Abstract
The genetic basis for combined pituitary hormone deficiency (CPHD) is complex, involving 30 genes in a variety of syndromic and nonsyndromic presentations. Molecular diagnosis of this disorder is valuable for predicting disease progression, avoiding unnecessary surgery, and family planning. We expect that the application of high throughput sequencing will uncover additional contributing genes and eventually become a valuable tool for molecular diagnosis. For example, in the last 3 years, six new genes have been implicated in CPHD using whole-exome sequencing. In this review, we present a historical perspective on gene discovery for CPHD and predict approaches that may facilitate future gene identification projects conducted by clinicians and basic scientists. Guidelines for systematic reporting of genetic variants and assigning causality are emerging. We apply these guidelines retrospectively to reports of the genetic basis of CPHD and summarize modes of inheritance and penetrance for each of the known genes. In recent years, there have been great improvements in databases of genetic information for diverse populations. Some issues remain that make molecular diagnosis challenging in some cases. These include the inherent genetic complexity of this disorder, technical challenges like uneven coverage, differing results from variant calling and interpretation pipelines, the number of tolerated genetic alterations, and imperfect methods for predicting pathogenicity. We discuss approaches for future research in the genetics of CPHD.
-
Introduction
Definition, clinical features, and genetic complexity of CPHD
Retrospective on gene discovery for CPHD in early years
Criteria for identifying pathogenic genes and variants for CPHD
-
Classification and Review of CPHD Causative Genes and Variants
Gene-level assessment to confirm CPHD causative genes
Adapted workflow for variant-level classification of reported variants in CPHD patients
-
Re-evaluation on Pathogenicity and Penetrance of Reported Genes and Variants for CPHD
Frequently screened and currently most relevant genes with CPHD
Less frequently reported genes in CPHD cases
Genes implicated in CPHD that require additional evidence about causality
CPHD candidate genes discovered by WES
Summary
-
Strategies to Improve Diagnostic Yield for CPHD and Future Directions
Incomplete coverage of WES
Genetic variants that are difficult to detect with WES (structural variants, splicing region, enhancers, and noncoding variants)
Large number of tolerated variants: filtering by expression profiles and pathways
Functional testing: genetic editing and animal models of human disease
Multifactorial disease
Conclusions
I. Introduction
A. Definition, clinical features, and genetic complexity of CPHD
Combined pituitary hormone deficiency (CPHD) (also called panhypopituitarism) is a condition classically characterized by a shortage of GH and at least one other pituitary hormone. If GH is the only deficient hormone, a diagnosis of isolated GH deficiency (IGHD) is made. About 45% of IGHD cases evolve to CPHD after a median follow-up time of 5.4 years (1), and CPHD is more likely to develop in patients with more severe idiopathic IGHD (2). The prevalence of CPHD is estimated to be 1 in 8000 individuals worldwide (Genetics Home Reference at NIH, https://www.ghr.nlm.nih.gov). Clinically, CPHD is often discovered when children exhibit reduced growth velocity, although hormone deficiency at birth can cause hypoglycemia and sudden death, emphasizing the necessity for early detection and treatment (3, 4). The clinical workup for growth insufficiency includes measurement of sitting and standing height, circulating hormone levels, hormone secretion in response to stimulation, imaging of the brain and pituitary gland, and determination of bone age with hand x-rays (3, 5, 6). Imaging sometimes reveals a pituitary mass, and genetic testing can often predict whether the mass is likely benign, thus avoiding unnecessary surgery (7, 8). Family history is also important because mean parental height is used to calculate the child's target height. If there is no family history of growth insufficiency, the case is termed sporadic, and it could be genetic or environmental (see Genetic Terminology, Box 1). In familial cases involving multiple affected individuals, the likelihood is increased that the cause is genetic, although environmental factors could play a role in the expressivity or severity of the features.
Box 1. Genetic Terminology.
- Inheritance patterns
- Sporadic: No family history
- Familial: Multiple related individuals are affected.
- Autosomal: Chromosomes other than the sex chromosomes X and Y
- X-linked: The disorder is due to lesion on the X chromosome; typically boys are affected, receiving the mutant allele from an unaffected mother.
- Recessive: Affected individuals have a lesion in both alleles.
- Dominant: Affected individuals have a lesion in only one allele.
- Effects of mutations
- LOF: Loss-of-function mutations result in lack of a gene product or an inactive one. Often recessive (see exception below).
- Dominant haploinsufficient: Loss of function of one allele is sufficient to cause the phenotype.
- Dominant negative: Mutant protein interferes with the function of the normal protein produced by the unaffected allele.
- GOF: Gain-of-function mutations cause over expression, mis-expression or excess function. Often dominant.
- Mutations
- Missense: Nucleotide change results in an amino acid substitution.
- Nonsense: Nucleotide change causes a premature stop codon.
- Indel: Insertion or deletion of nucleotides.
- Frame shift: An indel that changes the reading frame of the protein.
- CNV: Copy number variants are gene duplications or deletions that result in overdosage or underdosage of genes, respectively.
- Splicing: Nucleotide changes that render the donor or acceptor sequences ineffective for splicing are commonly identified in exome sequencing, but changes within introns can create novel splice sites that disrupt gene function, and these are often missed.
- De novo mutation: The affected individual has a lesion that is not detected in either parent (see mosaicism below).
- Splice enhancer: DNA sequence motif that directs or improves splice site utilization. Mutations in exonic splice enhancers may not change the encoded amino acid but may still be deleterious because of the failure to utilize the appropriate splice site.
- Phenotypic variability and penetrance
- Variable expressivity: Different individuals with the same genetic lesion can exhibit a range of symptoms from mild to severe.
- Incomplete penetrance: Not all individuals with the genetic lesion exhibit a clinical phenotype. Contrasts with fully penetrant, which means every individual with the lesion will be affected to some degree.
- Gonadal mosaicism: If a mutation occurs after fertilization, the individual may have germ cells that are normal as well as ones that carry the mutation. The mutation may or may not be detected in somatic tissue such as peripheral blood, which is often used to track mutations in a pedigree. If gonadal mosaicism is present but not detected, the mutation may be erroneously thought to be de novo, but there is a risk for additional affected children.
- Miscellaneous
- SNP: Single nucleotide polymorphism.
- Digenic disorder: Two different mutated genes are required to cause the clinical phenotype.
- Double heterozygote: An individual who is heterozygous for variants in two different genes.
- Compound heterozygote: An individual with a different variant in each allele of the same gene.
- Hemizygote: An individual with only one copy of the gene either because the other copy is deleted or the gene is on the X-chromosome and the individual is male.
- Genetic heterogeneity: A disorder that can be caused by mutations in a variety of different genes.
Conventions are to indicate a mutation in the predicted protein (p.) with the normal or reference amino acid, the position of the change, and the alteration. Asterisk indicates a nonsense codon ie. p.W194*. For cDNA (c.), the nucleotide position is followed by the reference and change, ie, c.334C>Tindicates a C to T change. Ins and del indicate insertions and deletions, respectively.
CPHD is caused by both genetic and nongenetic factors including trauma, brain surgery, tumor, infection, chronic heavy metal poisoning, irradiation, and autoimmune diseases (9–15). Genetic defects causing CPHD typically result in insufficient anterior pituitary gland development and hormone secretion manifesting in early childhood. Mutations in genes expressed in the developing head, hypothalamus, and/or pituitary cause CPHD. The earliest acting genes in head development are often associated with craniofacial abnormalities in addition to pituitary dysfunction (syndromic CPHD), whereas genes expressed in the hypothalamus or intrinsic to the pituitary cause nonsyndromic CPHD. The genetic complexity of CPHD can be explained by the heterogeneity of underlying factors, which contributes in part to the broad phenotypic spectrum. To date, 30 genes have been reported to be involved in the pathogenesis of CPHD (Figure 1). GLI2, HESX1, LHX3, LHX4, OTX2, POU1F1, PROP1, and SOX2 are the most studied ones (16–21).
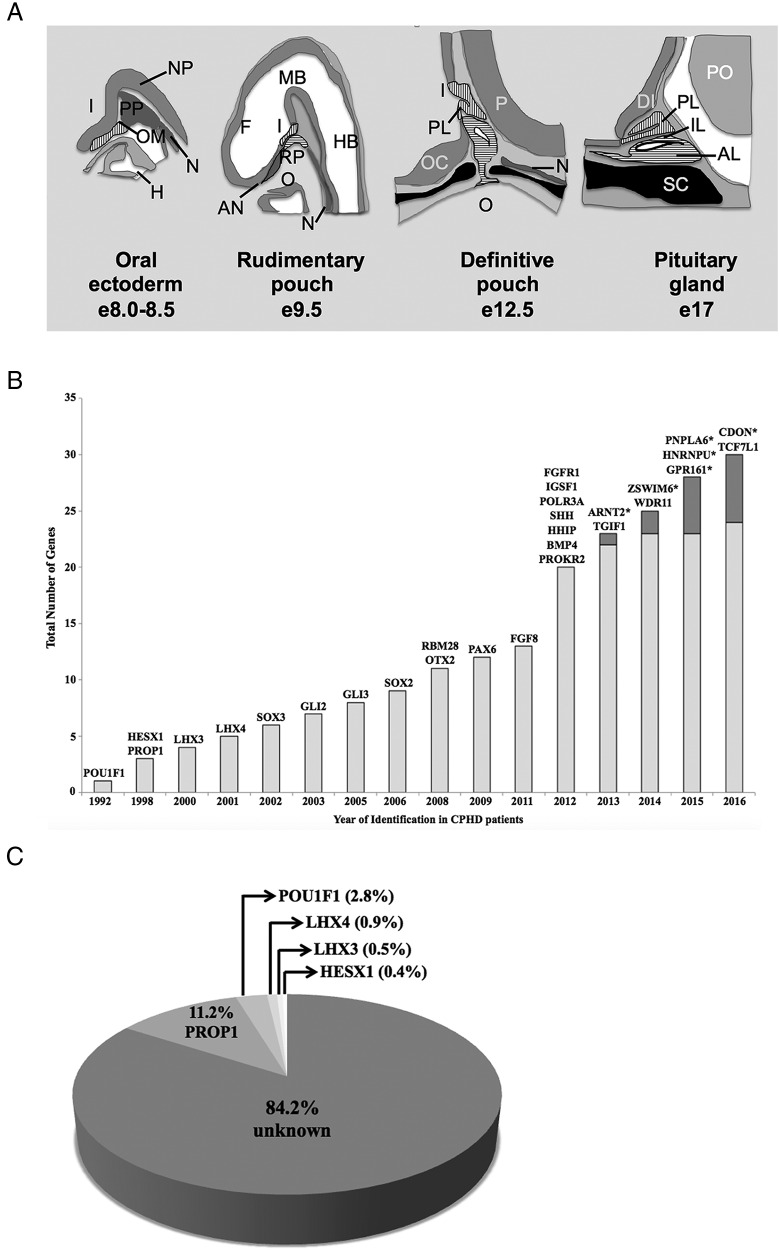
Figure 1.
Genetics of CPHD: timeline for discovery, cases explained, and developmental expression. A, Mouse pituitary development. Tissue fated to become Rathke's pouch is located at the anterior portion of the embryo. It is in constant contact with the neural ectoderm, which becomes the infundibulum. At e9.5, the oral ectoderm begins to invaginate to produce Rathke's pouch (horizontal hatching). At e12, the cartilage (black) begins to form, and Rathke's pouch pinches off. The posterior lobe (vertical hatching) is formed from evagination of neural ectoderm, and it produces FGF, which stimulates the growth of the pouch. The anterior and intermediate lobes are derived from Rathke's pouch and contain the major hormone-producing cell types by birth. The corresponding time in human development is weeks 4–9 (Carnegie stages 8–22) (418). Orientation: sagittal section with rostral to the left and dorsal at the top. I, infundibulum; H, heart; N, notochord; OM, oral membrane; PP, prechordal plate; F, forebrain; MB, midbrain; HB, hindbrain; AN, anterior neural pore; RP, Rathke's pouch; O, oral cavity; PL, posterior lobe; P, pontine flexure; OC, optic chiasma; IL, intermediate lobe; AL, anterior lobe; DI, diencephalon; PO, pons; SC, sphenoid cartilage. [Redrawn from H. Z. Sheng and H. Westphal: Early steps in pituitary organogenesis. Trends Genet. 1999;15(6):236–240 (166), with permission. © Elsevier.] B, The timeline of gene discovery for CPHD. The light gray bars represent the total number of genes identified by the candidate screening methods; the dark gray bars represent the total number of genes identified by the WES. For each year, the identified gene names are at the top of the bar. *, Gene identified by WES. C, Pie chart indicating the familial and sporadic cases of CPHD that were explained by mutations in the genes PROP1, POU1F1, LHX3, LHX4, and HESX1. Data are summarized from an extensive literature survey, which illustrates the variation in frequency of PROP1 mutations by ethnic groups (46). Because systematic screening has not yet been done for the other CPHD genes, it is not possible to estimate their frequency, but it is expected to be quite small.
Precise clinical phenotypic characterization of patients is a very important foundation for genetic studies. It is important to document the nature and progression of hormone deficiencies, take note of associated features, and eliminate cases that have a likely environmental etiology. Grouping of patients based on clinical phenotype can also help in identifying causal genes, which was done successfully with other endocrine disorders such as hypogonadotropic hypogonadism (HH), Kallmann syndrome (KS), and IGHD (22–25). As we learn more about the genetic etiology of CPHD, the data suggest that it is part of a spectrum disorder, with holoprosencephaly (HPE) and septo-optic dysplasia (SOD) at the severe end and HH and IGHD at the mild end of the spectrum (Table 1). The overlap derives from common developmental programs. For example, the eye, ear, nose, pituitary gland, and some cranial nerve ganglia are all derived from placodes in the developing head, and developmental regulation involves common genetic pathways (26). SOD features underdeveloped optic nerves, dysfunctional pituitary, and brain malformations. It results from altered craniofacial development in the midline and can be caused by mutations in HESX1, OTX2, SOX2, SOX3, and PAX6 (26, 27). Other examples of CPHD with craniofacial involvement are patients with HPE, which occurs when the forebrain of the embryo fails to separate into two cerebral hemispheres with or without other craniofacial midline structural anomalies or eye defects. Genes in the sonic hedgehog (SHH) signaling pathway can cause both HPE and CPHD (28, 29).
Table 1.
Genes Implicated in Cases of CPHD Can Present With Other Clinical Phenotypes
| Gene | Phenotypesa |
||||
|---|---|---|---|---|---|
| Craniofacialb | Pituitaryc | Peripheral Tissuesd | |||
| ARNT2 | Brain, eye | CPHD | Kidney, urinary tract | ||
| BMP4 | Eye, craniofacial, cleft lip, palate, spina bifida aperta | CPHD | Renal hypodysplasia, hypospadias, polydactyly | ||
| CDON | HPE | (CPHD) | None | ||
| CHD7 | Eye, craniofacial | CPHD | None | ||
| FGF8 | SOD, KS: anosmia | HHe | CPHD | IGHD | Vacterl, hypospadia |
| FGFR1 | Pfeiffer, Hartsfield, Jackson-Weiss syndromes | CPHD | None | ||
| GLI2 | HPE, cleft lip, palate | HH | CPHD | IGHD | Polydactyly, cryptorchidism |
| GLI3 | Pallister Hall syndrome, hypothalamic hamartoma, Greig cephalopolysyndactyly | CPHD | IGHD | Skeletal, polydactyly | |
| GPR161 | None | (CPHD) | (IGHD) | None | |
| HESX1 | SOD | CPHD | IGHD | None | |
| HHIP | HPE | (CPHD) | None | ||
| HNRNPU | Lennox-Gastaut syndrome, CNS, epilepsy, intellectual disability | (CPHD) | None | ||
| IGSF1 | None | CPHD, TSH only | Macro-orchidism | ||
| LHX3 | Hearing | CPHD | Skeletal | ||
| LHX4 | None | CPHD | None | ||
| OTX2 | Eye, craniofacial | CPHD | IGHD | None | |
| PAX6 | Eye | CPHD | None | ||
| PNPLA6 | Spastic paraplegia, Leber congenital amaurosis and other syndromes, eye, craniofacial | HH | CPHD | Muscle wasting | |
| POLR3A | Hypomyelination, hypodontia | HH | (IGHD) | None | |
| POU1F1 | None | CPHD | IGHD | None | |
| PROKR2 | KS: anosmia | HH | (CPHD) | (IGHD) | Hirschsprung disease |
| PROP1 | None | CPHD | None | ||
| RBM28 | Alopecia, neurological defects, intellectual disability, tooth defects | (CPHD) | None | ||
| SHH | HPE, eye, dental anomalies | (CPHD) | None | ||
| SOX2 | Eye, dental anomalies, hearing impairment | HH | CPHD | Micropenis | |
| SOX3 | Intellectual disability, abnormal facial features, speech difficulties, retrognathia, hearing impairment | HH | CPHD | IGHD | Sex reversal, digital anomalies, micropenis |
| TCF7L1 | SOD | CPHD | None | ||
| TGIF1 | HPE | CPHD | None | ||
| WDR11 | Cleft palate, hearing, brain and dental abnormalities, KS: anosmia | HH | (CPHD) | None | |
| ZSWIM6 | Acromelic frontonasal dysostosis, brain | CPHD | Skeletal, cryptorchidism | ||
Phenotypes listed represent the range of symptoms reported in various individuals. The pituitary hormone deficiency and associated craniofacial and peripheral tissue phenotypes can vary and/or be absent.
Craniofacial refers to all aspects of head development except the pituitary, which is listed separately.
The parentheses indicate need for additional evidence or patient examples to add certainty to the role of the gene with the indicated pituitary phenotype.
Peripheral tissue phenotypes are listed, except for those that are secondary to the pituitary hormone deficiencies.
The HH can be of hypothalamic or pituitary origin. Cases were arbitrarily assigned to the pituitary category for simplicity.
HH and CPHD have overlapping genetic etiologies. HH is characterized by gonadotropin insufficiency, reduced sex steroid production, and delayed puberty or infertility (30). It can be caused by reduced production of GnRH due to failed migration of GnRH neurons during development, as well as impaired regulation of GnRH secretion. Mutations in many genes can cause HH, and some of these genes are also implicated in CPHD (CHD7, PROKR2, WDR11, FGFR1, and FGF8) (30–33) (Figure 1 and Table 1). In addition to X-linked or autosomal recessive Mendelian cases of HH, panel sequencing of known genes in large cohorts of patients provided evidence that individuals with HH sometimes carry deleterious alleles within multiple HH genes (30). CPHD may prove to be a multifactorial disease as well because identical mutations can produce a spectrum of phenotypic severity in different CPHD patients, and incomplete penetrance is not uncommon in family pedigrees. This suggests the involvement of multiple genes and/or environment in enhancing or suppressing the severity of the features. Studies in mutant mice clearly document the effects of genetic and environmental factors, including maternal alcohol consumption on craniofacial development, and the fact that genetic lesions that are normally tolerated can sensitize the fetus to environmental challenges (34–36).
Height is a highly heritable trait, and it is estimated that genetic factors explain 80–90% of the variation (37). Numerous genome-wide association studies (GWAS) have been carried out on large population groups in an effort to identify the genetic factors that contribute to height (38–40). The loci identified to date explain about 30% of the heritability. Interestingly, a few genes implicated by GWAS are mutated in cases of GH insufficiency or resistance, including GLI2, GH1, GHR, and HHIP. In addition, association was found with HESX1 and POU1F1 in a pygmy population (41). Loci involved in skeletal dysplasia or intrauterine growth retardation are also implicated in height determination (42). Some of the missing heritability could be attributable to gene-gene interactions and epigenetic effects (43). For example, it is known that nutrition affects growth, and nutritional effects can be inherited trans generationally through incomplete erasure of epigenetic marks (44). It is notable that variation near the DNA methyltransferase gene, DNMT3A, is associated with height, and there are several well-known syndromes that are imprinted and affect height (45). Epigenetic heredity is a determinant of adult height, but more research is needed to identify the specific contributions of epigenetic factors.
Most patients with CPHD (∼84%) have no genetic diagnosis (Figure 1) (46–69). Gene identification methods for human diseases have evolved from positional cloning of single genes and Sanger sequencing to massively parallel sequencing. With reduced costs and increased application of next-generation sequencing (NGS) techniques, we expect that more novel genes and variants will be discovered. However, the assignment of pathogenicity and causality of genes and variants is not trivial, and there has not yet been a systematic review of this topic for CPHD. The purpose of this review is to document the inheritance mechanisms and pathogenicity of genes implicated in CPHD and its related diseases, to apply current criteria for classifying pathogenic variants, and to facilitate future novel gene discovery in CPHD. Other recent reviews have provided other perspectives on IGHD and CPHD (21, 25, 70–73).
B. Retrospective on gene discovery for CPHD in early years
There has been rapid progress in identifying genes that cause CPHD since 1992, and now a total of 30 genes are implicated, although the amount of evidence for individual genes is variable (Figure 1 and Table 2). We review the early discovery process here.
Table 2.
CPHD Genes Evaluated and Classified for the Causality
| Gene Name | Evidence 1: Multiple Unrelated Patients | Evidence 2: Functional Studies | Evidence 3: Gene Expression | Evidence 4: Interactions and Pathways | Total Supporting Evidence |
|---|---|---|---|---|---|
| POU1F1 | + | + | + | First gene discovered | 4 |
| PROP1 | + | + | + | + | 4 |
| HESX1 | + | + | + | + | 4 |
| LHX3 | + | + | + | + | 4 |
| LHX4 | + | + | + | + | 4 |
| OTX2 | + | + | + | + | 4 |
| GLI2 | + | + | + | + | 4 |
| SOX2 | + | + | + | + | 4 |
| SOX3 | + | + | + | + | 4 |
| FGF8 | + | + | + | + | 4 |
| FGFR1 | + | + | + | + | 4 |
| GLI3 | + | + | + | + | 4 |
| PAX6 | + | + | + | + | 4 |
| ARNT2 | + | + | + | 3 | |
| BMP4 | + | + | + | 3 | |
| IGSF1 | + | + | + | 3 | |
| PNPLA6 | + | + | + | 3 | |
| SHH | + | + | + | 3 | |
| TCF7L1 | + | + | + | 3 | |
| ZSWIM6 | + | + | + | 3 | |
| CHD7 | + | + | 2 | ||
| PROKR2 | + | + | 2 | ||
| TGIF1 | + | + | 2 | ||
| CDON | + | 1 | |||
| GPR161 | + | 1 | |||
| HHIP | + | 1 | |||
| HNRNPU | + | 1 | |||
| POLR3A | + | 1 | |||
| RBM28 | + | 1 | |||
| WDR11 | + | 1 |
Separation of groups of genes by line space indicate the groups of genes that are supported as CPHD candidates by different numbers of evidence.
1. Spontaneous mouse mutant strains reveal CPHD candidate genes
The first genes identified in CPHD patients were found after studies in mice implicated the genes in pituitary development and function. A spontaneous mutation causing dwarfism, infertility, and lethargy was reported in 1929. These mice, known as Snell dwarfs, were used to demonstrate that the pituitary gland regulates growth and the function of multiple target organs (74, 75). The molecular basis for the disorder was unknown until 1990 (76, 77). The pituitary transcription factor POU1F1 (previously known as PIT-1 and GHF-1) was discovered based on its ability to transactivate the GH and prolactin (PRL) genes (78, 79) and was tested as a candidate gene for the mutation. A recessive loss-of-function mutation in Pou1f1 (p.W251C) was shown to be responsible for the GH, PRL, and TSH deficiency in these mutants. Two years later, a homozygous nonsense mutation in POU1F1 gene was identified in a CPHD patient from consanguineous parents (80), which was the first report in humans of a defect in a transcription factor causing CPHD.
Similarly, the Ames dwarf mutation arose spontaneously in 1961, and in 1996 genetic mapping and positional cloning experiments demonstrated that these mice have panhypopituitarism caused by a p.S83P substitution in the paired-like homeodomain transcription factor, termed Prophet of Pit-1 (Prop1) (81–83). Two years later, four CPHD families with homozygosity or compound heterozygosity for inactivating mutations of PROP1 were identified (84).
The Ames and Snell dwarf mice are reasonable phenocopies of the human patients, although there are some differences. In humans, a differentiating diagnosis between PROP1 and POU1F1 patients can be the presence or absence of gonadotropin deficiency, respectively, in addition to low GH, TSH, and PRL (85). Both Prop1- and Pou1f1-deficient mice lack GH, TSH, and PRL, and both have low gonadotropins. However, supplementation with GH and T4 is sufficient to restore fertility in both mutants (86, 87). Mice require thyroid hormone to develop the feedback loops that regulate gonadotropin function (88). Patients with PROP1 mutations show progressive hormone deficiency that can eventually result in the life-threatening loss of ACTH, but adrenal function is preserved in Prop1 mutant mice (89). However, it is important to note that mice homozygous for loss-of-function mutations in either Prop1 or Pou1f1 can manifest quite different symptoms on different genetic backgrounds, ranging from newborn lethality to a vigorous long life, even when environmental parameters like food intake and endemic diseases are invariant (90, 91). This demonstrates that the effects of both PROP1 and POU1F1 deficiency are enhanced or suppressed by genetic variation inherent among inbred mouse strains. The mouse models provide excellent tools to identify such interacting genes (92, 93).
2. Genetically engineered mouse models identify CPHD candidate genes
The LIM homeodomain gene Lhx3 (p-Lim or Lim3) is expressed in the developing pituitary gland. In 1996, the Lhx3−/− mutant mice were generated by homologous recombination in embryonic stem cells and were discovered to lack both the anterior and intermediate lobes of the pituitary gland (94). The pituitary primordium, Rathke's pouch (Figure 1), formed in the mutants but it failed to grow, and only Pomc-expressing cells were detected. Four years later, the human gene was cloned and characterized, and a homozygous LHX3 defect was identified in patients with CPHD and rigid cervical spine with limited head rotation (95, 96).
Lhx4 is expressed in the pituitary primordium (Rathke's pouch) and persists in the anterior and intermediate lobes through adulthood (97). In 1997, genetically engineered Lhx4−/− mice were generated. They initially form Rathke's pouch normally, but very few cells undergo differentiation into the five anterior pituitary-specific cell lineages (97). The human LHX4 was cloned, and a germline splice-site mutation in LHX4 was found in a patient with CPHD and a small sella turcica in 2001 (98).
Hesx1 is expressed in embryonic stem cells and becomes restricted to the developing forebrain, hypothalamus, and Rathke's pouch (99). In 1998, mice homozygous for a Hesx1 disruption were generated and found to exhibit variable anterior central nervous system (CNS) defects and pituitary dysmorphology (100). Afterward, patients with SOD and/or CPHD were screened for HESX1 mutations, and siblings homozygous for a p.R160C substitution were identified (100).
3. Genes that cause severe craniofacial defects are associated with syndromic CPHD
The midline location of the pituitary gland and its close developmental association with the forebrain result in the association of CPHD with HPE. HPE can be caused by mutations in SHH, GLI2, PTCH1, TGIF, SIX3, ZIC2, NODAL, FOXH1, CDON, FGF8, and DISP1 (101–103). The SHH signaling pathway, which activates target genes under the control of the GLI family transcription factors, is the best studied cause of HPE. GLI2 mutations were first identified in a genetic screen of 390 HPE patients (104). Three of the four index families carrying GLI2 heterozygous mutations had hypopituitarism. Constitutive Gli2 knockout mice die embryonically. Conditional deletion of Gli2 in Rathke's pouch demonstrated that Gli2 is required for normal GH, PRL, and ACTH production (105). Shh and Gli2 signaling also control the diencephalic expression of Bmp4 and Fgf8, which are necessary for stimulating anterior pituitary development (106). Mutations in these genes were later identified in patients with hypopituitarism (see section III.B).
4. CPHD genes revealed through detection of cytogenetic abnormalities
The involvement of SOX3 gene with X-linked panhypopituitarism was discovered in 2002 through cytogenetic testing and subsequent mutation analysis in a patient with mental retardation and IGHD (107). The patient had an in-frame duplication of 33 bp that expanded a polyalanine tract by 11 alanines. Later, in 2004, Sox3 was studied in mice. Sox3 is highly expressed in ventral diencephalon, and targeted disruption of Sox3 leads to abnormal development of Rathke's pouch (108).
5. Genes identified through functional relationships with other CPHD genes
Heterozygous OTX2 mutations were identified in patients with anophthalmia and microphthalmia in 2005 using a candidate gene screening approach (109, 110). Mouse studies showed that Otx2 is required for activation of Hesx1 expression in the developing forebrain (111), and HESX1 mutations can cause eye and/or pituitary defects (100). Additionally, an OTX2 binding site was identified in the promoter of the Hesx1 gene (112). The connection between OTX2 and HESX1 led to screening and identification of OTX2 mutations in CPHD families with or without ocular abnormalities. A heterozygous frameshift mutation in OTX2 was identified in a patient with anophthalmia, short stature, and GH deficiency, and a missense mutation was found in two unrelated children with CPHD who presented with neonatal hypoglycemia and deficiencies of GH, TSH, LH, FSH, and ACTH (53, 113). As early as 1995, however, mouse fetuses heterozygous for a genetically engineered Otx2 knockout were discovered to have a highly variable phenotypic spectrum that is dependent upon the genetic background (35, 114). The phenotypes included pituitary aplasia, ocular malformations, facial clefting, and acephaly. Homozygotes had early embryonic lethality. The mechanism whereby Otx2 haploinsufficiency causes hypopituitarism was revealed by specifically deleting Otx2 in the neural ectoderm that gives rise to the ventral hypothalamus and posterior pituitary lobe. The hypoplasia of these regions led to reduced bone morphogenetic protein and fibroblast growth factor (FGF) signaling and, secondarily, reduced growth of the anterior lobe (115). Deletion in the anterior lobe had no consequences.
6. Summary
Early successes in identification of CPHD genes relied on phenotypic similarities between mouse models and human patients, the underlying knowledge of genetic pathways that regulate pituitary development, the occasional association of CPHD with genetic lesions that cause craniofacial anomalies, and cytogenetic findings. The emerging new paradigm is to conduct an unbiased search for new CPHD genes using NGS and utilize cell lines and animal models to demonstrate the functional relevance of the genetic changes and understand the mechanism of action of the novel genes. Criteria are being established to ensure the authentic pathogenicity and causality of genetic variation, and we will apply the current guidelines to genes and variants implicated in CPHD (116, 117).
C. Criteria for identifying pathogenic genes and variants for CPHD
To date, most human CPHD genes were identified by Sanger sequencing of individual candidate genes, which is not an efficient approach for diseases with a great deal of genetic heterogeneity (118). Moreover, gene-by-gene screening is unlikely to identify digenic or multigenic disease unless all known genes are investigated in each patient. Whole exome sequencing (WES), currently the most commonly used massively parallel sequencing technique, examines the exons of all known protein-coding genes simultaneously and offers a powerful approach to rapidly screen for candidate disease-causing mutations, with a current diagnosis rate of 25% (119). The sequencing technique and data analysis pipeline of WES are not the focus of this review and have been comprehensively discussed elsewhere (120–122). The first case of a CPHD causative gene identified by WES was ARNT2 (123). Since then, five other genes were implicated in CPHD by WES (Figure 1). We expect that application of WES and whole genome sequencing (WGS) will continue to increase our understanding of this disorder and provide valuable information about oligogenic disease.
The high rate of acquiring data by high throughput sequencing and the ambiguity in interpretation has prompted important dialog among genetic researchers and medical geneticists about assigning pathogenicity to genuine disease-causing genetic variants. In 2014, a group of 27 experts in genetic research, analysis, and clinical diagnostic sequencing published guidelines for investigating causality of sequence variants in human diseases (117). The challenges in assessing sequence variants in human disease are discussed in the guidelines, and a list of factors to consider at both the gene level and the variant level is provided for presumed monogenic diseases. One year later, the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) updated their standards and guidelines for the interpretation of sequence variants (116). With awareness of the increased complexity in sequence interpretation during clinical genetic testing, the ACMG-AMP guidelines provided a stringent and semiquantitative set of criteria for variant classification. A recent pilot evaluation of the performance of ACMG-AMP guidelines on 99 variants among nine laboratories showed that consensus discussion and detailed review among the researchers greatly improved the concordance of variant classifications (124). More effort is needed to increase application of a standardized approach for variant assessment and to achieve consistent concordance with variant interpretations. To date, there has not been a systematic overview in the literature of the pathogenicity of all the genes and variants that have already been reported to cause CPHD. It is timely and important to conduct such a survey before the field fully embraces the novel sequencing technologies for new gene identification in CPHD.
II. Classification and Review of CPHD Causative Genes and Variants
We present our methods for identifying genes implicated in CPHD and evaluating the pathogenicity using the currently accepted guidelines for genetic variation.
A. Gene-level assessment to confirm CPHD causative genes
Combining the search results from free (Online Mendelian Inheritance in Man [OMIM] and PubMed) and subscription (Human Gene Mutation Database [HGMD]) databases, we identified 30 genes that are associated with either nonsyndromic or syndromic CPHD in the literature by June 30, 2016. We applied the four categories of evidence suggested by MacArthur et al (117) to assess the causality of these genes for CPHD (Table 2). These lines of evidence include: 1) variants in the same gene and similar clinical phenotypes have been observed in multiple unrelated individuals; 2) functional studies, including biochemistry, cell culture, and/or animal models confirm the deleterious effects of detected variants in the genes; 3) the protein that the gene encodes is expressed in the appropriate developing tissue such as hypothalamus and/or pituitary, according to BioGPS (www.biogps.org), Gene Expression Database (GXD; http://www.informatics.jax.org/expression.shtml), and/or the internally maintained Embryonic Mouse Pituitary cDNA library (125) (https://sites.google.com/a/umich.edu/riken-database-2014/); and 4) the gene product interacts with other proteins or plays a role in the biological pathways that are implicated in the disease. Replication in unrelated individuals and functional studies are the most important types of evidence. We consider the genes that have at least two types of evidence, to date, that support their candidacy as causative genes for CPHD. These include the frequently screened and historically most relevant genes (POU1F1, PROP1, HESX1, LHX3, LHX4, SOX3, GLI2, SOX2, and OTX2) and the genes that are recently identified or less commonly reported in CPHD (BMP4, FGF8, FGFR1, GLI3, PAX6, IGSF1, SHH, ARNT2, TCF7L1, CHD7, PROKR2, TGIF1, PNPLA6, and ZSWIM6). Also, seven genes, CDON, GPR161, RBM28, POLR3A, HHIP, WDR11, and HNRNPU, are grouped together awaiting additional evidence to confirm causality. Among these, CDON, GPR161, and HNRNPU were discovered by WES, and the other four genes, RBM28, POLR3A, HHIP, WDR11, were from candidate gene screening.
B. Adapted workflow for variant-level classification of reported variants in CPHD patients
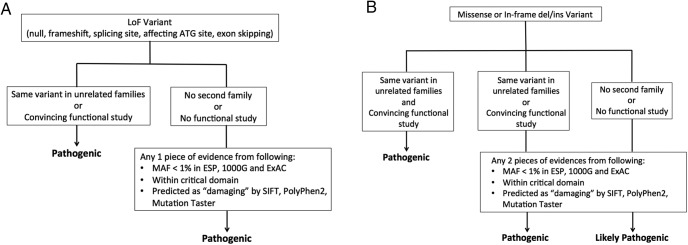
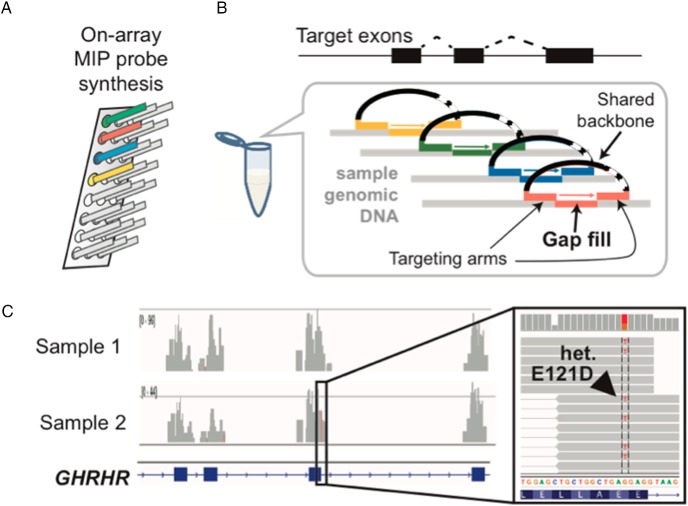
There are about 2,500 reported variants in the 30 CPHD-associated genes. For each variant, the classification criteria that we used are based on the ACMG-AMP guidelines, which is so far the most detailed and quantitative system for variant interpretation in genetic testing (116). Our retrospective review of variants implicated as causing CPHD focuses on specific criteria including minor allele frequency (MAF), which we expect to be rare in the case of a rare disease, computational prediction of a deleterious effect on function, and confirmation of reduced activity with functional assays. At the gene level, we weighed gene expression in the relevant tissues, gene function in pertinent biology pathways, variants in multiple families with similar phenotypes, and cosegregation of the clinical phenotype with the variant in a pedigree. We applied the weight and the rules for combining criteria in the ACMG-AMP guidelines to generate our workflow for the interpretation in loss-of-function variants and missense or in-frame indels (Figure 2). The MAF was determined from three public population databases: Exome Aggregation Consortium (ExAC; http://exac.broadinstitute.org/), Exome Variant Server (EVS; http://evs.gs.washington.edu/EVS/), and 1000 Genomes Project (1000G; http://browser.1000genomes.org/index.html). We used three in silico prediction programs for predicting the effects of missense variants: SIFT (http://sift.jcvi.org/), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), and MutationTaster (http://www.mutationtaster.org/). SIFT predictions are classified as: tolerated and damaging. PolyPhen-2 predictions are classified as: benign, possibly or probably damaging. MutationTaster predictions are classified as: probably deleterious, known to be deleterious, probably harmless, known to be harmless. None of the programs that predict pathogenicity are perfect, which is an ongoing challenge for interpretation of genome sequence information. Functional studies are critical because prediction programs have both false negatives and false positives (117, 126, 127). Improved accuracy in prediction programs would be invaluable for the field because functional testing is particularly time consuming, but functional testing is likely to continue to be necessary and critically important for interpretation. Functional testing also has challenges, which we discuss in Section IV.D.
Figure 2.
Work flow for assessment on the reported genetic variants of CPHD. A, Loss-of-function (LoF) variants; B, missense or in-frame insertion and deletion (in/del) variants.
III. Re-evaluation on Pathogenicity and Penetrance of Reported Genes and Variants for CPHD
A. Frequently screened and currently most relevant genes with CPHD
1. PROP1
PROP1 is the most frequently mutated gene known to cause CPHD. Patients with mutations in PROP1 are typically identified initially with short stature due to GH deficiency. Most patients also exhibit reduced TSH and PRL at the time of diagnosis. At the onset of puberty, many patients with PROP1 mutations also exhibit LH and FSH deficiency and fail to develop secondary sexual characteristics. The loss of gonadotropins may also present as an evolving characteristic identified in adulthood. Many human patients also develop ACTH deficiency as they age (128). We hypothesize that the role of PROP1 in establishing pituitary stem cell pools and promoting pituitary stem cells to migrate and transition to differentiation is the basis for progressive hormone failure in humans (129). Magnetic resonance imaging (MRI) is invaluable for assessing whether the pituitary insufficiency is associated with other brain abnormalities, disrupted pituitary stalk, or ectopic posterior pituitary, which is among the varying range of physical features of the pituitary gland in patients with PROP1 mutations. Most cases display pituitary hypoplasia or aplasia; however, there are reports of pituitary hyperplasia, a waxing and waning of pituitary hyperplasia to hypoplasia, as well as reports of pituitary masses (56, 128, 130–137).
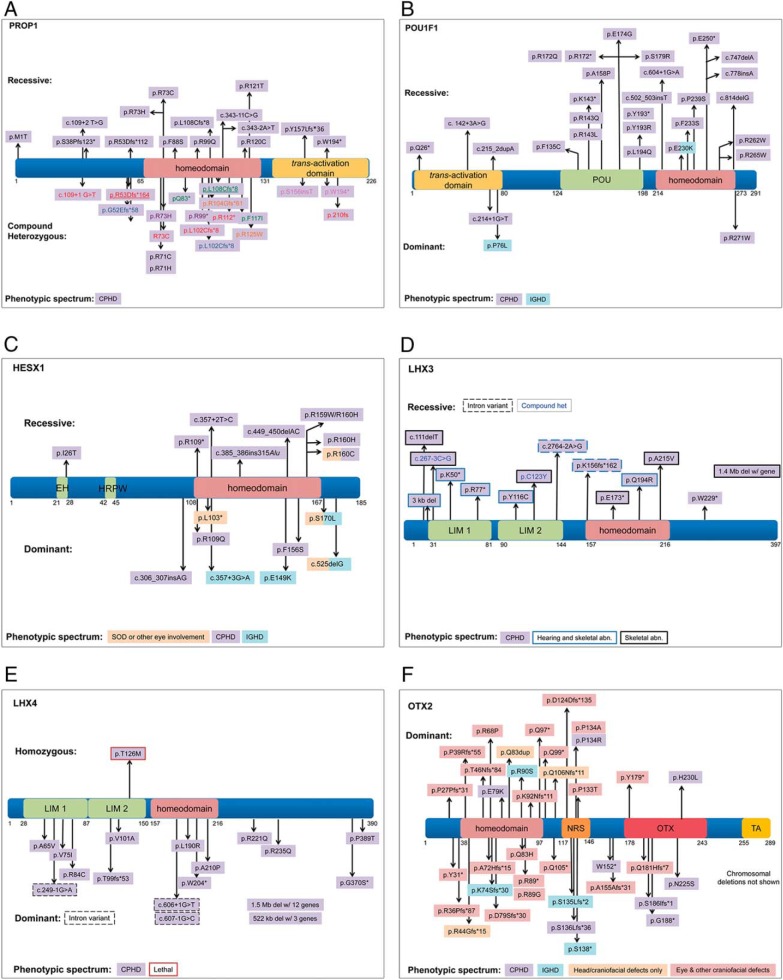
The frequency of PROP1 mutations varies among population groups and is highest in Eastern Europe and Russia (46–48, 64). Two of the most common variants identified are: a two base pair deletion, c.301_302delAG, which results in frameshift and early termination of the protein at amino acid 109; and c.150delA, which results in a frameshift at amino acid 53 and early termination at amino acid 164 (47). There are a couple of reports of heterozygous variants in PROP1 with insufficient data to support the pathogenicity (138, 139). All of the other cases are either homozygous or compound heterozygous, recessive, mostly loss-of-function variants in PROP1. There are no convincing examples of incomplete penetrance, meaning individuals with two loss-of-function alleles but no endocrine disorder, yet there are examples of variable hormone profiles and age of onset within a pedigree, including one unusual case in which the patient reached a normal height but acquired CPHD as an adult (140). Interestingly, in one case with a homozygous p.W194* variant, the patient initially presented with HH and developed progressive GH and TSH deficiency during adulthood (141). In another familial case, with two affected brothers containing a compound heterozygous variant (c.150_150delA; c.334C>T), the patients were initially diagnosed with hypothyroidism due to only a slight deviation from normal linear growth. In subsequent follow-up exams, deficiencies in GH, LH, FSH, and PRL were also detected (136). Twenty-nine PROP1 variants have been reported, including complete gene deletions (six variants), alterations in splicing (four variants), nonsense variants (eight variants), and missense variants (11 variants). Fourteen of the 29 variants are proven pathogenic through functional studies, analysis of multiple carriers within a pedigree, analysis of multiple pedigrees, and allele frequency (Figure 3). Five variants are assumed pathogenic because they cause protein truncations.
Figure 3.
Examples of pathogenic variants identified in CPHD genes.
2. POU1F1
POU1F1 is a pituitary-specific transcription factor necessary for expression of GH, TSH, and PRL. POU1F1 contains a POU-specific and POU-homeodomain, which are both necessary for DNA binding. A total of 30 different variants in POU1F1 have been described, and they are generally recessive with pituitary hypoplasia and CPHD. All patients with homozygous variants are familial, and all compound heterozygous cases but one are familial. About half of these variants have been proven to affect function with in vitro experiments. We determined that all of the homozygous and compound heterozygous variants are either likely pathogenic or pathogenic resulting in CPHD (Figure 3). There are three dominant-negative variants reported in POU1F1 that are pathogenic. The POU1F1 missense p.R271W acts as a dominant inhibitor of transcription resulting in CPHD (142). A few p.R271W cases exhibit cognitive impairment or hearing impairment, which may be due to hypothyroidism during pregnancy when the mother carries the same variant (143). Approximately half of the p.R271W variants are fully penetrant. Two cases are clearly incompletely penetrant, and the rest of the cases did not present enough family history to determine penetrance. A splicing variant at c.214+1G>T was also determined to be a dominant-negative variant, but it is incompletely penetrant (144). The third pathogenic dominant-negative variant identified is a missense p.P76L and results in IGHD. This variant is completely penetrant because all nine members of family carrying it exhibit the IGHD phenotype (145). Several other heterozygous variants in POU1F1 have been reported but, due to a lack of patient data, cannot be labeled as pathogenic with certainty (58, 64, 146–149). A single case with a homozygous deletion of POU1F1 together with the neighboring CHMP2B and VGLL3 genes presented with an unusual syndromic phenotype. The child exhibited CPHD and hypoplastic pituitary typical for POU1F1 mutations but had craniofacial defects and cognitive impairment and was initially unresponsive to GH therapy. The unusual phenotype may be due to the additive effects of deleting all three genes (150).
3. HESX1
HESX1 is a homeobox gene that plays an important role in early brain development. It is essential for the formation of the pituitary and the development of the forebrain, including optic nerves (151, 152). Both recessive and dominant variants in HESX1 can cause CPHD (Figure 3). All eight recessive mutations of HESX1 gene can be classified as pathogenic and have been studied by functional assays or animal models (100, 153–158). Most of the recessive variants cause CPHD, except that homozygous p.R160C causes SOD and hypopituitarism. Most variants affect the DNA binding homeodomain or the engrailed homology domain that binds corepressors of the TLE family (p.I26T). Among the 14 heterozygous variants found in HESX1, four are loss-of-function variants (c.307_308insAG; c.525delG; c.308T>A, p.L103*; c.357+3G>A) and can be classified as pathogenic. Only one heterozygous missense variant, p.S170L, was identified in two unrelated families, and one family had two affected children with mild SOD. The rest of the heterozygous missense variants reported in HESX1 are either incompletely penetrant or lack information on family segregation. More evidence is still needed to assign causality. Two heterozygous missense variants, p.H42Y and p.V75L, were identified in HH patients. The variant p.H42Y is rare in the population databases, but the prediction programs do not agree on whether it is likely damaging. The p.V75L variant is a novel one, not recorded in any population database, but MutationTaster predicts it would be tolerated. The functional effects of p.H42Y and p.V75L were not assessed. More evidence is needed to determine whether HESX1 mutations cause HH.
4. LHX3
LHX3 is a member of the Lin11, Isl-1, and Mec-3 (LIM) homeodomain protein family of transcription factors. It is expressed in Rathke's pouch and the neural tube and is important for pituitary development and the organization of spinal cord neurons (159, 160). Variants in LHX3 that cause CPHD are autosomal recessive and are mostly missense and nonsense mutations and include two complete gene deletions of LHX3 (161–163) (Figure 3). GH and TSH are consistently reduced in patients with LHX3 mutations, and in descending order PRL, LH/FSH, and ACTH production is also affected. Limited neck rotation is the most common accompanying symptom (164). In mice, Lhx3 and Lhx4 are expressed in the ventral motor neurons; therefore, the limited neck rotation may be due to abnormal innervation of the anterior neck muscles. Hyperplastic anterior pituitary gland, sensorineural hearing impairment, respiratory difficulties, and skeletal abnormalities were also seen in some cases (161–164). The variants found in LHX3 do not cluster in any particular region or domain of the protein. Two heterozygous variants in LHX3, p.T63M and p.A322T, were found in patients with CPHD (165). The p.A322T variant has a rare allele frequency of 0.003, but it is predicted to be benign using SIFT, which means it is unlikely to be causative. There was no allele frequency information for variant p.T63M, but it was also predicted to be benign.
5. LHX4
LHX4 also encodes a LIM homeodomain protein. Mutations in LHX4 mostly result in CPHD cases with autosomal dominant inheritance and incomplete penetrance (50, 66, 98, 167) (Figure 3). There is only one documented homozygous LHX4 variant (p.T126M) and it is associated with a lethal phenotype (168). Heterozygous LHX4 variants are generally loss-of-function alleles, and patients carrying these heterozygous variants present with CPHD, short stature, small sella turcica and cerebellar defects, hypoplastic anterior pituitary, respiratory disease, genital malformations, some craniofacial features, and hypothyroidism (50, 66, 165–167, 169–173). In some cases, one parent carries the putative causative allele and has significant short stature, but without CPHD or any other phenotypic abnormality. There are at least five reports of de novo deletions encompassing LHX4 (165, 170, 172). The clinical features of a patient with a de novo heterozygous 522-kb deletion including LHX4, CEP350, QSOX1, and ACBD6 are highly similar to other heterozygous missense LHX4 cases. Thus, the hemizygosity for the other genes is unlikely to contribute to this patient's phenotype (165). A 7.5-Mb duplication containing LHX4 is associated with tall stature, cognitive deficits, and craniofacial dysmorphism, leaving open the possibility that LHX4 overdosage could be problematic, but more studies are necessary to assess this definitively (174). Several LHX4 variants of uncertain significance were reported in individuals with CPHD (66, 173). The variants p.242delK (MAF = 0.000364), p.N271S (MAF = not reported (n.r.)), and p.Q346R (MAF = 8.2e−6) are predicted to be benign and do not alter LHX4 transactivation properties in transfected cells (173). Although p.G370S (MAF = n.r.) has normal transactivation properties, it was predicted to be deleterious (66). Additional studies are warranted to determine whether the p.G370S variant affects LHX4-interacting proteins or in vivo function in zebrafish or mice (175, 176).
6. OTX2
OTX2 encodes orthodenticle homeobox 2, a member of the bicoid class of homeobox transcription factors. OTX2 is involved in the development of the brain and other head structures, most notably the eyes and pituitary gland (35, 114, 177, 178). Variants in OTX2 account for approximately 2–3% of cases of anophthalmia or microphthalmia (179), and about one-third of these patients also have pituitary hormone deficiencies (180). Other symptoms, such as developmental delay, intellectual disabilities, and mouth and ear defects, can occur in variable combinations with eye and pituitary defects (180). All reported human OTX2 variants are heterozygous, and 19 of 55 currently reported cases of OTX2 mutations in humans present signs of pituitary disease (Figure 3). Of these, 37% are sporadic mutations, 42% are de novo, 16% are familial dominant with complete penetrance, and only 5% (one case) is familial dominant with incomplete penetrance, although incomplete penetrance is more common in OTX2 variants overall at 22% (53, 109, 113, 179–204). Eye, pituitary, and craniofacial defects are also observed in Otx2-heterozygous mutant mice (35). Otx2-homozygous knockout mice die during midgestation, and there are at least two strain-specific modifiers that alter the penetrance of the heterozygous phenotype in mice (114). Thus, it is likely that genetic and/or environmental factors influence the penetrance of OTX2 mutations in patients.
7. GLI2
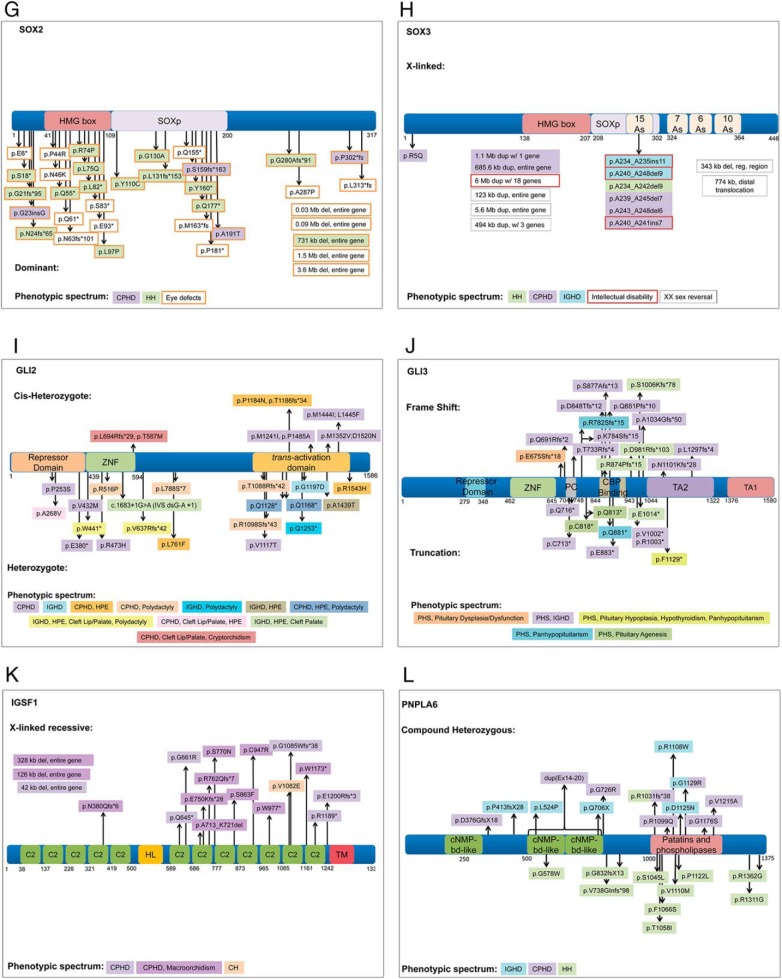
Heterozygous, loss-of-function mutations in GLI2 have been reported in 53 patients with a variety of clinical features with CPHD on the milder end of the spectrum (104, 205–218). There is no conclusive evidence of dominant-negative alleles. Ten of these variants can be classified as pathogenic based on functional studies and segregation in the pedigrees (Figure 3). In many cases, a pathogenic variant is associated with a variable phenotype and/or lack of penetrance in the family. For example, over 20% of the variants have one unaffected parent carrying the allele. GLI2 variants are commonly associated with HPE, cleft palate, CPHD, and polydactyly. One-third of the patients with GLI2 variants exhibit CPHD. Most of these CPHD patients (83%) are nonsyndromic in that they display no features characteristic of HPE, cleft palate, or polydactyly. IGHD (13%), hypopituitarism with unspecified hormone deficiencies (6%), and HH (2%) have also been associated with GLI2 variants, but most of these cases (85%) have some features of HPE, cleft palate, or polydactyly. Combinations of HPE, cleft palate, and polydactyly without pituitary hormone insufficiency represent < 30% of recorded GLI2 variants, with HPE presiding as the predominant phenotype within this group. Interestingly, two GLI2 variants suspected to be causal are found in patients who also have variants in ZIC2 and SHH, two genes that can cause HPE. The combined effect of the two heterozygous genes could be contributing to the disease penetrance in these cases.
8. SOX2
SOX2 is a member of the high-mobility group (HMG) transcription factors related to SRY (sex-determining region Y). Sox2 is important for embryonic development and is expressed in progenitor cells of many tissues including the developing brain, pituitary, and the otic and nasal placodes (219, 220). SOX2 variants are autosomal dominant with incomplete penetrance (Figure 3). In some cases, the unaffected carrier mother has been shown to be a gonadal mosaic (221–223). The clinical features associated with SOX2 mutations include eye disorders (anophthalmia and microphthalmia), brain malformations (usually hippocampal), intellectual disabilities, esophageal atresia, genital abnormalities, sensorineural hearing loss, and hypoplastic anterior pituitary. Pituitary masses have been observed in affected patients, but they do not progress (219). The hypothalamic-pituitary phenotypes caused by variants in SOX2 include HH or CPHD (224–226), but endocrine testing was not performed in most of these individuals (222, 226–231). Most SOX2 variants result in the absence or truncation of the gene affecting the HMG box domain (219, 225, 232–235). The eye and pituitary phenotype variability of SOX2 mutations is demonstrated in a pedigree with a mother and two children who share a heterozygous deletion resulting in a frameshift truncating the SOX2 protein, p.G280Afs91* (236). The mother presented with HH and normal eyes, whereas the two children presented with eye defects, one with clinically normal pituitary function and the other with IGHD.
9. SOX3
SOX3 is another member of the SRY-related HMG transcription factor family. In familial cases of SOX3-related hypothalamic-pituitary phenotypes, CPHD is X-linked with incomplete penetrance. SOX3 variants can cause CPHD (237–241), IGHD (107), or normosmic HH (32) with or without intellectual disability and anterior pituitary hypoplasia. Large duplications of Xq26 that included SOX3 were found in individuals with hypopituitarism (238, 240). Some other cases of X-linked hypopituitarism suspected to be caused by SOX3 dosage effects require further study to identify the precise lesions (242). Most mutations within the SOX3 gene are insertions and deletions in the first polyalanine tract where poly-A expansions are associated with reduced transactivation and poly-A deletions are associated with increased transactivation (32, 107, 237, 239–241) (Figure 3). There is one documented amino acid substitution in SOX3 in a male patient with CPHD (GH, LH, FSH deficiency) without intellectual disability (p.R5Q), which he inherited from his apparently unaffected mother (237). However, there was no difference in transcriptional activity when compared with the wild-type SOX3, so the significance of this variant is not clear. An example of the incomplete penetrance of SOX3 mutations is provided by a heterozygous female with CPHD and deletion of six alanines (237), with an unaffected carrier father. SOX3 regulatory region rearrangements are shown to cause XX male sex reversal in mice and humans, which can be accompanied with low gonadotropin serum levels and intellectual disability (243–246).
B. Less frequently reported genes in CPHD cases
1. BMP4
The 37 documented BMP4 mutations identified in patients are associated with a range of phenotypes that include cleft lip, cleft palate, eye and craniofacial defects, hypospadias, renal hypoplasia or dysplasia, spina bifida, and colon cancer. Most of the mutations are heterozygous (34 of 37), half are familial (17), and the other half are sporadic or de novo. Of the 17 familial mutations, only four are penetrant. The single reported case of a heterozygous BMP4 mutation with a pituitary phenotype (p.R300P) is in a sporadic case with CPHD and a hypoplastic pituitary gland (247). The mutation prediction software suggested that it is pathogenic, and it is positioned in a highly conserved region of the BMP4 gene, but no functional studies were carried out. Although we cannot conclude that this alteration is pathogenic, it is worth screening CPHD patients for lesions in BMP4 because it contributes to the formation of Rathke's pouch in the mouse (248, 249).
2. FGF8
There are 23 mutations reported in FGF8, and most of the patients have KS (HH and anosmia). About half of the cases (11 of 23) are heterozygous and sporadic. The penetrance is unclear in many cases due to the lack of pedigree information. There are eight cases that are heterozygous and familial, but of those mutations, only one is penetrant. Interestingly, three different FGF8 variants are reported to cause hypopituitarism (Supplemental Figure 1). A patient homozygous for a p.R189H variant had low ACTH, TSH, and PRL, along with HPE (103, 250). The parents of the patient are heterozygous and unaffected. The variant is predicted to be pathogenic according to mutation prediction software, and the MAF was 0.077. Based upon the above evidence, it is likely causative. A patient with CPHD and HH is heterozygous for an FGF8 variant that compromised an exonic splicing enhancer site (c.216G>A p.T72T). No family history was reported, but the nucleotide change resulted in higher FGF8 expression and higher activity in cell culture (33). It is predicted to be disease causing and has a MAF of 4.13e−5. We concluded that this variant is pathogenic. Finally, a patient with low GH, thyroid hormone, LH, and FSH, hyposmia, and cleft palate is heterozygous for deletion of a thymine at position 574, which results in protein truncation and likely affects receptor interaction (c.574delT) (251). This variant is likely causative. A heterozygous p.Q216E variant was reported in a patient with low GH and SOD (103, 250). The p.Q216E variant has not been reported in the public population databases, but it is not predicted to be disease causing. Without more evidence, this variant cannot be labeled as causative. FGF8 is expressed in the diencephalon and prospective hypothalamus during development in the human and mouse embryo (103, 250). Loss- and gain-of-function Fgf8 experiments in the mouse highlight the importance of Fgf8 in normal pituitary development. Ectopic expression of Fgf8 in the mouse pituitary gland leads to a dysmorphic and hyperplastic pituitary gland that lacks GH and TSH expression (106), whereas reduced FGF8 expression in an Fgf8 hypomorph causes pituitary hypoplasia (103, 250). The three patients with FGF8 mutations and hypopituitarism provide convincing evidence that FGF8 mutations are a cause of CPHD.
3. FGFR1
FGFR1 is one of four FGF receptors and plays an important role in the development of the nervous system. Human variants in FGFR1 cause multiple diseases, such as Pfeiffer syndrome, Hartsfield syndrome, Jackson-Weiss syndrome, etc. Pathogenic variants in FGFR1 are reported for HH with or without anosmia. There are several reports suggesting the oligogenic effects of FGFR1 and other GnRH deficiency genes, such as HS6ST1 (252) and SEMA3A (253). Heterozygous missense variants in the FGFR1 gene were identified in unrelated SOD and CPHD patients (31, 33, 254). Three variants, p.S450F, p.P483S, and c.336C>T or p.T112T, were identified in SOD probands (33). The p.S450F and p.P483S variants were proved to be loss-of-function mutations by functional studies. The synonymous change c.336C>T was not observed in healthy controls or public datasets. This variant is predicted to generate a new exonic splicing enhancer binding site for the SRp40 splicing factor and/or to disrupt an overlapping putative exonic splicing enhancer octamer. Further investigation is needed to assess the functional effects of p.T112T. The heterozygous variant c.1342C>T, p.R448W was found in two unrelated CPHD patients. It is rare (MAF = 0.0008 in ExAC; not found in EVS and 1000G databases), located in the juxtamembrane domain of FGFR1 protein, and predicted to be damaging by SIFT, PolyPhen-2, and MutationTaster. It has reduced transactivation activity in luciferase assays in transfected cells. Thus, FGFR1 p.R448W is a pathogenic variant. The variant c.320C>T, p.S107L is located in the Ig-like C2-type 1 domain of FGFR1 protein and was found in two unrelated Japanese families. The MAF of p.S107L in the East Asian population is 4%, and both SIFT and PolyPhen-2 predict that p.S107L is a benign change. Therefore, p.S107L is unlikely to be a pathogenic variant. Two unrelated CPHD patients carry p.P772S, but there was no change in the activity of the p.P772S protein, and the variant p.P772S is homozygous in several cases from ExAC and 1000G databases. Thus, the p.P772S is likely to be a benign variant. The p.V102I change is a variant of uncertain significance because all the programs predict it to be benign, and the functional study showed only a mild reduction in protein activity. More information is needed to further evaluate p.V102I. Thus, of four FGFR1 variants reported in CPHD patients, only p.R448W is clearly pathogenic.
4. GLI3
GLI3 is a zinc finger transcription factor and can act as a transcriptional activator or repressor of downstream targets of the SHH pathway. Heterozygous mutations in GLI3 are responsible for most cases of Greig cephalopolysyndactyly syndrome and Pallister Hall syndrome. Greig cephalopolysyndactyly syndrome is associated with abnormal development of the limbs, head, and face and includes polydactyly, macrocephaly, and hypertelorism. Pallister Hall syndrome has some of the same features but also includes hypothalamic hamartoma and bifid epiglottis. Pallister Hall syndrome can also be associated with pituitary phenotypes including IGHD (255–258) and CPHD (256, 259, 260) with pituitary hypoplasia or agenesis (Figure 3). Many of the mutations are de novo, although there are a few examples of familial cases. There is a very strong correlation between the location of the mutation within the gene and the phenotype. Greig cephalopolysyndactyly syndrome is associated with gene deletions and loss-of-function mutations in the amino terminal third of the protein, which affect DNA binding in the zinc finger domain, and in the carboxy-terminal portion, which affects the transactivation domain. Pallister Hall syndrome is associated with mutations within the middle third of the protein that cause truncation before the transactivation domains, rendering the protein a constitutive repressor. In mice, Gli2 and Gli3 have overlapping functions in hypothalamic development (261). Thus, GLI3 mutations may cause pituitary dysfunction that is secondary to hypothalamic defects.
5. IGSF1
The X-linked gene Ig superfamily member 1 (IGSF1) encodes a large, 150-kDa transmembrane protein. It contains 12 C2-type Ig-like loops in two groups of five and seven loops, respectively, separated by a hydrophobic linker (262, 263). It is thought that the nascent protein is cleaved in the hydrophobic linker to produce an N-terminal domain that is retained in the cytoplasm, whereas the larger C-terminal domain, containing a transmembrane protein, is translocated to the plasma membrane (264). IGSF1 is expressed in many different tissues, in particular the testes and pituitary gland (265–268). Accordingly, 18 different human IGSF1 variants have been reported, many with multiple affected family members across several generations, and some have also been found in unrelated families (265, 269–274) (Figure 3). Because the gene is X-linked, most reported patients have been males who are therefore hemizygous for IGSF1, presenting with central hypothyroidism (TSH deficiency) as the primary phenotype, with varying combinations of GH and/or PRL deficiencies and macroorchidism as a common symptom (Figure 3). Female carriers of IGSF1 mutations were initially reported as unaffected, although more extensive studies have now shown that a subset (30–60%) of female carriers can be affected with central hypothyroidism, PRL deficiency, delayed age at menarche, and obesity (273, 275, 276). The gene is overall highly likely to be pathogenic for CPHD, with strong evidence from the human variants, and cell- and animal-based models have recapitulated some, but not all, of the phenotypes observed in the human variants. Igsf1-knockout mice also display central hypothyroidism, with defective pituitary TRH receptor (TRHR) signaling appearing to be the cause, although the mutant mice do not display the variety of other hormone deficiencies and macroorchidism observed in patients with IGSF1 mutations (265).
6. PAX6
PAX6 is a highly conserved member of the paired homeodomain transcription factor family. Haploinsufficiency in PAX6 has long been known to cause aniridia (277, 278). PAX6 mutations provide a classic example of dosage-sensitive effects, in that homozygosity for loss-of-function mutations in mice cause anophthalmia, haploinsufficiency causes micro-ophthalmia, and overdosage also causes micro-ophthalmia (279, 280). During mouse pituitary development, Pax6 is transiently expressed on the dorsal side of Rathke's pouch between embryonic day (e) 9.0 and e12.5, and it is thought to be involved in establishing a sharp boundary between dorsal and ventral cell types (281). But strain background appears to influence the features associated with Pax6 loss of function (282, 283). A heterozygous deletion encompassing two genes and a PAX6 enhancer was identified in a patient with IGHD, cleft palate, and optic disc cupping (200). The patient's clinically unaffected father carried the mutation, but he was mosaic for the deletion. The patient's brother had HPE, but no DNA was available for analysis. A heterozygous variant p.N116S (in the 422 amino acid isoform, or p.N130S in the 436 amino acid isoform) was identified in a Japanese male with CPHD and cryptorchidism, without ocular malformation (200). The mother is also heterozygous for this variant, but she is clinically unaffected. The p.N116S variant is not reported in the public population databases, and it is predicted as damaging by MutationTaster. The p.N116S variant is located within the paired homeodomain of the protein and affects the transactivation function of PAX6, but not the DNA binding ability (200). Over 350 heterozygous mutations in PAX6 have been reported in individuals with ocular malformations, and pituitary hormone deficiency has only been reported for two patients. Therefore, PAX6 mutations rarely cause pituitary abnormalities.
7. PROKR2
Heterozygous and homozygous mutations in the prokineticin receptor 2 (PROKR2) have been reported in patients with HH or KS, and less frequently in CPHD, IGHD, or Hirschsprung's disease. PROKR2 encodes a G protein-coupled receptor (GPR73L1). Upon stimulation by the ligand PROK1, G proteins evoke an intracellular calcium release, and this feature is the basis for testing variant PROKR2 protein function in vitro. Functional links to causality in HH and KS are well established (284–290). Prokr2 is highly expressed in the olfactory placode and GnRH neurons (291), and Prokr2−/− mice recapitulate the hypogonadal phenotype of humans with HH (292–294). Several individuals with HH or KS are heterozygous or compound heterozygous for PROKR2 mutations and frequently carry one or more mutations in other genes mutated in HH or KS (digenic and multigenic cases) such as KAL1, FGFR1, GNRHR, GPR54, KISS1, and NELF (284, 295). Thus, HH and KS are excellent examples of oligogenic disease, and the involvement of PROKR2 is strongly supported for this condition.
A frameshift and nine missense variants in PROKR2 were found in patients with CPHD or IGHD (Supplemental Figure 1). It is not clear how mutations in PROKR2 cause CPHD or IGHD. Prokr2 is expressed in the developing brain and adult posterior pituitary gland, and variants in PROKR2 were proposed to affect neuronal migration and/or development of the posterior lobe, which acts as a signaling center for stimulation of anterior pituitary growth (33, 287, 296). However, in late gestation, Prokr2−/− mice have apparently normal expression of Ghrh, Gh, Prl, Tshb, and Pomc (288). Although Prokr2−/− mice were not examined postnatally, the existence of HH but not CPHD or IGHD suggests that mice do not model the human disorder perfectly. Alternatively, some individuals may have HH due to pathogenic variants in PROKR2 and may be GH deficient due to variants in another gene or genes, resulting in a diagnosis of CPHD. In support of this idea, a patient with PROKR2 p.R85H was carrying a heterozygous variant in HESX1 (287). Additional studies are necessary to resolve this issue for other patients with PROKR2 variants and a diagnosis of CPHD or IGHD.
8. SHH
In 2013, two heterozygous missense variants in the SHH gene were identified in CPHD patients (297, 298). Both variants are rare and exhibit incomplete penetrance. All three software programs predict that p.G427R is benign, and there was no evidence from functional studies to support the pathogenicity. The other variant, p.A226T, was originally reported in a patient with HPE (29). The software prediction programs list it as damaging/disease causing. Thus, “likely pathogenic” is a more accurate classification for p.A226T unless convincing functional studies are carried out.
9. TCF7L1
TCF7L1 gene encodes the transcription factor 7-like 1, which is a member of the T-cell factor (TCF)/lymphoid enhancer factor family. Within the nucleus, TCF/lymphoid enhancer factors interact with β-catenin to activate the expression of target genes (299, 300). The WNT signaling pathway and TCFs regulate proliferation of Rathke's pouch and differentiation of hormone-producing cells in the pituitary (301–306). In a recent study, TCF7L1 expression was reported in the developing forebrain and pituitary gland. Mouse studies showed that it is required in the prospective hypothalamus to maintain normal expression of the hypothalamic signals involved in the induction and subsequent expansion of Rathke's pouch progenitors (307). Two heterozygous variants, p.R92P and p.R400Q, were identified in a screen of SOD patients and normal family members for mutations in TCF7L1 by Sanger sequencing (307). The individual with the p.R92P variant has forebrain defects and normal pituitary function, and he inherited the variant from his unaffected father. The individual with the p.R400Q variant has SOD, absent posterior pituitary, and small anterior pituitary, and he is GH deficient. The variant was inherited from the unaffected mother, and two unaffected siblings also carried the variant. Although the p.R400Q is not completely penetrant, its pathogenicity was supported by: 1) reduced repressive function of TCF7L1 p.R400Q in vitro and in vivo; 2) the rare allele frequency (0.01% in ExAC); and 3) prediction that it is damaging by SIFT, PolyPhen-2, and MutationTaster. Thus, TCF7L1 is a candidate gene for other cases of CPHD with the likely contribution of other genes or environmental factors.
10. TGIF1
TGIF1 encodes a homeodomain protein that represses TGFβ and retinoic acid signaling. TGFβ signaling has been shown in the paracrine communication of thyrotropes and folliculostellate cells in the regulation of TSH secretion (308). Many heterozygous loss-of-function mutations in TGIF are reported for HPE, and there is one report of TGIF1 and CPHD with a central incisor, pituitary stalk disruption, and anterior lobe hypoplasia (p.Q267X), but no functional studies were carried out (297). Tgif1 and Tgif2 are expressed in the developing pituitary gland at e14.5 (125). The methods for testing TGIF1 function are well established (297, 309), and the nonsense mutation seems likely to be a loss-of-function allele. Mice homozygous for a Tgif null mutation that deletes the entire gene are viable on mixed genetic backgrounds, but on a more pure C57BL/6J background, a proportion of the Tgif1 null animals die perinatally (310). A different genetically engineered Tgif1 mutation deleted exon 3 and is predicted to produce a truncated 80-amino acid protein (311). Mice heterozygous for this mutation do exhibit HPE, microcephaly, and exencephaly with high penetrance on the C57BL/6 background but are protected on the 129 strain background. Affected animals have altered expression of both Pax6 and Shh. Mouse studies have demonstrated that Tgif1 and Tgif2 have overlapping function at gastrulation that is required for development of the anterior visceral endoderm and for normal Otx2 expression (312). Although additional evidence is needed to support TGIF1 as a cause of CPHD, the mouse studies strongly support the idea that it could have a role.
11. CHD7
Heterozygous loss of function in the chromodomain helicase DNA-binding protein 7 gene (CHD7) is the major causative factor for CHARGE syndrome (313, 314). Hormone defects include hypogonadism, growth failure with or without GH deficiency, and hypothyroidism (315). A CHARGE patient treated with GH exhibited improved growth rate and achieved target height (316). Two unrelated CHARGE syndrome children carrying rare variants in the CHD7 gene were reported to have congenital hypopituitarism due to structural abnormalities of the pituitary gland (317). However, functional studies on these two variants, p.P732A and c.IVS35+6T>C, have not been done. The missense variant p.P732A is rare, but both SIFT and PolyPhen-2 predict it as benign. CHD7 is expressed in the pituitary and the hypothalamus (318). Mice heterozygous for a Chd7 gene trap allele have HH due to decreased GnRH neurogenesis, and although they have growth insufficiency, GH and IGF-1 are normal (319). Additional studies are necessary to understand the growth insufficiency of individuals with CHD7 mutations.
C. Genes implicated in CPHD that require additional evidence about causality
1. HHIP
Several members of the hedgehog signaling pathway have been implicated in HPE and abnormal pituitary function in humans and mice (218, 320). Because of this, a candidate gene mutation screening for HHIP (hedgehog-interacting protein) variants was carried out for 93 CPHD patients, and one patient was identified as heterozygous for a de novo c.-1G>C variant in the 5′-untranslated region (298). Because the −1 position of the Kozak consensus is 28% G and 45% C in humans, this change seems unlikely to affect translation (321). Consistent with this, cell culture studies did not show a significant functional effect. The patient was born preterm at 33 weeks in a breech delivery, with asphyxia requiring resuscitation and neonatal jaundice. She needed thyroid and glucocorticoid hormone replacement as a neonate and was still prepubertal at 13 years. It is difficult to rule out perinatal hypoxia as a contributor to her CPHD (322). Although HHIP is an excellent candidate gene for CPHD based on the known role of SHH signaling, the c.-1G>C variant appears benign. Mice heterozygous for a null allele of Hhip are unaffected, and homozygotes die at birth due to respiratory distress arising from developmental defects of the lung (323). They have patterning defects in multiple organs including the CNS and the pancreas, where endocrine cell proliferation is reduced (324, 325).
2. POLR3A
Polymerase RNA 3 DNA directed polypeptide A (POLR3A) encodes a subunit of RNA polymerase 3. It is implicated in 4H syndrome (hypomyelination, hypogonadotropic hypogonadism, and hypodontia), one of the POL3-related leukodystrophies (326). There is one documented patient with 4H and late onset GH deficiency, a compound heterozygous variant (p.R1005H, p.A1331T) with recessive inheritance (327). The p.R1005H variant (MAF = 0.00019) was predicted deleterious using SIFT but benign using PolyPhen-2. The p.A1331T variant (MAF = 8.24e−6) was predicted deleterious with both SIFT and PolyPhen-2. Approximately 50% of POLR3A 4H patients have short stature, but to date, there are no reports of endocrine evaluation to support GH deficiency or hypothyroidism as the cause of short stature in these patients (326).
3. RBM28
RNA binding motif protein 28 is involved with RNA processing, being a component of the spliceosomal small nuclear ribonucleoprotein complexes (328, 329). RBM28 has been implicated in the development of alopecia, neurological defects, and endocrinopathy (ANE) syndrome. Individuals with mutations in this gene present with variable hair loss, intellectual disability, central adrenal insufficiency, and tooth abnormalities. There is one documented variant that is associated with recessive hypopituitarism: five siblings had a homozygous mutation, p.L351P (MAF = 6.59e−5), predicted to be deleterious using SIFT and PolyPhen-2. All five patients had CPHD and ANE syndrome (329, 330). The hormone deficiencies varied: all of the siblings had GH, LH, and FSH deficiencies, but only two had ACTH deficiencies, and one was PRL deficient. One of the five siblings also had anterior pituitary hypoplasia. Rbm28 is expressed in the developing mouse pituitary gland at e12.5 and e14.5 (125). Although RBM28 is an excellent candidate, the mechanism by which RBM28 leads to CPHD remains to be elucidated.
4. WDR11
Mutations in the WD40 repeat containing protein WDR11 have been implicated in autosomal dominant HH and KS, and occasionally with craniofacial abnormalities including cleft palate, sensorineural hearing loss, and brain and dental abnormalities and WDR11 is expressed in the developing hypothalamus and pituitary (125, 331). Six unrelated patients with HH or KS contained sporadic heterozygous variations in the WDR11 gene (331, 332). Four of the missense variants (p.A435T, p.H690Q, p.R448.5q, and p.R395W) were located in the EMX1 binding region, which has overlapping function with OTX2. Coimmunoprecipitation of EMX1 and WDR11 with these variants determined that p.A435T (MAF = 0.00047) and p.H690Q (MAF = 6.49e−5) completely disrupted EMX1 binding, p.R448.5q (MAF = 0.00045) reduced EMX1 binding, and p.R395W (MAF = 0.001) had no effect (331). These results do not align with SIFT and PolyPhen-2 predictions because p.R395W was predicted to be tolerated and possibly damaging by SIFT and PolyPhen-2, respectively. Variants p.A435T, p.R448.5q, and p.F1150L (MAF = 0.001) were predicted to be tolerated and benign, whereas p.H690Q was predicted to be deleterious and probably damaging by SIFT and PolyPhen-2. However, the researchers found that the three variants that altered EMX1 binding were predicted by SPPIDER (http://sppider.cchmc.org/) (333) to alter protein-protein interactions. Given the expression pattern, association with OTX2 and HH, WDR11 may be a candidate gene for CPHD.
5. IFT172
The intraflagellar transport (IFT) 172 gene encodes a subunit of the B subcomplex of IFT. The IFT complex is important for ciliary assembly and maintenance and plays a role in hedgehog signaling (334). A patient with pituitary hypoplasia, ectopic posterior lobe, growth retardation, facial abnormalities, intellectual delay, skeletal abnormalities, and degeneration of kidney and eye was found to be a compound heterozygote for mutations in IFT172: p.Cys1727Arg and a novel splice site mutation in intron 4, c.337–2AC (335). He had a normal GH stimulation test but persistently low IGF-1 and a positive response to recombinant human GH therapy. His features are similar to patients with Mainzer-Saldino syndrome, which can be caused by mutations in IFT140 or IFT172. These variants are predicted to be deleterious, but no functional studies were done. Although this patient did not have CPHD, his case suggests that IFT172 could be another candidate gene for CPHD, and it would likely be associated with other developmental anomalies. Mice homozygous for null allele die during gestation with multiple anomalies including neural tube defects, VACTERL-like features, and homeotic transformations of the skeleton (336, 337). A conditional allele exists and could be used to study the effects of IFT172 on hypothalamic and pituitary development (338).
D. CPHD candidate genes discovered by WES
1. ARNT2
The ARNT2 gene encodes a basic helix-loop-helix transcription factor: the aryl hydrocarbon receptor nuclear translocator. The only ARNT2 mutation described to date is the c.1372_1373dupTC mutation, which occurred in a family of six generations with consanguineous marriages (123). Because of the consanguinity, autosomal recessive inheritance was predicted, and single nucleotide polymorphism (SNP) analysis was carried out on four affected individuals to identify regions of homozygosity. WES was carried out on one affected individual, and the segregation of the mutation was confirmed in the pedigree. The clinical presentation included CPHD, brain, eye, kidney, and urinary tract abnormalities in all individuals homozygous for the c.1372_1373dupTC mutation, and none of the heterozygotes. Two SNPs in the promoter region of ARNT2 affect transcription, but their significance in human disease is not known (339). Arnt2 has a broad expression pattern in the mouse embryo, including the CNS, spinal cord, hypothalamus, pituitary, eye, lung, and kidney, which correlates with the embryonic human expression (340). The patient phenotype is well connected to the developmental expression pattern. Mice homozygous for a null allele of Arnt2 die shortly after birth of unknown cause, and they lack hypothalamic secretory neurons including vasopressin, oxytocin, CRH, TRH, and somatostatin (341, 342). This phenotype is similar to that of mice with a knockout of Sim1, which encodes a related protein that is known to dimerize with ARNT2, providing further support for the role of ARNT2 in CPHD (343, 344).
2. ZSWIM6
ZSWIM6 is a transcription factor containing a zinc finger domain and a SWIM domain, which are important in DNA binding and protein-protein interactions, respectively. A heterozygous mutation in the sin-3 like domain of ZSWIM6 (p.R1163W) has been found in eight unrelated individuals with acromelic frontonasal dysostosis (345, 346). This disorder is characterized by severe frontonasal dysplasia, neurocognitive and motor delays, and preaxial polydactyly. Other associated variable features include hypopituitarism, absent olfactory bulbs, cryptorchidism, hypoplastic corpus callosum, and patellar hypoplasia. There was no detailed endocrine analysis of hypothalamic-pituitary function. Exome sequencing was carried out on two unrelated affected individuals and a trio with a mildly affected father, unaffected mother and affected child (345). A dominant mode of inheritance was initially considered, but no likely candidate variants were shared by the father and the affected child, and no variants were detected in unrelated affected individuals. An apparent de novo variant was found in the affected child from the trio, and the same de novo mutation was found in the two unrelated individuals that were sequenced. An unrelated adoptee carried the same mutation and may have been mosaic. Mosaicism could explain the mildly affected father as well, although the mutation was not detected in his DNA sample. Recently, four additional examples of unrelated individuals with acromelic frontonasal dysostosis were found to have the same p.R1163W variant (346). MutationTaster and in silico structural and atomic-resolution modeling predict that the p.R1163W variant is pathogenic, and primary osteoblast cell lines from patients, but not controls, had alteration in hedgehog signaling transcripts (345). This variant has not been identified in the normal population, but heterozygous deletions of ZSWIM6 are found in unaffected individuals, suggesting that the p.R1163W change is a gain-of-function mutation.
3. GPR161
G protein-coupled receptor 161 (GPR161) gene is located on 1q24.2. Activation of the receptor elevates cAMP, which leads to proteolytic conversion of Gli transcription factors (GLI2 and GLI3) into Gli repressor forms and inhibition of SHH signaling (347). There is a single report of CPHD caused by a homozygous missense GPR161 mutation c.56T>A; p.L19Q in a consanguineous family. WES was carried out on two affected individuals and an unaffected sibling, and variants were filtered considering autosomal recessive inheritance. The parents were confirmed to be heterozygous for the p.L19Q variant, and segregation in the pedigree was consistent with autosomal recessive inheritance. The two affected individuals both have pituitary stalk interruption syndrome, other pituitary radiographic abnormalities, and GH deficiency. One sister has IGHD, but the other also has diabetes insipidus and central hypothyroidism. PolyPhen-2 predicts that the p.L19Q variant, in a highly conserved residue, is probably damaging. Structure prediction programs suggest that the variant is an extracellular loop that could affect ligand binding, but the ligand is not known, and SHH signaling is difficult to assess in fibroblasts. Gpr161 is expressed in the developing mouse pituitary gland, but detailed expression studies have not been carried out (125). Without further information, it is difficult to be certain that GPR161 p.L19Q causes hypopituitarism because: 1) this variant has a MAF of 2% among Asians and 3% overall, and there are seven homozygous individuals; 2) there is no functional study to prove that the p.L19Q variant is deleterious; and 3) we would expect a variant in GPR161 to be gain of function in order to enhance the suppression of SHH signaling, which seems unlikely for a recessive inheritance pattern. However, GPR161 also regulates WNT and retinoic acid signaling, which are important for normal pituitary development, and the effects of GPR161 p.L19Q on these signaling pathways was not assessed (348–350). Identification of additional individuals with CPHD or IGHD and variants in GPR161 could enhance the confidence that this gene has a role in pituitary development and function.
Studies in mice reveal that Gpr161 is important for development of multiple organs, and genetic background can have a major effect on the penetrance of the phenotype (348). Homozygotes for a hypomorphic (reduced function) C-terminal truncation allele are generally not viable and have extensive craniofacial abnormalities, spina bifida, and limb defects. On certain genetic backgrounds the mutants are viable and have a belly spot and vacuolated lens. SHH signaling is not affected in these mutants, but retinoic acid and WNT signaling are, and retinoic acid injection can rescue the neural tube defects. Homozygotes for a null allele have fully penetrant lethality, craniorachischisis, and craniofacial abnormalities (351). These data suggest that if GPR161 p.L19Q is causal, the allele may be hypomorphic and influence retinoic acid signaling.
4. HNRNPU
HNRNPU (heterogeneous nuclear ribonucleoprotein U) is a member of the group of proteins that form complexes with nascent pre-mRNA transcripts in the nucleus of the cell. It is implicated in CNS abnormalities, with patients displaying learning disabilities and seizures. Some individuals are diagnosed with Lennox-Gastaut syndrome, which is characterized by epilepsy and occasionally impaired intellectual development (352). HNRNPU was first implicated as a causal gene for CPHD when found in a patient during WES of 119 trios with undiagnosed disorders who were referred for genetic testing (353). A splice site variant, c.1615 –1 G>A, was found in a child with CPHD and epilepsy. It is predicted to be deleterious using SIFT and probably damaging by PolyPhen-2. HNRNPU interacts with PITX2 and may be involved in the regulation of heterogeneous nuclear RNA stability (354). HNRNPU can also inhibit GNRH transcription (355, 356). Functional studies are needed to assess the effects of this variant on HNRNPU function. Mouse HNRNPU differs from human by only a single amino acid. Mice homozygous for a hypomorphic mutation in Hnrnpu exhibit early embryonic lethality at e9.5, growth retardation, and defects of the allantois (357). Mice homozygous for selective deletion of Hnrnpu have lethal cardiomyopathy and extensive defects in splicing mRNAs that are important for heart development (358).
5. PNPLA6
PNPLA6 encodes the patatin-like phospholipase domain containing protein 6, which is responsible for the de-esterification of membrane phosphatidylcholine into fatty acids and glycerophosphocholine. The first PNPLA6 mutation implicated in a human disease was discovered in 2008 in a linkage study focused on a progressive spastic paraplegia and a distal muscle-wasting phenotype (359). Since then, autosomal recessive, compound heterozygous mutations have been found in individuals with a variety of neurodegenerative conditions including childhood blindness (360), Leber congenital amaurosis (360), Oliver MacFarlane syndrome, Laurence-Moon syndrome, Gordon Holmes syndrome, and Boucher-Neuhäuser syndrome (361). The distinction between the syndromes may be artificial, arising from variable clinical presentations. Individuals with Oliver MacFarlane or Laurence-Moon syndrome have congenital hypopituitarism including GH, TSH, and gonadotropin deficiency and chorioretinal atrophy. Craniofacial dysmorphology, ataxia, and paraplegia may also be evident. Boucher-Neuhäuser and Gordon Holmes syndromes have cerebellar ataxia and HH, but chorioretinal dystrophy is present only in the former (361, 362). Childhood blindness results from widespread photoreceptor neuron death (360). Homozygous and compound heterozygous mutations affecting the patatin-like phospholipase domain are fully penetrant. The mechanism underlying pituitary dysfunction may be impaired vesicular transport because PNPLA6 mutant cells exhibit reduced secretion in response to stimulation (363). PNPLA6 is expressed in the developing pituitary gland, and loss-of-function mutations have been identified in six unrelated individuals with pituitary dysfunction. Thus, PNPLA6 should be considered in cases of CPHD or HH with features characteristic of PNPLA6 associated syndromes. In mice, conditional inactivation of Pnpla6 in the CNS causes neurodegeneration (364). However, fetuses homozygous for a null allele exhibit failed placental development, impaired blood vessel development, growth retardation by e7.5, and death by e9 (365). These results suggest that the human mutations in PNPLA6 may be partial loss-of-function alleles.
6. CDON
CDON (cell adhesion associated, oncogene regulated) is a cell surface SHH-binding protein that promotes SHH signaling activity (366). Heterozygous loss-of-function mutations in CDON have been reported in HPE patients, including some with GH deficiency and absent pituitary (p.P689A and p.T790A, respectively) (102). Cdon−/− mice recapitulate the range of HPE phenotypes from unaffected or mildly affected to severe, depending on genetic background (367). Interestingly, maternal ethanol exposure synergizes dramatically with Cdon loss of function to produce highly penetrant HPE (36). On a 129S6 genetic background, Cdon−/− mice are unaffected, but if exposed to ethanol, 75% have HPE, providing strong evidence for gene-environment interaction. Cell-based assays using Cdon−/− mouse embryonic fibroblasts showed that the p.P689A variant is defective in conferring SHH-dependent induction of Ptch1 and Gli1, but the p.T790A was not tested. By WES, a novel nonsense variant p.E922* was recently identified in a patient with pituitary stalk interruption syndrome and panhypopituitarism (368). The mother also carried this variant, and she had convergent strabismus that required surgical correction but normal height. This variant has never been reported in the public population database. The mutation of amino acid 922 to a stop codon will result in a truncated CDON protein without the transmembrane and intracellular domain, which is expected to destroy function. Taken together, the GH-deficient HPE patient with p.689A, the p.E922* CPHD patient, and the studies in genetically engineered mice suggest that CDON is a candidate gene for other cases of CPHD.
E. Summary
We assessed the pathogenicity of reported variants in CPHD genes according to the current standards in the field. It is not surprising that most of the criteria were met by most of the earlier report and more established genes. More analysis is required to prove causality of some of the most recently reported candidates. Overall, WES has identified six new candidate genes for CPHD. To date, only CDON, ZSWIM6, and PNPLA6 mutations have been found in multiple unrelated individuals. For disorders that are highly heterogeneous like CPHD, it can be helpful to take advantage of the online platform MatchMaker Exchange (http://www.matchmakerexchange.org/) to facilitate identifying cases with similar phenotypic and genotypic profiles among different research groups. In addition, functional studies are critical for testing the pathogenicity of the detected variants both in vitro and in vivo.
IV. Strategies to Improve Diagnostic Yield for CPHD and Future Directions
NGS tools have identified genetic variants responsible for many human diseases, and in some cases the molecular diagnosis has effectively guided physicians to effective therapeutic intervention (369). We expect that rigorous application of these methods will result in better molecular diagnosis of CPHD and provide new insight into pituitary organogenesis. However, the diagnostic yield for WES in a research setting is currently between 10 and 30% (370). The potential reasons for this and some strategies to improve the discovery of new genes and variants are discussed below.
A. Incomplete coverage of WES
WES is designed to identify coding variants and splice variants at intron-exon borders in all known genes, but in practice, the whole exome is not assayed because some areas of the genome are difficult to capture and/or sequence. For example, SOX3 is 75% guanine and cytosine GC, and more than half of the SOX3 coding region has < 20 fold coverage in the ExAC database (http://exac.broadinstitute.org/gene/ENSG00000134595). We also experienced gaps in covering this gene in our WES analysis of CPHD patients (unpublished data). To avoid a false-negative result, such problematic areas should be screened by Sanger sequencing with modifications (371, 372), but this can be tedious. Improvements in high throughput capture and sequencing are emerging and are likely to continue to evolve, resulting in improved diagnostic yield (373). Some coverage gaps can be avoided using exome sequencing methods targeted to specific collections of candidate genes (Figure 4). This enriches the capture of a region of interest, reducing the likelihood of incomplete coverage. Targeted candidate gene sequencing methods can be applied to many samples at a time, providing an economical way to obtain deep coverage of genes in the panel (374). This can be a useful first step in determining causes of disease within a large sample set. Although the targeted gene panels only provide information about the chosen genes or regions, they provide an economical, high throughput way to screen large sample sets for candidate genes at greater read depth than would be obtained with WES.
Figure 4.
Molecular inversion probe (MIP) capture. A, MIP probe panels are synthesized in parallel on a microarray. B, In a single-well reaction, hundreds to thousands of exons are targeted in parallel by capture probes. As each probe anneals, the target is enzymatically copied and ligated to the probe backbone to yield a sequencing library. C, Example of MIPS sequencing data at GHRHR showing coverage for two samples, including a heterozygous missense SNV.
B. Genetic variants that are difficult to detect with WES (structural variants, splicing region, enhancers, and noncoding variants)
Rearrangements and copy number variations (CNVs), including insertions and deletions (indels) and duplications, are often missed by WES. Chromosomal microarray (SNP array) is a first-tier screen for genetic defects caused by CNVs and is recommended as a complement to exome sequencing (375). SNP arrays cover a wide area of the genome, including intergenic and nongenic regions. Increasingly dense arrays, up to several million probes, offer high-resolution mapping of CNVs. SNP arrays typically detect a pathogenic CNV in 20% of the cases (376). WGS is another approach to identify lesions missed by WES, and as the cost of WGS drops, we expect it to replace WES. In many cases, the bioinformatics analysis must be specifically directed at examining mismatched paired-end reads to detect structural variation (377–379). If cytogenetic abnormalities or CNVs are known to be present from karyotype or chromosomal microarray analysis, jumping libraries can be carried out as an alternative to WGS to identify precise, gene-level information about the genetic lesion (380, 381).
Splice enhancers are sequences that enhance the proper usage of weak splice sites by serine-arginine splice regulators. Variations in exonic splice enhancers that do not change the encoded amino acid are an important cause of disease that is missed by most software prediction programs (382). Indeed, silent mutations in the exonic splice enhancers of the GH gene are an important contribution to autosomal dominant IGHD, which can progress to cause CPHD (383). Skipping exon 3 results in production of the 17.5-kDa GH isoform that blocks secretion of the 22-kDa biologically active form. Pharmaceuticals that improve the use of weaker splice sites are being explored as a treatment for this form of IGHD, and for prevention of the progression to CPHD (384). Improvements in software may make it easier to predict exonic splice enhancers and identify silent, pathogenic mutations.
Variation in regulatory elements is likely to be an important cause of disease (385), and 88% of the SNPs identified in GWAS are not in the exome (386). Taken together, these observations suggest that identifying regulatory regions of the genome (regulome) is an important area of future investigation. Many of the critical regulatory elements are species, developmental, and/or cell type specific. Analysis of the pituitary or hypothalamic regulome has not yet been included in the ENCODE project. Thus, more information is needed about the location of hypothalamic-pituitary regulatory elements for effective utilization of CNV and WGS information. We have recently identified the binding sites for PROP1 in rodent cell lines, which is an important step toward defining the pituitary regulome (129). A concerted effort to identify regulatory sites in the developing hypothalamus and pituitary gland would be valuable information as the field moves toward WGS analysis of CPHD.
C. Large number of tolerated variants: filtering by expression profiles and pathways
A challenge in interpreting WES data is the large number of tolerated, rare variants, which makes it difficult to filter the genes and nominate the appropriate candidate for functional studies. Often, WES is applied to trios including the unaffected parents and a single affected child. If the disorder is caused by a recessive mutation or de novo mutation, the list of candidate genes will usually be manageably short. If the disorder is caused by a dominant mutation with incomplete penetrance, then the list of candidate genes will be much longer, and it could be intractable. If there are multiple affected individuals in a pedigree, it is advisable to apply WES to the most distantly related affected individuals because they will have fewer rare variants in common that require filtering.
We reviewed the gene expression patterns of genes that are known to cause CPHD to determine whether gene expression profiles would be an appropriate way to prioritize candidate genes. Gene expression data from BioGPS (http://biogps.org) (387, 388) and MGI-GXD (Mouse Genome Informatics, http://www.informatics.jax.org; Gene Expression Database [GXD]) (389) were used to determine the enrichment of the 30 CPHD candidate genes in mouse and human hypothalamus and pituitary at different development stages. We considered gene expression enriched if its expression in the pituitary or hypothalamus was greater than three times the median in BioGPS or was present in the indicated tissue in MGI-GXD. For many genes, numerous embryonic and fetal developmental ages have been examined in the mouse hypothalamus and pituitary. Such extensive human gene expression analysis has yet to be performed. Human 16-week fetal pituitary expression and hypothalamic and pituitary expression for specific genes at various Carnegie stages were used to determine the developmental human gene expression for the 30 candidate genes (103, 123, 218, 232, 307, 362, 390, 391).
We found that the known genes have high brain and pituitary specificity for expression, and notably, expression is skewed to embryonic, but not adult stages (Table 3). Based on the expression patterns of these known genes, future candidate genes for CPHD causality should include genes that affect early head development, including anterior structures at gastrulation, craniofacial placode development, and the organogenesis of the hypothalamus and pituitary gland (21, 392–394). Expression profiling has been carried out in developing mouse pituitary gland at e12.5 and e14.5 (125), and in migrating neural crest and craniofacial placodes (e8.5 to e10.5) (395, 396). Numerous fetal-specific, novel expressed sequence tags were identified by expression analysis of fetal (male, 16 weeks) and adult human pituitary glands (391). A more comprehensive database on head, hypothalamic, and pituitary gene expression with spatiotemporal information would be valuable for advancing CPHD research.
Table 3.
CPHD Gene Expression
| Gene | Fetala |
Adult |
||||||
|---|---|---|---|---|---|---|---|---|
| Mouse |
Human |
Mouse |
Human |
|||||
| Hypothalamus | Pituitary | Hypothalamus | Pituitary | Hypothalamus | Pituitary | Hypothalamus | Pituitary | |
| POU1F1 | − | x | ? | x | − | x | − | x |
| PROP1 | − | x | ? | − | − | − | − | − |
| HESX1 | − | x | x | x | − | − | − | − |
| LHX3 | x | x | ? | x | − | x | − | x |
| LHX4 | x | x | ? | − | − | − | − | − |
| OTX2 | x | x | ? | − | − | − | − | − |
| GLI2 | x | x | x | x | − | x | − | − |
| SOX2 | x | x | x | x | x | x | − | − |
| SOX3 | x | − | ? | − | − | − | − | − |
| FGF8 | x | x | x | − | − | − | − | − |
| FGFR1 | x | x | x | x | − | − | − | − |
| GLI3 | x | − | ? | − | − | − | − | − |
| PAX6 | x | x | ? | − | − | − | − | − |
| ARNT2 | x | x | x | − | x | − | x | − |
| BMP4 | x | x | x | − | − | − | − | − |
| IGSF1 | x | x | ? | x | x | x | x | x |
| PNPLA6 | x | x | ? | x | − | − | − | − |
| SHH | x | x | ? | − | − | − | − | − |
| TCF7L1 | x | x | x | x | − | − | − | − |
| ZSWIM6 | x | x | ? | − | − | − | − | − |
| CHD7 | x | x | ? | − | − | x | − | − |
| PROKR2 | x | ? | ? | − | x | − | − | − |
| TGIF1 | x | x | ? | − | − | − | − | − |
| CDON | x | ? | ? | − | − | − | − | − |
| GPR161 | ? | x | ? | − | x | − | − | − |
| HHIP | ? | x | ? | − | − | − | − | − |
| HNRNPU | ? | ? | ? | x | − | − | − | − |
| POLR3A | − | − | ? | − | − | − | x | x |
| RBM28 | x | x | ? | − | − | − | − | − |
| WDR11 | ? | x | ? | − | − | − | − | − |
For many genes, numerous embryonic and fetal developmental ages have been examined in mouse. For human genes, information is available for 16-week gestation in Ref. 391. “x” menas enriched expression, “−” means the expression is not enriched, and “?” means unknown.
Identifying genes in the CPHD interactome could be useful in filtering candidate genes for functional studies (397, 398). For example, several genes that encode members of the SHH signaling pathway are implicated in CPHD, and as more is learned about the mechanism of action of other known CPHD genes, pathway analysis could assist in prioritizing candidate genes. Knowledge of protein interactions with other proteins, DNA, RNA, and metabolites will also be valuable in understanding the mechanisms of disease.
D. Functional testing: genetic editing and animal models of human disease
As an increasing number of rare variants are documented in both diseased and healthy populations, understanding which genetic changes are benign and which increase disease risk is the new frontier of translational research. Some of the most common functional characterizations involve cell transfection studies, in which the mutated gene of interest is transfected into a population of cells, and the function, expression, and stability of the mutated protein is characterized. Although imperfect, these assays can be scaled up to test many variants. Transfection studies can yield false negatives. For example, a transcription factor variant that does not influence DNA binding or transactivation of a consensus reporter gene in transient transfection may still have a negative effect on protein-protein interactions that are necessary for function and/or specificity in intact animals because these interactions and chromatin remodeling properties are not assayed in the cell culture system. For example, OTX2 interactions with LHX1, FOXA2, and LDB1 are important for its function (399–401). Protein interactions critical for LIM homeodomain proteins and POU1F1 are emerging, but many critical interactions are still unknown (142, 402).
Animal models are the “gold standard” for functional studies and can give invaluable insight into the mechanism of action. For example, mutations in OTX2 fail to activate transcription of POU1F1 and several other promoters in transfected cells, which clearly documented the deleterious effect of the variants on function. However, the mechanism of action does not involve regulation of Pou1f1 because deletion of Otx2 in the anterior pituitary has no effect, and the primary region of Otx2 expression is in the ventral diencephalon (115). Disruption of Otx2 in early head development and in the developing hypothalamus affects FGF signaling, which secondarily affects anterior pituitary growth. Another advantage of animal models is that they provide an opportunity to standardize the genetic background, which makes it easier to determine whether specific mutations are loss-of-function or hypomorphic alleles (390).
The time and expense associated with generating mouse models of human disease has led to the implementation of zebrafish as a high throughput, economical method of assessing function of human genetic variants. This approach is particularly relevant for CPHD studies. Pituitary development is conserved among vertebrates, and many of the genes that are critical for mammalian hypothalamic pituitary development have orthologs that function the same way in zebrafish (307, 403, 404). Zebrafish were used successfully to test OTX2 mutations discovered in patients with the otocephaly-dysgnathia complex, and prrx1, pgap1, and msx1 knockdown enhanced the severity of the phenotype (192). The use of morpholinos to disrupt expression has been complicated by off-target effects and nonreproducible results, however (405). Genome-editing technology, using CRISPR/Cas9, is an alternative to morpholino studies in fish and has also facilitated generating mouse models of disease.
CRISPR is an endonuclease that can be targeted to a region of the genome in a highly specific manner. Briefly, CRISPR is transfected into a one-cell embryo with a single-guide RNA, targeting the endonuclease to the gene of interest, and the digestion and repair process disrupts it (406). In the presence of a donor DNA plasmid containing regions of homology to the cut region, knock-ins and floxed alleles are also possible (407–410). Technological improvements have increased specificity and efficiency and reduced off-target effects (411). This technology will allow us to understand how genes function in a physiological context in a variety of model organisms because it obviates the need for stem cells; all of the necessary materials can be injected into fertilized eggs or cells. Gene editing may have value for treating some genetic disorders (412), in addition to its role in generating animal models of disease.
E. Multifactorial disease
Systematic genetic screening for CPHD has only been reported for a few genes, and it is likely that there are additional genes to discover because approximately 84% of the cases are undiagnosed (46). The work summarized here makes it is clear that CPHD is a genetically complex disorder involving 30 or more genes. In addition, the responsible genes, range of syndromic features, and common occurrence of incomplete penetrance associated with CPHD suggest that it is an oligogenic disease and part of a spectrum disorder that includes craniofacial abnormalities. CPHD has genetic and phenotypic overlap with SOD, HPE, and HH, and there is evidence for multifactorial genetic contributions to these disorders (33, 413–415).
Our experience in screening a collection of sporadic and familial CPHD patients for mutations by WES is consistent with this hypothesis (unpublished results). In our screen of 69 exomes from 25 unrelated families, we were able to make a clear molecular diagnosis for only three of the probands. We found mutations in HESX1 (416) and GHRHR (unpublished data). We did not identify another major, simple Mendelian gene like PROP1, which accounts for approximately 11% of cases worldwide (46). We did identify rare heterozygous variants that are predicted to be deleterious in six genes that have been implicated as PROP1 targets (129) or in HH, HPE, and SOD (unpublished data). Six probands have heterozygous, rare, likely deleterious variants in two or more genes, suggesting digenic or oligogenic disease. These results suggest that CPHD, like HH, can arise from compounding effects of multiple genes and requires analysis of genetic load in cases and controls (353).
Given the large sample sizes that are necessary to identify modifier genes, it is possible that mice could be used to identify loci and genetic pathways that enhance and suppress incompletely penetrant CPHD phenotypes (92, 417). For example, two loci have been mapped that modify the craniofacial features of Otx2+/− mice, but the underlying genes have not been identified yet (114).
V. Conclusions
Over the past 24 years, 30 CPHD genes have been identified, which has resulted in a remarkable improvement in our basic understanding of the development of the hypothalamic-pituitary axis, the cellular and molecular regulation of hormone production and secretion, and the genetic basis for endocrine disorders (see Main Points, Box 2). The emergence of cheaper and efficient NGS technologies promises to advance both basic and clinical CPHD research. The benefits will extend beyond CPHD because many of the genes identified also affect craniofacial development, which is not uncommon and can have serious consequences for quality of life. As we enter the era of genomic medicine, we anticipate faster and more accessible genetic testing, enormous datasets, and a more complicated decision-making process to define the genetic causes for human diseases. We envision that the current NGS technologies will continue to improve in accuracy and diagnostic yield. The focus of this review has been to assess the candidates of CPHD at both the gene and variant levels to ensure that reporting of “causal” genetic defects has sufficient validation and supporting evidence, with the hope that this will be instructive for genetic analyses of other endocrine system disorders. We expect this review will provide a foundation for endocrinologists and other clinicians as they expand their work into precision medicine. Finally, we encourage collaborations and data sharing among the clinical genetic-testing laboratories and academic research groups in order to empower the identification of genes underlying rare genetic endocrine disorders.
Box 2. Main Points.
Pituitary gland and hypothalamic development are dependent upon the expression of critical transcription factors and cell-cell signaling.
Mutations in these transcription factor and signaling pathway genes are responsible for some cases of pituitary hormone deficiency.
Thirty genes have been implicated in CPHD, but 84–90% of the cases still have unknown etiology.
The most commonly mutated gene in patients with combined hormone deficiency is the pituitary transcription factor PROP1, which accounts for 11% of cases.
The clinical presentation can vary among patients with the same mutation because of incomplete penetrance, variable expressivity, and/or the involvement of other genetic or environmental factors.
The genetics of CPHD place this phenotype in spectrum of disorders ranging from craniofacial defects such as holoprosencephaly to IGHD.
Advanced genome sequencing technology and bioinformatics hold promise for a better understanding of the genetic causes of this disorder.
Functional studies are necessary to assess the pathogenicity of novel genetic variants.
Acknowledgments
We thank our local and international collaborators, including Drs. I. Arnhold, L. Carvalho, F. Correa, and B. Mendonça at the University of São Paulo, Brazil; Drs. F. Castinetti, R. Reynaud, and T. Brue at the University of Marseille, France; Dr. M. Dattani at University College London, United Kingdom; Drs. I. Bergada, D. Braslavsky, and A. Keselman from Centro de Investigaciones Endocrinológicas, C. Bergadá, Consejo Nacional de Investigaciones Científicas y Técnicas, Division of Endocrinology, Buenos Aires, Argentina; and Dr. Gutiérrez, Children's Hospital, Buenos Aires, Argentina. We also thank the patients and their families for their participation in genetic studies.
This work was supported by the University of Michigan (to S.A.C., J.Z.L., J.O.K., R.E.M.), Human Genetics Training Grant GM007544 (to A.Z.D.), and the National Institutes of Health (HD30428, HD34283; to S.A.C.).
Current address for M.I.P.M.: Instituto de Investigaciones Biomédicas, UBA-CONICET, Buenos Aires, Argentina.
Current address for Q.F.: Regeneron Pharmaceuticals, Tarrytown, New York 10591.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- CNS
- central nervous system
- CNV
- copy number variation
- CPHD
- combined pituitary hormone deficiency
- e
- embryonic day
- FGF
- fibroblast growth factor
- GWAS
- genome-wide association studies
- HH
- hypogonadotropic hypogonadism
- HHIP
- hedgehog-interacting protein
- HMG
- high-mobility group
- HPE
- holoprosencephaly
- IFT
- intraflagellar transport
- IGHD
- isolated GH deficiency
- KS
- Kallmann syndrome
- MAF
- minor allele frequency
- NGS
- next-generation sequencing
- PRL
- prolactin
- PROKR2
- prokineticin receptor 2
- SHH
- sonic hedgehog
- SNP
- single nucleotide polymorphism
- SOD
- septo-optic dysplasia
- TCF
- T-cell factor
- WES
- whole exome sequencing
- WGS
- whole genome sequencing.
Reference
- 1. Otto AP, França MM, Correa FA, et al. Frequent development of combined pituitary hormone deficiency in patients initially diagnosed as isolated growth hormone deficiency: a long term follow-up of patients from a single center. Pituitary. 2015;18(4):561–567. [DOI] [PubMed] [Google Scholar]
- 2. Blum WF, Deal C, Zimmermann AG, et al. Development of additional pituitary hormone deficiencies in pediatric patients originally diagnosed with idiopathic isolated GH deficiency. Eur J Endocrinol. 2014;170(1):13–21. [DOI] [PubMed] [Google Scholar]
- 3. Higham CE, Johannsson G, Shalet SM. Hypopituitarism [published online March 31, 2016]. Lancet. 10.1016/s0140-6736(16)30053-8. [DOI] [PubMed] [Google Scholar]
- 4. Sadeghi-Nejad A, Senior B. A familial syndrome of isolated “aplasia” of the anterior pituitary. Diagnostic studies and treatment in the neonatal period. J Pediatr. 1974;84(1):79–84. [DOI] [PubMed] [Google Scholar]
- 5. Manzoor Mughal A, Hassan N, Ahmed A. Bone age assessment methods: a critical review. Pak J Med Sci. 2014;30(1):211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deal C, Hasselmann C, Pfäffle RW, et al. Associations between pituitary imaging abnormalities and clinical and biochemical phenotypes in children with congenital growth hormone deficiency: data from an international observational study. Horm Res Paediatr. 2013;79(5):283–292. [DOI] [PubMed] [Google Scholar]
- 7. Obermannova B, Pfaeffle R, Zygmunt-Gorska A, et al. Mutations and pituitary morphology in a series of 82 patients with PROP1 gene defects. Horm Res Paediatr. 2011;76(5):348–354. [DOI] [PubMed] [Google Scholar]
- 8. Zygmunt-Górska A, Starzyk J, Adamek D, et al. Pituitary enlargement in patients with PROP1 gene inactivating mutation represents cystic hyperplasia of the intermediate pituitary lobe. Histopathology and over 10 years follow-up of two patients. J Pediatr Endocrinol Metab. 2009;22(7):653–660. [DOI] [PubMed] [Google Scholar]
- 9. Acerini CL, Tasker RC. Neuroendocrine consequences of traumatic brain injury. J Pediatr Endocrinol Metab. 2008;21(7):611–619. [DOI] [PubMed] [Google Scholar]
- 10. de Bree R, Lips P, Leemans CR. The need for patients' endocrine function vigilance following treatment of head and neck cancer. Curr Opin Otolaryngol Head Neck Surg. 2008;16(2):154–157. [DOI] [PubMed] [Google Scholar]
- 11. Medic-Stojanoska M, Pekic S, Curic N, Djilas-Ivanovic D, Popovic V. Evolving hypopituitarism as a consequence of traumatic brain injury (TBI) in childhood - call for attention. Endocrine. 2007;31(3):268–271. [DOI] [PubMed] [Google Scholar]
- 12. Melo ME, Marui S, Carvalho LR, et al. Hormonal, pituitary magnetic resonance, LHX4 and HESX1 evaluation in patients with hypopituitarism and ectopic posterior pituitary lobe. Clin Endocrinol (Oxf). 2007;66(1):95–102. [DOI] [PubMed] [Google Scholar]
- 13. Molitch ME, Gillam MP. Lymphocytic hypophysitis. Horm Res. 2007;68(suppl 5):145–150. [DOI] [PubMed] [Google Scholar]
- 14. Popovic V, Pekic S, Pavlovic D, et al. Hypopituitarism as a consequence of traumatic brain injury (TBI) and its possible relation with cognitive disabilities and mental distress. J Endocrinol Invest. 2004;27(11):1048–1054. [DOI] [PubMed] [Google Scholar]
- 15. Sharma V, O'Connor RA. Acute presentation of Sheehan's syndrome. J Obstet Gynaecol. 2006;26(5):471–472. [DOI] [PubMed] [Google Scholar]
- 16. Castinetti F, Reynaud R, Quentien MH, et al. Combined pituitary hormone deficiency: current and future status. J Endocrinol Invest. 2015;38(1):1–12. [DOI] [PubMed] [Google Scholar]
- 17. Kelberman D, Dattani MT. Hypothalamic and pituitary development: novel insights into the aetiology. Eur J Endocrinol. 2007;157(suppl 1):S3–S14. [DOI] [PubMed] [Google Scholar]
- 18. Davis SW, Castinetti F, Carvalho LR, et al. Molecular mechanisms of pituitary organogenesis: In search of novel regulatory genes. Mol Cell Endocrinol. 2010;323(1):4–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Castinetti F, Reynaud R, Saveanu A, Barlier A, Brue T. Genetic causes of combined pituitary hormone deficiencies in humans. Ann Endocrinol (Paris). 2012;73(2):53–55. [DOI] [PubMed] [Google Scholar]
- 20. Davis SW, Ellsworth BS, Peréz Millan MI, et al. Pituitary gland development and disease: from stem cell to hormone production. Curr Top Dev Biol. 2013;106:1–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kelberman D, Rizzoti K, Lovell-Badge R, Robinson IC, Dattani MT. Genetic regulation of pituitary gland development in human and mouse. Endocr Rev. 2009;30(7):790–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alatzoglou KS, Dattani MT. Genetic forms of hypopituitarism and their manifestation in the neonatal period. Early Hum Dev. 2009;85(11):705–712. [DOI] [PubMed] [Google Scholar]
- 23. Dattani MT. Growth hormone deficiency and combined pituitary hormone deficiency: does the genotype matter? Clin Endocrinol (Oxf). 2005;63(2):121–130. [DOI] [PubMed] [Google Scholar]
- 24. Quinton R, Duke VM, Robertson A, et al. Idiopathic gonadotrophin deficiency: genetic questions addressed through phenotypic characterization. Clin Endocrinol Oxf. 2001;55(2):163–174. [DOI] [PubMed] [Google Scholar]
- 25. Alatzoglou KS, Webb EA, Le Tissier P, Dattani MT. Isolated growth hormone deficiency (GHD) in childhood and adolescence: recent advances. Endocr Rev. 2014;35(3):376–432. [DOI] [PubMed] [Google Scholar]
- 26. Singh S, Groves AK. The molecular basis of craniofacial placode development. Wiley Interdiscip Rev Dev Biol. 2016;5(3):363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kelberman D, Dattani MT. Genetics of septo-optic dysplasia. Pituitary. 2007;10(4):393–407. [DOI] [PubMed] [Google Scholar]
- 28. Kauvar EF, Muenke M. Holoprosencephaly: recommendations for diagnosis and management. Curr Opin Pediatr. 2010;22(6):687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roessler E, Belloni E, Gaudenz K, et al. Mutations in the C-terminal domain of sonic hedgehog cause holoprosencephaly. Hum Mol Genet. 1997;6(11):1847–1853. [DOI] [PubMed] [Google Scholar]
- 30. Stamou MI, Cox KH, Crowley WF., Jr Discovering genes essential to the hypothalamic regulation of human reproduction using a human disease model: adjusting to life in the “-omics” era. Endocr Rev. 2015;36(6):603–621. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31. Fukami M, Iso M, Sato N, et al. Submicroscopic deletion involving the fibroblast growth factor receptor 1 gene in a patient with combined pituitary hormone deficiency. Endocr J. 2013;60(8):1013–1020. [DOI] [PubMed] [Google Scholar]
- 32. Izumi Y, Suzuki E, Kanzaki S, et al. Genome-wide copy number analysis and systematic mutation screening in 58 patients with hypogonadotropic hypogonadism. Fertil Steril. 2014;102(4):1130–1136.e3. [DOI] [PubMed] [Google Scholar]
- 33. Raivio T, Avbelj M, McCabe MJ, et al. Genetic overlap in Kallmann syndrome, combined pituitary hormone deficiency, and septo-optic dysplasia. J Clin Endocrinol Metab. 2012;97(4):E694–E699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sulik KK, Johnston MC, Webb MA. Fetal alcohol syndrome: embryogenesis in a mouse model. Science. 1981;214(4523):936–938. [DOI] [PubMed] [Google Scholar]
- 35. Matsuo I, Kuratani S, Kimura C, Takeda N, Aizawa S. Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev. 1995;9(21):2646–2658. [DOI] [PubMed] [Google Scholar]
- 36. Hong M, Krauss RS. Cdon mutation and fetal ethanol exposure synergize to produce midline signaling defects and holoprosencephaly spectrum disorders in mice. PLoS Genet. 2012;8(10):e1002999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Visscher PM, Medland SE, Ferreira MA, et al. Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full siblings. PLoS Genet. 2006;2(3):e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bakshi A, Zhu Z, Vinkhuyzen AA, et al. Fast set-based association analysis using summary data from GWAS identifies novel gene loci for human complex traits. Sci Rep. 2016;6:32894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wood AR, Esko T, Yang J, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet 2014;46(11):1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schierding W, Antony J, Cutfield WS, Horsfield JA, O'Sullivan JM. Intergenic GWAS SNPs are key components of the spatial and regulatory network for human growth [published online June 10, 2016]. Hum Mol Genet. 10.1093/hmg/ddw165. [DOI] [PubMed] [Google Scholar]
- 41. Lachance J, Vernot B, Elbers CC, et al. Evolutionary history and adaptation from high-coverage whole-genome sequences of diverse African hunter-gatherers. Cell. 2012;150(3):457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chan Y, Salem RM, Hsu YH, et al. Genome-wide analysis of body proportion classifies height-associated variants by mechanism of action and implicates genes important for skeletal development. Am J Hum Genet. 2015;96(5):695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Trerotola M, Relli V, Simeone P, Alberti S. Epigenetic inheritance and the missing heritability. Hum Genomics. 2015;9:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Simeone P, Alberti S. Epigenetic heredity of human height. Physiol Rep. 2014;2(6):e12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tatton-Brown K, Seal S, Ruark E, et al. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat Genet. 2014;46(4):385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. De Rienzo F, Mellone S, Bellone S, et al. Frequency of genetic defects in combined pituitary hormone deficiency: a systematic review and analysis of a multicentre Italian cohort. Clin Endocrinol (Oxf). 2015;83(6):849–860. [DOI] [PubMed] [Google Scholar]
- 47. Dusatkova P, Pfäffle R, Brown MR, et al. Genesis of two most prevalent PROP1 gene variants causing combined pituitary hormone deficiency in 21 populations. Eur J Hum Genet. 2016;24(3):415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Navardauskaite R, Dusatkova P, Obermannova B, et al. High prevalence of PROP1 defects in Lithuania: phenotypic findings in an ethnically homogenous cohort of patients with multiple pituitary hormone deficiency. J Clin Endocrinol Metab. 2014;99(1):299–306. [DOI] [PubMed] [Google Scholar]
- 49. Kandemir N, Vurallı D, Tasķıran E, et al. Frequency of mutations in PROP-1 gene in Turkish children with combined pituitary hormone deficiency. Turk J Pediatr. 2012;54(6):570–575. [PubMed] [Google Scholar]
- 50. Takagi M, Ishii T, Inokuchi M, et al. Gradual loss of ACTH due to a novel mutation in LHX4: comprehensive mutation screening in Japanese patients with congenital hypopituitarism. PLoS One. 2012;7(9):e46008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. de Graaff LC, Argente J, Veenma DC, et al. PROP1, HESX1, POU1F1, LHX3 and LHX4 mutation and deletion screening and GH1 P89L and IVS3+1/+2 mutation screening in a Dutch nationwide cohort of patients with combined pituitary hormone deficiency. Horm Res Paediatr. 2010;73(5):363–371. [DOI] [PubMed] [Google Scholar]
- 52. Mehta A, Hindmarsh PC, Mehta H, et al. Congenital hypopituitarism: clinical, molecular and neuroradiological correlates. Clin Endocrinol (Oxf). 2009;71(3):376–382. [DOI] [PubMed] [Google Scholar]
- 53. Diaczok D, Romero C, Zunich J, Marshall I, Radovick S. A novel dominant negative mutation of OTX2 associated with combined pituitary hormone deficiency. J Clin Endocrinol Metab. 2008;93(11):4351–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Coya R, Vela A, Pérez de Nanclares G, et al. Panhypopituitarism: genetic versus acquired etiological factors. J Pediatr Endocrinol Metab. 2007;20(1):27–36. [DOI] [PubMed] [Google Scholar]
- 55. Reynaud R, Gueydan M, Saveanu A, et al. Genetic screening of combined pituitary hormone deficiency: experience in 195 patients. J Clin Endocrinol Metab. 2006;91(9):3329–3336. [DOI] [PubMed] [Google Scholar]
- 56. Lemos MC, Gomes L, Bastos M, et al. PROP1 gene analysis in Portuguese patients with combined pituitary hormone deficiency. Clin Endocrinol (Oxf). 2006;65(4):479–485. [DOI] [PubMed] [Google Scholar]
- 57. Halász Z, Toke J, Patócs A, et al. High prevalence of PROP1 gene mutations in Hungarian patients with childhood-onset combined anterior pituitary hormone deficiency. Endocrine. 2006;30(3):255–260. [DOI] [PubMed] [Google Scholar]
- 58. Rainbow LA, Rees SA, Shaikh MG, et al. Mutation analysis of POUF-1, PROP-1 and HESX-1 show low frequency of mutations in children with sporadic forms of combined pituitary hormone deficiency and septo-optic dysplasia. Clin Endocrinol (Oxf). 2005;62(2):163–168. [DOI] [PubMed] [Google Scholar]
- 59. Lebl J, Vosáhlo J, Pfaeffle RW, et al. Auxological and endocrine phenotype in a population-based cohort of patients with PROP1 gene defects. Eur J Endocrinol. 2005;153(3):389–396. [DOI] [PubMed] [Google Scholar]
- 60. Turton JP, Mehta A, Raza J, et al. Mutations within the transcription factor PROP1 are rare in a cohort of patients with sporadic combined pituitary hormone deficiency (CPHD). Clin Endocrinol (Oxf). 2005;63(1):10–18. [DOI] [PubMed] [Google Scholar]
- 61. McLennan K, Jeske Y, Cotterill A, et al. Combined pituitary hormone deficiency in Australian children: clinical and genetic correlates. Clin Endocrinol (Oxf). 2003;58(6):785–794. [DOI] [PubMed] [Google Scholar]
- 62. Fofanova OV, Takamura N, Kinoshita E, et al. A mutational hot spot in the Prop-1 gene in Russian children with combined pituitary hormone deficiency. Pituitary. 1998;1(1):45–49. [DOI] [PubMed] [Google Scholar]
- 63. Deladoëy J, Flück C, Büyükgebiz A, et al. “Hot spot” in the PROP1 gene responsible for combined pituitary hormone deficiency. J Clin Endocrinol Metab. 1999;84(5):1645–1650. [DOI] [PubMed] [Google Scholar]
- 64. Fofanova OV, Takamura N, Kinoshita E, et al. Rarity of PIT1 involvement in children from Russia with combined pituitary hormone deficiency. Am J Med Genet. 1998;77(5):360–365. [DOI] [PubMed] [Google Scholar]
- 65. Pfaeffle RW, Savage JJ, Hunter CS, et al. Four novel mutations of the LHX3 gene cause combined pituitary hormone deficiencies with or without limited neck rotation. J Clin Endocrinol Metab. 2007;92(5):1909–1919. [DOI] [PubMed] [Google Scholar]
- 66. Castinetti F, Saveanu A, Reynaud R, et al. A novel dysfunctional LHX4 mutation with high phenotypical variability in patients with hypopituitarism. J Clin Endocrinol Metab. 2008;93(7):2790–2799. [DOI] [PubMed] [Google Scholar]
- 67. Turton JP, Reynaud R, Mehta A, et al. Novel mutations within the POU1F1 gene associated with variable combined pituitary hormone deficiency. J Clin Endocrinol Metab. 2005;90(8):4762–4770. [DOI] [PubMed] [Google Scholar]
- 68. Vieira TC, Boldarine VT, Abucham J. Molecular analysis of PROP1, PIT1, HESX1, LHX3, and LHX4 shows high frequency of PROP1 mutations in patients with familial forms of combined pituitary hormone deficiency. Arq Bras Endocrinol Metabol. 2007;51(7):1097–1103. [DOI] [PubMed] [Google Scholar]
- 69. Kim SS, Kim Y, Shin YL, Kim GH, Kim TU, Yoo HW. Clinical characteristics and molecular analysis of PIT1, PROP1,LHX3, and HESX1 in combined pituitary hormone deficiency patients with abnormal pituitary MR imaging. Horm Res. 2003;60(6):277–283. [DOI] [PubMed] [Google Scholar]
- 70. Castinetti F, Reynaud R, Saveanu A, et al. Mechanisms in Endocrinology: an update in the genetic aetiologies of combined pituitary hormone deficiency. Eur J Endocrinol. 2016;174(6):R239–R247. [DOI] [PubMed] [Google Scholar]
- 71. Pfäffle R, Klammt J. Pituitary transcription factors in the aetiology of combined pituitary hormone deficiency. Best Pract Res Clin Endocrinol Metab. 2011;25(1):43–60. [DOI] [PubMed] [Google Scholar]
- 72. Tajima T, Ishizu K, Nakamura A. Molecular and clinical findings in patients with LHX4 and OTX2 mutations. Clin Pediatr Endocrinol. 2013;22(2):15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cohen LE. Genetic disorders of the pituitary. Curr Opin Endocrinol Diabetes Obes. 2012;19(1):33–39. [DOI] [PubMed] [Google Scholar]
- 74. Carsner RL, Rennels EG. Primary site of gene action in anterior pituitary dwarf mice. Science. 1960;131(3403):829. [DOI] [PubMed] [Google Scholar]
- 75. Snell GD. Dwarf, a new Mendelian recessive character of the house mouse. Proc Natl Acad Sci USA. 1929;15(9):733–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Camper SA, Saunders TL, Katz RW, Reeves RH. The Pit-1 transcription factor gene is a candidate for the murine Snell dwarf mutation. Genomics. 1990;8(3):586–590. [DOI] [PubMed] [Google Scholar]
- 77. Li S, Crenshaw EB, 3rd, Rawson EJ, Simmons DM, Swanson LW, Rosenfeld MG. Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU-domain gene pit-1. Nature. 1990;347(6293):528–533. [DOI] [PubMed] [Google Scholar]
- 78. Dollé P, Castrillo JL, Theill LE, Deerinck T, Ellisman M, Karin M. Expression of GHF-1 protein in mouse pituitaries correlates both temporally and spatially with the onset of growth hormone gene activity. Cell. 1990;60(5):809–820. [DOI] [PubMed] [Google Scholar]
- 79. Mangalam HJ, Albert VR, Ingraham HA, et al. A pituitary POU domain protein, Pit-1, activates both growth hormone and prolactin promoters transcriptionally. Genes Dev. 1989;3(7):946–958. [DOI] [PubMed] [Google Scholar]
- 80. Tatsumi K, Miyai K, Notomi T, et al. Cretinism with combined hormone deficiency caused by a mutation in the PIT1 gene. Nat Genet. 1992;1(1):56–58. [DOI] [PubMed] [Google Scholar]
- 81. Buckwalter MS, Katz RW, Camper SA. Localization of the panhypopituitary dwarf mutation (df) on mouse chromosome 11 in an intersubspecific backcross. Genomics. 1991;10(3):515–526. [DOI] [PubMed] [Google Scholar]
- 82. Sornson MW, Wu W, Dasen JS, et al. Pituitary lineage determination by the Prophet of Pit-1 homeodomain factor defective in Ames dwarfism. Nature. 1996;384(6607):327–333. [DOI] [PubMed] [Google Scholar]
- 83. Schaible R, Gowen JW. A new dwarf mouse. Genetics. 1961;46(896):349–356. [Google Scholar]
- 84. Wu W, Cogan JD, Pfäffle RW, et al. Mutations in PROP1 cause familial combined pituitary hormone deficiency. Nat Genet. 1998;18(2):147–149. [DOI] [PubMed] [Google Scholar]
- 85. Parks JS, Brown MR, Hurley DL, Phelps CJ, Wajnrajch MP. Heritable disorders of pituitary development. J Clin Endocrinol Metab. 1999;84(12):4362–4370. [DOI] [PubMed] [Google Scholar]
- 86. O'Hara BF, Bendotti C, Reeves RH, Oster-Granite ML, Coyle JT, Gearhart JD. Genetic mapping and analysis of somatostatin expression in Snell dwarf mice. Molec. Brain Res. 1988;464(4):283–292. [DOI] [PubMed] [Google Scholar]
- 87. Bartke A. The response of two types of dwarf mice to growth hormone, thyrotropin, and thyroxine. Gen Comp Endocrinol. 1965;5(4):418–426. [DOI] [PubMed] [Google Scholar]
- 88. Stahl JH, Kendall SK, Brinkmeier ML, et al. Thyroid hormone is essential for pituitary somatotropes and lactotropes. Endocrinology. 1999;140(4):1884–1892. [DOI] [PubMed] [Google Scholar]
- 89. Nasonkin IO, Ward RD, Bavers DL, et al. Aged PROP1 deficient dwarf mice maintain ACTH production. PLoS One. 2011;6(12):e28355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Eicher EM, Beamer WG. New mouse dw allele: genetic location and effects on lifespan and growth hormone levels. J Hered. 1980;71(3):187–190. [DOI] [PubMed] [Google Scholar]
- 91. Nasonkin IO, Ward RD, Raetzman LT, et al. Pituitary hypoplasia and respiratory distress syndrome in Prop1 knockout mice. Hum Mol Genet. 2004;13(22):2727–2735. [DOI] [PubMed] [Google Scholar]
- 92. Nadeau JH. Modifier genes in mice and humans. Nat Rev Genet. 2001;2(3):165–174. [DOI] [PubMed] [Google Scholar]
- 93. Fang Q, Longo-Guess C, Gagnon LH, et al. A modifier gene alleviates hypothyroidism-induced hearing impairment in Pou1f1dw dwarf mice. Genetics. 2011;189(2):665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sheng HZ, Zhadanov AB, Mosinger B, Jr, et al. Specification of pituitary cell lineages by the LIM homeobox gene Lhx3. Science. 1996;272(5264):1004–1007. [DOI] [PubMed] [Google Scholar]
- 95. Netchine I, Sobrier ML, Krude H, et al. Mutations in LHX3 result in a new syndrome revealed by combined pituitary hormone deficiency. Nat Genet. 2000;25(2):182–186. [DOI] [PubMed] [Google Scholar]
- 96. Sloop KW, Showalter AD, Von Kap-Herr C, Pettenati MJ, Rhodes SJ. Analysis of the human LHX3 neuroendocrine transcription factor gene and mapping to the subtelomeric region of chromosome 9. Gene. 2000;245(2):237–243. [DOI] [PubMed] [Google Scholar]
- 97. Sheng HZ, Moriyama K, Yamashita T, et al. Multistep control of pituitary organogenesis. Science. 1997;278(5344):1809–1812. [DOI] [PubMed] [Google Scholar]
- 98. Machinis K, Pantel J, Netchine I, et al. Syndromic short stature in patients with a germline mutation in the LIM homeobox LHX4. Am J Hum Genet. 2001;69(5):961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hermesz E, Mackem S, Mahon KA. Rpx: a novel anterior-restricted homeobox gene progressively activated in the prechordal plate, anterior neural plate and Rathke's pouch of the mouse embryo. Development. 1996;122(1):41–52. [DOI] [PubMed] [Google Scholar]
- 100. Dattani MT, Martinez-Barbera JP, Thomas PQ, et al. Mutations in the homeobox gene HESX1/Hesx1 associated with septo-optic dysplasia in human and mouse. Nat Genet. 1998;19(2):125–133. [DOI] [PubMed] [Google Scholar]
- 101. Roessler E, Muenke M. The molecular genetics of holoprosencephaly. Am J Med Genet C Semin Med Genet. 2010;154C(1):52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bae GU, Domené S, Roessler E, et al. Mutations in CDON, encoding a hedgehog receptor, result in holoprosencephaly and defective interactions with other hedgehog receptors. Am J Hum Genet. 2011;89(2):231–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. McCabe MJ, Gaston-Massuet C, Tziaferi V, et al. Novel FGF8 mutations associated with recessive holoprosencephaly, craniofacial defects, and hypothalamo-pituitary dysfunction. J Clin Endocrinol Metab. 2011;96(10):E1709–E1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Roessler E, Du YZ, Mullor JL, et al. Loss-of-function mutations in the human GLI2 gene are associated with pituitary anomalies and holoprosencephaly-like features. Proc Natl Acad Sci USA. 2003;100(23):13424–13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wang Y, Martin JF, Bai CB. Direct and indirect requirements of Shh/Gli signaling in early pituitary development. Dev Biol. 2010;348(2):199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Treier M, Gleiberman AS, O'Connell SM, et al. Multistep signaling requirements for pituitary organogenesis in vivo. Genes Dev. 1998;12(11):1691–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Laumonnier F, Ronce N, Hamel BC, et al. Transcription factor SOX3 is involved in X-linked mental retardation with growth hormone deficiency. Am J Hum Genet. 2002;71(6):1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Rizzoti K, Brunelli S, Carmignac D, Thomas PQ, Robinson IC, Lovell-Badge R. SOX3 is required during the formation of the hypothalamo-pituitary axis. Nat Genet. 2004;36(3):247–255. [DOI] [PubMed] [Google Scholar]
- 109. Ragge NK, Brown AG, Poloschek CM, et al. Heterozygous mutations of OTX2 cause severe ocular malformations. Am J Hum Genet. 2005;76(6):1008–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Henderson RA, Williamson K, Cumming S, et al. Inherited PAX6, NF1 and OTX2 mutations in a child with microphthalmia and aniridia. Eur J Hum Genet. 2007;15(8):898–901. [DOI] [PubMed] [Google Scholar]
- 111. Rhinn M, Dierich A, Le Meur M, Ang S. Cell autonomous and non-cell autonomous functions of Otx2 in patterning the rostral brain. Development. 1999;126(19):4295–4304. [DOI] [PubMed] [Google Scholar]
- 112. Spieler D, Bäumer N, Stebler J, et al. Involvement of Pax6 and Otx2 in the forebrain-specific regulation of the vertebrate homeobox gene ANF/Hesx1. Dev Biol. 2004;269(2):567–579. [DOI] [PubMed] [Google Scholar]
- 113. Dateki S, Fukami M, Sato N, Muroya K, Adachi M, Ogata T. OTX2 mutation in a patient with anophthalmia, short stature, and partial growth hormone deficiency: functional studies using the IRBP, HESX1, and POU1F1 promoters. J Clin Endocrinol Metab. 2008;93(10):3697–3702. [DOI] [PubMed] [Google Scholar]
- 114. Hide T, Hatakeyama J, Kimura-Yoshida C, et al. Genetic modifiers of otocephalic phenotypes in Otx2 heterozygous mutant mice. Development. 2002;129(18):4347–4357. [DOI] [PubMed] [Google Scholar]
- 115. Mortensen AH, Schade V, Lamonerie T, Camper SA. Deletion of OTX2 in neural ectoderm delays anterior pituitary development. Hum Mol Genet. 2014;24(4):939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. MacArthur DG, Manolio TA, Dimmock DP, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508(7497):469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Neveling K, Feenstra I, Gilissen C, et al. A post-hoc comparison of the utility of sanger sequencing and exome sequencing for the diagnosis of heterogeneous diseases. Hum Mutat. 2013;34(12):1721–1726. [DOI] [PubMed] [Google Scholar]
- 119. Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312(18):1870–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Koboldt DC, Larson DE, Sullivan LS, et al. Exome-based mapping and variant prioritization for inherited Mendelian disorders. Am J Hum Genet. 2014;94(3):373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Goldstein DB, Allen A, Keebler J, et al. Sequencing studies in human genetics: design and interpretation. Nat Rev Genet. 2013;14(7):460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Bamshad MJ, Ng SB, Bigham AW, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12(11):745–755. [DOI] [PubMed] [Google Scholar]
- 123. Webb EA, AlMutair A, Kelberman D, et al. ARNT2 mutation causes hypopituitarism, post-natal microcephaly, visual and renal anomalies. Brain. 2013;136(Pt 10):3096–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Amendola LM, Jarvik GP, Leo MC, et al. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet. 2016;98(6):1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Brinkmeier ML, Davis SW, Carninci P, et al. Discovery of transcriptional regulators and signaling pathways in the developing pituitary gland by bioinformatic and genomic approaches. Genomics 2009;93(5):449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Niroula A, Vihinen M. Variation interpretation predictors: principles, types, performance, and choice. Hum Mutat. 2016;37(6):579–597. [DOI] [PubMed] [Google Scholar]
- 127. Dong C, Wei P, Jian X, et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet. 2015;24(8):2125–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Böttner A, Keller E, Kratzsch J, et al. PROP1 mutations cause progressive deterioration of anterior pituitary function including adrenal insufficiency: a longitudinal analysis. J Clin Endocrinol Metab. 2004;89(10):5256–5265. [DOI] [PubMed] [Google Scholar]
- 129. Pérez Millán MI, Brinkmeier ML, Mortensen AH, Camper SA. PROP1 triggers epithelial-mesenchymal transition-like process in pituitary stem cells. Elife 2016;5:e14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Kelberman D, Turton JP, Woods KS, et al. Molecular analysis of novel PROP1 mutations associated with combined pituitary hormone deficiency (CPHD). Clin Endocrinol (Oxf). 2009;70(1):96–103. [DOI] [PubMed] [Google Scholar]
- 131. Akcay A, Ulucan K, Taskin N, et al. Suprasellar mass mimicking a hypothalamic glioma in a patient with a complete PROP1 deletion. Eur J Med Genet. 2013;56(8):445–451. [DOI] [PubMed] [Google Scholar]
- 132. Vallette-Kasic S, Barlier A, Teinturier C, et al. PROP1 gene screening in patients with multiple pituitary hormone deficiency reveals two sites of hypermutability and a high incidence of corticotroph deficiency. J Clin Endocrinol Metab. 2001;86(9):4529–4535. [DOI] [PubMed] [Google Scholar]
- 133. Riepe FG, Partsch CJ, Blankenstein O, Mönig H, Pfäffle RW, Sippell WG. Longitudinal imaging reveals pituitary enlargement preceding hypoplasia in two brothers with combined pituitary hormone deficiency attributable to PROP1 mutation. J Clin Endocrinol Metab. 2001;86(9):4353–4357. [DOI] [PubMed] [Google Scholar]
- 134. Voutetakis A, Maniati-Christidi M, Kanaka-Gantenbein C, et al. Prolonged jaundice and hypothyroidism as the presenting symptoms in a neonate with a novel Prop1 gene mutation (Q83X). Eur J Endocrinol. 2004;150(3):257–264. [DOI] [PubMed] [Google Scholar]
- 135. Avbelj Stefanija M, Kotnik P, Bratanič N, et al. Novel mutations in HESX1 and PROP1 genes in combined pituitary hormone deficiency. Horm Res Paediatr. 2015;84(3):153–158. [DOI] [PubMed] [Google Scholar]
- 136. Ziemnicka K, Budny B, Drobnik K, et al. Two coexisting heterozygous frameshift mutations in PROP1 are responsible for a different phenotype of combined pituitary hormone deficiency. J Appl Genet. 2016;57(3):373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Baş F, Uyguner ZO, Darendeliler F, et al. Molecular analysis of PROP1, POU1F1, LHX3, and HESX1 in Turkish patients with combined pituitary hormone deficiency: a multicenter study. Endocrine. 2015;49(2):479–491. [DOI] [PubMed] [Google Scholar]
- 138. 1000 Genomes Project Consortium, Abecasis GR, Altshuler D, Auton A, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Abrão MG, Billerbeck AE, Nishi MY, Marui S, Mendonça BB. Standardization of DNA extraction with NaCl from oral mucosa cells: application in PROP1 gene study [in Portuguese]. Arq Bras Endocrinol Metabol. 2005;49(6):978–982. [DOI] [PubMed] [Google Scholar]
- 140. Nogueira CR, Sabacan L, Jameson JL, Medeiros-Neto G, Kopp P. Combined pituitary hormone deficiency in an inbred Brazilian kindred associated with a mutation in the PROP-1 gene. Mol Genet Metab. 1999;67(1):58–61. [DOI] [PubMed] [Google Scholar]
- 141. Reynaud R, Barlier A, Vallette-Kasic S, et al. An uncommon phenotype with familial central hypogonadism caused by a novel PROP1 gene mutant truncated in the transactivation domain. J Clin Endocrinol Metab. 2005;90(8):4880–4887. [DOI] [PubMed] [Google Scholar]
- 142. Skowronska-Krawczyk D, Ma Q, Schwartz M, et al. Required enhancer-matrin-3 network interactions for a homeodomain transcription program. Nature. 2014;514(7521):257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Pine-Twaddell E, Romero CJ, Radovick S. Vertical transmission of hypopituitarism: critical importance of appropriate interpretation of thyroid function tests and levothyroxine therapy during pregnancy. Thyroid. 2013;23(7):892–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Blankenstein O, Mühlenberg R, Kim C, Wüller S, Pfäffle R, Heimann G. A new C-terminal located mutation (V272ter) in the PIT-1 gene manifesting with severe congenital hypothyroidism. Possible functionality of the PIT-1 C-terminus. Horm Res. 2001;56(3–4):81–86. [DOI] [PubMed] [Google Scholar]
- 145. Sobrier ML, Tsai YC, Pérez C, et al. Functional characterization of a human POU1F1 mutation associated with isolated growth hormone deficiency: a novel etiology for IGHD. Hum Mol Genet. 2016;25(3):472–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Ohta K, Nobukuni Y, Mitsubuchi H, et al. Characterization of the gene encoding human pituitary-specific transcription factor, Pit-1. Gene. 1992;122(2):387–388. [DOI] [PubMed] [Google Scholar]
- 147. Malvagia S, Poggi GM, Pasquini E, et al. The de novo Q167K mutation in the POU1F1 gene leads to combined pituitary hormone deficiency in an Italian patient. Pediatr Res. 2003;54(5):635–640. [DOI] [PubMed] [Google Scholar]
- 148. Gat-Yablonski G, Klar A, Hirsch D, et al. Three novel mutations in POU1F1 in Israeli patients with combined pituitary hormone deficiency. J Pediatr Endocrinol Metab. 2005;18(4):385–393. [DOI] [PubMed] [Google Scholar]
- 149. Cohen LE. A “hot spot” in the Pit-1 gene responsible for combined pituitary hormone deficiency: clinical and molecular correlates. J Clin Endocrinol Metab. 1995;80(2):679–684. [DOI] [PubMed] [Google Scholar]
- 150. Gat-Yablonski G, Frumkin-Ben David R, Bar M, Potievsky O, Phillip M, Lazar L. Am J Med Genet A. 2011;155A(9):2242–2246. [DOI] [PubMed] [Google Scholar]
- 151. Martinez-Barbera JP, Rodriguez TA, Beddington RS. The homeobox gene Hesx1 is required in the anterior neural ectoderm for normal forebrain formation. Dev Biol. 2000;223(2):422–430. [DOI] [PubMed] [Google Scholar]
- 152. McCabe MJ, Alatzoglou KS, Dattani MT. Septo-optic dysplasia and other midline defects: the role of transcription factors: HESX1 and beyond. Best Pract Res Clin Endocrinol Metab. 2011;25(1):115–124. [DOI] [PubMed] [Google Scholar]
- 153. Brickman JM, Clements M, Tyrell R, et al. Molecular effects of novel mutations in Hesx1/HESX1 associated with human pituitary disorders. Development. 2001;128(24):5189–5199. [DOI] [PubMed] [Google Scholar]
- 154. Carvalho LR, Woods KS, Mendonca BB, et al. A homozygous mutation in HESX1 is associated with evolving hypopituitarism due to impaired repressor-corepressor interaction. J Clin Invest. 2003;112(8):1192–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Sobrier ML, Netchine I, Heinrichs C, et al. Alu-element insertion in the homeodomain of HESX1 and aplasia of the anterior pituitary. Hum Mutat. 2005;25(5):503. [DOI] [PubMed] [Google Scholar]
- 156. Sobrier ML, Maghnie M, Vié-Luton MP, et al. Novel HESX1 mutations associated with a life-threatening neonatal phenotype, pituitary aplasia, but normally located posterior pituitary and no optic nerve abnormalities. J Clin Endocrinol Metab. 2006;91(11):4528–4536. [DOI] [PubMed] [Google Scholar]
- 157. Durmaz B, Cogulu O, Dizdarer C, Stobbe H, Pfaeffle R, Ozkinay F. A novel homozygous HESX1 mutation causes panhypopituitarism without midline defects and optic nerve anomalies. J Pediatr Endocrinol Metab. 2011;24(9–10):779–782. [DOI] [PubMed] [Google Scholar]
- 158. Reynaud R, Albarel F, Saveanu A, et al. Pituitary stalk interruption syndrome in 83 patients: novel HESX1 mutation and severe hormonal prognosis in malformative forms. Eur J Endocrinol. 2011;164(4):457–465. [DOI] [PubMed] [Google Scholar]
- 159. Bach I, Rhodes SJ, Pearse RV, 2nd, et al. P-Lim, a LIM homeodomain factor, is expressed during pituitary organ and cell commitment and synergizes with Pit-1. Proc Natl Acad Sci USA. 1995;92(7):2720–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Tsuchida T, Ensini M, Morton SB, et al. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79(6):957–970. [DOI] [PubMed] [Google Scholar]
- 161. Bechtold-Dalla Pozza S, Hiedl S, Roeb J, et al. A recessive mutation resulting in a disabling amino acid substitution (T194R) in the LHX3 homeodomain causes combined pituitary hormone deficiency. Horm Res Paediatr. 2012;77(1):41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Bhangoo AP, Hunter CS, Savage JJ, et al. Clinical case seminar: a novel LHX3 mutation presenting as combined pituitary hormonal deficiency. J Clin Endocrinol Metab. 2006;91(3):747–753. [DOI] [PubMed] [Google Scholar]
- 163. Kriström B, Zdunek AM, Rydh A, et al. A novel mutation in the LHX3 gene is responsible for combined pituitary hormone deficiency, hearing impairment, and vertebral malformations. Endocrinology. 2009;150(2):1069–1069. [DOI] [PubMed] [Google Scholar]
- 164. Bonfig W, Krude H, Schmidt H. A novel mutation of LHX3 is associated with combined pituitary hormone deficiency including ACTH deficiency, sensorineural hearing loss, and short neck-a case report and review of the literature. Eur J Pediatr. 2011;170(8):1017–1021. [DOI] [PubMed] [Google Scholar]
- 165. Dateki S, Fukami M, Uematsu A, et al. Mutation and gene copy number analyses of six pituitary transcription factor genes in 71 patients with combined pituitary hormone deficiency: identification of a single patient with LHX4 deletion. J Clin Endocrinol Metab. 2010;95(8):4043–4047. [DOI] [PubMed] [Google Scholar]
- 166. Sheng HZ, Westphal H. Early steps in pituitary organogenesis. Trends Genet. 1999;15(6):236–240. [DOI] [PubMed] [Google Scholar]
- 167. Pfaeffle RW, Hunter CS, Savage JJ, et al. Three novel missense mutations within the LHX4 gene are associated with variable pituitary hormone deficiencies. J Clin Endocrinol Metab. 2008;93(3):1062–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Gregory LC, Humayun KN, Turton JP, McCabe MJ, Rhodes SJ, Dattani MT. Novel lethal form of congenital hypopituitarism associated with the first recessive LHX4 mutation. J Clin Endocrinol Metab. 2015;100(6):2158–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Tajima T, Hattori T, Nakajima T, Okuhara K, Tsubaki J, Fujieda K. A novel missense mutation (P366T) of the LHX4 gene causes severe combined pituitary hormone deficiency with pituitary hypoplasia, ectopic posterior lobe and a poorly developed sella turcica. Endocr J. 2007;54(4):637–641. [DOI] [PubMed] [Google Scholar]
- 170. Filges I, Bischof-Renner A, Röthlisberger B, et al. Panhypopituitarism presenting as life-threatening heart failure caused by an inherited microdeletion in 1q25 including LHX4. Pediatrics. 2012;129(2):e529–e534. [DOI] [PubMed] [Google Scholar]
- 171. Braslavsky D, Saveanu A, Keselman A, et al. Genetic screening in children with congenital combined pituitary hormone deficiency. Horm Res Paediatr. 2012;78(suppl 2):27. [Google Scholar]
- 172. Tajima T, Yorifuji T, Ishizu K, Fujieda K. A novel mutation (V101A) of the LHX4 gene in a Japanese patient with combined pituitary hormone deficiency. Exp Clin Endocrinol Diabetes. 2010;118(7):405–409. [DOI] [PubMed] [Google Scholar]
- 173. Rochette C, Jullien N, Saveanu A, et al. Identifying the deleterious effect of rare LHX4 allelic variants, a challenging issue. PLoS One 2015;10(5):e0126648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Kehrer M, Liehr T, Benkert T, et al. Interstitial duplication of chromosome region 1q25.1q25.3: report of a patient with mild cognitive deficits, tall stature and facial dysmorphisms. Am J Med Genet A. 2015;167A(3):653–656. [DOI] [PubMed] [Google Scholar]
- 175. Howard PW, Maurer RA. A point mutation in the LIM domain of Lhx3 reduces activation of the glycoprotein hormone alpha-subunit promoter. J Biol Chem. 2001;276(22):19020–19026. [DOI] [PubMed] [Google Scholar]
- 176. de Bruijn DR, van Dijk AH, Willemse MP, van Kessel AG. The C terminus of the synovial sarcoma-associated SSX proteins interacts with the LIM homeobox protein LHX4. Oncogene. 2008;27(5):653–662. [DOI] [PubMed] [Google Scholar]
- 177. Simeone A, Acampora D, Gulisano M, Stornaiuolo A, Boncinelli E. Nested expression domains of four homeobox genes in developing rostral brain. Nature. 1992;358(6388):687–690. [DOI] [PubMed] [Google Scholar]
- 178. Ang SL, Conlon RA, Jin O, Rossant J. Positive and negative signals from mesoderm regulate the expression of mouse Otx2 in ectoderm explants. Development. 1994;120(10):2979–2989. [DOI] [PubMed] [Google Scholar]
- 179. Wyatt A, Bakrania P, Bunyan DJ, et al. Novel heterozygous OTX2 mutations and whole gene deletions in anophthalmia, microphthalmia and coloboma. Hum Mutat. 2008;29(11):E278–E283. [DOI] [PubMed] [Google Scholar]
- 180. Schilter KF, Schneider A, Bardakjian T, et al. OTX2 microphthalmia syndrome: four novel mutations and delineation of a phenotype. Clin Genet. 2011;79(2):158–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Ashkenazi-Hoffnung L, Lebenthal Y, Wyatt AW, et al. A novel loss-of-function mutation in OTX2 in a patient with anophthalmia and isolated growth hormone deficiency. Hum Genet. 2010;127(6):721–729. [DOI] [PubMed] [Google Scholar]
- 182. Gorbenko Del Blanco D, Romero CJ, Diaczok D, de Graaff LC, Radovick S, Hokken-Koelega AC. A novel OTX2 mutation in a patient with combined pituitary hormone deficiency, pituitary malformation, and an underdeveloped left optic nerve. Eur J Endocrinol. 2012;167(3):441–452. [DOI] [PubMed] [Google Scholar]
- 183. Dateki S, Kosaka K, Hasegawa K, et al. Heterozygous orthodenticle homeobox 2 mutations are associated with variable pituitary phenotype. J Clin Endocrinol Metab. 2010;95(2):756–764. [DOI] [PubMed] [Google Scholar]
- 184. Tajima T, Ohtake A, Hoshino M, et al. OTX2 loss of function mutation causes anophthalmia and combined pituitary hormone deficiency with a small anterior and ectopic posterior pituitary. J Clin Endocrinol Metab. 2009;94(1):314–319. [DOI] [PubMed] [Google Scholar]
- 185. Nolen LD, Amor D, Haywood A, et al. Deletion at 14q22–23 indicates a contiguous gene syndrome comprising anophthalmia, pituitary hypoplasia, and ear anomalies. Am J Med Genet A. 2006;140(16):1711–1718. [DOI] [PubMed] [Google Scholar]
- 186. You T, Lv Y, Liu S, et al. Novel OTX2 mutation associated with congenital anophthalmia and microphthalmia in a Han Chinese family. Acta Ophthalmol. 2012;90(6):e501–e502. [DOI] [PubMed] [Google Scholar]
- 187. Gerth-Kahlert C, Williamson K, Ansari M, et al. Clinical and mutation analysis of 51 probands with anophthalmia and/or severe microphthalmia from a single center. Mol Genet Genomic Med. 2013;1(1):15–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Vincent A, Forster N, Maynes JT, et al. OTX2 mutations cause autosomal dominant pattern dystrophy of the retinal pigment epithelium. J Med Genet. 2014;51(12):797–805. [DOI] [PubMed] [Google Scholar]
- 189. Slavotinek AM, Garcia ST, Chandratillake G, et al. Exome sequencing in 32 patients with anophthalmia/microphthalmia and developmental eye defects. Clin Genet. 2015;88(5):468–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Henderson RH, Williamson KA, Kennedy JS, et al. A rare de novo nonsense mutation in OTX2 causes early onset retinal dystrophy and pituitary dysfunction. Mol Vis. 2009;15:2442–2447. [PMC free article] [PubMed] [Google Scholar]
- 191. Gonzalez-Rodriguez J, Pelcastre EL, Tovilla-Canales JL, et al. Mutational screening of CHX10, GDF6, OTX2, RAX and SOX2 genes in 50 unrelated microphthalmia-anophthalmia-coloboma (MAC) spectrum cases. Br J Ophthalmol. 2010;94(8):1100–1104. [DOI] [PubMed] [Google Scholar]
- 192. Chassaing N, Sorrentino S, Davis EE, et al. OTX2 mutations contribute to the otocephaly-dysgnathia complex. J Med Genet. 2012;49(6):373–379. [DOI] [PubMed] [Google Scholar]
- 193. Sergouniotis PI, Urquhart JE, Williams SG, et al. Agnathia-otocephaly complex and asymmetric velopharyngeal insufficiency due to an in-frame duplication in OTX2. J Hum Genet. 2015;60(4):199–202. [DOI] [PubMed] [Google Scholar]
- 194. Aradhya S, Lewis R, Bonaga T, et al. Exon-level array CGH in a large clinical cohort demonstrates increased sensitivity of diagnostic testing for Mendelian disorders. Genet Med. 2012;14(6):594–603. [DOI] [PubMed] [Google Scholar]
- 195. Patat O, van Ravenswaaij-Arts CM, Tantau J, et al. Otocephaly-dysgnathia complex: description of four cases and confirmation of the role of OTX2. Mol Syndromol. 2013;4(6):302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Delahaye A, Bitoun P, Drunat S, et al. Genomic imbalances detected by array-CGH in patients with syndromal ocular developmental anomalies. Eur J Hum Genet. 2012;20(5):527–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Prasov L, Masud T, Khaliq S, et al. ATOH7 mutations cause autosomal recessive persistent hyperplasia of the primary vitreous. Hum Mol Genet. 2012;21(16):3681–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Balikova I, de Ravel T, Ayuso C, et al. High frequency of submicroscopic chromosomal deletions in patients with idiopathic congenital eye malformations. Am J Ophthalmol. 2011;151(6):1087–1094.e45. [DOI] [PubMed] [Google Scholar]
- 199. Takenouchi T, Nishina S, Kosaki R, et al. Concurrent deletion of BMP4 and OTX2 genes, two master genes in ophthalmogenesis. Eur J Med Genet. 2013;56(1):50–53. [DOI] [PubMed] [Google Scholar]
- 200. Takagi M, Nagasaki K, Fujiwara I, et al. Heterozygous defects in PAX6 gene and congenital hypopituitarism. Eur J Endocrinol. 2015;172(1):37–45. [DOI] [PubMed] [Google Scholar]
- 201. Ballesta-Martínez MJ, López-González V, Dulcet LA, Rodríguez-Santiago B, Garcia-Miñaúr S, Guillen-Navarro E. Autosomal dominant oculoauriculovertebral spectrum and 14q23.1 microduplication. Am J Med Genet A. 2013;161A(8):2030–2035. [DOI] [PubMed] [Google Scholar]
- 202. Zielinski D, Markus B, Sheikh M, et al. OTX2 duplication is implicated in hemifacial microsomia. PLoS One. 2014;9(5):e96788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Gergics P, Fang Q, George AS, et al. OTX2 deficiency and combined pituitary hormone deficiency: interacting genes affect penetrance. In: Proceedings of the Endocrine Society's 98th Annual Meeting and Expo; April 1–4, 2016; Boston, MA Abstract SUN-520. [Google Scholar]
- 204. Moreira M, Franca MM, Otto AP, et al. A novel heterozygous OTX2 deleterious variant (p.H230L) in a patient with hypopituitarism and ectopic posterior pituitary without eye malformation. In: Proceedings of the Endocrine Society's 94th Annual Meeting and Expo; June 23–26, 2012; Houston, TX Abstract SUN-592. [Google Scholar]
- 205. Arnhold IJ, França MM, Carvalho LR, Mendonca BB, Jorge AA. Role of GLI2 in hypopituitarism phenotype. J Mol Endocrinol. 2015;54(3):R141–R150. [DOI] [PubMed] [Google Scholar]
- 206. Bear KA, Solomon BD, Antonini S, et al. Pathogenic mutations in GLI2 cause a specific phenotype that is distinct from holoprosencephaly. J Med Genet. 2014;51(6):413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Bertolacini CD, Ribeiro-Bicudo LA, Petrin A, Richieri-Costa A, Murray JC. Clinical findings in patients with GLI2 mutations–phenotypic variability. Clin Genet. 2012;81(1):70–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Flemming GM, Klammt J, Ambler G, et al. Functional characterization of a heterozygous GLI2 missense mutation in patients with multiple pituitary hormone deficiency. J Clin Endocrinol Metab. 2013;98(3):E567–E575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. França MM, Jorge AA, Carvalho LR, et al. Novel heterozygous nonsense GLI2 mutations in patients with hypopituitarism and ectopic posterior pituitary lobe without holoprosencephaly. J Clin Endocrinol Metab. 2010;95(11):E384–E391. [DOI] [PubMed] [Google Scholar]
- 210. Fromer M, Pocklington AJ, Kavanagh DH, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506(7487):179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Kevelam SH, van Harssel JJ, van der Zwaag B, Smeets HJ, Paulussen AD, Lichtenbelt KD. A patient with a mild holoprosencephaly spectrum phenotype and deletion encompassing GLI2. Am J Med Genet A. 2012;158A(1):166–173. [DOI] [PubMed] [Google Scholar]
- 212. Rahimov F, Ribeiro LA, de Miranda E, Richieri-Costa A, Murray JC. GLI2 mutations in four Brazilian patients: how wide is the phenotypic spectrum? Am J Med Genet A. 2006;140(23):2571–2576. [DOI] [PubMed] [Google Scholar]
- 213. Richieri-Costa A, Ribeiro LA. Holoprosencephaly-like phenotype: clinical and genetic perspectives. Am J Med Genet A. 2006;140(23):2587–2593. [DOI] [PubMed] [Google Scholar]
- 214. Roessler E, Ermilov AN, Grange DK, et al. A previously unidentified amino-terminal domain regulates transcriptional activity of wild-type and disease-associated human GLI2. Hum Mol Genet. 2005;14(15):2181–2188. [DOI] [PubMed] [Google Scholar]
- 215. Solomon BD, Pineda-Alvarez DE, Gropman AL, Willis MJ, Hadley DW, Muenke M. High intellectual function in individuals with mutation-positive microform holoprosencephaly. Mol Syndromol. 2012;3(3):140–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Vieira AR, Avila JR, Daack-Hirsch S, et al. Medical sequencing of candidate genes for nonsyndromic cleft lip and palate. PLoS Genet. 2005;1(6):e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Wannasilp N, Solomon BD, Warren-Mora N, et al. Holoprosencephaly in a family segregating novel variants in ZIC2 and GLI2. Am J Med Genet A. 2011;155A(4):860–864. [DOI] [PubMed] [Google Scholar]
- 218. Gregory LC, Gaston-Massuet C, Andoniadou CL, et al. The role of the sonic hedgehog signalling pathway in patients with midline defects and congenital hypopituitarism. Clin Endocrinol (Oxf). 2015;82(5):728–738. [DOI] [PubMed] [Google Scholar]
- 219. Alatzoglou KS, Andoniadou CL, Kelberman D, et al. SOX2 haploinsufficiency is associated with slow progressing hypothalamo-pituitary tumours. Hum Mutat. 2011;32(12):1376–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Wood HB, Episkopou V. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech Dev. 1999;86(1–2):197–201. [DOI] [PubMed] [Google Scholar]
- 221. Schneider A, Bardakjian TM, Zhou J, et al. Familial recurrence of SOX2 anophthalmia syndrome: phenotypically normal mother with two affected daughters. Am J Med Genet A. 2008;146A(21):2794–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Faivre L, Williamson KA, Faber V, et al. Recurrence of SOX2 anophthalmia syndrome with gonosomal mosaicism in a phenotypically normal mother. Am J Med Genet A. 2006;140(6):636–639. [DOI] [PubMed] [Google Scholar]
- 223. Chassaing N, Gilbert-Dussardier B, Nicot F, et al. Germinal mosaicism and familial recurrence of a SOX2 mutation with highly variable phenotypic expression extending from AEG syndrome to absence of ocular involvement. Am J Med Genet A. 2007;143A(3):289–291. [DOI] [PubMed] [Google Scholar]
- 224. Kelberman D, Rizzoti K, Avilion A, et al. Mutations within Sox2/SOX2 are associated with abnormalities in the hypothalamo-pituitary-gonadal axis in mice and humans. J Clin Invest. 2006;116(9):2442–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Macchiaroli A, Kelberman D, Auriemma RS, et al. A novel heterozygous SOX2 mutation causing congenital bilateral anophthalmia, hypogonadotropic hypogonadism and growth hormone deficiency. Gene. 2014;534(2):282–285. [DOI] [PubMed] [Google Scholar]
- 226. Schneider A, Bardakjian T, Reis LM, Tyler RC, Semina EV. Novel SOX2 mutations and genotype-phenotype correlation in anophthalmia and microphthalmia. Am J Med Genet A. 2009;149A(12):2706–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Schilter KF, Reis LM, Schneider A, et al. Whole-genome copy number variation analysis in anophthalmia and microphthalmia. Clin Genet. 2013;84(5):473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Hagstrom SA, Pauer GJ, Reid J, et al. SOX2 mutation causes anophthalmia, hearing loss, and brain anomalies. Am J Med Genet. 2005;138A(2):95–98. [DOI] [PubMed] [Google Scholar]
- 229. Bakrania P, Robinson DO, Bunyan DJ, et al. SOX2 anophthalmia syndrome: 12 new cases demonstrating broader phenotype and high frequency of large gene deletions. Br J Ophthalmol. 2007;91(11):1471–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Williamson KA, Hever AM, Rainger J, et al. Mutations in SOX2 cause anophthalmia-esophageal-genital (AEG) syndrome. Hum Mol Genet. 2006;15(9):1413–1422. [DOI] [PubMed] [Google Scholar]
- 231. Ragge NK, Quaghebeur G, Stewart H. SOX2 anophthalmia syndrome in adulthood - a neurodegenerative picture? Clin Genet. 2013;83(5):482–484. [DOI] [PubMed] [Google Scholar]
- 232. Kelberman D, de Castro SC, Huang S, et al. SOX2 plays a critical role in the pituitary, forebrain, and eye during human embryonic development. J Clin Endocrinol Metab. 2008;93(5):1865–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Jayakody SA, Andoniadou CL, Gaston-Massuet C, et al. SOX2 regulates the hypothalamic-pituitary axis at multiple levels. J Clin Invest. 2012;122(10):3635–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Sato N, Kamachi Y, Kondoh H, et al. Hypogonadotropic hypogonadism in an adult female with a heterozygous hypomorphic mutation of SOX2. Eur J Endocrinol. 2007;156(2):167–171. [DOI] [PubMed] [Google Scholar]
- 235. Takagi M, Narumi S, Asakura Y, et al. A novel mutation in SOX2 causes hypogonadotropic hypogonadism with mild ocular malformation. Horm Res Paediatr. 2014;81(2):133–138. [DOI] [PubMed] [Google Scholar]
- 236. Stark Z, Storen R, Bennetts B, Savarirayan R, Jamieson RV. Isolated hypogonadotropic hypogonadism with SOX2 mutation and anophthalmia/microphthalmia in offspring. Eur J Hum Genet. 2011;19(7):753–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Alatzoglou KS, Kelberman D, Cowell CT, et al. Increased transactivation associated with SOX3 polyalanine tract deletion in a patient with hypopituitarism. J Clin Endocrinol Metab. 2011;96(4):E685–E690. [DOI] [PubMed] [Google Scholar]
- 238. Bauters M, Frints SG, Van Esch H, et al. Evidence for increased SOX3 dosage as a risk factor for X-linked hypopituitarism and neural tube defects. Am J Med Genet A. 2014;164A(8):1947–1952. [DOI] [PubMed] [Google Scholar]
- 239. Takagi M, Ishii T, Torii C, Kosaki K, Hasegawa T. A novel mutation in SOX3 polyalanine tract: a case of Kabuki syndrome with combined pituitary hormone deficiency harboring double mutations in MLL2 and SOX3. Pituitary. 2014;17(6):569–574. [DOI] [PubMed] [Google Scholar]
- 240. Woods KS, Cundall M, Turton J, et al. Over- and underdosage of SOX3 is associated with infundibular hypoplasia and hypopituitarism. Am J Hum Genet. 2005;76(5):833–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Burkitt Wright EM, Perveen R, Clayton PE, et al. X-linked isolated growth hormone deficiency: expanding the phenotypic spectrum of SOX3 polyalanine tract expansions. Clin Dysmorphol. 2009;18(4):218–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Solomon NM, Ross SA, Forrest SM, et al. Array comparative genomic hybridisation analysis of boys with X-linked hypopituitarism identifies a 3.9 Mb duplicated critical region at Xq27 containing SOX3. J Med Genet. 2007;44(4):e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Sutton E, Hughes J, White S, et al. Identification of SOX3 as an XX male sex reversal gene in mice and humans. J Clin Invest. 2011;121(1):328–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Vetro A, Dehghani MR, Kraoua L, et al. Testis development in the absence of SRY: chromosomal rearrangements at SOX9 and SOX3. Eur J Hum Genet. 2015;23(8):1025–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Moalem S, Babul-Hirji R, Stavropolous DJ, et al. XX male sex reversal with genital abnormalities associated with a de novo SOX3 gene duplication. Am J Med Genet A. 2012;158A(7):1759–1764. [DOI] [PubMed] [Google Scholar]
- 246. Haines B, Hughes J, Corbett M, et al. Interchromosomal insertional translocation at Xq26.3 alters SOX3 expression in an individual with XX male sex reversal. J Clin Endocrinol Metab. 2015;100(5):E815–E820. [DOI] [PubMed] [Google Scholar]
- 247. Breitfeld J, Martens S, Klammt J, et al. Genetic analyses of bone morphogenetic protein 2, 4 and 7 in congenital combined pituitary hormone deficiency. BMC Endocr Disord. 2013;13:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Dattani MT, Robinson IC. The molecular basis for developmental disorders of the pituitary gland in man. Clin Genet. 2000;57(5):337–346. [DOI] [PubMed] [Google Scholar]
- 249. Takuma N, Sheng HZ, Furuta Y, et al. Formation of Rathke's pouch requires dual induction from the diencephalon. Development. 1998;125(23):4835–4840. [DOI] [PubMed] [Google Scholar]
- 250. Ericson J, Norlin S, Jessell TM, Edlund T. Integrated FGF and BMP signaling controls the progression of progenitor cell differentiation and the emergence of pattern in the embryonic anterior pituitary. Development. 1998;125(6):1005–1015. [DOI] [PubMed] [Google Scholar]
- 251. Suzuki E, Yatsuga S, Igarashi M, et al. De novo frameshift mutation in fibroblast growth factor 8 in a male patient with gonadotropin deficiency. Horm Res Paediatr. 2014;81(2):139–144. [DOI] [PubMed] [Google Scholar]
- 252. Tornberg J, Sykiotis GP, Keefe K, et al. Heparan sulfate 6-O-sulfotransferase 1, a gene involved in extracellular sugar modifications, is mutated in patients with idiopathic hypogonadotrophic hypogonadism. Proc Natl Acad Sci USA. 2011;108(28):11524–11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Hanchate NK, Giacobini P, Lhuillier P, et al. SEMA3A, a gene involved in axonal pathfinding, is mutated in patients with Kallmann syndrome. PLoS Genet. 2012;8(8):e1002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Correa FA, Trarbach EB, Tusset C, et al. FGFR1 and PROKR2 rare variants found in patients with combined pituitary hormone deficiencies. Endocr Connect. 2015;4(2):100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Démurger F, Ichkou A, Mougou-Zerelli S, et al. New insights into genotype-phenotype correlation for GLI3 mutations. Eur J Hum Genet. 2015;23(1):92–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Johnston JJ, Olivos-Glander I, Killoran C, et al. Molecular and clinical analyses of Greig cephalopolysyndactyly and Pallister-Hall syndromes: robust phenotype prediction from the type and position of GLI3 mutations. Am J Hum Genet. 2005;76(4):609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Johnston JJ, Sapp JC, Turner JT, et al. Molecular analysis expands the spectrum of phenotypes associated with GLI3 mutations. Hum Mutat. 2010;31(10):1142–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Galasso C, Scirè G, Fabbri F, et al. Long-term treatment with growth hormone improves final height in a patient with Pallister-Hall syndrome. Am J Med Genet. 2001;99(2):128–131. [DOI] [PubMed] [Google Scholar]
- 259. Ng D, Johnston JJ, Turner JT, et al. Gonadal mosaicism in severe Pallister-Hall syndrome. Am J Med Genet A. 2004;124A(3):296–302. [DOI] [PubMed] [Google Scholar]
- 260. Narumi Y, Kosho T, Tsuruta G, et al. Genital abnormalities in Pallister-Hall syndrome: report of two patients and review of the literature. Am J Med Genet A. 2010;152A(12):3143–3147. [DOI] [PubMed] [Google Scholar]
- 261. Haddad-Tóvolli R, Paul FA, Zhang Y, et al. Differential requirements for Gli2 and Gli3 in the regional specification of the mouse hypothalamus. Front Neuroanat. 2015;9:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Frattini A, Faranda S, Redolfi E, Allavena P, Vezzoni P. Identification and genomic organization of a gene coding for a new member of the cell adhesion molecule family mapping to Xq25. Gene. 1998;214(1–2):1–6. [DOI] [PubMed] [Google Scholar]
- 263. Mazzarella R, Pengue G, Jones J, Jones C, Schlessinger D. Cloning and expression of an immunoglobulin superfamily gene (IGSF1) in Xq25. Genomics. 1998;48(2):157–162. [DOI] [PubMed] [Google Scholar]
- 264. Robakis T, Bak B, Lin SH, Bernard DJ, Scheiffele P. An internal signal sequence directs intramembrane proteolysis of a cellular immunoglobulin domain protein. J Biol Chem. 2008;283(52):36369–36376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Sun Y, Bak B, Schoenmakers N, et al. Loss-of-function mutations in IGSF1 cause an X-linked syndrome of central hypothyroidism and testicular enlargement. Nat Genet. 2012;44(12):1375–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Chong H, Pangas SA, Bernard DJ, et al. Structure and expression of a membrane component of the inhibin receptor system. Endocrinology. 2000;141(7):2600–2607. [DOI] [PubMed] [Google Scholar]
- 267. Bernard DJ, Woodruff TK. Inhibin binding protein in rats: alternative transcripts and regulation in the pituitary across the estrous cycle. Mol Endocrinol. 2001;15(4):654–667. [DOI] [PubMed] [Google Scholar]
- 268. Bernard DJ, Burns KH, Haupt B, Matzuk MM, Woodruff TK. Normal reproductive function in InhBP/p120-deficient mice. Mol Cell Biol. 2003;23(14):4882–4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Nakamura A, Bak B, Silander TL, et al. Three novel IGSF1 mutations in four Japanese patients with X-linked congenital central hypothyroidism. J Clin Endocrinol Metab. 2013;98(10):E1682–E1691. [DOI] [PubMed] [Google Scholar]
- 270. Tajima T, Nakamura A, Ishizu K. A novel mutation of IGSF1 in a Japanese patient of congenital central hypothyroidism without macroorchidism. Endocr J. 2013;60(2):245–249. [DOI] [PubMed] [Google Scholar]
- 271. Asakura Y, Abe K, Muroya K, et al. Combined growth hormone and thyroid-stimulating hormone deficiency in a Japanese patient with a novel frameshift mutation in IGSF1. Horm Res Paediatr. 2015;84(5):349–354. [DOI] [PubMed] [Google Scholar]
- 272. Joustra SD, Roelfsema F, Endert E, et al. Pituitary hormone secretion profiles in IGSF1 deficiency syndrome. Neuroendocrinology. 2016;103(3–4):408–416. [DOI] [PubMed] [Google Scholar]
- 273. Hughes JN, Aubert M, Heatlie J, et al. Identification of an IGSF1-specific deletion in a five-generation pedigree with X-linked central hypothyroidism without macroorchidism. Clin Endocrinol (Oxf). 2016;85(4):609–615. [DOI] [PubMed] [Google Scholar]
- 274. Tenenbaum-Rakover Y, Turgeon MO, London S, et al. Familial central hypothyroidism caused by a novel IGSF1 gene mutation [published online July 25, 2016]. Thyroid. 10.1089/thy.2015.0672. [DOI] [PubMed] [Google Scholar]
- 275. Joustra SD, Schoenmakers N, Persani L, et al. The IGSF1 deficiency syndrome: characteristics of male and female patients. J Clin Endocrinol Metab. 2013;98(12):4942–4952. [DOI] [PubMed] [Google Scholar]
- 276. Joustra SD, Heinen CA, Schoenmakers N, et al. IGSF1 deficiency: lessons from an extensive case series and recommendations for clinical management. J Clin Endocrinol Metab. 2016;101(4):1627–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Jordan T, Hanson I, Zaletayev D, et al. The human PAX6 gene is mutated in two patients with aniridia. Nat Genet. 1992;1(5):328–332. [DOI] [PubMed] [Google Scholar]
- 278. Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7(4):463–471. [DOI] [PubMed] [Google Scholar]
- 279. Favor J, Gloeckner CJ, Neuhäuser-Klaus A, et al. Relationship of Pax6 activity levels to the extent of eye development in the mouse, Mus musculus. Genetics. 2008;179(3):1345–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Manuel M, Pratt T, Liu M, Jeffery G, Price DJ. Overexpression of Pax6 results in microphthalmia, retinal dysplasia and defective retinal ganglion cell axon guidance. BMC Dev Biol. 2008;8:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Kioussi C, O'Connell S, St-Onge L, et al. Pax6 is essential for establishing ventral-dorsal cell boundaries in pituitary gland development. Proc Natl Acad Sci USA. 1999;96(25):14378–14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Chanas SA, Collinson JM, Ramaesh T, et al. Effects of elevated Pax6 expression and genetic background on mouse eye development. Invest Ophthalmol Vis Sci. 2009;50(9):4045–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Bentley CA, Zidehsarai MP, Grindley JC, Parlow AF, Barth-Hall S, Roberts VJ. Pax6 is implicated in murine pituitary endocrine function. Endocrine. 1999;10(2):171–177. [DOI] [PubMed] [Google Scholar]
- 284. Cole LW, Sidis Y, Zhang C, et al. Mutations in prokineticin 2 and prokineticin receptor 2 genes in human gonadotrophin-releasing hormone deficiency: molecular genetics and clinical spectrum. J Clin Endocrinol Metab. 2008;93(9):3551–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Caronia LM, Martin C, Welt CK, et al. A genetic basis for functional hypothalamic amenorrhea. N Engl J Med. 2011;364(3):215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Abreu AP, Noel SD, Xu S, Carroll RS, Latronico AC, Kaiser UB. Evidence of the importance of the first intracellular loop of prokineticin receptor 2 in receptor function. Mol Endocrinol. 2012;26(8):1417–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Reynaud R, Jayakody SA, Monnier C, et al. PROKR2 variants in multiple hypopituitarism with pituitary stalk interruption. J Clin Endocrinol Metab. 2012;97(6):E1068–E1073. [DOI] [PubMed] [Google Scholar]
- 288. McCabe MJ, Gaston-Massuet C, Gregory LC, et al. Variations in PROKR2, but not PROK2, are associated with hypopituitarism and septo-optic dysplasia. J Clin Endocrinol Metab. 2013;98(3):E547–E557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Libri DV, Kleinau G, Vezzoli V, et al. Germline prokineticin receptor 2 (PROKR2) variants associated with central hypogonadism cause differential modulation of distinct intracellular pathways. J Clin Endocrinol Metab. 2014;99(3):E458–E63. [DOI] [PubMed] [Google Scholar]
- 290. Sbai O, Monnier C, Dodé C, Pin JP, Hardelin JP, Rondard P. Biased signaling through G-protein-coupled PROKR2 receptors harboring missense mutations. FASEB J. 2014;28(8):3734–3744. [DOI] [PubMed] [Google Scholar]
- 291. Dodé C, Teixeira L, Levilliers J, et al. Kallmann syndrome: mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genet. 2006;2(10):e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Matsumoto S, Yamazaki C, Masumoto KH, et al. Abnormal development of the olfactory bulb and reproductive system in mice lacking prokineticin receptor PKR2. Proc Natl Acad Sci USA. 2006;103(11):4140–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Prosser HM, Bradley A, Caldwell MA. Olfactory bulb hypoplasia in Prokr2 null mice stems from defective neuronal progenitor migration and differentiation. Eur J Neurosci. 2007;26(12):3339–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Xiao L, Zhang C, Li X, et al. Signaling role of prokineticin 2 on the estrous cycle of female mice. PLoS One 2014;9(3):e90860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Abreu AP, Trarbach EB, de Castro M, et al. Loss-of-function mutations in the genes encoding prokineticin-2 or prokineticin receptor-2 cause autosomal recessive Kallmann syndrome. J Clin Endocrinol Metab. 2008;93(10):4113–4118. [DOI] [PubMed] [Google Scholar]
- 296. Puverel S, Nakatani H, Parras C, Soussi-Yanicostas N. Prokineticin receptor 2 expression identifies migrating neuroblasts and their subventricular zone transient-amplifying progenitors in adult mice. J Comp Neurol. 2009;512(2):232–242. [DOI] [PubMed] [Google Scholar]
- 297. Tatsi C, Sertedaki A, Voutetakis A, et al. Pituitary stalk interruption syndrome and isolated pituitary hypoplasia may be caused by mutations in holoprosencephaly-related genes. J Clin Endocrinol Metab. 2013;98(4):E779–E784. [DOI] [PubMed] [Google Scholar]
- 298. Gorbenko del Blanco D, de Graaff LC, Visser TJ, Hokken-Koelega AC. Single-nucleotide variants in two Hedgehog genes, SHH and HHIP, as genetic cause of combined pituitary hormone deficiency. Clin Endocrinol (Oxf). 2013;78(3):415–423. [DOI] [PubMed] [Google Scholar]
- 299. Roose J, Molenaar M, Peterson J, et al. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395(6702):608–612. [DOI] [PubMed] [Google Scholar]
- 300. Sharpe C, Lawrence N, Martinez Arias A. Wnt signalling: a theme with nuclear variations. Bioessays. 2001;23(4):311–318. [DOI] [PubMed] [Google Scholar]
- 301. Cha KB, Douglas KR, Potok MA, Liang H, Jones SN, Camper SA. WNT5A signaling affects pituitary gland shape. Mech Dev. 2004;121(2):183–194. [DOI] [PubMed] [Google Scholar]
- 302. Potok MA, Cha KB, Hunt A, et al. WNT signaling affects gene expression in the ventral diencephalon and pituitary gland growth. Dev Dyn. 2008;237(4):1006–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Brinkmeier ML, Potok MA, Cha KB, et al. TCF and Groucho-related genes influence pituitary growth and development. Mol Endocrinol. 2003;17(11):2152–2161. [DOI] [PubMed] [Google Scholar]
- 304. Brinkmeier ML, Potok MA, Davis SW, Camper SA. TCF4 deficiency expands ventral diencephalon signaling and increases induction of pituitary progenitors. Dev Biol. 2007;311(2):396–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Olson LE, Tollkuhn J, Scafoglio C, et al. Homeodomain-mediated β-catenin-dependent switching events dictate cell-lineage determination. Cell. 2006;125(3):593–605. [DOI] [PubMed] [Google Scholar]
- 306. Kioussi C, Briata P, Baek SH, et al. Identification of a Wnt/Dvl/β-catenin → Pitx2 pathway mediating cell-type-specific proliferation during development. Cell. 2002;111(5):673–685. [DOI] [PubMed] [Google Scholar]
- 307. Gaston-Massuet C, McCabe MJ, Scagliotti V, et al. Transcription factor 7-like 1 is involved in hypothalamo-pituitary axis development in mice and humans. Proc Natl Acad Sci USA. 2016;113(5):E548–E557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. García M, Fernández A, Moreno JC. Central hypothyroidism in children. Endocr Dev. 2014;26:79–107. [DOI] [PubMed] [Google Scholar]
- 309. El-Jaick KB, Powers SE, Bartholin L, et al. Functional analysis of mutations in TGIF associated with holoprosencephaly. Mol Genet Metab. 2007;90(1):97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Bartholin L, Melhuish TA, Powers SE, et al. Maternal Tgif is required for vascularization of the embryonic placenta. Dev Biol. 2008;319(2):285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Kuang C, Xiao Y, Yang L, et al. Intragenic deletion of Tgif causes defects in brain development. Hum Mol Genet. 2006;15(24):3508–3519. [DOI] [PubMed] [Google Scholar]
- 312. Powers SE, Taniguchi K, Yen W, et al. Tgif1 and Tgif2 regulate Nodal signaling and are required for gastrulation. Development. 2010;137(2):249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Jongmans MC, Admiraal RJ, van der Donk KP, et al. CHARGE syndrome: the phenotypic spectrum of mutations in the CHD7 gene. J Med Genet. 2006;43(4):306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Lalani SR, Safiullah AM, Fernbach SD, et al. Spectrum of CHD7 mutations in 110 individuals with CHARGE syndrome and genotype-phenotype correlation. Am J Hum Genet. 2006;78(2):303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Asakura Y, Toyota Y, Muroya K, et al. Endocrine and radiological studies in patients with molecularly confirmed CHARGE syndrome. J Clin Endocrinol Metab. 2008;93(3):920–924. [DOI] [PubMed] [Google Scholar]
- 316. Esposito A, Tufano M, Di Donato I, et al. Effect of long-term GH treatment in a patient with CHARGE association. Ital J Pediatr. 2014;40:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Gregory LC, Gevers EF, Baker J, et al. Structural pituitary abnormalities associated with CHARGE syndrome. J Clin Endocrinol Metab. 2013;98(4):E737–E743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Hurd EA, Capers PL, Blauwkamp MN, et al. Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mamm Genome. 2007;18(2):94–104. [DOI] [PubMed] [Google Scholar]
- 319. Layman WS, Hurd EA, Martin DM. Reproductive dysfunction and decreased GnRH neurogenesis in a mouse model of CHARGE syndrome. Hum Mol Genet. 2011;20(16):3138–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Paulo SS, Fernandes-Rosa FL, Turatti W, et al. Sonic Hedgehog mutations are not a common cause of congenital hypopituitarism in the absence of complex midline cerebral defects. Clin Endocrinol (Oxf). 2015;82(4):562–569. [DOI] [PubMed] [Google Scholar]
- 321. Grzegorski SJ, Chiari EF, Robbins A, Kish PE, Kahana A. Natural variability of Kozak sequences correlates with function in a zebrafish model. PLoS One. 2014;9(9):e108475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Craft WH, Underwoood LE, Van Wyk JJ. High incidence of perinatal insult in children with idiopathic hypopituitarism. J Pediatr. 1980;96(3 Pt 1):397–402. [DOI] [PubMed] [Google Scholar]
- 323. Chuang PT, Kawcak T, McMahon AP. Feedback control of mammalian Hedgehog signaling by the Hedgehog-binding protein, Hip1, modulates Fgf signaling during branching morphogenesis of the lung. Genes Dev. 2003;17(3):342–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324. Jeong J, McMahon AP. Growth and pattern of the mammalian neural tube are governed by partially overlapping feedback activities of the hedgehog antagonists patched 1 and Hhip1. Development. 2005;132(1):143–154. [DOI] [PubMed] [Google Scholar]
- 325. Kawahira H, Ma NH, Tzanakakis ES, McMahon AP, Chuang PT, Hebrok M. Combined activities of hedgehog signaling inhibitors regulate pancreas development. Development. 2003;130(20):4871–4879. [DOI] [PubMed] [Google Scholar]
- 326. Wolf NI, Vanderver A, van Spaendonk RM, et al. Clinical spectrum of 4H leukodystrophy caused by POLR3A and POLR3B mutations. Neurology. 2014;83(21):1898–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327. Potic A, Brais B, Choquet K, Schiffmann R, Bernard G. 4H syndrome with late-onset growth hormone deficiency caused by POLR3A mutations. Arch Neurol. 2012;69(7):920–923. [DOI] [PubMed] [Google Scholar]
- 328. Damianov A, Kann M, Lane WS, Bindereif A. Human RBM28 protein is a specific nucleolar component of the spliceosomal snRNPs. Biol Chem. 2006;387(10–11):1455–1460. [DOI] [PubMed] [Google Scholar]
- 329. Nousbeck J, Spiegel R, Ishida-Yamamoto A, et al. Alopecia, neurological defects, and endocrinopathy syndrome caused by decreased expression of RBM28, a nucleolar protein associated with ribosome biogenesis. Am J Hum Genet. 2008;82(5):1114–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330. Spiegel R, Shalev SA, Adawi A, Sprecher E, Tenenbaum-Rakover Y. ANE syndrome caused by mutated RBM28 gene: a novel etiology of combined pituitary hormone deficiency. Eur J Endocrinol. 2010;162(6):1021–1025. [DOI] [PubMed] [Google Scholar]
- 331. Kim HG, Ahn JW, Kurth I, et al. WDR11, a WD protein that interacts with transcription factor EMX1, is mutated in idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am J Hum Genet. 2010;87(4):465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332. Quaynor SD, Kim HG, Cappello EM, et al. The prevalence of digenic mutations in patients with normosmic hypogonadotropic hypogonadism and Kallmann syndrome. Fertil Steril. 2011;96(6):1424–1430.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333. Porollo A, Meller J. Prediction-based fingerprints of protein-protein interactions. Proteins. 2007;66(3):630–645. [DOI] [PubMed] [Google Scholar]
- 334. Ngan ES, Kim KH, Hui CC. Sonic hedgehog signaling and VACTERL association. Mol Syndromol. 2013;4(1–2):32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335. Lucas-Herald AK, Kinning E, Iida A, et al. A case of functional growth hormone deficiency and early growth retardation in a child with IFT172 mutations. J Clin Endocrinol Metab. 2015;100(4):1221–1224. [DOI] [PubMed] [Google Scholar]
- 336. Friedland-Little JM, Hoffmann AD, Ocbina PJ, et al. A novel murine allele of intraflagellar transport protein 172 causes a syndrome including VACTERL-like features with hydrocephalus. Hum Mol Genet. 2011;20(19):3725–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337. Gorivodsky M, Mukhopadhyay M, Wilsch-Braeuninger M, et al. Intraflagellar transport protein 172 is essential for primary cilia formation and plays a vital role in patterning the mammalian brain. Dev Biol. 2009;325(1):24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338. Howard PW, Howard TL, Maurer RA. Generation of mice with a conditional allele for Ift172. Transgenic Res. 2010;19(1):121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339. Bersten DC, Bruning JB, Peet DJ, Whitelaw ML. Human variants in the neuronal basic helix-loop-helix/Per-Arnt-Sim (bHLH/PAS) transcription factor complex NPAS4/ARNT2 disrupt function. PLoS One. 2014;9(1):e85768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340. Aitola MH, Pelto-Huikko MT. Expression of Arnt and Arnt2 mRNA in developing murine tissues. J Histochem Cytochem. 2003;51(1):41–54. [DOI] [PubMed] [Google Scholar]
- 341. Hosoya T, Oda Y, Takahashi S, et al. Defective development of secretory neurones in the hypothalamus of Arnt2-knockout mice. Genes Cells. 2001;6(4):361–374. [DOI] [PubMed] [Google Scholar]
- 342. Keith B, Adelman DM, Simon MC. Targeted mutation of the murine arylhydrocarbon receptor nuclear translocator 2 (Arnt2) gene reveals partial redundancy with Arnt. Proc Natl Acad Sci USA. 2001;98(12):6692–6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343. Michaud JL, DeRossi C, May NR, Holdener BC, Fan CM. ARNT2 acts as the dimerization partner of SIM1 for the development of the hypothalamus. Mech Dev. 2000;90(2):253–261. [DOI] [PubMed] [Google Scholar]
- 344. Michaud JL, Rosenquist T, May NR, Fan CM. Development of neuroendocrine lineages requires the bHLH-PAS transcription factor SIM1. Genes Dev. 1998;12(20):3264–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345. Smith JD, Hing AV, Clarke CM, et al. Exome sequencing identifies a recurrent de novo ZSWIM6 mutation associated with acromelic frontonasal dysostosis. Am J Hum Genet. 2014;95(2):235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346. Twigg SR, Ousager LB, Miller KA, et al. Acromelic frontonasal dysostosis and ZSWIM6 mutation: phenotypic spectrum and mosaicism. Clin Genet. 2016;90(3):270–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347. Karaca E, Buyukkaya R, Pehlivan D, et al. Whole-exome sequencing identifies homozygous GPR161 mutation in a family with pituitary stalk interruption syndrome. J Clin Endocrinol Metab. 2015;100(1):E140–E147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348. Li BI, Matteson PG, Ababon MF, et al. The orphan GPCR, Gpr161, regulates the retinoic acid and canonical Wnt pathways during neurulation. Dev Biol. 2015;402(1):17–31. [DOI] [PubMed] [Google Scholar]
- 349. Rhodes SJ, Chen R, DiMattia GE, et al. A tissue-specific enhancer confers Pit-1-dependent morphogen inducibility and autoregulation on the pit-1 gene. Genes Dev. 1993;7(6):913–932. [DOI] [PubMed] [Google Scholar]
- 350. Cohen LE, Zanger K, Brue T, Wondisford FE, Radovick S. Defective retinoic acid regulation of the Pit-1 gene enhancer: a novel mechanism of combined pituitary hormone deficiency. Mol Endocrinol. 1999;13(3):476–484. [DOI] [PubMed] [Google Scholar]
- 351. Mukhopadhyay S, Wen X, Ratti N, et al. The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell. 2013;152(1–2):210–223. [DOI] [PubMed] [Google Scholar]
- 352. Epi4K Consortium, Epilepsy Phenome/Genome Project, Allen AS, Berkovic SF, Cossette P, et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501(7466):217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353. Zhu X, Petrovski S, Xie P, et al. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet Med. 2015;17(10):774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354. Huang Y, Huang K, Boskovic G, et al. Proteomic and genomic analysis of PITX2 interacting and regulating networks. FEBS Lett. 2009;583(4):638–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355. Jin JM, Yang WX. Molecular regulation of hypothalamus-pituitary-gonads axis in males. Gene 2014;551(1):15–25. [DOI] [PubMed] [Google Scholar]
- 356. Zhao S, Korzan WJ, Chen CC, Fernald RD. Heterogeneous nuclear ribonucleoprotein A/B and G inhibits the transcription of gonadotropin-releasing-hormone 1. Mol Cell Neurosci. 2008;37(1):69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357. Roshon MJ, Ruley HE. Hypomorphic mutation in hnRNP U results in post-implantation lethality. Transgenic Res. 2005;14(2):179–192. [DOI] [PubMed] [Google Scholar]
- 358. Ye J, Beetz N, O'Keeffe S, et al. hnRNP U protein is required for normal pre-mRNA splicing and postnatal heart development and function. Proc Natl Acad Sci USA. 2015;112(23):E3020–E3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359. Rainier S, Bui M, Mark E, et al. Neuropathy target esterase gene mutations cause motor neuron disease. Am J Hum Genet. 2008;82(3):780–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360. Kmoch S, Majewski J, Ramamurthy V, et al. Mutations in PNPLA6 are linked to photoreceptor degeneration and various forms of childhood blindness. Nat Commun. 2015;6:5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361. Synofzik M, Gonzalez MA, Lourenco CM, et al. PNPLA6 mutations cause Boucher-Neuhauser and Gordon Holmes syndromes as part of a broad neurodegenerative spectrum. Brain. 2014;137(Pt 1):69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362. Hufnagel RB, Arno G, Hein ND, et al. Neuropathy target esterase impairments cause Oliver-McFarlane and Laurence-Moon syndromes. J Med Genet. 2015;52(2):85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363. Topaloglu AK, Lomniczi A, Kretzschmar D, et al. Loss-of-function mutations in PNPLA6 encoding neuropathy target esterase underlie pubertal failure and neurological deficits in Gordon Holmes syndrome. J Clin Endocrinol Metab. 2014;99(10):E2067–E2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364. Akassoglou K, Malester B, Xu J, Tessarollo L, Rosenbluth J, Chao MV. Brain-specific deletion of neuropathy target esterase/swisscheese results in neurodegeneration. Proc Natl Acad Sci USA. 2004;101(14):5075–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365. Moser M, Li Y, Vaupel K, et al. Placental failure and impaired vasculogenesis result in embryonic lethality for neuropathy target esterase-deficient mice. Mol Cell Biol. 2004;24(4):1667–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366. Kang JS, Mulieri PJ, Miller C, Sassoon DA, Krauss RS. CDO, a robo-related cell surface protein that mediates myogenic differentiation. J Cell Biol. 1998;143(2):403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367. Zhang W, Kang JS, Cole F, Yi MJ, Krauss RS. Cdo functions at multiple points in the Sonic Hedgehog pathway, and Cdo-deficient mice accurately model human holoprosencephaly. Dev Cell. 2006;10(5):657–665. [DOI] [PubMed] [Google Scholar]
- 368. Bashamboo A, Bignon-Topalovic J, Rouba H, McElreavey K, Brauner R. A nonsense mutation in the hedgehog receptor CDON associated with pituitary stalk interruption syndrome. J Clin Endocrinol Metab. 2016;101(1):12–15. [DOI] [PubMed] [Google Scholar]
- 369. Koboldt DC, Steinberg KM, Larson DE, Wilson RK, Mardis ER. The next-generation sequencing revolution and its impact on genomics. Cell. 2013;155(1):27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370. Chong JX, Buckingham KJ, Jhangiani SN, et al. The genetic basis of Mendelian phenotypes: discoveries, challenges, and opportunities. Am J Hum Genet. 2015;97(2):199–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371. Prasov L, Brown NL, Glaser T. A critical analysis of Atoh7 (Math5) mRNA splicing in the developing mouse retina. PLoS One. 2010;5(8):e12315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372. Kieleczawa J. Fundamentals of sequencing of difficult templates–an overview. J Biomol Tech. 2006;17(3):207–217. [PMC free article] [PubMed] [Google Scholar]
- 373. Patwardhan A, Harris J, Leng N, et al. Achieving high-sensitivity for clinical applications using augmented exome sequencing. Genome Med. 2015;7:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374. Abulí A, Boada M, Rodríguez-Santiago B, et al. NGS-based assay for the identification of individuals carrying recessive genetic mutations in reproductive medicine. Hum Mutat. 2016;37(6):516–523. [DOI] [PubMed] [Google Scholar]
- 375. Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86(5):749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376. Lim EC, Brett M, Lai AH, et al. Next-generation sequencing using a pre-designed gene panel for the molecular diagnosis of congenital disorders in pediatric patients. Hum Genomics. 2015;9:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377. Geister KA, Brinkmeier ML, Cheung LY, et al. LINE-1 mediated insertion into Poc1a (protein of centriole 1 A) causes growth insufficiency and male infertility in mice. PLoS Genet. 2015;11(10):e1005569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378. Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12(5):363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379. Sudmant PH, Rausch T, Gardner EJ, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526(7571):75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380. Talkowski ME, Ordulu Z, Pillalamarri V, et al. Clinical diagnosis by whole-genome sequencing of a prenatal sample. N Engl J Med. 2012;367(23):2226–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381. Brand H, Pillalamarri V, Collins RL, et al. Cryptic and complex chromosomal aberrations in early-onset neuropsychiatric disorders. Am J Hum Genet. 2014;95(4):454–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382. Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3(4):285–298. [DOI] [PubMed] [Google Scholar]
- 383. Lee MS, Wajnrajch MP, Kim SS, et al. Autosomal dominant growth hormone (GH) deficiency type II: the Del32–71-GH deletion mutant suppresses secretion of wild-type GH. Endocrinology. 2000;141(3):883–890. [DOI] [PubMed] [Google Scholar]
- 384. Miletta MC, Petkovic V, Eblé A, Flück CE, Mullis PE. Rescue of isolated GH deficiency type II (IGHD II) via pharmacologic modulation of GH-1 splicing. Endocrinology. 2016;157(10):3972–3982. [DOI] [PubMed] [Google Scholar]
- 385. Kellis M, Wold B, Snyder MP, et al. Defining functional DNA elements in the human genome. Proc Natl Acad Sci USA. 2014;111(17):6131–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386. Edwards SL, Beesley J, French JD, Dunning AM. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet. 2013;93(5):779–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387. Wu C, Orozco C, Boyer J, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10(11):R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388. Wu C, Jin X, Tsueng G, Afrasiabi C, Su AI. BioGPS: building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res. 2016;44(D1):D313–D316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389. Smith CM, Finger JH, Hayamizu TF, et al. The mouse Gene Expression Database (GXD): 2014 update. Nucleic Acids Res. 2014;42(Database issue):D818–D824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390. Sajedi E, Gaston-Massuet C, Signore M, et al. Analysis of mouse models carrying the I26T and R160C substitutions in the transcriptional repressor HESX1 as models for septo-optic dysplasia and hypopituitarism. Dis Model Mech. 2008;1(4–5):241–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391. Ma Y, Qi X, Du J, et al. Identification of candidate genes for human pituitary development by EST analysis. BMC Genomics. 2009;10:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392. Mortensen AH, MacDonald JW, Ghosh D, Camper SA. Candidate genes for panhypopituitarism identified by gene expression profiling. Physiol Genomics. 2011;43(19):1105–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393. Chen J, Meng Y, Zhou J, et al. Identifying candidate genes for type 2 diabetes mellitus and obesity through gene expression profiling in multiple tissues or cells. J Diabetes Res. 2013;2013:970435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394. Bancalari RE, Gregory LC, McCabe MJ, Dattani MT. Pituitary gland development: an update. Endocr Dev. 2012;23:1–15. [DOI] [PubMed] [Google Scholar]
- 395. Brunskill EW, Potter AS, Distasio A, et al. A gene expression atlas of early craniofacial development. Dev Biol. 2014;391(2):133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396. Hochheiser H, Aronow BJ, Artinger K, et al. The FaceBase Consortium: a comprehensive program to facilitate craniofacial research. Dev Biol. 2011;355(2):175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397. Sahni N, Yi S, Zhong Q, et al. Edgotype: a fundamental link between genotype and phenotype. Curr Opin Genet Dev. 2013;23(6):649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398. Sahni N, Yi S, Taipale M, et al. Widespread macromolecular interaction perturbations in human genetic disorders. Cell. 2015;161(3):647–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399. Costello I, Nowotschin S, Sun X, et al. Lhx1 functions together with Otx2, Foxa2, and Ldb1 to govern anterior mesendoderm, node, and midline development. Genes Dev. 2015;29(20):2108–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400. Yasuoka Y, Suzuki Y, Takahashi S, et al. Occupancy of tissue-specific cis-regulatory modules by Otx2 and TLE/Groucho for embryonic head specification. Nat Commun. 2014;5:4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401. Nakano T, Murata T, Matsuo I, Aizawa S. OTX2 directly interacts with LIM1 and HNF-3β. Biochem Biophys Res Commun. 2000;267(1):64–70. [DOI] [PubMed] [Google Scholar]
- 402. Bach I. The LIM domain: regulation by association. Mech Dev. 2000;91(1–2):5–17. [DOI] [PubMed] [Google Scholar]
- 403. Nica G, Herzog W, Sonntag C, Hammerschmidt M. Zebrafish pit1 mutants lack three pituitary cell types and develop severe dwarfism. Mol Endocrinol. 2004;18(5):1196–1209. [DOI] [PubMed] [Google Scholar]
- 404. Pogoda HM, Hammerschmidt M. How to make a teleost adenohypophysis: molecular pathways of pituitary development in zebrafish. Mol Cell Endocrinol. 2009;312(1–2):2–13. [DOI] [PubMed] [Google Scholar]
- 405. Morcos PA, Vincent AC, Moulton JD. Gene editing versus morphants. Zebrafish. 2015;12(5):319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406. Wang H, Yang H, Shivalila CS, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407. Chu VT, Weber T, Graf R, et al. Efficient generation of Rosa26 knock-in mice using CRISPR/Cas9 in C57BL/6 zygotes. BMC Biotechnol. 2016;16:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408. Qi LS, Larson MH, Gilbert LA, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409. Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154(6):1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410. Hilton IB, D'Ippolito AM, Vockley CM, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33(5):510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411. Cho SW, Kim S, Kim Y, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24(1):132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412. Tebas P, Stein D, Tang WW, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370(10):901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413. Pitteloud N, Quinton R, Pearce S, et al. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J Clin Invest. 2007;117(2):457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414. Mitchell AL, Dwyer A, Pitteloud N, Quinton R. Genetic basis and variable phenotypic expression of Kallmann syndrome: towards a unifying theory. Trends Endocrinol Metab. 2011;22(7):249–258. [DOI] [PubMed] [Google Scholar]
- 415. Ming JE, Muenke M. Multiple hits during early embryonic development: digenic diseases and holoprosencephaly. Am J Hum Genet. 2002;71(5):1017–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416. Fang Q, Benedetti AF, Ma Q, et al. HESX1 mutations in patients with congenital hypopituitarism: variable phenotypes with the same genotype. Clin Endocrinol (Oxf). 2016;85(3):408–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417. Génin E, Feingold J, Clerget-Darpoux F. Identifying modifier genes of monogenic disease: strategies and difficulties. Hum Genet. 2008;124(4):357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418. Xue L, Cai JY, Ma J, et al. Global expression profiling reveals genetic programs underlying the developmental divergence between mouse and human embryogenesis. BMC Genomics 2013;14:568. [DOI] [PMC free article] [PubMed] [Google Scholar]