Abstract
Progress on understanding how genome structure evolves is accelerating with the arrival of new genomic, comparative, and theoretical approaches. This article reviews progress in understanding how chromosome inversions and sex chromosomes evolve, and how their evolution affects species’ ecology. Analyses of clines in inversion frequencies in flies and mosquitoes imply strong local adaptation, and roles for both over- and under dominant selection. Those results are consistent with the hypothesis that inversions become established when they capture locally adapted alleles. Inversions can carry alleles that are beneficial to closely related species, causing them to introgress following hybridization. Models show that this “adaptive cassette” scenario can trigger large range expansions, as recently happened in malaria mosquitoes. Sex chromosomes are the most rapidly evolving genome regions of some taxa. Sexually antagonistic selection may be the key force driving transitions of sex determination between different pairs of chromosomes and between XY and ZW systems. Fusions between sex-chromosomes and autosomes most often involve the Y chromosome, a pattern that can be explained if fusions are mildly deleterious and fix by drift. Sexually antagonistic selection is one of several hypotheses to explain the recent discovery that the sex determination system has strong effects on the adult sex ratios of tetrapods. The emerging view of how genome structure evolves invokes a much richer constellation of forces than was envisioned during the Golden Age of research on Drosophila karyotypes.
Key words: chromosome inversion, chromosome fusion, sex chromosome, sexually antagonistic selection, species range, sex ratio

Mark Kirkpatrick is the Painter Professor of Genetics at the University of Texas. He received a BA from Harvard and a PhD from the University of Washington. Kirkpatrick uses models and statistics to study several topics in evolutionary genetics, including the evolution of chromosome rearrangements, sex determination, speciation, quantitative genetics, and species ranges.
In 2005, our picture of the evolution of the human genome was upended. Previously, we believed that the chromosomes of humans and chimpanzees differed by 9 inversions and one fusion that had been established since our most recent common ancestor. With the arrival of the chimp genome, we learned that the 2 species differ by some 1500 inversions (Feuk et al. 2005). In one fell swoop, the amount of structural evolution to be explained exploded by a factor of 150.
This is one of several dramatic recent discoveries that are drawing our attention back to questions that were at the center of evolutionary genetics in the mid-20th Century. Do new chromosomal rearrangements become fixed mainly by selection or drift? Are rearrangement polymorphisms maintained by coadapted gene complexes or some other form of selection? Are differences in chromosome structure often important to isolation between species? Leading geneticists of the last century, notably Theodosius Dobzhansky and MJD White, debated these questions endlessly but without clear resolution (Kirkpatrick 2010). The arrival of the genomic era holds the prospect that we may be able to solve some of those classic puzzles.
This article is fruit of the generous invitation to give the Wilhelmina Key Distinguished Lecture to the 2015 meeting of the American Genetic Association. It reviews some of the recent advances that have been made in understanding how the genome structure evolves. The topic is vast, and so here we will prune the scope by focusing largely on work from our lab. The first topic is the evolution of chromosome inversions. We will see how inversion polymorphisms can be established by local adaptation, and how genomic data can provide tests of that hypothesis against alternatives. We then consider how the introgression of inversions between species can trigger range expansions, which may have been a key to the evolution of virulent malaria vectors in the recent evolutionary past. We then turn to the evolution of sex chromosomes, which in many groups of animals and plants are the most rapidly evolving regions of the genome. Theory and data suggest that sexual selection can trigger evolutionary shifts of sex determination from 1 chromosome pair to another, and transitions between XY (male heterogamety) and ZW (female heterogamety) determination. Sex chromosomes also provide a unique perspective on the forces responsible for chromosome fusions. A meta-analysis of fusions in vertebrates and population genetic models suggest that many fusions may be mildly deleterious and become fixed by drift. The article closes by suggesting that the sex chromosomes may have a surprising feedback on the sex ratios and hence social environment of many animals.
Chromosome Rearrangements and Local Adaptation
Much of our early understanding of inversion polymorphisms came from Dobzhansky’s studies of Drosophila pseudoobscura (Lewontin et al. 1981). Other species (particularly other dipterans) have also been discovered to have abundant inversion polymorphism. Famous examples include additional species of Drosophila, notably D. subobscura (Prevosti et al. 1988) and Anopheles mosquitoes (Ayala et al. 2014). Several of these inversions show striking geographical variation in frequency that are correlated with climatic variables (Huey et al. 2000, Ayala et al. 2011).
What could maintain these clines in the face of the strong gene flow expected in these mobile species? The obvious hypothesis is local adaptation. Schaeffer (2008) studied this idea using the data on the frequencies of 5 inversions in D. pseudoobscura that had been collected by a team of researchers across the Southwestern US over a span of 4 decades. He clustered the data from different localities into 6 geographical regions, then estimated the fitnesses of all 15 genotypes in each region by numerically fitting migration-selection models to those data. The results suggest strong selection that varies between regions.
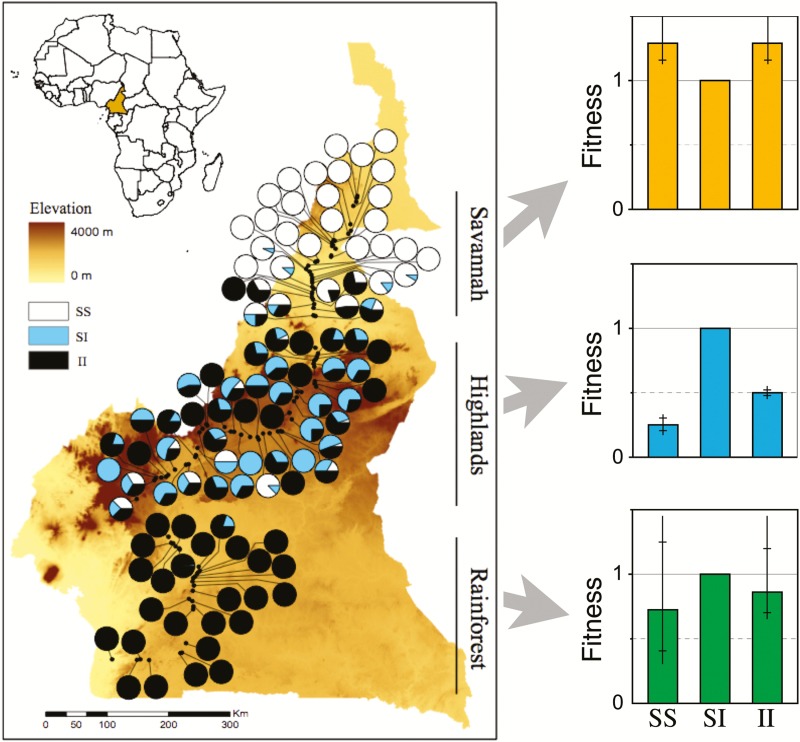
In a similar spirit, we estimated fitnesses in different parts of the range of the malaria mosquito Anopheles funestus by fitting a spatially explicit model to a dramatic cline in the frequency of inversion 3Ra (Ayala et al. 2013). Again, the results point to very strong selection that varies in space (Figure 1). Further, the model suggests the inversion mediates strong assortative mating. In one part of the species’ range, the combination of pre- and postzygotic mechanisms may result in 95% reproductive isolation between homozygotes for the alternative chromosomal arrangements. A final observation is that the inversion experiences overdominant selection in some parts of its range, and underdominant selection in others. This result casts an amusing light on a long-running controversy about inversions. Dobzhansky (1970) argued that inversion polymorphisms are maintained by balancing selection, while White (1978) argued that inversions are generally underdominant. The mosquito results support both views.
Figure 1.
Left: Inversion 2La in the malaria mosquito Anopheles funestus has a strong frequency cline in Cameroon. The pie diagrams show the frequencies of the 3 genotypes (S = standard chromosome, I = inverted chromosome). Samples are from 105 locations along a highway that traverses 3 ecological zones: savannah (north), highlands (center), and lowland tropical rainforest (south). Right: Estimates of relative viabilities for the 3 genotypes in the 3 zones estimated by fitting a spatially explicit genetic model to the data using likelihood. The bars and whiskers show the 95% and 99% confidence limits. From Ayala et al. (2013).
There are 3 evolutionary paths by which an inversion can become locally adapted. First, it can be locally adapted when it first appears because its breakpoints fortuitously generate a favorable mutation. Second, an inversion can capture locally adapted alleles within the inverted chromosome segment. A simple model shows that this event can cause a new inversion to become established (Kirkpatrick and Barton 2006). The inversion gets an evolutionary advantage in this case because it binds the locally adapted alleles together and prevents them from recombining onto unfavorable genetic backgrounds. Other chromosome rearrangements, such as fusions and translocations, can also be established by this mechanism (Guerrero and Kirkpatrick 2014). (Rearrangements can also be established by the same mechanism when species hybridize. In effect, the alleles captured by the rearrangements are ‘locally adapted’ to the genetic background of their own species, and so decreased recombination is again favored.) A third hypothesis is that locally adapted alleles accumulate within the inversion after it is established by some other mechanism (Noor et al. 2001; Navarro and Barton 2003).
The first and second hypotheses are expected to leave contrasting signatures in neutral polymorphism that could be detected with genomic data (Guerrero et al. 2012, Guerrero and Kirkpatrick 2014). When heterozygous, large inversions admit a very small amount of recombination via gene conversion and double recombination (Andolfatto et al. 2001). This causes a genetic flux between inverted and uninverted chromosomes that is strongest near the center of the inversion and virtually absent near the breakpoints. As a result, divergence in neutral polymorphism between the 2 chromosomal arrangements will vary as we move along the chromosome. The pattern resembles the cables of a suspension bridge, with peaks of divergence at the breakpoints (Figure 2). That pattern is modified if the inversion carries locally adapted alleles at loci between the breakpoints. In that case, we expect additional peaks of divergence between inverted and uninverted chromosomes centered on the locally adapted loci.
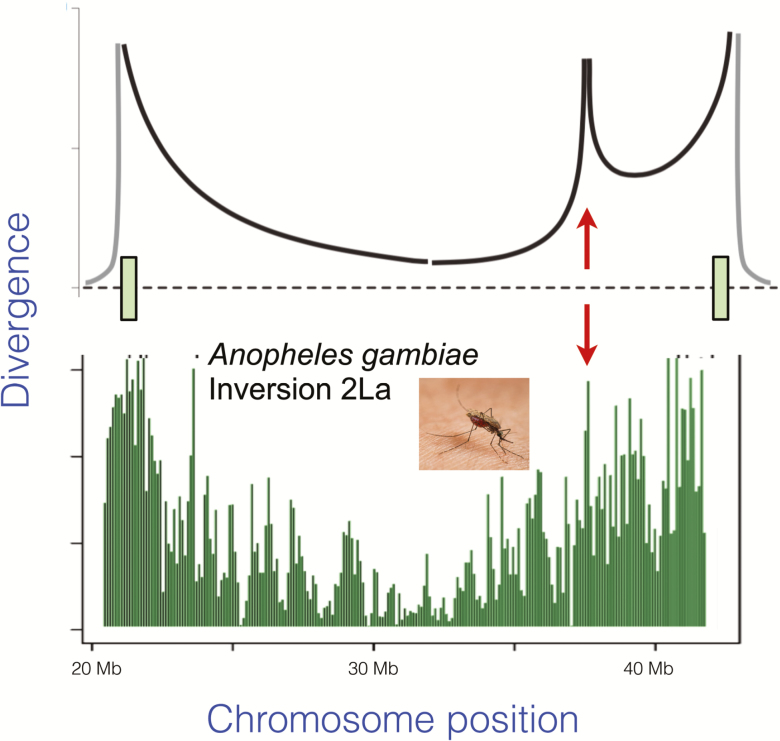
Figure 2.
Top: Coalescent simulations show that divergence between uninverted and inverted chromosomes will show a distinctive pattern. Divergence (measured as F ST or d xy) is highest at the breakpoints (indicated by the rectangles on the chromosome map at bottom). Locally adapted loci within the inversion (indicated by the arrow) will show additional divergence peaks. From Guerrero et al. (2012). Bottom: Divergence between standard and inverted chromosomes in the region spanned by inversion 2La in the mosquito Anopheles gambiae. The data are from pooled sequences of mosquitoes sampled from Cameroon from Cheng et al. (2012).
What do the data in fact show? To date, sequences from inverted and uninverted chromosomes needed for this analysis are only available from 2 species: Anopheles gambiae (Cheng et al. 2012) and D. melanogaster (Corbett-Detig and Hartl 2012). Figure 2 shows that the pattern of divergence does indeed show the predicted suspension bridge pattern. Even more exciting is that there are peaks of divergence inside the inversion, suggestive of locally adapted loci.
But before we conclude that these inversions do carry locally adapted loci, we must rule out the null hypothesis that the peaks of divergence are simply sampling noise. That could be done using coalescent models (Guerrero et al. 2012; Peischl et al. 2013; Guerrero and Kirkpatrick 2014; Rousset et al. 2014), but no one has yet tackled that job. If the inversions do in fact show signals of carrying locally adapted loci, a second issue is whether those loci were responsible for establishing the inversion. Testing that hypothesis will be difficult, because locally adapted variants that accumulate after an inversion establishes can leave much the same pattern of neutral polymorphism. The discovery of locally adapted polymorphisms still segregating in the ancestral, uninverted population of chromosomes might suggest the inversion captured pre-existing alleles that were locally adapted when it first appeared.
Inversions, Adaptive Cassettes and Species Ranges
Many discussions of inversions emphasize their importance as a mechanism that genetically isolates species (White 1978; King 1993). Inversions may also play the opposite role: they carry beneficial alleles that can drive introgression between species.
Malaria is transmitted to humans in Africa by species of Anopheles mosquitoes. Several of them share inversion polymorphisms, the result of trans-specific introgression (White et al. 2011; Fontaine et al. 2015). A similar situation may have occurred in Heliconius butterflies. These are mimetic species that rely on adaptation at several loci that mediate wing color patterns to successfully resemble their model species. Locally adapted inversions that enable mimicry in H. numata may have introgressed from another species (M. Joron, personal communication).
These examples show that inversions can play the role of “adaptive cassettes” that carry positively selected alleles which have already stood the test of natural selection. Indeed, it would be surprising if inversions don’t play this role more often than the 2 examples cited above might suggest. Horizontal gene transfer of adaptive alleles between distantly related microbes is ubiquitous (Ochman et al. 2000; Darmon and Leach 2014). Mitochondria in animals (Ballard and Whitlock 2004) and chloroplasts in plants (Rieseberg and Soltis 1991) can rapidly introgress between species with little or no nuclear gene flow, suggesting the introgression is driven by positive selection. As the adaptive introgression of genes in microbes and organelles in eukaryotes is apparently widespread, by analogy it is plausible that a similar process is common with chromosome inversions.
The introgression of inversions may have had important impacts on the geographical ranges of Anopheles mosquitoes (Besansky et al. 2003). A little less than 1 million years ago, inversion 2La introgressed from Anopheles gambiae into A. arabensis, where it completely replaced the alternative chromosomal arrangement (Fontaine et al. 2015). This inversion is known to confer resistance to desiccation (Fouet et al. 2012). It is highly plausible that the spread of 2La into A. arabensis expanded that species’ ecological tolerance and enabled it to colonize new habitats.
To refine this intuition, we developed a model of how a species’ geographical range evolves following establishment of a locally adapted inversion (Kirkpatrick and Barrett 2015). We considered 2 scenarios (Figure 3). In the first, the inversion spreads because it captures locally adapted alleles that are already present in a peripheral population. As the inversion is established, the density of the peripheral population increases, but not by much. That is because the inversion increases the mean fitness of the peripheral population by suppressing the production of low fitness recombinants. They are not common to begin with, however, and so their elimination has only a slight impact on population density.
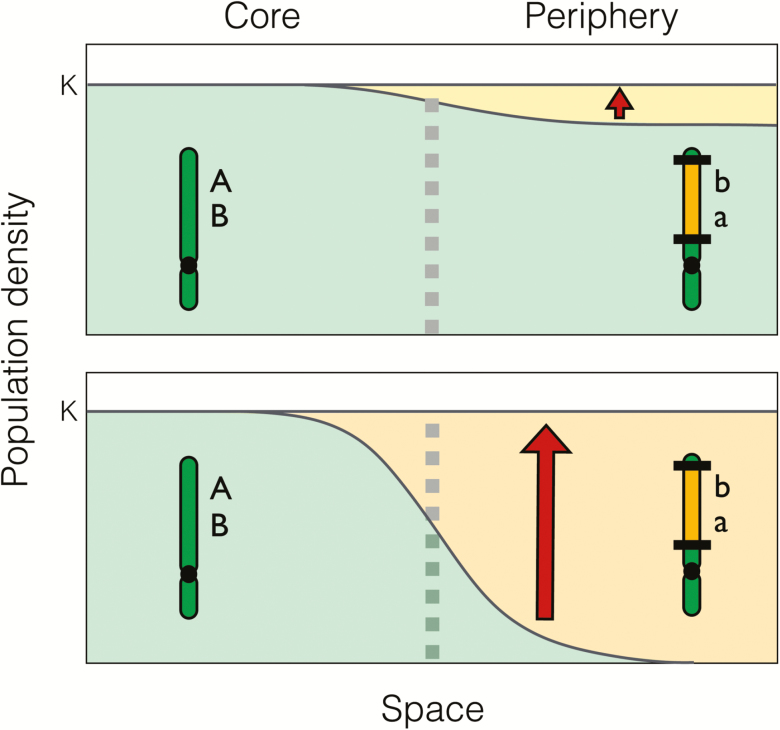
Figure 3.
A model shows that peripheral populations of a species can expand with the establishment of a locally adapted inversion. Top: In the capture scenario, alleles adapted to the range periphery (to the right of the vertical dashed line) are present before the inversion appears. The equilibrium population density (shown in dark gray) is slightly depressed below carrying capacity by recombinant genotypes. The inversion then captures those alleles and becomes established at the periphery, which causes the population density to increase to the carrying capacity, K (small arrow). Bottom: In the adaptive cassette scenario, the locally adapted alleles are initially absent. Populations in the periphery are maladapted and far below carrying capacity. With the introgression of an inversion that carries the locally adapted alleles, the densities increase greatly and the range expands outwards (large arrow). From Kirkpatrick and Barrett (Kirkpatrick and Barrett 2015).
A second way that a locally adapted inversion can establish is by the adaptive cassette scenario, which leads to quite different results (Figure 3). In this case, the alleles adapted to conditions at the periphery are initially absent, so the population there can be very poorly adapted and its density low. The introgression of an inversion carrying 2 locally adapted alleles has exactly the same effect as a single beneficial mutation of large effect. As it becomes established, the mean fitness of the peripheral populations can increase substantially, and their densities increase correspondingly.
Our conclusion is that an inversion carrying alleles adapted to conditions at the periphery can indeed trigger a range expansion, as may have happened in the recent evolutionary past of Anopheles mosquitoes. Establishment of the inversion will typically have only modest impacts on population densities if it spreads by capturing locally adapted alleles that are already segregating. In contrast, the inversion can have very large effects on the species range if it first appears with novel beneficial alleles, as can happen when the inversion introgresses following hybridization with another species.
Evolution of the Genome by Sexual Selection
While natural selection is responsible for many features of the genome, sexual selection can play an important role too. The most obvious place for it to intervene in genomic affairs is in the sex chromosomes. Animals such as mammals, fruitflies, and higher birds have a genetic sex determination system in which one of the chromosomes is largely nonrecombining and highly degenerate. In the classical narrative, heteromorphic sex chromosomes like these evolve in 3 steps (Charlesworth et al. 2005). Initially, the sex chromosomes are freely recombining and largely undifferentiated. One or more loci on these chromosomes then segregate for alleles under sexually antagonistic selection. This is the situation in which one allele is beneficial to males and detrimental to females, while the other allele has the converse fitness effects. Many of the genes that fit this bill are likely to be those for sexually selected traits that increase male mating success but decrease the survival of both sexes.
In the second step, an inversion on the proto-Y chromosome (say) captures the male determining region and the male-beneficial allele at the sexually antagonistic locus. The inversion spreads by blocking recombination, which guarantees that the proto-Y will always carry the high fitness allele. An unfortunate side-effect is that the Y is now unable to recombine with any other chromosome (unlike the X). That condition triggers the third step, in which the Y degenerates by several processes that afflict nonrecombining parts of the genome (Bachtrog 2008). Additional inversions can expand the nonrecombining segment down almost the entire length of the Y, eventually leading to the situation we now see in eutherian mammals. An analogous evolutionary path has been followed by the W chromosome in some taxa with female heterogamety, such as many birds. Genomic data is consistent with this narrative: moving along the Y and W chromosomes, we see “strata” of different ages corresponding to a series of chromosome inversions (Bachtrog 2013; Zhou et al. 2014).
While this scenario explains the evolution of highly diverged sex chromosomes, it does not explain the remarkable diversity of sex chromosomes we see in other groups. A dramatic example comes from stickleback fishes (Kitano et al. 2009; Ross et al. 2009). Every one of the 7 species in this clade has a unique sex chromosome system (Figure 4). In some species, an autosome has fused with a Y chromosome to form a neo-Y. In others, the sex determination system has jumped to an entirely different linkage group. The 4 spine stickleback (Apeltes quadracus) has made an even more radical transition: it has evolved a ZW sex determination system.
Figure 4.
Stickleback fishes show a remarkable diversity of sex chromosomes. At left is a phylogeny for 7 species. The first column to the right of the species names shows the type of sex chromosome system. X1X2Y indicates that an autosome has fused with the ancestral Y, which results in a neo-X (the former partner of the fused autosome) that is unlinked to but segregates with the ancestral X. The second column shows the linkage group that determines sex. After Hendry et al. (2009).
What evolutionary forces might be driving this evolutionary frenzy? Hypotheses include random genetic drift, pleiotropic selection of sex-determination alleles, sex ratio selection, and transmission distortion (Beukeboom and Perrin 2014). Another possibility is sexually antagonistic selection, the very force thought to be key to the evolution of nonrecombining sex chromosomes (van Doorn and Kirkpatrick 2007, van Doorn and Kirkpatrick 2010). In many animals, mutations at numerous autosomal loci cause sex reversal (Beukeboom and Perrin 2014). If a dominant male-determining mutation occurs near a locus under sexually antagonistic selection, the resulting can be a neo-Y chromosome that hijacks sex determination (Figure 5). By virtue of carrying a male-beneficial allele, the neo-Y can have higher fitness than the ancestral Y. As it spreads, the ancestral Y is lost. The evolutionary endpoint is conversion of the ancestral sex determining chromosomes to autosomes (the former X chromosomes), while the new linkage group assumes full responsibility for sex determination. This process can drive both transitions of sex determination from one linkage group to another, and transitions between XY and ZW systems.
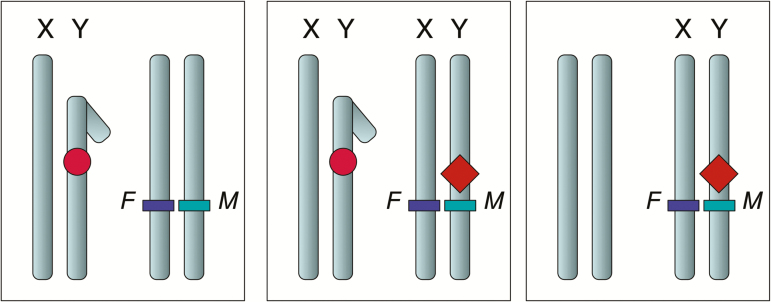
Figure 5.
Sexually antagonistic selection can cause an autosome to hijack sex determination from the ancestral sex chromosomes. Left panel: In the ancestral sex chromosomes, a dominant male-determining factor (the circle) is carried by the Y chromosome. On a pair of autosomes, a locus under sexually antagonistic selection segregates for 2 alleles, 1 beneficial to females (F) and the other to males (M). Middle panel: A dominant sex-reversal mutation that makes all carriers develop as male (the diamond) appears on the autosome near to the sexually antagonistic locus. Selection causes the mutation to become associated (in linkage disequilibrium) with the male-beneficial allele (M). During the transition, both linkage groups act as sex chromosomes, but individual males carry only 1 of the 2 Y chromosomes. Right panel: If sexually antagonistic selection is sufficiently strong, the chromosome with the new sex-determining mutant spreads. It becomes a neo-Y, its homolog becomes a neo-X, and the ancestral Y is lost. The linkage group that previously determined sex is now a pair of autosomes consisting of the ancestral X chromosomes. Details of the model are given in van Doorn and Kirkpatrick (2007).
Tantalizing hints support the idea that sexual antagonism might underlie evolutionary transitions in sex determination. In stickleback fishes, the neo-sex chromosomes that have evolved in the Japan Sea species carry quantitative trait loci (QTL) for 2 male-limited traits that are candidates for sexually antagonistic selection (Kitano et al. 2009). While most cichlids in Lake Malawi have XY sex determination, several species have a new ZW system (Ser et al. 2010). The neo-W chromosome is linked to a novel color pattern thought to be under sexually antagonistic selection because it enhances female crypsis but is detrimental to male breeding success (Roberts et al. 2009). These data strongly implicate sexually antagonistic selection in the origin of a new sex determination system.
It may be possible to glean further evidence for this process from genomic data. Coalescent modeling shows that a locus under sexually antagonistic selection in the pseudoautosomal (recombining) region of the sex chromosomes leaves a characteristic signature in patterns of neutral polymorphism (Kirkpatrick and Guerrero 2014). Moving away from the sex-determining region, divergence between the X and Y is expected to decline. Loci under sexually antagonistic selection, however, will generate peaks of divergence (Figure 6). It is no coincidence that these peaks are reminiscent of those we saw earlier at locally adapted loci within chromosome inversions (Figure 2). The alleles at a locus under sexually antagonistic selection are locally adapted, but to a sex rather than to a locality. These models can be used to estimate the strength of sexually antagonistic selection from the divergence between X and Y chromosomes (Kirkpatrick and Guerrero 2014).
Figure 6.
Divergence between the X and Y chromosomes (e.g. measured as F ST) can be used to scan for loci under sexually antagonistic selection in the recombining (or pseudoautosomal) region of sex chromosomes. Coalescent models show that divergence declines as we move down the chromosomes, away from the sex-determining region (SDR, at left). Divergence will again peak in the vicinity of a locus under sexually antagonistic selection (marked as SA). After Kirkpatrick and Guerrero (2014).
The first report of data consistent with the pattern seen in Figure 6 comes from the recombining sex chromosomes of the plant Silene latifolia. Qiu et al. (2013) found a gene in the pseudoautosomal region that shows different allele frequencies on X and Y chromosomes. They interpreted this as evidence for sexually antagonistic selection. No phenotype has yet been associated with this chromosome region, however, so the target of selection remains elusive. New genomic methods that will give us phased sequences of the pseudoautosomal regions may soon enable us to scan for loci under sexually antagonistic selection, while QTL studies could be used to associate those regions with phenotypes.
Y do Chromosomes Fuse?
Karyotypes show bewildering variety across the Tree of Life. The jack jumper ant has only a single pair of chromosomes (Crosland and Crozier 1986), while the protozoan Oxytricha trifallax has some 16 000 pairs (Swart et al. 2013). One process that contributes to this variation is chromosome fusion. Sex chromosomes offer a unique window onto how fusions evolve. Fusions between sex chromosomes and autosomes are surprisingly common. Hundreds of cases have been detected cytologically by their distinctive signature: they cause one sex to have an odd number of chromosomes (The Tree of Sex Consortium 2014).
X, Y, Z and W chromosomes differ in several key regards: their effective population sizes, their mutation rates, and the fractions of their evolutionary lives spent in each sex. Differences between their propensities to fuse therefore hold clues about the evolutionary forces that drive fusions. Three have been proposed. Sexually antagonistic selection can fix a fusion between an autosome and a sex chromosome, for example when it links a male beneficial allele on autosome with the Y chromosome (Charlesworth and Charlesworth 1980). Fusions can generate beneficial changes to gene expression patterns (Chang et al. 2013). Third, meiotic drive in females can give a transmission advantage to fused chromosomes (Pardo-Manuel de Villena and Sapienza 2001; Yoshida and Kitano 2012).
To gain insight about general patterns, we performed a metaanalysis of sex chromosome fusions in vertebrates (Pennell et al. 2015). Three striking patterns appear in the data (Table 1). Among fish and squamate reptiles, there are many more Y-autosome fusions than X-autosome fusions. Second, many more fusions occur in species with XY sex determination than those with ZW sex determination. Third, mammals have much more equitable numbers of X-autosome and Y-autosome fusions than do fish and reptiles, suggesting that fusions in mammals may evolve differently than in the other 2 clades.
Table 1.
Numbers of species with different sex chromosome systems known in vertebrates. The 4 columns following the taxa are systems with a fusion between a sex chromosome and an autosome: Y-autosome fusions (X1X2Y systems), X-autosome fusions (XY1Y2 systems), W-autosome fusions (Z1Z2W systems), and Z-autosome fusions (ZW1W2 systems). From Pennell et al. (2015)
| Taxa | Y-A | X-A | W-A | Z-A | XY | ZW |
|---|---|---|---|---|---|---|
| Fish | 42 | 3 | 0 | 2 | 109 | 38 |
| Amphibians | 1 | 0 | 0 | 0 | 29 | 16 |
| Reptiles | 40 | 0 | 2 | 4 | 120 | 240 |
| Birds | — | — | 0 | 3 | 0 | 192 |
| Mammals | 18 | 24 | — | — | 467 | 0 |
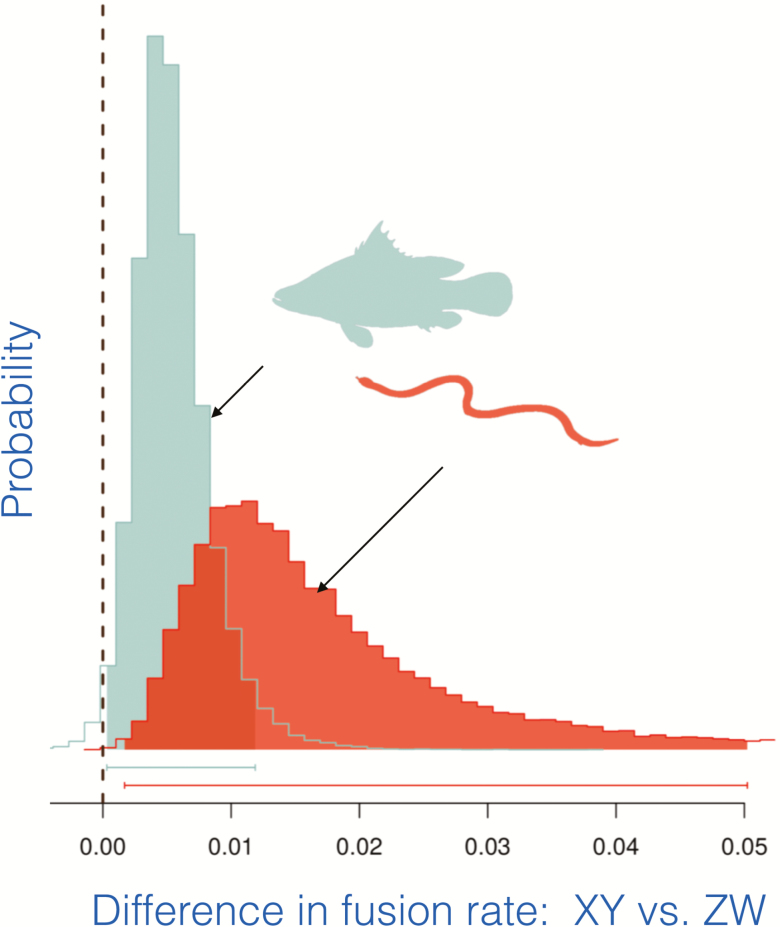
We next asked if the 2 strong trends seen in fish and reptiles reflect differences between the fixation rates of fusions with the 4 types of sex chromosomes. We developed Markov chain Monte Carlo models for the evolutionary transitions between fused and unfused sex chromosomes on the phylogeny of the species in our dataset. The results strongly indicate that among fish and reptiles, fusions fix at a higher rate in XY than in ZW systems (Figure 7). Further analysis shows that this pattern results because Y-autosome fusions establish more frequently than any of the 3 other kinds.
Figure 7.
Posterior probability densities of the difference in fixation rates of fusions between autosomes and sex chromosomes (rates in XY species minus in ZW species). These results are based on MCMC models that allow transitions between fused and unfused states to occur along the phylogeny. From Pennell et al. (2015).
What is it about the biology of Y chromosomes that gives them a predilection to fuse? We tackled that question by developing classic population genetic models the broad range of evolutionary forces that might play a role: positive and negative selection, sexually antagonistic selection, sex-biased mutation, meiotic drive, and random genetic drift. The models account for differences in the rates that the 4 types of fusions originate (for example, because there are more X than Y chromosomes in a population), and for differences in the probability that a fusion will fix once it has appeared.
It is less easy than one might think to explain the high rate at which Y-autosome fusions establish. Consider the hypothesis that fusions are typically deleterious and become fixed by random genetic drift. In that case, Y-autosome fusions will fix more frequently than X-autosome fusions, because the Y has a smaller effective population size than the X. But for the same reason, if all else is equal, then W-autosome fusions will also fix at an equally high rate, which is not consistent with the data.
To break the symmetry between the Y and W chromosomes, the models show that combinations of 2 or more evolutionary forces must be in play. The most plausible hypothesis is that they are deleterious and further that mutation rates for fusions are strongly male-biased and/or that the reproductive sex ratio is strongly female-biased. Other combinations of evolutionary forces can also lead to elevated rates of Y-A fusions. One is female meiotic drive that consistently favors unfused chromosomes in fishes and reptiles. That does not seem a very likely hypothesis, however, since female meiotic drive in mammals (where it has been best studied) sometimes favors fused and sometimes unfused chromosomes (Pardo-Manuel de Villena and Sapienza 2001; Yoshida and Kitano 2012). Another hypothesis suggested by the models is the action of sexually antagonistic selection in combination with male-biased mutation and male-biased operational sex ratios. Again, these conditions (particularly the last one) do not seem particularly plausible.
In sum, the data and models suggest that in fish and reptiles, many fusions between sex chromosomes and autosomes are weakly deleterious and were fixed by random genetic drift. It is plausible that this nearly neutral theory of fusions also applies to autosomes, and so plays an important role in the evolution of the entire karyotype.
The Genome’s Revenge: Sex Chromosomes and Sex Ratios
To here, our discussion has explored how the genome’s structure evolves in response to evolutionary forces acting on phenotypes. Can the arrow of causality ever point in the other direction—can the structure of the genome itself have important consequences for phenotypes?
Recent comparative analyses suggest that the sex chromosomes have unexpected and surprisingly strong effects on the adult sex ratio (or ASR), which in turn affects demography and social behavior. It has long been known that mammals typically have female-biased ASRs, while most birds have male-biased ASRs (Szekely et al. 2014). Another well-known difference between mammals and birds is sex determination: mammals have an XY system, while birds have a ZW system. But of course by themselves, these data give no reason to think there is a connection between sex chromosomes and the ASR, since it is a comparison between just 2 clades.
To see if the correlation between ASR and sex determination is general, we carried out a metaanalysis of 344 species of tetrapods whose sex determination system is known (Pipoly et al. 2015). Crucially, the dataset includes groups such as frogs and skinks that have variation in their sex determination system. Figure 8 shows the results in a phylogenetic context. Visually, it seems that species with XY sex determination tend to have a female-biased ASR, just as seen in mammals. Conversely, species with ZW sex determination tend to have a male-biased ASR.
Figure 8.
The adult sex ratio (ASR) in tetrapods is strongly influenced by the sex chromosome system. Groups like mammals with XY sex determination (red in the inner band) tend to have female-biased ASR (red in the outer band). Groups like birds with ZW sex determination (blue in the inner band) tend to have male-biased ASR (blue in the outer band). This correlation remains statistically significant after correcting for the phylogenetic relatedness (phylogeny shown at center). From Pipoly et al. (2015).
That impression is confirmed by phylogenetically corrected statistical analyses. Further, the correlation between sex chromosome system and ASR remains intact after controlling for several potentially confounding variables: body size, sexual size dimorphism, breeding latitude, and sex-biased dispersal. The effect is strong: the genetic sex determination system explains 27% of the variance in the ASRs of amphibians, and 25% of the variance in reptiles.
How do sex chromosomes affect the adult sex ratio? One possibility is through the sex ratio at conception, for example by different types of meiotic drive. That hypothesis is not supported in mammals and birds, at least, because the sex ratio at birth those groups tend to be balanced. The most plausible explanation therefore seems to be differential mortality after birth, which could result from several causes. The most obvious is degeneration of the Y and W chromosomes that follows when they stop recombining (Bachtrog 2013). There are several other hypotheses, including sexually antagonistic selection acting on sex-linked loci, and the physiological deterioration of Y and W chromosomes (e.g. by telomere shortening). Population genetic models, however, suggest that none of these hypotheses can easily explain the strength and consistency of the pattern (Pipoly et al. 2015). The puzzle of how sex chromosomes affect adult sex ratios is ripe for further investigation using experimental and theoretical, as well as comparative, approaches.
A Second Golden Age
The mid-Twentieth Century was a Golden Age for the study of karyotype evolution. The world of genetics had not yet coalesced around a small number of model species. Dobzhansky and colleagues did pioneering work on polymorphism in Drosophila pseudoobscura. The team at the University of Texas and others painstakingly worked out the evolutionary relations between the chromosomes of other species of flies (Kohler 1994). Meanwhile, a vast database of karyotypes in additional groups of insects, mammals, and other taxa accumulated (White 1973; King 1993).
This trove has been largely neglected since the advent of molecular biology, but it is ready to be exploited using modern phylogenetic methods (see the Tree of Sex Consortium (2014)). The genomic revolution is now extending outward across a taxonomically rich range of species. With the arrival of sequencing technologies that can detect structural variation, we will soon have an entirely new level of genetic resolution on diverse forms of life. Both classic forward-time and modeling approaches are now being brought to bear on questions about how and why genome structure evolves. Experimental evolution and field experiments can now be used to test hypotheses in some systems. Opportunities for functional tests of genetic effects are vastly expanded by the recent arrival of the CRISPR-Cas9 technology. This confluence of new data and analyses places us at the dawn of the second Golden Age.
Funding
This work was supported by grants from the National Science Foundation (DEB-0819901) and the National Institutes of Health (R01GM116853-01).
Acknowledgments
I am grateful to D. Ayala, R. Guerrero, K. Peichel, and 2 anonymous reviewers for discussion and useful comments on the manuscript. Nil Rahola generously provided the photo of Anopheles gambiae.
References
- Andolfatto P, Depaulis F, Navarro A. 2001. Inversion polymorphisms and nucleotide variability in Drosophila. Genet Res. 77:1–8. [DOI] [PubMed] [Google Scholar]
- Ayala D, Fontaine MC, Cohuet A, Fontenille D, Vitalis R, Simard F. 2011. Chromosomal inversions, natural selection and adaptation in the malaria vector Anopheles funestus. Mol Biol Evol. 28:745–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala D, Guerrero RF, Kirkpatrick M. 2013. Reproductive isolation and local adaptation quantified for a chromosome inversion in a malaria mosquito. Evolution. 67:946–958. [DOI] [PubMed] [Google Scholar]
- Ayala D, Ullastres A, González J. 2014. Adaptation through chromosomal inversions in Anopheles. Front Genet. 5:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. 2008. The temporal dynamics of processes underlying Y chromosome degeneration. Genetics. 179:1513–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet. 14:113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard JWO, Whitlock MC. 2004. The incomplete natural history of mitochondria. Mol Ecol. 13:729–744. [DOI] [PubMed] [Google Scholar]
- Besansky NJ, Krzywinski J, Lehmann T, Simard F, Kern M, Mukabayire O, Fontenille D, Toure Y, Sagnon NF. 2003. Semipermeable species boundaries between Anopheles gambiae and Anopheles arabiensis: Evidence from multilocus DNA sequence variation. Proc Natl Acad Sci USA 100:10818–10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beukeboom LW, Perrin N. 2014. The evolution of sex determination. Oxford, UK: Oxford University Press. [Google Scholar]
- Chang SL, Lai HY, Tung SY, Leu JY. 2013. Dynamic large-scale chromosomal rearrangements fuel rapid adaptation in yeast populations. Plos Genetics 9:e31003232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. 1980. Sex-differences in fitness and selection for centric fusions between sex-chromosomes and autosomes. Genet Res 35:205–214. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Marais G. 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity (Edinb). 95:118–128. [DOI] [PubMed] [Google Scholar]
- Cheng C, White BJ, Kamdem C, Mockaitis K, Costantini C, Hahn MW, Besansky NJ. 2012. Ecological genomics of Anopheles gambiae along a latitudinal cline: a population-resequencing approach. Genetics. 190:1417–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig RB, Hartl DL. 2012. Population genomics of inversion polymorphisms in Drosophila melanogaster . Plos Genetics 8:e1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosland MWJ, Crozier RH. 1986. Myrmecia pilosula, an ant with only one pair of chromosomes. Science 231:1278–1278. [DOI] [PubMed] [Google Scholar]
- Darmon E, Leach DRF. 2014. Bacterial genome instability. Microbiol Mol Biol Rev 78:1–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky TG. 1970. Genetics of the Evolutionary Process. New York: Columbia University Press. [Google Scholar]
- Feuk L, MacDonald JR, Tang T, Carson AR, Li M, Rao G, Khaja R, Scherer SW. 2005. Discovery of human inversion polymorphisms by comparative analysis of human and chimpanzee DNA sequence assemblies. Plos Genet. 1:489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine MC, Pease JB, Steele A, Waterhouse RM, Neafsey DE, Sharakhov IV, Jiang X, Hall AB, Catteruccia F, Kakani E, et al. 2015. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347:1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouet C, Gray E, Besansky NJ, Costantini C. 2012. Adaptation to aridity in the malaria mosquito Anopheles gambiae: chromosomal inversion polymorphism and body size influence resistance to desiccation. PLoS One. 7:e34841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero RF, Kirkpatrick M. 2014. Local adaptation and the evolution of chromosome fusions. Evolution 68:2747–2756. [DOI] [PubMed] [Google Scholar]
- Guerrero RF, Rousset F, Kirkpatrick M. 2012. Coalescence patterns for chromosomal inversions in divergent populations. Philos Trans Roy Soc B Biol Sci. 367:430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry AP, Bolnick DI, Berner D, Peichel CL. 2009. Along the speciation continuum in sticklebacks. J Fish Biol. 75:2000–2036. [DOI] [PubMed] [Google Scholar]
- Huey RB, Gilchrist GW, Carlson ML, Berrigan D, Serra L. 2000. Rapid evolution of a geographic cline in size in an introduced fly. Science 287:308–309. [DOI] [PubMed] [Google Scholar]
- King M. 1993. Species evolution: the role of chromosomal change. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Kirkpatrick M. 2010. How and why chromosome inversions evolve. Plos Biol. 8:e1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Barrett B. 2015. Chromosome inversions, adaptive cassettes and the evolution of species’ ranges. Mol Ecol. 24:2046–2055. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M, Barton N. 2006. Chromosome inversions, local adaptation and speciation. Genetics. 173:419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Guerrero RF. 2014. Signatures of sex-antagonistic selection on recombining sex chromosomes. Genetics. 197:531–U147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano J, Ross JA, Mori S, Kume M, Jones FC, Chan YF, Absher DM, Grimwood J, Schmutz J, Myers RM, et al. 2009. A role for a neo-sex chromosome in stickleback speciation. Nature 461:1079–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler RE. 1994. Lords of the fly: Drosophila genetics and the experimental life. Chicago: University of Chicago Press. [Google Scholar]
- Lewontin RC, Moore JA, Provine WB, Wallace B. 1981. Dobzhansky’s Genetics of Natural Populations, I - XLIII. New York: Columbia University Press. [Google Scholar]
- Navarro A, Barton NH. 2003. Accumulating postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evolution. 57:447–459. [DOI] [PubMed] [Google Scholar]
- Noor MAF, Grams KL, Bertucci LA, Reiland J. 2001. Chromosomal inversions and the reproductive isolation of species. Proc Natl Acad Sci USA. 98:12084–12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature. 405:299–304. [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F, Sapienza C. 2001. Female meiosis drives karyotypic evolution in mammals. Genetics. 159:1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peischl S, Koch E, Guerrero RF, Kirkpatrick M. 2013. A sequential coalescent algorithm for chromosomal inversions. Heredity (Edinb). 111:200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell MW, Kirkpatrick M, Otto SP, Vamosi JC, Peichel CL, Valenzuela N, Kitano J. 2015. Y fuse? sex chromosome fusions in fishes and reptiles. Plos Genet. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipoly I, Bokony V, Kirkpatrick M, Donald PF, Szekely T, Liker A. 2015. The genetic sex-determination system predicts adult sex ratios in tetrapods. Nature. 527:91–94. [DOI] [PubMed] [Google Scholar]
- Prevosti A, Ribo G, Serra L, Aguade M, Balana J, Monclus M, Mestres F. 1988. Colonization of America by Drosophila subobscura - experiment in natural populations that supports the adaptive role of chromosome inversion polymorphism. Proc Natl Acad Sci USA 85:5597–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu S, Bergero R, Charlesworth D. 2013. Testing for the footprint of sexually antagonistic polymorphisms in the pseudoautosomal region of a plant sex chromosome pair. Genetics. 194:663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Soltis DE. 1991. Phylogenetic consequences of cytoplasmic gene flow in plants. Evol Trends Plants. 5:65–84. [Google Scholar]
- Roberts RB, Ser JR, Kocher TD. 2009. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid Fishes. Science. 326:998–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JA, Urton JR, Boland J, Shapiro MD, Peichel CL. 2009. Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae). Plos Genet. 5:e1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F, Kirkpatrick M, Guerrero RF. 2014. Matrix inversions for chromosomal inversions: a method to construct summary statistics in complex coalescent models. Theor Popul Biol. 97:1–10. [DOI] [PubMed] [Google Scholar]
- Schaeffer SW. 2008. Selection in heterogeneous environments maintains the gene arrangement polymorphism of Drosophila pseudoobscura . Evolution. 62:3082–3099. [DOI] [PubMed] [Google Scholar]
- Ser JR, Roberts RB, Kocher TD. 2010. Multiple interacting loci control sex determination in lake Malawi cichlid fish. Evolution. 64:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart EC, Bracht JR, Magrini V, Minx P, Chen X, Zhou Y, Khurana JS, Goldman AD, Nowacki M, Schotanus K, et al. 2013. The Oxytricha trifallax macronuclear genome: A complex eukaryotic genome with 16,000 tiny chromosomes. Plos Biol. 11:e1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekely T, Weissing FJ, Komdeur J. 2014. Adult sex ratio variation: implications for breeding system evolution. J Evol Biol. 27:1500–1512. [DOI] [PubMed] [Google Scholar]
- The Tree of Sex Consortium 2014. Tree of Sex: A database of sexual systems. Scientific Data 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn GS, Kirkpatrick M. 2007. Turnover of sexual chromosomes induced by sexual conflict. Nature. 449:909–912. [DOI] [PubMed] [Google Scholar]
- van Doorn GS, Kirkpatrick M. 2010. Transitions between male and female heterogamety caused by sex-antagonistic selection. Genetics. 186:629–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White BJ, Collins FH, Besansky NJ. 2011. Evolution of Anopheles gambiae in relation to humans and malaria. Annu Rev Ecol Evol Syst. 42:111–132. [Google Scholar]
- White MJD. 1973. Animal cytology and evolution. 3rd edn Cambridge: Cambridge University Press. [Google Scholar]
- White MJD. 1978. Modes of speciation. San Francisco: W.H. Freeman. [Google Scholar]
- Yoshida K, Kitano J. 2012. The contribution of female meiotic drive to the evolution of neo-sex chromosomes. Evolution. 66:3198–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Zhang JL, Bachtrog D, An N, Huang QF, Jarvis ED, Gilbert MTP, Zhang GJ. 2014. Complex evolutionary trajectories of sex chromosomes across bird taxa. Science. 346:1246338. [DOI] [PMC free article] [PubMed] [Google Scholar]