Summary
N-glycans from invertebrates and protists have often unusual structures which present analytical challenges. Both core and antennal modifications can be quite different from the more familiar vertebrate glycan motifs; thereby, contrary to the concept that ‘simple’ organisms have ‘simple’ N-glycans, rather complex oligosaccharides structures, including zwitterionic and anionic ones, have been found in a range of species. Thus, to facilitate the optimised elucidation of the maximal possible range of structures, the analytical workflow for glycomics of these organisms should include sequential release and fractionation steps. Peptide:N-glycosidase F is sufficient to isolate N-glycans from fungi and some protists, but in most invertebrates core α1,3-fucose is present, so release of the glycans from glycopeptides by peptide:N-glycosidases A is required. Subsequent solid-phase extraction with graphitised carbon and reversed phase resins enables different classes of N-glycans to be separated prior to high-pressure liquid chromatography (HPLC) and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). Depending on the types and numbers of glycans present, either reversed- or normal-phase HPLC (or both in series) enable even single isomeric or isobaric structures to be separated prior to MALDI-TOF MS and MS/MS. The use of enzymatic or chemical treatments allows further insights to be gained, although some glycan modifications (especially methylation) are resistant. Using a battery of methods, sometimes up to 100 structures from a single organism can be assigned, a complexity which raises evolutionary questions regarding the function of these glycans.
Keywords: glycosylation, glycome, mass spectrometry, phosphorylated glycans, sulphated glycans
1. Introduction
The modification of asparagine residues of proteins by glycans (N-glycosylation) occurs in almost all eukaryotes as well as in some prokaryotes. While the N-glycans of higher organisms often have defined functions in terms of organismal viability or protein function, the biology of those of lower organisms is less well understood, partly due to restricted glycomic knowledge or limitations in the possibilities for genetic manipulation of many species. Nevertheless, it is known that in flies the formation of the neural system depends on proper N-glycan processing, whereas for nematodes some glyco-modifications play a role in inter-organismal interactions(1,2). For parasites, whether unicellular or multicellular, glycans are involved in binding to host cells or modulation of the host immune system(3). Certainly, even now, a detailed glycomic exploration is required before we can even begin to understand phylogenetic distribution of glycomotifs and their function.
Although the basic principles of N-glycan processing in eukaryotes (formation of a lipid-linked precursor, followed by transfer and further processing) are similar, the exact structures differ greatly between species. While prokaryotic N-glycans are quite unusual, only a few parasitic eukaryotes do not N-glycosylate their proteins at all. One source of variation in eukaryotes is that a number of unicellular species do not synthesise the Glc3Man9GlcNAc2 dolichol-linked precursor familiar from, e.g., yeast and mammalian systems(4); instead Giardia and Plasmodium only transfer GlcNAc1-2 to proteins, Trichomonas vaginalis Man5GlcNAc2, Tetrahymena Glc3Man5GlcNAc2 and trypanosomatids Man5-9GlcNAc2. This source of variation is due to the absence of different alg genes encoding glycosyltransferases of the endoplasmic reticulum; also, the oligosaccharyltransferase complexes necessary for transfer of the glycan precursor to asparagine residues often have a lower number of subunits in some protists.
After removal/transfer of terminal glucose residues to the protein-bound glycan (dependent on the species), transport of the folded glycoproteins from the endoplasmic reticulum, a large variety of genus-, species- and cell-specific processing events take place in the Golgi apparatus. Residues are removed and others added; not just monosaccharides such as xylose, fucose, glucuronic acid, N-acetylglucosamine, galactopyranose and galactofuranose are transferred, but also other moieties such as phosphate, sulphate, phosphorylcholine and phosphoethanolamine. On the other hand, sialic acid tends to be rare outside the vertebrates. While our knowledge of vertebrate and plant N-glycosylation is extensive, the wide variability, accompanied by a lack of predictability, of invertebrate, fungal and protist N-glycans (5) means that the release, fractionation and analysis methods must be ‘open’ to unexpected outcomes. Here we summarise methods successfully applied to N-glycans from a number of quite different organisms, including protist, fungal, molluscan, insect and nematode species (6–17). A particular focus is on sample preparation, solid-phase extraction, HPLC, MALDI-TOF MS and chemical or enzymatic treatments.
2. Materials
2.1. Reagents, Buffers and Columns (see Note 1)
2.1.1. Cell/Tissue Disruption, Proteolytic Digestion and Glycopeptide Purification
Tight fitting glass homogenizer (e.g., glass Dounce-type), porcelain mortar and pestle, or sonifier.
Pepsin A from porcine gastric mucosa (Sigma).
Thermolysin (Promega).
Sequencing grade modified trypsin dissolved to 1 mg/ml in 1 mM HCl (Promega or Roche).
Dowex 50W×8, 200-400 mesh, H+ form (Sigma-Aldrich), pre-equilibrated with 2% acetic acid.
0.5%, 2% and 10% (v/v) acetic acid.
0.5 M ammonium acetate (pH 6.0; prepared from 0.5 M acetic acid adjusted to pH 6 with ammonia)
20-ml polypropylene columns (Bio-Rad, EconoPac).
Sephadex™ G25 (medium; GE Healthcare).
Glass columns (BioRad) of 1.5 cm diameter and 50 cm length.
Orcinol monohydrate (Sigma), 200 mg dissolved in 100 ml of 20% (v/v) H2SO4, suitable for spraying or pre-treatment of TLC plates (Silica Gel 60 F254 plates (Merck)).
2.1.2. Glycan Release
N-glycosidase F (PNGase F) from Flavobacterium meningosepticum (Roche, now sold through Sigma-Aldrich; recombinant).
N-glycosidase A (PNGase A) from almond meal (Roche, now sold through Sigma-Aldrich). Recombinant PNGase A (prepared in-house) can also be used.
50 mM ammonium hydrogen carbonate (pH 8) or ammonium acetate (pH 5).
Chromatography media as in 2.1.2.
2.1.3. Solid-Phase Extraction for Glycan Purification and Subfractionation
Acetonitrile (MeCN; e.g., VWR, LC-MS grade).
Methanol (MeOH; e.g., Roth, HPLC gradient grade)
C18-SepPak (100 mg) cartridge or C18 material placed into a 1 ml solid-phase extraction (SPE) column (e.g., Supelco). Pre-equilibrate by sequential application of 100% MeOH and then water.
Non-porous graphitized carbon (PGC) column: 250 mg ENVI™Carb bulk material (Sigma-Aldrich) per 1-ml SPE tube. Pre-equilibrate by sequential application of 100% MeCN and then water.
Syringe mounted on an adaptor for expelling solutions as well as clamp for holding column.
2.1.4. Glycan Derivatization or modification
2-aminopyridine (2AP or PA; ≥99%, Sigma-Aldrich); prepare fresh PA-solution of 100 mg of PA in a mixture of 76 µL of concentrated HCl and 152 µL of water.
Sodium cyanoborohydride (95%, Sigma-Aldrich); prepare fresh cyanoborohydride/PA solution with 4.5 mg of sodium cyanoborohydride in a mixture of 9 µL of the PA solution and 13.5 µL of water (consider bubble formation).
Concentrated hydrochloric acid (37% HCl; Roth)
96-well F black plates (Nunc).
PA-labeled partial dextran hydrolysate, 2-20 glucose units (bought in non-pyridylaminated form from Sigma-Aldrich and labeled as described below).
Sephadex™ G15 (GE Healthcare).
Glass columns (BioRad) of 1 cm diameter and 50 cm length
2.1.5. HPLC
Tosoh Amide-80 column (4.6 × 250 mm, 5 µm; stored in 95% acetonitrile) with a guard column, for normal phase (NP)-HPLC. Pre-equilibrate in a 1:3 mixture of 10 mM ammonium formate (pH 7.0; i.e., 0.1 M formic acid adjusted to pH 7 with ammonia) and 95% acetonitrile in water. Alternative columns are available from other suppliers (e.g., Waters or Phenomenex).
Agilent Hypersil ODS (4 mm × 250 mm, 5 µm; stored in 30% MeOH) with a guard column, for reverse phase (RP)-HPLC. Pre-equilibrate in 0.1 M ammonium acetate, pH 4.0 (i.e., 0.1 M acetic acid adjusted to pH 4 with ammonia). Alternatives are fused core RP-HPLC columns with a superior, but also subtly different, resolution: e.g., Ascentis® Express RP-Amide (150 × 4.6 mm, 2.7μm; Supelco) or Kinetex™ 5µm XB-C18 (250 × 4.6mm; Phenomenex).
Thermo Dionex IonPac AS11 (4 × 250 mm) with guard column, for hydrophilic interaction/anion exchange (HIAX); stored in 95% acetonitrile; pre-equilibrate with 80% acetonitrile in ammonium acetate, pH 3.85 (i.e., 800 mM ammonia adjusted with acetic acid to pH 3.85).
Acetonitrile and methanol should be of LC-MS and HPLC gradient grade respectively.
2.1.6. Mass spectrometry
- MALDI matrices
- 3 mg/ml 6-aza-2-thiothymine (ATT; Sigma-Aldrich) in 50% EtOH.
- 20 mg/ml (i.e., 2% (w/v)) 2,5-dihydroxybenzoic acid (DHB; Sigma-Aldrich) in 30% (v/v) MeCN/water.
- 10 mg/ml (i.e., 1% (w/v)) α-cyano-4-hydroxycinnamic acid (ACH; Sigma-Aldrich) in 0.1% trifluoroacetic acid, 50% MeCN/water.
Seven-component peptide Mr standard mixture (Bruker Daltonics).
Appropriate steel MALDI plate.
2.1.7. Enzymatic and chemical treatments
Mannosidases: jack bean α-mannosidase from Sigma, Aspergillus (α1,2-specific) from Prozyme, Xanthomonas (α1,2/3-specific or α1,6-specific) from New England Biolabs (NEB), Helix pomatia β-mannosidase from Sigma or bacterial endo-α-mannosidase expressed in-house(9).
Fucosidases: bovine kidney (α1,2/6-preferring) from Sigma, almond (α1,3-specific) from Calbiochem, Xanthomonas (α1,2-specific) from NEB, Corynebacterium (α1,2-specific) from Takara or microbial (α1,2-specific) from Megazyme.
Galactosidases: recombinant Aspergillus niger or oryzae β-galactosidases (prepared in-house (18)), Xanthomonas (β1,3-specific) from NEB, Bacillus fragilis (β1,4-specific) from NEB or green coffee bean α-galactosidase from Sigma.
Hexosaminidases: recombinant Apis mellifera FDL linkage-specific β-N-acetylglucosaminidase or C. elegans HEX-4 β-N-acetylgalactosaminidase (both prepared in-house(19)), jack bean β-N-acetylhexosaminidase from Sigma, Streptomyces chitinase (β1,3/4/6-N-acetylhexosaminidase from NEB) or chicken liver (α1,3/4-N-acetylgalactosaminidase from Sigma).
Glucuronidase: E. coli β-glucuronidase from Megazyme (should be subject to desalting, e.g., with a 10000 MWCO ultrafiltration device and dilution/re-concentration).
Hydrofluoric acid (48%; e.g., from Sigma-Aldrich).
2.1.8. Western blotting
Standard SDS-PAGE electrophoresis apparatus (e.g., BioRad) and standard electrophoresis buffers.
Trans blot SD-semi dry transfer cell (e.g., BioRad), standard nitrocellulose membrane and transfer buffers.
Antibody dilution and blocking solution (also used for antibody dilution): 0.5% (w/v) crystalline bovine serum albumin (Roth) in 100 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.05% (v/v) Tween-20.
Anti-horseradish peroxidase (anti-HRP; Sigma-Aldrich) from rabbit; use at 1 µg/ml (1:20,000) in blocking solution.
Goat anti-rabbit IgG, conjugated with alkaline phosphatase (Vector Labs); use at 1:2000 in blocking solution.
Anti-phosphorylcholine (TEPC-15; Sigma-Aldrich) from mouse; use at 1:200 in blocking solution.
Goat anti-mouse IgA, conjugated with horseradish peroxidase or alkaline phosphate (Sigma-Aldrich); use at 1:1000 in blocking solution.
Biotinylated lectins (Vector Labs); typically use at 1:2000 in blocking solution.(20)
Anti-Biotin from goat conjugated with alkaline phosphatase (Sigma); use at 1:10,000 in blocking solution.
2.1.9. Glycan Data Analysis
Glycoworkbench 2.1 (free download at www.glycoworkbench.org/).
flexAnalysis (Bruker Daltonics).
Calculator for manual interpretation.
2.2. Equipment
Vacuum centrifuge (e.g., Speedvac, Thermo).
MALDI-TOF-TOF-MS: Autoflex Speed or UltrafleXtreme MALDI-TOF-TOF (Bruker Daltonics, Billerica, MA). Alternatives are available commercially from Shimadzu or Applied Biosystems.
Liquid chromatograph with fluorescence detector; e.g., LC-30 AD with RF 20 AXS (Shimadzu).
Fraction collector, such as BioRad model 2110.
Multifunctional microplate reader (such as Infinite M200 monochromator based instrument; Tecan).
Micro-centrifuge, such as Heraeus (Thermo).
Probe sonifier, e.g., model 250 (Branson).
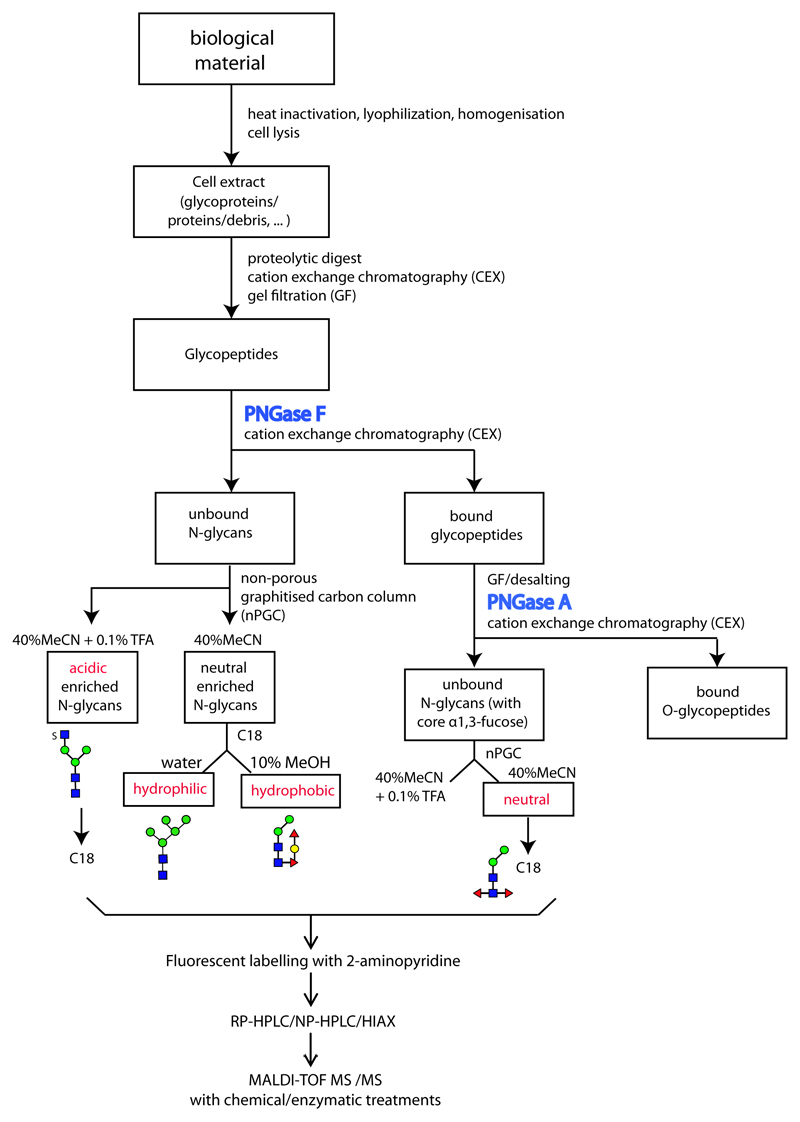
3. Methods (see flow chart in Figure 1)
Figure 1. Glycomic workflow for the analysis of N-glycans.
Initially, samples are proteolysed, the glycopeptides enriched by cation exchange (CEX) and gel filtration (GF) and the glycans released enzymatically, whereby PNGase A (and not PNGase F) is capable of releasing the core α1,3-fucosylated N-glycans. Subsequent sub-fractionation by non-porous graphitised carbon (NPGC) and/or reversed-phase (C18) resins result in pools differing in terms of anionic and zwitterionic modifications. Finally, all N-glycans are analysed by different types of HPLC (reversed or normal phase or hydrophilic interaction/anion exchange; RP, NP or HIAX) in combination with MALDI-TOF MS/MS and chemical/enzymatic treatments. The remaining O-glycopeptides can be subject to β-elimination and LC-MS. The example structures, shown according to the nomenclature of the Consortium for Functional Glycomics, are from a marine snail.(12)
3.1. Cell/Tissue Disruption, Proteolytic Digestion and Glycopeptide Purification
Biological material (see Note 2) can be whole organisms, tissues, cells, (semi-)purified proteins, media/buffer containing secreted proteins, etc., stored at -80 °C after collection; large volumes of sample should be reduced by precipitation using 5 volumes of methanol.
Suspend cells or other biological material (1-6 g wet weight) in 10 ml of boiling water for 5-10 min.
After cooling, disperse cells or tissue using a tight fitting glass homogenizer (for cellular samples, a probe sonifier may be suitable to generate subcellular particles as determined using phase contrast microscopy). For fungi/marine organisms: biological material (2-5 g) were heat denatured for 10 min then lyophilised over night. Lyophilized samples were ground in liquid nitrogen in a mortar and pestle to produce a powder which was suspended in a minimal volume of water (wait briefly to avoid freezing of the added water) and transferred into 150 ml round-bottomed flask.
Add formic acid [5% (v/v) final concentration] and 1 mg of pepsin (per 3 g wet weight). Incubate for 1 day at 37°C (final volume of 5-10 ml) and centrifuge to remove insoluble material. Alternatively, add 100 mM ammonium carbonate:ammonium hydrogen carbonate buffer to a final concentration of 20-50 mM (pH 8), followed by CaCl2 to a final concentration of 0.5 mM and finally thermolysin (1 mg protease per 1g wet weight) and incubate for 2 h at 70°C. Another alternative is trypsin, but this results in rather large glycopeptides which may not be efficiently enzymatically deglycosylated by PNGase A.
As required (e.g., when using thermolysin), acidify the sample with an aliquot of 10% acetic acid. Then incubate the proteolytic supernatant in a beaker with 10 packed ml of prewashed Dowex-50W×8 for 1 h at 23°C. Pour into a column (e.g., a BioRad Econo-Pac polypropylene column). Wash the column with 2% (v/v) acetic acid to remove unbound material, and elute glycopeptides with 0.5 M ammonium acetate (pH 6). Collect 1.5 ml fractions and assess for carbohydrate by orcinol reactivity (e.g., by spotting onto TLC plates pre-treated with orcinol and then developed at 90°C for 5-10 mins (see Note 3); lyophilize selected fractions and resuspend in not more than 3 ml of water.
Subject the sample to gel filtration on an 80-ml Sephadex G25 column (1.5 × 45 cm) in 0.5% (v/v) acetic acid and collect 4 ml fractions (see Note 4). Pool the orcinol-positive fractions (these should contain glycopeptides) and lyophilize.
3.2. Release of N-Glycans (see Note 5)
3.2.1. Sequential Release by PNGase F and PNGase A (6,12)
Resuspend the glycopeptides in water, heat for 5 min at 95°C (to denature remaining proteases), and subject to digestion overnight at 37°C with 3 µL PNGase F (Roche) in a final concentration of 20-50 mM NH4HCO3 (pH 8.0) and a volume of 200-300 µl.
Acidify the sample (Note 6) and repeat the Dowex-50W×8 chromatography step (see step 4 of 3.1), collect 1.5 ml fractions and lyophilize the unbound (free N-glycans, lacking core α1,3-fucose, ready for further purification) and the bound fractions eluted with 0.5 M ammonium acetate pH 6 (remaining glycopeptides).
Desalt the remaining glycopeptides on Sephadex G25 (as in step 5 of 3.1). Lyophilize orcinol-positive fractions and dissolve together in 50 mM ammonium acetate, pH 5.0. Incubate with 3 µL of PNGase A (Roche) overnight at 37°C.
Repeat the Dowex-50 chromatography step (step 4 of 3.1). The unbound fraction contains core α1,3-fucosylated glycans which are also ready for further purification below, whereas the remaining glycopeptides can be subject to β-elimination for O-glycan analysis(12).
3.3. Solid-Phase Extraction for Glycan Purification and Subfractionation
3.3.1. Solid-phase Extraction and Enrichment of Hydrophobic N-Glycans (12)
Dilute glycan samples in 100 µl water and apply to a C18-SepPak (see 2.1.3.3).
Elute most N-glycans with water.
Elute further hydrophobic N-glycans with 15% MeOH.
Wash column with 40% MeOH and 100% MeOH.
Dry by vacuum centrifugation or lyophilise.
Assess quality of pools by mass spectrometry.
3.3.2. Enrichment of Anionic N-Glycans by Non-porous Graphitized Carbon (8,12)
Dissolve glycans in water (100-200 µl) and apply them to graphitised carbon pre-equilibrated as above (2.1.3.4).
Wash the column or cartridge with water (1-2 ml).
Elute primarily neutral N-glycans with 40% MeCN (1 ml).
Elute further N-glycans with 0.1 % TFA in 40% MeCN (1 ml).
Dry neutral and acidic-enriched pools by vacuum centrifugation or lyophilization before analysis or derivatization.
Assess quality of pools by mass spectrometry.
3.4. Derivatization of N-glycans (see Note 7)
3.4.1. Reductive Amination of Glycans
Fluorophores are conjugated at the reducing terminus of the glycan, which is uniquely constituted by a reactive carbonyl moiety. Many fluorophores are available including 2-aminobenzamide (2AB), AEAB and 2-aminopyridine (2AP or PA), the latter which is described here.
Transfer 80 µL of fresh 2-aminopyridine (PA) solution (see 2.1.4.1) to the glycan sample dried in a 1.5-ml polypropylene microcentrifuge tube, apply safety cap to avoid popping, and incubate in boiling water for 15 min.
Continue the reaction by transferring 4 µL of a fresh cyanoborohydride/PA solution (see 2.1.4.2) to the sample and incubating overnight at 90°C in an oven.
Dilute the sample in 1.5 ml of 0.5% acetic acid, apply to a 30 ml Sephadex G15 column (1 × 40 cm) equilibrated in 0.5% acetic acid, and collect 1.5 ml fractions. Transfer aliquots of fractions (80 µl) to a 96 F black plate and detect fluorescence in a microtiter plate reader (excitation/emission: 320/400nm). Pool fluorescent glycans eluting before the excess labeling reagent and lyophilize.
3.5. HPLC separation methods
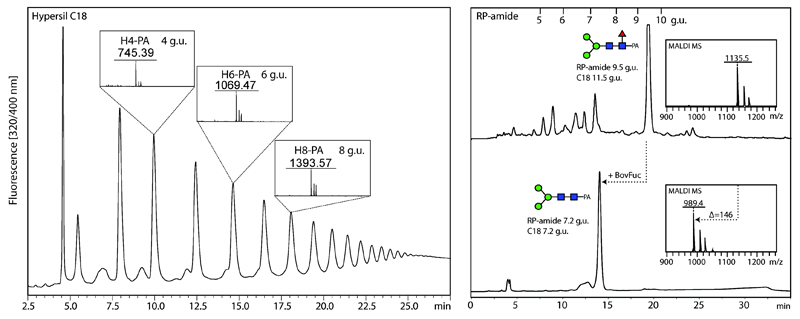
Pyridylaminated N-glycans can be analyzed by either NP- or RP-HPLC or HIAX using an HPLC system equipped with a fluorescence detector. NP- and RP-columns are calibrated daily in terms of glucose units, using PA-labeled forms of partial dextran hydrolysates (see Note 8 and Figure 2).
Figure 2. Examples of RP-HPLC separations.
RP-HPLC columns can be calibrated in terms of glucose units for day-to-day comparisons; the order of elution is the same as for NP-HPLC with the lowest molecular weight gluco-oligomers coming first as verified by MALDI-TOF MS of three of the standard peaks. Some modifications have also a major effect on elution times on RP-HPLC, as exemplified by the elution of Manα1,6(Manα1,3)Manβ1,4GlcNAcβ1,4(Fucα1,6)GlcNAc-PA before and after bovine α-fucosidase digestion, which results in a reduction in elution time from 9.5 to 7.2 glucose units on an RP-amide column (11.5 to 7.2 g.u. in the case of a standard C18 column); the example glycan shown is from Trichuris suis.(17)
3.5.1. Option 1: NP-HPLC
Dissolve dried sample in 50 μL of a 1:3 mixture of solvent A (10 mM ammonium formate, pH 7.0) and solvent B (95% acetonitrile in water).
Inject a portion of the sample into a Tosoh-80 column equilibrated in the same 1:3 mixture.
Elute column at 1 ml/min as follows: 0-5 mins, 75% solvent B; 5-15 mins, 75-65% B; 15-40 mins, 65% B; 40-55 mins, 65-57% B; followed by a return to the starting conditions. Other variant: 1-5min: 75%B; 5-10 min, 75-70% B; 10-15 min, 70-65% B; 15-55 min; 65-55% B; 55-56 min; 55-75%B; 56-60 min, 75% B. The remaining % (up to 100 % (v/v)) derive from delivery of solvent A by the HPLC system.
Detect glycans by fluorescence using excitation at 310 nm and emission at 380 nm. Collect fractions based on fluorescence intensity and lyophilize prior to later analyses.
3.5.2. Option 2: RP-HPLC (see also Fig. 2)
Dissolve dried sample in 50 µL of water.
Inject portion of sample onto an RP-HPLC column (e.g., Agilent Hypersil ODS or Phenomenex Hyperclone) pre-equilibrated with 100 mM ammonium acetate (pH 4; solvent C).
Elute at 1.5 ml/min using a linear gradient from the starting buffer to a solution composed of 30% (v/v) MeOH (solvent D), at 1% per min (in the case of fungal samples(10), a shallower gradient was applied: 0-35 min, 0-15% D; 35-36 min, 15-35% D; 36-46 min, 35-0% D).
Detect glycans by fluorescence using excitation at 320 nm and emission at 400 nm. Collect fractions based on fluorescence intensity and lyophilize prior to later analyses. For fused core RP columns, the gradients may have to be adapted and the flow rate reduced to 0.8 ml/min if manually collecting fractions.
3.5.3. Option 3: 2-D HPLC (see Note 9)
First fractionate by NP-HPLC (see Subheading 3.5.1.).
Collect fractions, lyophilize and identify fractions of interest by MALDI-TOF MS (see Subheading 3.6).
Subject desired fractions to RP-HPLC (see Subheading 3.5.2) and analyze by MALDI-TOF MS (see Subheading 3.6).
3.5.4. Option 4: HIAX (8,21)
Dissolve sample in 10 µl water, before adding 40 µl acetonitrile; pre-equilibrate AS11 column (Dionex) with 4:1 80% acetonitrile/800 mM ammonium acetate, pH 3.85 (solvent E is 800 mM ammonia, pH 3.85, and solvent F is 80% acetonitrile).
Check column by injecting a mixture of PA-labeled oligomannosidic glycans e.g., Man3-9GlcNAc2).
Inject portion of sample onto pre-equilibrated column.
Elute with the following gradient: 0-5 min, 99% F; 5-50 min, 99-90% F, 50-65 min; 90-80% F, 65-85 min, 80-7% F; 85-110 min, 75-65% F; 110-112 min, 65% F; 112-120 min, 65-99% F; 120-130 min, 99% F.
Collect (either based on fluorescence intensity or time) and dry fractions for further analyses.
3.6. MALDI-TOF and TOF-TOF Analysis (see Note 10)
Dried native or chemically-treated (see below) samples are resuspended in 5-20 µl water; exoglycosidase digested samples can normally be used directly.
Prepare a method blank (without analyte) to differentiate signals from background contamination (especially for exoglycosidase digests).
Spot 0.5 µL of the sample onto a steel MALDI target plate and vacuum dry; thereafter 0.5 µL of the matrix ATT or DHB is added and drying repeated; sometimes the signals can be improved or shifted to [M+H]+ if 0.25 µl 20 mM ammonium sulphate pH 5 is co-spotted with the sample. Spot a peptide mixture (using ACH as matrix) as an external mass calibrant.
Analyze glycans using an MALDI-TOF-TOF in reflectron positive or negative ion modes. Acquire MALDI spectra at laser frequency of 1000 Hz and sum 1000-2000 individual spectra of each sample. Typically, glycans require higher laser power and/or higher detector gain settings than peptides. Decreasing the lens voltage, as compared to factory settings, may also improve detection of some glycans.
For glycan fragmentation, perform TOF-TOF MS/MS experiments by selecting precursor ions for laser-induced dissociation using the LIFT apparatus in Bruker instruments (see Note 11).
3.7. Enzymatic and chemical treatments
3.7.1. Exoglycosidase digestions (see Note 12)
Resuspend pmol quantities of 2 PA-labeled glycans in an appropriate amount of water.
Typically 1 µl pyridylaminated glycans and 0.5 µl 50 mM ammonium acetate (pH 5.0) are mixed with 0.2 µl enzyme (regardless of U/ml) and 0.8 µl water in a PCR tube. Incubate tubes at 37°C overnight in an oven. If performing a number of reactions with the same enzyme, the glycosidase can be pre-mixed with buffer and water for immediate use to reduce pipetting errors. Some glycosidases have other pH or cation requirements: e.g., β-glucuronidase requires pH 7 and α1,2/3-mannosidase requires 0.5 mM CaCl2 (refer to suppliers’ information as required); other glycosidases may need to be diluted before use due to contaminants.
3.7.2. Chemical cleavage to remove phosphoesters, galactofuranose or fucose residues (see Note 13)
Incubate dried glycan fractions with 3 µl 48% hydrofluoric acid (HF; caution) on ice for 24-48 hours in plastic microcentrifuge tubes, and then dry under a stream of dry N2 gas or in a Speedvac.
Re-evaporate from water to remove traces of HF.
3.8. Western blotting (see Note 14)
Analyze crude whole cell extracts (25 µg total protein material) or purified glycoproteins of interest by Western blotting after separation by SDS-PAGE and transfer to nitrocellulose membrane using a semi-dry blotting apparatus.
Use standard Western blot procedures to probe the membrane with anti-carbohydrate antibody or lectin, such as rabbit anti-HRP (IgG) for α3-linked core fucose or β1,2-xylose, mouse anti-phosphorylcholine (TEPC-15; IgA) for phosphorylcholine, or biotin-conjugated lectins.
Use a relevant secondary antibody: e.g., alkaline phosphatase–conjugated forms of goat anti-rabbit antibody (for anti-HRP), goat-anti-mouse IgA (for TEPC-15) or anti-biotin (or streptavidin; for lectins).
Colour detection is with 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium or 3,3´-diaminobenzidine for alkaline phosphatase or peroxidase conjugates respectively; SigmaFAST tablets are available for both.
3.9. Glycan Data Analysis
-
1.
Analyze MS spectra using the relevant software (e.g., flexAnalysis for Bruker instruments). Ions are typically singly charged under the MALDI conditions described, and only exact monoisotopic m/z values (i.e., the 12C-only ion) should be examined. If possible, select [M+H]+ ions for peak picking in positive ion mode, or [M-H]– in negative ion mode. Spectra can be exported as mzXML files if supporting information is required for publications.
-
2.
Predict glycan compositions based on mass/charge (m/z) matching with potential glycan compositions using manual calculations or GlycoWorkbench software. Typically MALDI is accurate to +/- 0.3 daltons; N-glycans will contain varying numbers of hexose (Δm/z 162), N-acetylhexosamine (Δm/z 203), deoxyhexose (e.g., fucose; Δm/z 146), pentose (e.g., xylose; Δm/z 132), sulphate/phosphate (Δm/z 80), methylphosphate (Δm/z 94) or zwitterionic residues (phosphorylcholine, phosphoethanolamine, N-methyl-2-aminoethylphosphonate or aminoethylphosphonate with respectively Δm/z 165, 123, 121 or 107). Apply corrections for reducing terminal derivatization or permethylation. The nature of the hexose and N-acetylhexosamine residues depends on the species; indeed, even in fungi, hexose residues are not automatically mannose(10). Sialic acids are rare in non-vertebrate species; their presence in invertebrate or protist samples may suggest contamination with food or media components. Western blotting data can additionally aid interpretation.
-
4.
Predict structural models based on known biosynthetic rules, and confirm using exoglycosidase digestions (Subheading 3.7.1), chemical cleavage (Subheading 3.7.2), and MS/MS studies (Subheading 3.6). These models can be converged with findings from elution times in 1D- and 2D-HPLC studies (Subheading 3.5).
4. Notes
A high quality source of deionized water, to dissolve reagents and samples, is required; in some cases, the water should be bought to avoid ionic or microbial contaminants.
Care must be taken to consider potential contamination from food sources or media components; for instance, foetal calf serum often used in parasite cultivation contains fetuin (a sialylated glycoprotein), fungal growth media contain milk glycoproteins (casein) or mosquito larvae are reared on ground cat or fish food. This can mean that cells or unicellular organisms are kept briefly in a serum-free medium or at least adequately washed or that buffer be exchanged in order to reduce media contaminants. Also, contamination by a polyhexose series is common; to reduce the relevant signals, samples (e.g., glycopeptides) can be pre-reduced prior to N-glycan release with 100 µL of 0.5% (w/v) sodium borohydride/2.5% (v/v) ammonia at room temperature for 2 h followed by addition of 2.5 µL of glacial acetic acid and lyophilization(22). The reduced material will then be inert to derivatization by reductive amination and the normal glycan release procedure then followed after adjustment to the pH required by the applied enzyme. Endogenous endo- and exoglycosidases are another potential problem, but heat inactivation before disruption of cell integrity should be sufficient to prevent glycan degradation.
Care should be taken when diluting or handling sulphuric acid required for the orcinol reagent (do not pour water directly into concentrated acid); for pre-treating TLC plates, apply a thin layer of orcinol reagent and then dry with a hair drier.
Whereas sample volume is not a problem for Dowex chromatography, in the case of gel filtration, the sample volume should be no more than 5% of that of the gel filtration column.
PNGase F is the standard N-glycosidase for releasing N-glycans, but does not release N-glycans modified by Fuc α3-linked to the core GlcNAc(23). PNGase A has broader specificity to include core α3-fucosylated N-glycans, but requires small peptides for optimal activity, is more expensive and differs in its pH optimum as compared to PNGase F. Newer broad specificity enzymes are now described, but are not yet commercially available. Endoglycosidases are more specific and so are less suited for whole organism glycomics unless pretreatment to remove, e.g., oligomannosidic forms is desired. Enzymatic deglycosylation is most effective after proteolysis, especially for PNGase A. However, PNGase F is also capable of deglycosylating intact proteins, which have been denatured by SDS or urea, if SDS is first diluted in the presence of NP40, or urea is sufficiently diluted; deglycosylation can be followed by glycan recovery or by Western blotting.(24) As appropriate, glycopeptides samples may be treated with PNGase A or F alone (e.g., with PNGase A alone if core α1,3-fucosylated glycans are a major component or with PNGase F alone if, as in the case of fungi, no core α1,3-fucosylated glycans are expected); the described approach of sequential release is, however, enabling a partial sub-fractionation of complex N-glycomes. Chemical release with hydrazine is also an option, but requires thorough purification to remove contaminants interfering with mass spectrometry.
To prevent failure of pH-sensitive steps, such as Dowex chromatography or digestion with glycosidases, confirm proper pH by dispensing 1 µL on a strip of appropriate pH indicator paper.
The simplest method is to analyze released N-glycans directly by MS. However, derivatization of the reducing terminus with a fluorophore improves MS sensitivity, and allows detection of glycans by RP or NP chromatography for identification based on co-chromatography with known standards. PA is commonly used in our own laboratory and in Japan(25); otherwise, 2AB or AA are another commonly used fluorophores with different properties in terms of hydrophobicity and HPLC/MS sensitivity. AEAB is of interest for preparation of natural glycan arrays(26).
Calibrated HPLC separations can allow structure prediction based on elution times, because compositional isoforms may be differentially retained on selected stationary phases. For both normal and reversed phase columns, the lowest glucose units elute first (Figure 2). In case of doubt, collected fractions containing the standards can be analysed by MALDI-TOF MS. Anionic glycans tend to elute earlier than the corresponding neutral structures on NP- and RP-HPLC columns(8,11); the elution of zwitterionic phosphorylcholine-containing glycans is either relatively late on standard RP-HPLC or relatively earlier on fused core RP-HPLC columns(27); core α1,3-fucose results in early elution and core α1,6-fucose in late elution on RP-HPLC regardless of the type of column. HPLC buffers are made from the individual components (acetic acid and ammonia) and not by merely adjusting the pH of an ammonium acetate solution prepared from the commercially-available salt; the stock solutions should also be filtered before use.
Typically, 2D-HPLC is generally performed with NP-HPLC in the first dimension and RP-HPLC in the second(6), however, we have occasionally done RP-HPLC followed by HIAX (10).
Here we focus on MALDI-TOF MS, but LC-ESI-MS is also a commonly-used method in glycomic analyses and its MSn capabilities are of interest in the case of unusual structures as well as for cross-ring cleavages; also permethylation or perdeuteromethylation can be performed, but care must be taken with clean-up in order to avoid loss of hydrophilic sulphated or phosphoester-modified glycans. We have found ATT to be a rather robust matrix for positive and negative MALDI-TOF MS and is also suitable for glycopeptides(24); DHB tends to favour formation of sodiated adducts of glycans. ACH is used as a matrix for the peptide standard; its chlorinated form can be also used for glycans and glycopeptides(28).
In our hands, best results for MS/MS are obtained with the [M+H]+ form in positive mode as sodiated adducts tend to show losses from the parent ion (B-fragments), but are relatively poor in terms of Y-fragments useful for determining core modifications. High-energy collision-induced decay using argon is also possible, but reduces the parent ion signal. If a neutral glycan sample is rather concentrated, then cross-ring cleavage in negative-ion mode is possible(17).
Although jack bean α-mannosidase is considered non-specific with respect to the linkage position on the underlying sugar, reaction rates vary considerably and thus times need to be extended to achieve removal of sterically constrained linkages. Some exoglycosidases are extremely sensitive to steric hindrance; furthermore, not all residues occurring in non-vertebrate glycans are susceptible to exoglycosidase digestion due either to the types of linkages or the presence of substitutions such as methylation or sulphate. If there is sufficient material, shifts in elution time by RP-HPLC offer extra information in addition to MALDI-TOF MS alone; e.g., removal of core α1,6-fucose results in a reduction in retention time (Figure 2). On-plate digestions in a humid chamber can also be performed. Volatile buffers (e.g., ammonium acetate) are to be preferred over manufacturer-supplied buffers with citrate, phosphate, etc.
Hydrofluoric acid removes phosphoesters (phosphate, methylphosphate, phosphorylcholine, phosphoethanolamine, 2-aminoethylphosphonate and its N-methylated form), galactofuranose and α1,3-fucose; α1,2- and α1,4-fucose is also partially susceptible, but α1,6-fucose and α-mannose is resistant(8,10,12,14). Hydrofluoric acid is to be treated with caution; a tube of calcium gluconate gel should be to hand in case of skin contact. As for exoglycosidase digestions, if there is sufficient material, shifts in elution time by RP-HPLC offer extra information in addition to MALDI-TOF MS alone.
Antibodies and lectins can help screen types of glycan modifications, such as core α3- vs. α6-linked fucose when using anti-HRP or phosphorylcholine when using TEPC-15, in the samples at the glycoprotein level.(7,22) Western blotting is readily adapted to the use of any lectin or antibody; enzyme-linked, fluorescence or ECL detection systems can be used. Concentrations of the antibody/lectin should be optimized by comparison with appropriate negative and positive controls; especially when using biotin-based systems, controls without the lectin (i.e., applying only the anti-biotin or streptavidin conjugate) are required. Nevertheless, interaction with a lectin is not a structural proof and can be misleading as many non-mammalian glyco-modifications have not been assessed for lectin reactivity.
Acknowledgment
This work was supported by the Austrian Fonds zur Förderung der wissenschaftlichen Forschung (FWF; grants P26662, P25058 and P23922 to A.H., K.P and I.B.H.W.).
References
- 1.Schachter H. Paucimannose N-glycans in Caenorhabditis elegans and Drosophila melanogaster. Carbohydr Res. 2009;344:1391–1396. doi: 10.1016/j.carres.2009.04.028. [DOI] [PubMed] [Google Scholar]
- 2.Sabotič J, Ohm RA, Künzler M. Entomotoxic and nematotoxic lectins and protease inhibitors from fungal fruiting bodies. Appl Microbiol Biotechnol. 2015;100:91–111. doi: 10.1007/s00253-015-7075-2. [DOI] [PubMed] [Google Scholar]
- 3.Rodrigues JA, Acosta-Serrano A, Aebi M, Ferguson MA, Routier FH, Schiller I, Soares S, Spencer D, Titz A, Wilson IB, Izquierdo L. Parasite Glycobiology: A Bittersweet Symphony. PLoS Pathog. 2015;11:e1005169. doi: 10.1371/journal.ppat.1005169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samuelson J, Banerjee S, Magnelli P, Cui J, Kelleher DJ, Gilmore R, Robbins PW. The diversity of dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc Natl Acad Sci U S A. 2005;102:1548–1553. doi: 10.1073/pnas.0409460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schiller B, Hykollari A, Yan S, Paschinger K, Wilson IBH. Complicated N-linked glycans in simple organisms. Biol Chem (Hoppe Seyler) 2012;393:661–673. doi: 10.1515/hsz-2012-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paschinger K, Hykollari A, Razzazi-Fazeli E, Greenwell P, Leitsch D, Walochnik J, Wilson IBH. The N-glycans of Trichomonas vaginalis contain variable core and antennal modifications. Glycobiology. 2012;22:300–313. doi: 10.1093/glycob/cwr149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schiller B, Makrypidi G, Razzazi-Fazeli E, Paschinger K, Walochnik J, Wilson IBH. Exploring the unique N-glycome of the opportunistic human pathogen Acanthamoeba. J Biol Chem. 2012;287:43191–43204. doi: 10.1074/jbc.M112.418095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hykollari A, Balog CI, Rendić D, Braulke T, Wilson IBH, Paschinger K. Mass spectrometric analysis of neutral and anionic N-glycans from a Dictyostelium discoideum model for human congenital disorder of glycosylation CDG IL. J Proteome Res. 2013;12:1173–1187. doi: 10.1021/pr300806b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hykollari A, Dragosits M, Rendić D, Wilson IBH, Paschinger K. N-glycomic profiling of a glucosidase II mutant of Dictyostelium discoideum by “off-line” liquid chromatography and mass spectrometry. Electrophoresis. 2014;35:2116–2129. doi: 10.1002/elps.201300612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hykollari A, Eckmair B, Voglmeir J, Jin C, Yan S, Vanbeselaere J, Razzazi-Fazeli E, Wilson IBH, Paschinger K. More than just oligomannose: An N-glycomic comparison of Penicillium species. Mol Cell Proteomics. 2016;15:73–92. doi: 10.1074/mcp.M115.055061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurz S, Jin C, Hykollari A, Gregorich D, Giomarelli B, Vasta GR, Wilson IBH, Paschinger K. Haemocytes and plasma of the eastern oyster (Crassostrea virginica) display a diverse repertoire of sulphated and blood group A-modified N-glycans. J Biol Chem. 2013;288:24410–24428. doi: 10.1074/jbc.M113.478933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckmair B, Jin C, Abed-Navandi D, Paschinger K. Multi-step fractionation and mass spectrometry reveals zwitterionic and anionic modifications of the N- and O-glycans of a marine snail. Mol Cell Proteomics. 2016 doi: 10.1074/mcp.M1115.051573. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurz S, Aoki K, Jin C, Karlsson NG, Tiemeyer M, Wilson IB, Paschinger K. Targetted release and fractionation reveal glucuronylated and sulphated N- and O-glycans in larvae of dipteran insects. J Proteomics. 2015;126:172–188. doi: 10.1016/j.jprot.2015.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan S, Brecker L, Jin C, Titz A, Dragosits M, Karlsson N, Jantsch V, Wilson IBH, Paschinger K. Bisecting galactose as a feature of N-glycans of wild-type and mutant Caenorhabditis elegans. Mol Cell Proteomics. 2015;14:2111–2125. doi: 10.1074/mcp.M115.049817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan S, Jin C, Wilson IBH, Paschinger K. Comparisons of Caenorhabditis fucosyltransferase mutants reveal a multiplicity of isomeric N-glycan structures. J Proteome Res. 2015;14:5291–5305. doi: 10.1021/acs.jproteome.5b00746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paschinger K, Wilson IBH. Two types of galactosylated fucose motifs are present on N-glycans of Haemonchus contortus. Glycobiology. 2015;25:585–590. doi: 10.1093/glycob/cwv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson IBH, Paschinger K. Sweet secrets of a therapeutic worm: Mass spectrometric N-glycomic analysis of Trichuris suis. Anal Bioanal Chem. 2016;408:461–471. doi: 10.1007/s00216-015-9154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dragosits M, Pflugl S, Kurz S, Razzazi-Fazeli E, Wilson IBH, Rendić D. Recombinant Aspergillus β-galactosidases as a robust glycomic and biotechnological tool. Appl Microbiol Biotechnol. 2014;98:3553–3567. doi: 10.1007/s00253-013-5192-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dragosits M, Yan S, Razzazi-Fazeli E, Wilson IBH, Rendić D. Enzymatic properties and subtle differences in the substrate specificity of phylogenetically distinct invertebrate N-glycan processing hexosaminidases. Glycobiology. 2015;25:448–464. doi: 10.1093/glycob/cwu132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iskratsch T, Braun A, Paschinger K, Wilson IBH. Specificity analysis of lectins and antibodies using remodeled glycoproteins. Anal Biochem. 2009;386:133–146. doi: 10.1016/j.ab.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 21.Neville DC, Dwek RA, Butters TD. Development of a single column method for the separation of lipid- and protein-derived oligosaccharides. J Proteome Res. 2009;8:681–687. doi: 10.1021/pr800704t. [DOI] [PubMed] [Google Scholar]
- 22.Pöltl G, Kerner D, Paschinger K, Wilson IBH. N-Glycans of the porcine nematode parasite Ascaris suum are modified with phosphorylcholine and core fucose residues. FEBS J. 2007;274:714–726. doi: 10.1111/j.1742-4658.2006.05615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tretter V, Altmann F, März L. Peptide-N4-(N-acetyl-β-glucosaminyl)asparagine amidase F cannot release glycans with fucose attached α1→3 to the asparagine-linked N-acetylglucosamine residue. Eur J Biochem. 1991;199:647–652. doi: 10.1111/j.1432-1033.1991.tb16166.x. [DOI] [PubMed] [Google Scholar]
- 24.Paschinger K, Gonzalez-Sapienza GG, Wilson IBH. Mass spectrometric analysis of the immunodominant glycan epitope of Echinococcus granulosus antigen Ag5. Int J Parasitol. 2012;42:279–285. doi: 10.1016/j.ijpara.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hase S, Ibuki T, Ikenaka T. Reexamination of the pyridylamination used for fluorescence labelling of oligosaccharides and its application to glycoproteins. J Biochem (Tokyo) 1984;95:197–203. doi: 10.1093/oxfordjournals.jbchem.a134585. [DOI] [PubMed] [Google Scholar]
- 26.Song X, Xia B, Stowell SR, Lasanajak Y, Smith DF, Cummings RD. Novel fluorescent glycan microarray strategy reveals ligands for galectins. Chem Biol. 2009;16:36–47. doi: 10.1016/j.chembiol.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan S, Wilson IBH, Paschinger K. Comparison of RP-HPLC modes to analyse the N-glycome of the free-living nematode. Pristionchus pacificus Electrophoresis. 2015;36:1314–1329. doi: 10.1002/elps.201400528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selman MH, Hoffmann M, Zauner G, McDonnell LA, Balog CI, Rapp E, Deelder AM, Wuhrer M. MALDI-TOF-MS analysis of sialylated glycans and glycopeptides using 4-chloro-alpha-cyanocinnamic acid matrix. Proteomics. 2012;12:1337–1348. doi: 10.1002/pmic.201100498. [DOI] [PubMed] [Google Scholar]