Abstract
Pre-mRNA splicing proceeds by two consecutive trans-esterification reactions via a lariat-intron intermediate. We present the 3.8Å cryoEM structure of the spliceosome immediately after lariat formation. The 5’-splice site is cleaved but remains close to the catalytic Mg2+ site in the U2/U6 snRNA triplex, and the 5’-phosphate of the intron nucleotide G(+1) is linked to the branch adenosine 2’OH. The 5’-exon is held between the Prp8 N-terminal and Linker domains, and base-pairs with U5 snRNA loop 1. Non-Watson-Crick interactions between the branch helix and 5’-splice site dock the branch adenosine into the active site, while intron nucleotides +3 to +6 base-pair with the U6 snRNA ACAGAGA sequence. Isy1 and the step one factors Yju2 and Cwc25 stabilise docking of the branch helix. The intron downstream of the branch site emerges between the Prp8 RT and Linker domains and extends towards Prp16 helicase, suggesting a plausible mechanism of remodelling before exon ligation.
Introduction
The spliceosome is a dynamic molecular machine1,2 that catalyzes pre-mRNA splicing in two sequential trans-esterifications analogous to group II intron self-splicing3. The major spliceosomal components - U1, U2, U4/U6, and U5 small nuclear ribonucleoprotein particles (snRNPs), and the two large Nineteen and Nineteen Related (NTC and NTR) protein complexes - assemble de novo on pre-mRNA substrates in an ordered manner4–6. Initially U1 and U2 snRNPs recognise the 5’-splice site (5’SS) and branch point (BP) sequences of pre-mRNA: subsequently the pre-assembled U4/U6.U5 tri-snRNP is recruited to form the fully assembled spliceosome (complex B). During catalytic activation Prp28 helicase displaces the 5’SS from U1 snRNP and allows it to base-pair with the U6 snRNA ACAGAGA sequence7,8. Brr2 helicase unwinds the U4/U6 snRNA duplex to release U4 snRNA and its associated proteins9,10, allowing recruitment of the NTC and NTR complexes. The resulting complex Bact is then remodelled to complex B*, which recruits step one-specific factors Yju2 and Cwc25. These factors stabilise a network of RNA interactions comprising U2, U5, and U6 snRNAs, which position the pre-mRNA 5’SS and BP sequences for catalysis of the first trans-esterification (branching) producing 5’-exon and lariat intron-3’exon intermediates. The resulting complex C is further remodelled to complex C* in which the 5’- and 3’-exons are aligned on U5 snRNA loop 1 to produce spliced mRNA and lariat intron products via the second trans-esterification (exon ligation)11,12. The spliced mRNA is released and the remaining Intron Lariat Spliceosome (ILS) is disassembled, recycling the snRNPs for new rounds of splicing.
During this splicing cycle DExD/H box helicases are recruited to the spliceosome at specific steps to remodel RNA-RNA interactions and induce binding or release of auxiliary factors13,14. Specifically, after branching, the step one factors Yju2 and Cwc25 are released by Prp16 helicase and Prp18-Slu7 and Prp22 are recruited to produce catalytically active complex C*(ref 13). Following exon ligation, the spliced mRNA is released by Prp22 helicase15 and the residual ILS is disassembled by Prp43 helicase16,17.
Here we describe the cryoEM structure of the spliceosome captured immediately after branching. This structure provides insight into recognition and positioning of the 5’SS and branch point at the active site, elucidates how proteins stabilise the architecture of the catalytic RNA core, and provides a molecular basis to understand the functions of RNA helicases and auxiliary factors in remodelling the spliceosome.
Overview of the structure
Spliceosomes from the yeast Saccharomyces cerevisiae were assembled on UBC4 pre-mRNA substrate18 with a mutation of the 3’-splice site (3’SS) sequence UAG|AG to UACAC, and purified via an affinity-tag on Slu7 or Prp18 (Methods). The purified spliceosomes contained predominantly lariat intron-3’exon intermediates (Extended Data Fig. 1), indicating that the purified spliceosomes represent complex C. We obtained a cryoEM reconstruction at 3.8Å overall resolution (Methods; Extended Data Figs. 1-6; Extended Data Table 1) into which 44 components have been modelled (Fig. 1; Extended Data Table 2). The U5 snRNP forms the core of the complex, which cradles the active site (Fig. 1a). Assembling onto this core, the NTC and NTR act as a multipronged clamp that stabilizes binding of the U2 snRNP core, the substrate, and auxiliary splicing factors to the U5 snRNP (Fig. 1a-c). The helicase module containing Brr2 and Prp16 protrudes from the U5 snRNP core (Fig. 1a,b).
Figure 1. Subunit architecture of the spliceosomal complex C.
a-c, three orthogonal views of the complex coloured according to the subunit identity. d, a list of all 44 modelled subunits of the complex grouped into functional sub-complexes.
As in U4/U6.U5 tri-snRNP19,20, the Large domain of Prp8 (ref. 21) forms the foundation of the assembly together with the stable foot unit, comprising GTP-bound Snu114 and the N-terminal domain of Prp8, firmly gripping the U5 snRNA (Fig. 2a,b). Prp8 has undergone a large structural change including a 30° rotation of the foot with respect to the Large domain when compared to U4/U6.U5 tri-snRNP19 (Extended Data Fig. 7). U4 snRNA and its associated proteins have been released upon unwinding of the U4/U6 duplex by Brr2 (ref 6). The 3’-domain of U2 snRNP comprising Msl1(U2B”), Lea1(U2A’) and the Sm core domain bridges the Prp8 RNaseH-like domain and the N-terminal HAT (Half-a-TPR)-repeat domain of Syf1 (Fig. 2a). Isy1 and Cef1 dock with the N-terminal and reverse transcriptase(RT)-like domains of Prp8 (ref. 21), respectively, and anchor the N-terminal end of Cfl1 together with Prp45/Prp46 (Fig. 2c,d). These interactions support the HAT-repeat arches of Syf1 and Cfl1 suspended over the Large domain of Prp8. The 5’-part of U2 snRNA and the 3’-part of U6 snRNA run side-by-side from the active site forming nine consecutive base-pairs extending towards the centre of the Syf1 HAT-repeat arch (Fig. 2a-e). Bud31 anchors the 5’-stem of U6 snRNA to the N-terminal domain of Prp8 (Fig. 2c). Cwc2 is wedged between Bud31, Ecm2 and Prp45 and guides the path of U6 snRNA22 (Fig. 2c). U2 snRNA downstream of the branch helix extends from the active site towards the 3’-domain of U2 snRNP, forming two stems bridging the U2 Sm ring with Ecm2/Cwc2 and the main body of the complex (Fig. 2d,e). Density for two RNA helices emanating from the U2 Sm ring is consistent with a stem-loop IIb/stem IIc arrangement and the catalytically competent conformation of the active site23,24 (Fig. 2f). The C-terminal region of Cwc21 forms a coiled-coil that interacts with Snu114 (ref. 25) (Fig. 2a) while the N-terminal half of Cwc21 extends towards Prp8 and points into the U5 snRNA stem minor groove.
Figure 2. Overview of the core structure.
a, Prp8 and its central role in organizing the entire assembly (SII denotes U2/U6 stem II). b, RNA only in the same orientation as in a (ISL, U6 snRNA Internal Stem-Loop; 5’SL, U6 snRNA 5’ Stem-Loop; SL1, U5 snRNA Stem-Loop 1; VSL, U5 snRNA Variable Stem-Loop; S3, U5 snRNA Stem III). c, Ecm2, Cwc2 and Bud31 binding to the 5’-end of the U6 snRNA. d, top view of the complex. e, RNA only in the same orientation as in d. f, Secondary structure diagram for the 3'-end of U2 snRNA.
Two large regions of weak density extend from the well-ordered core of the complex (Extended Data Fig. 1e). Focused classification allowed us to select subsets of particles (core+helicase, core+Prp19) (Extended Data Fig. 2), in which less well-ordered components can be more clearly visualised. The weak density observed in the latter class is readily attributable to Prp19, Cef1 and Snt309 based on its distinct shape first observed in ILS26 but the weaker density in complex C suggests these proteins are more loosely attached to the core than in ILS. A large lobe corresponding to a DEAH helicase in contact with Cwc25 is observed near the intron exit channel, downstream of the BP. Although its limited resolution does not allow us to build a model de novo, the density is of sufficient quality to fit a DEAH box helicase model unambiguously (Extended Data Fig. 6; Extended Data Table 2) and it has been interpreted as Prp16 as it contacts Cwc25. An even larger domain is observed in contact with the DEAH helicase domain. The structure of Brr2 helicase coupled to the Jab1/MPN domain of Prp8 (ref. 27) can be docked into this density, consistent with an interaction between Prp16 and Brr2 (ref. 28).
Active site
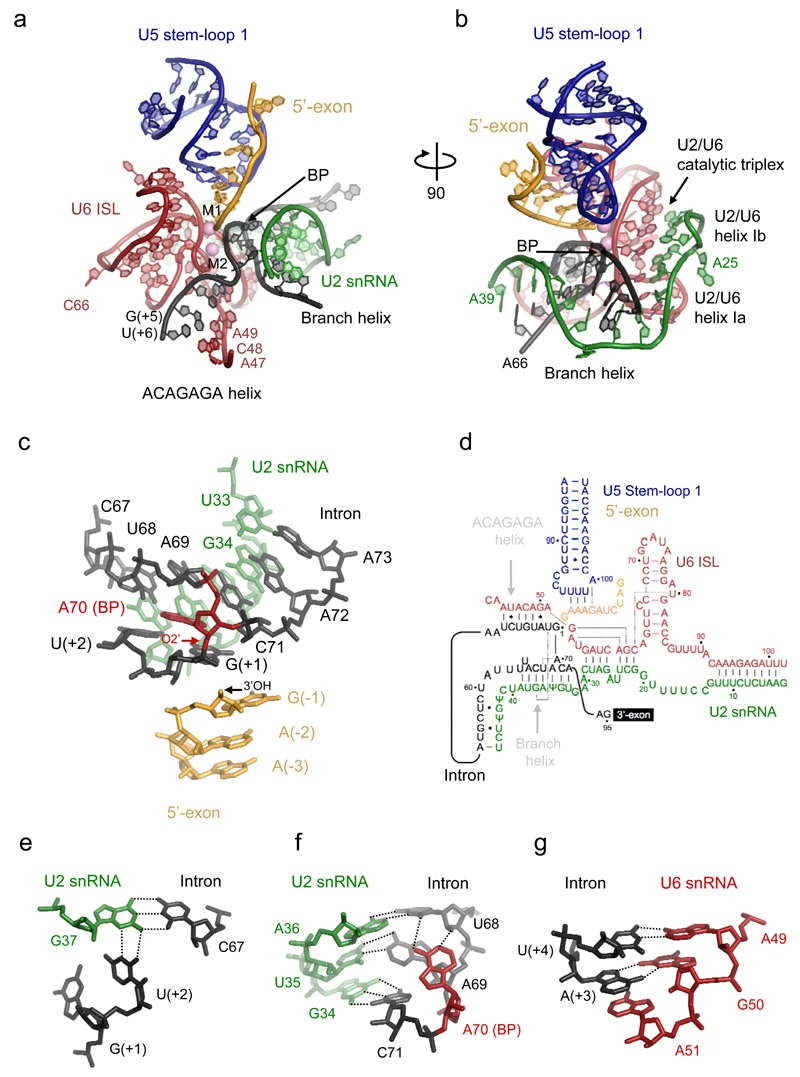
The map shows that the phosphodiester bond at the 5’SS is cleaved and the 5’-phosphate of the first intron nucleotide G(+1) forms a 2’-5’ phosphodiester linkage with the branch point adenosine (A70), in agreement with the RNA analysis (Extended Data Fig. 1b and 4b). The key RNA elements assemble around the active site harbouring the magnesium ion binding sites (Fig 3). The 3’OH of the 5’-exon remains close to the 5’-phosphate of G(+1) such that the normal 5’-3’ phosphodiester linkage at the 5’SS could be restored with minimal structural alteration (Fig. 3c). The adenine base of BP A70 is bulged out from the branch helix and its N1 and 6-amino group are hydrogen-bonded to the 2’OH and O2 of U68 creating a unique backbone conformation which enables the 2’OH of A70 to project towards the 5’-phosphate of intron G(+1) (Fig. 3f). In yeast the intron sequence following the 5’SS is stringently conserved as GUAUGU2. The G(+1) base is partially packed against the A70 base while the U(+2) base is within hydrogen-bonding distance of U2 snRNA G37 suggesting a possible base-triple interaction with intron C67 (Fig. 3e). Mutation of G(+1) to C, or of the branch A70 to C, would disrupt these interactions, consistent with the strong branching defects observed for these mutations29. Four conserved intron nucleotides A(+3)U(+4)G(+5)U(+6) form sequence-specific base-pairs with part of the ACAGAGA sequence of U6 snRNA7,8,30,31. The three 5’-exon nucleotides A(-2)A(-3)A(-4) form Watson-Crick base-pairs with loop 1 of U5 snRNA11(Fig. 3b, 4). Interestingly, the 5’-exon winds through a narrow channel between the Large and N-terminal domains of Prp8 formed during spliceosome activation (via 30° foot rotation) (Extended Data Fig. 7c) and stabilised by Cwc21 and the C-terminal domain of Cwc22 (Fig. 4a,b). Cwc22 consists of two HEAT repeat-containing domains that straddle the 5’-exon tunnel, providing insight into exon-junction complex deposition in higher eukaryotes32 (Extended Data Fig. 8).
Figure 3. Structure of the RNA catalytic core.
a, key RNA elements at the active site. ISL denotes Internal Stem-Loop. b, orthogonal view illustrating the branch helix and helices Ia and Ib of U2/U6 snRNA duplex. c, the branch helix and 5’-exon with the 2’-5’ phosphodiester linkage (red arrow). d, intricate RNA interactions at the active site (dotted lines indicate base triples; dot and star indicate G-U wobble and other non-canonical base-pairs). e, base-triple interaction between the branch helix and 5’-splice site. f, a network of interactions in the branch helix. g, Hoogsteen base-pair between intron A(+3) and G50 of U6 snRNA.
Figure 4. Proteins at the active site.
a, 5’exon channel formed between the Large and N-terminal domains of Prp8, Cwc21 and Cwc22. b, 5’exon:U5 loop 1 interaction surrounded by Prp8. Th/X denotes Thumb/domain X of Prp8 (residues 1300-1375). c, interactions between the 5’-exon, the N-terminal (purple) and Large (blue) domains of Prp8, and Yju2 (green). Interactions involving protein main and side chains are shown by solid and dotted lines. d, components surrounding U6 Internal Stem-Loop. e, Prp8 and Cef1 (myb1 domain) stabilise the catalytic triplex. HB denotes helix bundle of the RT domain (residues 750-870). f, structure of the catalytic triplex.
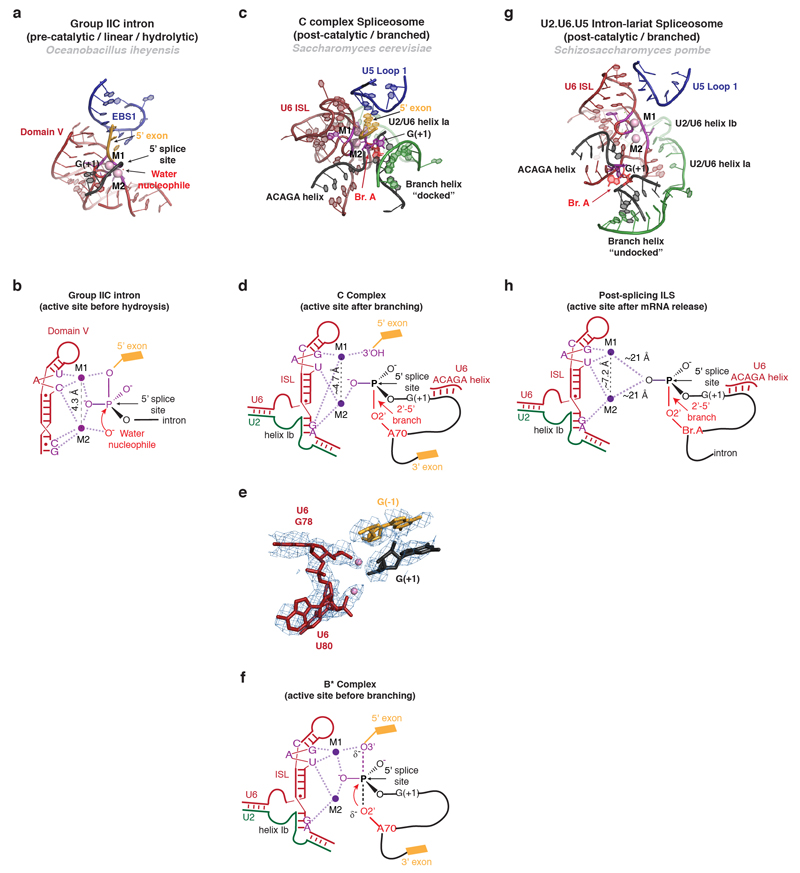
U6 snRNA following the ACAGAGA sequence forms Helices Ia and Ib by base-pairing with U2 snRNA and folds back to form an intramolecular stem loop (ISL), in agreement with the structure inferred from genetics33 (Fig. 3b,d). Helices Ia and Ib show continuous base-stacking and the bulged U2 snRNA nucleotides U24 and A25 protrude from Helix I and bind to the Prp8 RT domain (Fig. 3d,4d,e,5a). The Watson-Crick faces of U6 snRNA nucleotides G52 and A53 interact with the Hoogsteen faces of G60 and A59, respectively, forming two consecutive base triples as inferred from genetics34 (Fig. 4e,f). C66 and A79 bulge out from the ISL (Fig. 3a,b), allowing continuous base-stacking of the bulged U80 with G52 and A53 and stabilizing the catalytic triplex. It has been proposed that pre-mRNA splicing reactions are catalysed by a two-metal-ion mechanism35. Indeed ligands for the two divalent metal ions have been identified by stereo-specific phosphorothioate substitutions and metal rescue experiments36 and density attributable to Mg2+ ions is observed adjacent to these ligands (Extended Data Fig. 5). The 5’-exon 3’OH and the 5’ phosphate of G(+1) remain close to M1, while U6 snRNA metal ligands have repositioned slightly, in agreement with the previously observed repositioning of the branch in structures of a branched group II intron37. Nonetheless, the branch helix remains “docked” at the catalytic Mg2+ site, in striking contrast to its “undocked” configuration observed in the ILS structure, where it swings away from the ACAGAGA helix by 90º (ref 26; Extended Data Fig. 5).
Figure 5. Step 1 factors and branch site positioning.
a, interaction between the RNA catalytic core and Prp8. b, positioning of the branch helix by step 1 factors. c, corresponding view in S.pombe post splicing ILS complex26, showing dramatic repositioning of the branch helix and its further stabilisation by debranching co-factor Cwf19. d, a close-up view of step 1 factors interacting with the branch helix.
The intron downstream of the 5’SS GUAUGU sequence exits the active site near Cwc2, Ecm2, Clf1, Cef1 and Isy1 (Fig. 2), re-enters the spliceosome and runs side-by-side with U2 snRNA in the opposite direction through a channel between the Prp8 Endonuclease and RNaseH-like domains (Extended Data Fig. 7). The intron then forms the branch helix with the GΨAGUA sequence of U2 snRNA in proximity to the catalytic Mg2+ site (Fig. 3b, d) and exits the active site through a channel made by the Linker and RT-like domains of Prp8 (Fig. 2).
Roles of proteins around the active site
The RNA network at the active centre, comprising U2, U5 and U6 snRNAs and RNA substrate, is stabilised by a number of proteins (Figs 1,2,4). The catalytic RNA core is surrounded by the Linker and the helix bundle (HB) domains of Prp8 (ref.19,21) on one side and by NTC proteins (Prp45, Prp46, Isy1 and Cef1) and step one factors (Yju2 and Cwc25) on the other side, which together stabilise the catalytic RNA core for branching. Remarkable stacking of Prp8 Tyr671 and Tyr1620 against bases at positions G(-5) and A(-6) stabilises the 5’-exon:U5 snRNA loop 1 pairing (Fig. 4b,c). The linker between the N-terminal and Large domains of Prp8 runs across the major groove of U6 ISL, which is positioned in a pocket formed by Prp8 and Clf1, and the interactions are sealed by the extended N-terminus of Cwc15 (Fig. 4d). Cef1 stabilises the U2/U6 catalytic triplex34 (Fig. 4e,f).
Step one-specific factors probe the branch helix and stabilise its docking at the catalytic core (Fig. 5). A long α-helix of Cwc25 contacts the RNaseH-like domain and α-finger of Prp8 and its N-terminus is inserted into the widened major groove of the bulged branch helix (Fig. 5b,d). The N-terminus of Yju2 wraps around the branch helix (Fig. 5d) and its Arg4 makes a base-specific contact with the intron U(+2) while its main chain amide group contacts the backbone phosphate of the 5’-exon A(-2) (Fig. 4c). Isy1 projects its N-terminus deep into the active site forming contacts with the phosphate backbone of intron U68. Ser2 of Isy1 forms a hydrogen-bond with the O2 carbonyl group of U(+2) of the intron. One of the Isy1 helices inserts into the minor groove of the ACAGAGA/5’SS helix. Cwc25 forms multiple contacts with the branch site, consistent with cross-linking experiments38 and its role in juxtaposition of the 5’SS and BP for branching39,40,41. These spliceosomal factors are reminiscent of ribosomal proteins L27 and L16, which penetrate into the peptidyl transferase active site and stabilise tRNA binding42.
Remodelling of the spliceosome
The intron downstream of the BP emerges from the exit channel formed by the Prp8 RT and Linker domains and the α-finger, and projects towards Prp16 (Fig. 6a). Twelve nucleotides could span the distance between the last ordered intron nucleotide (BP+6) and the substrate RNA entry site of Prp16, consistent with Prp16 crosslinking to 4-thiouridine introduced 18 nucleotides downstream of the BP43. Prp16 translocates 3’→5’ towards the BP along the intron upon ATP hydrolysis43–45. Prp16 would thus pull the branch helix out of its pocket and hence destabilise the binding of Yju2 and Cwc25 (Fig 6b). The undocked branch helix would allow the 3’-exon to enter the active site31,45 and bind to U5 snRNA loop 1 (ref 11,12). Consistent with this, destabilisation of the branch helix by Isy1 deletion suppresses splicing defects caused by Prp16 mutations46. The step two factors, Prp18 and Slu7 are likely to dock into the space vacated by the branch helix/Yju2/Cwc25 to stabilise the 3’SS into the active site as Slu7 and Prp18 are in direct contact with the 3’SS bound to U5 snRNA loop 1 prior to exon ligation47 (Fig. 6b). Prp22 binds the 3’-exon at position +17 (ref. 15). Translocation of Prp22 on the 3’-exon in the 3’→5’ direction towards the active centre15,43 would displace Prp18-Slu7, releasing the mRNA. In our structure density assigned to Prp16 is in direct contact with Cwc25 (Fig. 6a), consistent with Cwc25 stabilising Prp16 binding to the spliceosome prior to branching44. We propose that the branch helix and 3’-exon confer specificity for auxiliary factors such as Cwc25-Yju2, Slu7-Prp18, which may act as adaptors that determine the identity of the next DEAH box helicase to remodel the active site.
Figure 6. The role of helicases in active site remodelling.
a, the intron sequence downstream from the branch site exits the spliceosome via a channel in Prp8 and extends towards Prp16. Translocation of Prp16 towards the branch helix would destabilise step 1 factors and displace the branch helix from its pocket. b, schematic illustrating how step 1 or step 2 specific factors can determine the specificity of the helicase recruited to the spliceosome at particular stages of splicing.
The structure of the S. pombe spliceosomal complex26,48 contains a lariat intron but not 5’-exon or the spliced mRNA. The catalytic RNA core is surrounded by a similar set of NTC and NTR proteins but the structure lacks step one or step two factors26,48, suggesting this corresponds to a post-splicing Intron Lariat Spliceosome (ILS)49. Instead Cwf19, a homolog of the debranching enzyme co-factor Drn150, intrudes between the Large and RNaseH-like domains of Prp8, occupying the binding sites for Isy1, Cwc25, and Yju2 found in our complex C. Cwf19 marks the ILS complex for disassembly by displacing the branch helix, which rotates by 90° in ILS with respect to complex C (Fig. 5c, Extended Data Fig. 7).
A pronounced conformational change between ILS and complex C is a large rotation of the NTC (Extended Data Fig. 7d). In ILS the N-terminus of Syf1 moves away from the core, promoting undocking of U2 snRNP. In complex C, the position of U2 snRNP is stabilised by the formation of stem IIc and binding of Prp19. U2 snRNP is in direct contact with the RNaseH domain of Prp8, which holds Cwc25 in place. This network of interactions suggests that binding of Prp19 and formation of stem IIc in U2 snRNA may have an allosteric effect on the positioning of the branch helix via step one factors. Extended arches of Syf1 and Clf1 may have a role in communicating the signal over long distance.
Our spliceosomal complex C structure reveals the active configuration of the catalytic core, elucidating the arrangement of the RNA substrate and its interaction with proteins. The structure accounts for a large body of biochemical and genetic data and provides crucial insights into substrate docking and catalysis and the role of DEAH helicases and auxiliary factors in spliceosome remodelling.
Methods
Prp18-HA and Slu7-TAPS tagging
SLU7-TAPS homology recombination cassettes were generated by PCR from pFA6a-TAPS-kanMX6, a modified version of pFA6a-TAP-kanMX6 in which the Calmodulin-binding peptide tag is replaced by two tandem copies of the StrepII tag51. The PCR product was used to transform yeast strain YSCC1 (MATa prc1 prb1 pep4 leu2 trp1 ura3 PRP19-HA)4 selecting for G418-resistance. Prp18_3xHA kanMX6 cassette was transformed into BY4741 strain (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and selected as above. Integration of the cassettes was confirmed by PCR and Western blotting.
Sample preparation
The Prp18-HA or Slu7-TAPS yeast strains were grown in a 120 L fermenter, and splicing extract was prepared using liquid nitrogen method36 essentially as previously described52. A DNA template for in vitro transcription was generated by addition of 2xMS2 stem loops53 to the 5’-end of the UBC4 pre-mRNA sequence18, in which the 3’-splice site sequence UAGAG was mutated to UACAC. Pre-mRNA substrate was generated by run-off transcription from a plasmid DNA template and labelled at the 3’-end with fluorescein-5-thiosemicarbazide54. In vitro splicing reactions were assembled using pre-mRNA substrate pre-bound to MS2-MBP fusion protein as previously described6,53. The resulting spliceosomes were bound by amylose-resin in HE-75 (20 mM HEPES KOH pH 7.8, 75 mM KCl, 0.25 mM EDTA, 5% glycerol, 0.01% NP-40) and eluted with 12 mM maltose. The sample was subsequently immobilised on either anti-HA-agarose (for Prp18-HA yeast extract) or Streptactin resin (for Slu7-TAPS yeast extract) in HE-100 (20 mM HEPES KOH pH 7.8, 100 mM KCl, 0.25 mM EDTA, 5% glycerol, 0.01% NP-40) and eluted with either HA peptide (for anti-HA-agarose) or desthiobiotin (for Streptactin resin), essentially as described55. The eluate was finally dialysed against HE-75 buffer (without glycerol and NP-40) for EM sample preparation. Analysis of fluorescently labelled RNA showed that pre-mRNA is converted to the lariat intron-3’-exon intermediate in our sample and hence it is referred to as complex C (Extended data Fig. 1b). Our experimental set-up was designed to purify step 2 complexes after Prp16 action, however the presence of step 1 factors in the structure and configuration of the active site clearly indicate that the complex has not undergone Prp16-mediated remodelling. It has been shown previously13 that in low salt conditions Prp18, Slu7 and Prp16 associate with complex B* and C. Analysis of protein components by gel electrophoresis and subsequent mass spectrometry shows that Prp16 as well as Prp22 are present, in agreement with the previous results (Extended Data Fig. 1a; Extended Data Table 2)6,13,43.
Electron microscopy
For cryo-EM analysis, Quantifoil R2/2 Cu 400 mesh grids were coated with a 5 – 7 nm-thick layer of homemade carbon film and glow discharged. After applying 3 mL of the sample, the grids were blotted for 2.5 – 3 s and vitrified in liquid ethane in FEI Vitrobot MKIII, at 100% humidity at 4 °C. Grids were loaded into an FEI Titan Krios transmission electron microscope operated at 300 kV and imaged using a Gatan K2 summit direct electron detector and a GIF Quantum energy filter (slit width 20 eV). Images were collected in super-resolution counting mode at 1.25 frames s-1 and a calibrated pixel size of 1.43 Å. A total dose of 40 e Å-2 over 16 s and a defocus range of 0.5 – 4 μm were used.
Image processing
A total of 2213 micrographs were subjected to whole-frame drift correction in MOTIONCORR56 followed by contrast transfer function (CTF) parameter estimation in CTFFIND4 (ref. 57). All subsequent processing steps were done using RELION58 unless otherwise stated. An initial subset of 5000 particles was selected manually and subjected to reference-free 2D classification. Resulting 2D class averages were low-pass filtered to 20 Å and used as templates for subsequent automated particle picking within RELION59. A total of 247,603 particles were selected after initial reference-free 2D classification and subjected to 3D classification (Extended Data Figure 2). An initial 3D reference was prepared by scaling and low pass-filtering (60 Å) the reconstruction of the Intron-Lariat complex (EMD-6413). A subset of 93,106 particles was selected after 3D classification. Particle-based beam-induced motion correction and radiation-damage weighting (particle polishing) followed by 3D Refinement resulted in a final reconstruction at 3.8 Å overall resolution and estimated accuracies of rotations of 1.1° (Extended Data Fig. 3).
Very weak density observed at two peripheral regions of the map corresponds to Brr2/Prp16 (helicase module) and Prp19/Cef1/Snt309 (Prp19 module). We used focused classification with signal subtraction to improve the resolution of these regions60. The region of interest was masked out and the projection of the remaining map was subtracted from the experimental particles using angular assignment from the last iteration of the 3D auto-refine run. Subtracted particles were 3D classified without image alignment and the best classes were selected for further refinement of the original (not subtracted) particles. This resulted in a smaller subset of the original particles, in which Brr2/Prp16 and Prp19/Cef1/Snt309 are more homogeneous and consequently the density is significantly improved in those regions (Extended Data Figure 2 and 3). 3D refinement of the selected 29210 Prp19-selected particles resulted in a map at overall 5.1Å resolution, while 15872 of the helicase-containing particles yielded a map at 10 Å resolution. For the global classification approach we generated a soft mask around the core of the complex and classified polished particles with finer angular sampling of 1.8° and local searches of 10°. The resulting two major classes of 37K and 47K particles were refined to 4.1Å and 3.9 Å respectively. They revealed a subtle conformational change of the U2 snRNP and Syf1 HAT arch correlated with the presence of WD40 domain near the stem IIc and IIb region of U2 snRNA. This WD40 domain belongs to Prp17 or Prp19, but the local resolution did not allow us to make an unambiguous assignment. All reported resolutions are based on the gold-standard Fourier shell correlation (FSC) = 0.143 criterion61. FSC curves were calculated using soft spherical masks and high-resolution noise substitution was used to correct for convolution effects of the masks on the FSC curves62. Prior to visualization, all maps were corrected for the modulation transfer function of the detector. Local resolution was estimated using Resmap63.
Model building
A list of protein and RNA components included in the model is given in Extended Data Table 2. Building started by docking known structures of S. cerevisiae Prp8, Snu114, U5 Sm ring, U5 snRNA19, Cwc2 (ref. 64) and Bud31 (ref. 65) into the map. Homology models for Cef1, Prp45, Prp46, Ecm2 and Cwc15 were built with SWISS-MODEL66, using structures from the S. pombe intron-lariat spliceosome26 as templates, and were docked into the map. This accounted for the majority of the protein density in the core, allowing building of the intron, U6 snRNA and U2 snRNA. RNA extending from the loop 1 of U5 snRNA was assigned to nucleotides -1 to -16 of the 5’ exon as previously predicted11. A model for the NTD of Cwc22 was built using SWISS-MODEL based on the structure of the human Cwc22:eIF4AIII complex32 and docked near Snu114. Clear density near the NTD of Cwc22 was interpreted as the MA3 domain at the C-terminus of Cwc22; this domain was built de novo. A coiled-coil was found contacting domain IV of Snu114. Based on an unpublished NMR structure from Arabidopsis thaliana (PDB ID: 2E62) and biochemical data25 we assigned this density to the CTD of Cwc21. Weak density was observed connecting this coiled-coil to a peptide contacting the 5’-exon. We therefore assigned this peptide as the N-terminus of Cwc21. Unassigned density remained near the branch-point helix. Based on secondary structure prediction67 we assigned a portion of this density to Yju2 and were able to build its NTD de novo; our assignment was supported by clear density for a zinc atom coordinated by four conserved cysteines. The remainder of the density could then be assigned to the N-termini of Cwc25 and Isy1.
The majority of the model building described above was for the core of the spliceosome where the resolution was uniformly between 3.5 – 4.5 Å (Extended Data Figure 4). For the periphery of the complex, the resolution was more heterogeneous, ranging from 4 to 20 Å. Clear features of the periphery were two large proteins with extended architectures. One of these proteins started in the core and projected outwards to the periphery. At the core, side-chains were easily visible for this protein and allowed assignment as the N-terminus of Clf1. Towards the C-terminus of Clf1 the resolution only allowed building of idealised poly-Ala helices, which were then assigned sequence based on secondary structure predictions67. For the other extended protein, few side-chains were visible but helices could be distinguished. This protein was generally built as poly-Ala helices, and based on secondary structure predictions67 was assigned as Syf1. A second Sm ring at medium-resolution was found in the map and was assigned as the U2 snRNA Sm ring. Homology models for the U2 snRNP proteins Lea1 and Msl1 were generated using SWISS-MODEL66 based on the structure of the human U2B”-U2A’-U2 snRNA complex68 and were docked into the adjacent density. The portion of the U2 snRNA in contact with Msl1 was most consistent with the previously proposed stem IV + stem V architecture and was built based on the secondary structure prediction69. Two RNA double helices were observed bridging the U2 Sm ring to Ecm2 and were assigned as stems IIb and IIc of the U2 snRNA. Using 3D classification, we found that some of the particles contained a large lobe of extra density connected to the reverse transcriptase and RNase H domains of Prp8 (see above). Although we could not resolve secondary structure in this region, we could perfectly dock the crystal structure of Brr2 and the Jab1/MPN domain of Prp8 (ref. 27). The remainder of the density could then well accommodate an I-TASSER70 homology model of Prp16 based on the crystal structure of Prp43 (ref. 71). Weak density connected to Clf1 and Syf1 had the characteristic shape of Prp19-Snt309-Cef1 (ref. 26). Focused classification in this region could improve the density enough to resolve the U-box dimers and thus dock a homology model of these proteins. Finally, three copies of the Prp19 WD40 domain crystal structure could be docked into very weak density adjacent to the Prp19 coiled-coils. With the exception of the helicase and Prp19 modules all models were manually rebuilt in order to obtain the best fit to the cryo-EM density. The model was refined using REFMAC 5.8 (ref. 72) with secondary structure restraints generated in PROSMART73 and RNA base-pair and stacking restraints generated in LIBG74. Extended Data Table 1 summarizes refinement statistics and PBD and EMDB accession codes.
Map visualisation
Maps were visualised in Chimera84 and figures were prepared using PyMOL (http://www.pymol.org).
Extended Data
Extended Data Figure 1. Biochemical characterisation of the complex and initial cryo-EM analysis.
a, SDS-PAGE analysis of the purified sample. Protein identities were confirmed by mass spectrometry analysis. Protein labels are coloured according to sub-complex identity (dark blue, U5 snRNP; light blue, helicase module; orange, NTC; yellow, NTR; green, U2 snRNP; purple, splicing factors; grey, not found in density) b, analysis of the fluorescently labelled substrate in the sample by denaturing PAGE, showing conversion of linear pre-mRNA (time point 0’) into branched lariat-intron intermediate (time point 30’), which is a predominant species in the purified sample (C complex). The two hairpins on the right depict the 2xMS2 stem-loops attached to the 5’end of the UBC4 pre-mRNA substrate for affinity purification. c, a typical cryo-EM micrograph collected on an FEI Titan Krios microscope operated at 300 kV and detected with a Gatan K2 Summit camera. d, reference-free 2D classification results. e, detail of a single class average with major domains labelled.
Extended Data Figure 2. Overview of the data processing scheme used in this study.
Iterative 2D classification, template selection and automated particle picking resulted in 248K particles which were classified in 3D with a scaled and low-pass filtered model of ILS (EMDB-6413) as a reference. The best class was refined to 3.8 Å resolution overall. Focused classification allowed us to obtain two other maps with improved quality of the peripheral regions (Prp19 and helicase modules, EMD-4056 and EMD-4057). Classification of the core complex with fine angular sampling and local searches revealed a subtle movement of the U2 snRNP which correlates with the appearance of the extra density, interpreted as a WD40 domain which belongs to Prp17 or Prp19.
Extended Data Figure 3. Global and local resolution analysis.
a, two orthogonal sections through the map showing variation in the local resolution as estimated by Resmap. b, an overall map of the core complex c, Gold-standard FSC plots for three maps used in this study. d, map of the core complex with a helicase module. e, a map of the core complex with Prp19 module.
Extended Data Figure 4. Examples of cryo-EM density at the core of the complex with atomic models built in.
a, U5 snRNA loop 1 with 5’-exon bound. b, the active site with exon, intron, U2 and U6 snRNAs. c, two helices of the Prp8 Reverse Transcriptase Thumb/X domain, showing a clear helical pitch and excellent densities for the side chains. d, Fourier Shell Correlation between model and the map and cross-validation of the model fitting. (The original atom positions have been randomly displaced up to 0.5Å and refined with restraints against the half1 map only. FSC was calculated for two half maps. Excellent correlation up to the high resolution between the model and the half2 map (which was not used in refinement) cross-validates the model for overfitting.
Extended Data Figure 5. Metal binding by the catalytic core of C complex.
a,b, Structure (a) and schematic representation (b) of the active site of a group IIC intron trapped in the pre-catalytic state in the presence of Ca2+ (PDB 4FAQ, ref. 75). The 5’ splice site scissile phosphate is aligned with the two metals bound at the core in a catalytic configuration, as shown in b. Note that, in this pre-catalytic structure, the group II domain VI is not present and therefore the structure does not contain the bulged adenosine nucleophile required for the branching reaction. As a result, the nucleophile is a water molecule, rather than the 2’-OH of the branch site adenosine found in spliceosomal introns. c-d, Structure of the RNA at the active site of spliceosomal C complex, showing the overall architecture (c), schematic of metal binding (d), and comparison of the model with the EM density (e). Note conservation of the metal binding residues compared to the group II intron (c.f. ref. 36) and proximity of the cleaved G(-1)-G(+1) bond to putative M1. f, Proposed interactions between U6 snRNA and the two catalytic Mg2+ during the transition state for branching, as inferred from biochemistry36. g, h, Structure (g) and schematic (h) of the RNA core of the U2.U6.U5 ILS complex in a post-catalytic configuration (PDB 3JB9, ref. 26), likely following release of the mRNA. The two Mg2+ are shown as modelled in the coordinates deposited by the authors of the ILS structure (PDB 3JB9, ref. 26). In the ILS structure M1 and M2 are further apart (7.2 A) than in most other structures of RNAs that coordinate catalytic metals (usually 3.9-5 A); nonetheless the ligands modeled for M1 and M2 are consistent with the ligands identified biochemically for the two catalytic Mg2+ necessary for splicing (compare PDB 3JB9 and 4R0D with the data in refs. 34 and 36). Note that the branch helix is undocked from the U6 snRNA metal binding site and G(+1) is far away from the two Mg2+ at the core. The substrate and snRNAs are colour-coded while residues that position the catalytic metals are shown in magenta.
Extended Data Figure 6. Examples of the structures of isolated components.
De novo built proteins are shown in cartoon form, along with a secondary structure diagram for the novel zinc finger fold of Yju2. Proteins that were modelled into low-resolution regions by rigid-body docking of crystal structures or homology models (Prp19 module, Brr2, Prp16, Prp8Jab1/MPN) are shown in their cryo-EM densities.
Extended Data Figure 7. Conformational changes between U4/U6.U5 tri-snRNP, Complex C and Intron-Lariat Spliceosome.
a, rearrangement of the RNaseH-like domain with respect to the main body of Prp8 in all three complexes. b, α-finger (1575-1598) contacting the key RNA and proteins in a context-dependent manner. c, Prp8 N-terminal domain movements along with Prp8 residues 1406-1436 transiently docking on top of the 5’-exon and Cwc21 in complex C, stabilising the 5’-exon and interdomain contacts in Prp8. d, conformational rearrangements between complex C and S.pombe ILS26 showing a coupled movement of the U2 snRNP, Syf1 and Prp19.
Extended Data Figure 8. Implications for deposition of the Exon-Junction Complex.
In higher eukaryotes exon-junction complexes (EJCs) are deposited 20 – 24 nt upstream of splice junctions, and form a binding platform for factors involved in nuclear export, translation, alternative splicing and nonsense-mediated mRNA decay76. The core EJC components eIF4AIII, MAGOH and Y14 are found in human B and C complexes77. Cwc22 is required for eIF4AIII recruitment to spliceosomes78–80 and holds it in an open, inactive conformation32. a, Crystal structure of the eIF4AIII:Cwc22 complex32 docked onto the spliceosomal C complex via superposition on Cwc22. b, Crystal structure of the core EJC81,82 superimposed on the previous model via the second RecA domain of eIF4AIII. c, The 5’-exon exiting the channel at the interface between the Prp8 Large and N-terminal domains is positioned perfectly for the deposition of the EJC, explaining how the Cwc22 MIF4G domain is involved in determining the distance of EJC deposition from the splice junction.
Extended Data Table 1. Cryo-EM data collection and refinement statistics.
| Core | Core+Prp191 | Core+helicase1 | |
|---|---|---|---|
| Data collection | |||
| Microscope | FEI Titan Krios | FEI Titan Krios | FEI Titan Krios |
| Voltage (kV) | 300 | 300 | 300 |
| Electron dose (e Å -2) | 40 | 40 | 40 |
| Detector | Gatan K2 Summit | Gatan K2 Summit | Gatan K2 Summit |
| Pixel size (Å) | 1.43 | 1.43 | 1.43 |
| Defocus Range (μm) | 0.5-4.0 | 0.5-4.0 | 0.5-4.0 |
| Reconstruction (Relion) | |||
| Particles | 93 106 | 29 210 | 15 872 |
| Box size (pix) | 412 | 412 | 412 |
| Accuracy of rotations (°) | 1.13 | 1.13 | 1.51 |
| Accuracy of translations (pix) | 0.64 | 0.96 | 1.30 |
| Map sharpening В-factor (Å2) | -57 | -17 | -350 |
| Final resolution (Å) | 3.75 | 5.08 | 9.78 |
| Model composition |  |
||
| Protein Residues | 7447 | 119783 | |
| RNA bases | 458 | 458 | |
| Ligands | 10 | 10 | |
| Refinement (Refmac) | |||
| Resolution (Å) | 3.8 | ||
| FSCaverage | 0.82 | ||
| R factor | 0.32 | ||
| R.m.s deviations | |||
| Bond lengths (Å) | 0.007 | ||
| Bong angles (°) | 1.25 | ||
| Validation2 | |||
| Molprobity score | 2.5 (98th percentile) | ||
| Clashscore, all atoms | 5.3 (100th percentile) | ||
| Good rotamers (%) | 80 | ||
| Ramachandran plot | |||
| Favoured (%) | 90.84 | ||
| Outliers (%) | 1.16 | ||
| RNA validation2 | |||
| Correct sugar puckers (%) | 95 | ||
| Good backbone conformations (%) | 60 | ||
| Deposition | |||
| PDB ID | 5LJ3 | 5LJ53 | 5LJ53 |
| EMDB ID | EMDB-4055 | EMDB-4056 | EMDB-4057 |
represents a sub-set of the whole dataset (Core).
determined by Molprobity83.
overall model including Prp19 and helicase modules.
Extended Data Table 2. Summary of model building for spliceosomal complex C.
| Proteins and RNA included in the model | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sub-complexes | Protein/RNA | Domains | Total residues | M.W. (Da) |
Modelled | Modelling template (PDB ID) |
Modelling | Resolution1 | Chain ID | Human/S. pombe names |
| U5 snRNP | Prp8 | N-terminal | 1-870 | 101,767 | 128-870 | 5GAN | Docked & rebuilt | 3.4 - 5.8 | A | 220K/Spp42 |
| Large | 871-1827 | 111,525 | 871-1827 | 5GAN | Docked & rebuilt | 3.6 - 6.2 | ||||
| RNaseH | 1828-2085 | 29,453 | 1837-2085 | 5GAN | Docked & rebuilt | 4.2 - 6.6 | ||||
| Jab1/MPN | 2086-2413 | 36,812 | 2148-2396 | 4BGD | Rigid docking | ~15 - 20 | ||||
| Snu114 | 1008 | 114,041 | 67-998 | 5GAN | Docked & rebuilt | 3.8 - 7.2 | C | 116K/Cwf10 | ||
| SmB | 196 | 22,403 | 4-102 | 5GAN | Docked | 4.6 - 7.2 | b | SmB/SmB | ||
| SmD3 | 110 | 11,229 | 4-85 | 5GAN | Docked | 4.4 - 7.8 | d | SmD3/SmD3 | ||
| SmD1 | 146 | 16,288 | 1-109 | 5GAN | Docked | 4.8 - 7.8 | h | SmD1/SmD1 | ||
| SmD2 | 110 | 12,856 | 15-108 | 5GAN | Docked | 5.2 - 8.0 | j | SmD2/SmD2 | ||
| SmF | 94 | 10,373 | 12-83 | 5GAN | Docked | 5.2 - 8.0 | f | SmF/SmF | ||
| SmE | 96 | 9,659 | 10-92 | 5GAN | Docked | 5.4 - 8.0 | e | SmE/SmE | ||
| SmG | 77 | 8,479 | 2-76 | 5GAN | Docked | 5.0 - 7.8 | g | SmG/SmG | ||
| U5 snRNA-L | 214 | 68,847 | 4-144 | De novo | 3.8 - 7.6 | U | ||||
| U2 snRNP | Msl1 | 111 | 12,830 | 28-111 | 1A9N | Homology modelled | 6.6 - 8.8 | Y | U2-B″ | |
| Lea1 | 238 | 27,193 | 1-167 | 1A9N | Homology modelled | 5.6 - 8.6 | W | U2-A′ | ||
| SmB | 196 | 22,403 | 4-102 | 5GAN | Docked | 5.4 - 8.2 | k | SmB/SmB | ||
| SmD3 | 110 | 11,229 | 4-85 | 5GAN | Docked | 6.0 - 8.2 | n | SmD3/SmD3 | ||
| SmD1 | 146 | 16,288 | 1-118 | 5GAN | Docked | 5.0 - 8.0 | l | SmD1/SmD1 | ||
| SmD2 | 110 | 12,856 | 15-108 | 5GAN | Docked | 5.0 - 7.6 | m | SmD2/SmD2 | ||
| SmF | 94 | 10,373 | 12-83 | 5GAN | Docked | 5.2 - 7.4 | q | SmF/SmF | ||
| SmE | 96 | 9,659 | 10-92 | 5GAN | Docked | 5.4 - 8.0 | p | SmE/SmE | ||
| SmG | 77 | 8,479 | 2-76 | 5GAN | Docked | 5.8 -8.2 | r | SmG/SmG | ||
| U2 snRNA | 1175 | 363,824 | 3-150; 1089-1169 |
De novo | 3.8 - 6.0 | Z | ||||
| U6 | U6 snRNA | 112 | 36,088 | 1-102 | De novo | 3.6 - 6.4 | V | |||
| NTC | Prp19 | U-box | 1-51 | 5,713 | 1-51 | 3JB9 | Homology modelled | ~20 | t,u,v,w | PRPF19/Cwf8 |
| Coiled-coil | 52-143 | 10,247 | 78-143 | 3JB9 | Homology modelled | ˜20 | ||||
| WD40 | 144-503 | 40,646 | 171-501 | 3LRV | Docked | ˜25-30 | ||||
| Snt309 | 175 | 20,709 | 12-174 | 3JB9 | Homology modelled | ˜20 | s | BCAS2/Cwf7 | ||
| Syf1 | 859 | 100,229 | 21-790 | Idealised alpha helices | 4.8 - 8 | T | SYF1/Cwf3 | |||
| Clf1 | Core | 1-271 | 32,396 | 1-271 | 3JB9 | Homology modelled & rebuilt | 3.8 - 6.4 | S | CRNKLl/Cwf4 | |
| Periphery | 272-687 | 50,067 | 277-556 | Idealised alpha helices | 5.2 - 8.8 | |||||
| Cef1 | N-terminal | 1-191 | 21,868 | 12-191 | 3JB9 | Homology modelled & rebuilt | 3.8 - 6.2 | O | CDC5L/Cdc5 | |
| Middle | 192-505 | 65,905 | - | Not modelled | - | |||||
| C-terminal | 506-590 | 9,994 | 506-590 | 3JB9 | Homology modelled | ~20 | ||||
| Isy1 | 235 | 32,992 | 1-96 | De novo | 3.8 - 6.2 | G | ISY1/Cwf12 | |||
| NTR | Prp45 | 379 | 42,483 | 32-224 | 3JB9 | Homology modelled & rebuilt | 4 - 8.4 | K | SNW1/Prp45 | |
| Prp46 | 451 | 50,700 | 111-445 | 3JB9 | Homology modelled & rebuilt | 3.4 - 6.6 | J | PLRG1/Prp5 | ||
| Ecm2 | 364 | 40,925 | 6-324 | 3JB9 | Homology modelled & rebuilt | 4.0 - 7.0 | N | RBM22/Cwf5 | ||
| Cwc2 | 339 | 38,431 | 3-252 | 3U1L | Docked & rebuilt | 3.6 - 6.0 | M | RBM22/Cwf2 | ||
| Cwcl5 | 175 | 19,935 | 7-40 | 3JB9 | Homology modelled & rebuilt | 3.6 - 7.6 | P | CNC15/Cwf15 | ||
| Bud31 | 157 | 18,447 | 2-156 | 2MY1 | Docked & rebuilt | 3.6 - 6.8 | L | BUD31/Cwf14 | ||
| Splicing factors | Yju2 | 278 | 32,312 | 2-115 | De novo | 3.8 - 5.4 | D | CCDC94/Cwf16 | ||
| Cwc21 | N-terminal | 1-64 | 7,057 | 2-50 | De novo | 3.8 - 7.4 | R | SRRM2/Cwf21 | ||
| Coiled-coil | 65-135 | 8,724 | 64-111 | 2E62 | Homology modelled | 4.4 - 7.6 | ||||
| Cwc22 | MIF4G | 1-288 | 33,187 | 11-262 | 4C9B | Homology modelled & adjusted | 4.6 - 8.2 | H | CWC22/Cwf22 | |
| MA3 | 289-577 | 34,125 | 289-481 | De novo | 3.8 - 7.0 | |||||
| Cwc25 | 179 | 20,374 | 3-48 | De novo | 3.8 - 7.0 | F | CWC25/Cwf25 | |||
| Helicases | Brr2 | 2,163 | 246,185 | 442-2163 | 4BGD | Docked | ˜13 - 20 | B | 200K/Brr2 | |
| Prp16 | 1,071 | 121,653 | 338-978 | 2XAU | Homology modeled & domains fitted | ˜12 - 15 | Q | DHX38/Prp16 | ||
| Substrate | 5′-exon | 20 | 6,683 | (-16) - (-1) | De novo | 3.4 - 6.4 | E | |||
| Intron | 95 | 30,405 | 1-10; 54-76 | De novo | 3.4 - 7.2 | I | ||||
Resolution was calculated by averaging ResMap-calculated resolution voxels over each residue using Chimera. The resolution of residues at the 5th and 95th percentile for each chain then gave the resolution range for that chain.
Acknowledgements
We thank Kelly Nguyen, Xiao-chen Bai and Sjors Scheres for their help and advice on data collection and processing; Christos Savva, Shaoxia Chen, Vinothkumar Kutti Ragunath, Greg McMullan, Jake Grimmett and Toby Darling for smooth running of the EM and computing facilities; the mass spec facility for help with protein identification, Alan Brown, Paul Emsley, Garib Murshudov and Jose Llacer for help and advice with model building and refinement; Lukas Langer and the members of the spliceosome group for help and advice throughout the project. We thank Jan Löwe, Venki Ramakrishnan, David Barford and Richard Henderson for their continuing support, Sjors Scheres, Clemens Plaschka and Lisa Strittmatter for critical reading of the manuscript and Josep Vilardell for a generous gift of reagent. The project was supported by the Medical Research Council (MC_U105184330). M.E.W was supported by a Rutherford Memorial Cambridge Scholarship, S.M.F. by EMBO and Marie Skłodowska-Curie fellowships.
Footnotes
Author contributions: W.P.G., M.E.W. & S.M.F. established experimental procedures, W.P.G. & M.E.W prepared the sample and grids, and processed EM data. W.P.G., M.E.W. & S.M.F. collected EM data. W.P.G., M.E.W., C.O. carried out model building and refinement. W.P.G., M.E.W., S.M.F., C.O. & K.N. analysed the structure. A.J.N. contributed to the project through his knowledge and experience on yeast splicing. Manuscript was written by W.P.G., M.E.W. & K.N. and finalised with input from all authors. K.N. initiated and orchestrated the spliceosome project.
Author Information: The cryo-EM maps have been deposited in the Electron Microscopy Data Bank with accession codes EMD-4055, EMD-4056, EMD-4057, EMD-4058, EMD-4059. The coordinates of the atomic models have been deposited in the Protein Data Bank under accession code 5LJ3 (core of the complex) and 5LJ5 (overall structure).
The authors declare no competing financial interests.
References
- 1.Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3:a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burge CB, Tuschl T, Sharp PA. In: The RNA World II. Gesteland RF, Cech TR, Atkins JF, editors. Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- 3.Lambowitz AM, Zimmerly S. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol. 2011;3:a003616. doi: 10.1101/cshperspect.a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan S-P, Kao D-I, Tsai W-Y, Cheng S-C. The Prp19p-associated complex in spliceosome activation. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- 5.Ohi MD, Gould KL. Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA. 2002;8:798–815. doi: 10.1017/s1355838202025050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fabrizio P, et al. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell. 2009;36:593–608. doi: 10.1016/j.molcel.2009.09.040. [DOI] [PubMed] [Google Scholar]
- 7.Lesser CF, Guthrie C. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science. 1993;262:1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- 8.Kandels-Lewis S, Séraphin B. Involvement of U6 snRNA in 5' splice site selection. Science. 1993;262:2035–2039. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- 9.Raghunathan PL, Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol. 1998;8:847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- 10.Laggerbauer B, Achsel T, Lührmann R. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc Natl Acad Sci USA. 1998;95:4188–4192. doi: 10.1073/pnas.95.8.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newman AJ, Norman C. U5 snRNA interacts with exon sequences at 5' and 3' splice sites. Cell. 1992;68:743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- 12.Sontheimer EJ, Steitz JA. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993;262:1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- 13.Ohrt T, et al. Molecular dissection of step 2 catalysis of yeast pre-mRNA splicing investigated in a purified system. RNA. 2013;19:902–915. doi: 10.1261/rna.039024.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cordin O, Hahn D, Beggs JD. Structure, function and regulation of spliceosomal RNA helicases. Current Opinion in Cell Biology. 2012;24:431–438. doi: 10.1016/j.ceb.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Schwer B. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol Cell. 2008;30:743–754. doi: 10.1016/j.molcel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Company M, Arenas J, Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 17.Tsai R-T, et al. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 2005;19:2991–3003. doi: 10.1101/gad.1377405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abelson J, et al. Conformational dynamics of single pre-mRNA molecules during in vitro splicing. Nature Struct Mol Biol. 2010;17:504–512. doi: 10.1038/nsmb.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen THD, et al. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Å resolution. Nature. 2016;530:298–302. doi: 10.1038/nature16940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan R, et al. The 3.8 Å structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis. Science. 2016;351:466–475. doi: 10.1126/science.aad6466. [DOI] [PubMed] [Google Scholar]
- 21.Galej WP, Oubridge C, Newman AJ, Nagai K. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature. 2013;493:638–643. doi: 10.1038/nature11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rasche N, et al. Cwc2 and its human homologue RBM22 promote an active conformation of the spliceosome catalytic centre. EMBO J. 2012;31:1591–1604. doi: 10.1038/emboj.2011.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hilliker AK, Mefford MA, Staley JP. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes Dev. 2007;21:821–834. doi: 10.1101/gad.1536107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perriman RJ, Ares M. Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes Dev. 2007;21:811–820. doi: 10.1101/gad.1524307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grainger RJ, Barrass JD, Jacquier A, Rain J-C, Beggs JD. Physical and genetic interactions of yeast Cwc21p, an ortholog of human SRm300/SRRM2, suggest a role at the catalytic center of the spliceosome. RNA. 2009;15:2161–2173. doi: 10.1261/rna.1908309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan C, et al. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015;349:1182–1191. doi: 10.1126/science.aac7629. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen THD, et al. Structural basis of Brr2-Prp8 interactions and implications for U5 snRNP biogenesis and the spliceosome active site. Structure. 2013;21:910–919. doi: 10.1016/j.str.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Nues RW, Beggs JD. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics. 2001;157:1451–1467. doi: 10.1093/genetics/157.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith DJ, Query CC, Konarska MM. ‘Nought may endure but mutability’: spliceosome dynamics and the regulation of splicing. Mol Cell. 2008;30:657–666. doi: 10.1016/j.molcel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konarska MM, Vilardell J, Query CC. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol Cell. 2006;21:543–553. doi: 10.1016/j.molcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 31.Kim CH, Abelson J. Site-specific crosslinks of yeast U6 snRNA to the pre-mRNA near the 5' splice site. RNA. 1996;2:995–1010. [PMC free article] [PubMed] [Google Scholar]
- 32.Buchwald G, Schüssler S, Basquin C, Le Hir H, Conti E. Crystal structure of the human eIF4AIII-CWC22 complex shows how a DEAD-box protein is inhibited by a MIF4G domain. Proc Natl Acad Sci USA. 2013;110:E4611–8. doi: 10.1073/pnas.1314684110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madhani HD, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71:803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- 34.Fica SM, Mefford MA, Piccirilli JA, Staley JP. Evidence for a group II intron–like catalytic triplex in the spliceosome. Nature Struct Mol Biol. 2014;21:464–471. doi: 10.1038/nsmb.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fica SM, et al. RNA catalyses nuclear pre-mRNA splicing. Nature. 2013;503:229–234. doi: 10.1038/nature12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robart AR, Chan RT, Peters JK, Rajashankar KR, Toor N. Crystal structure of a eukaryotic group II intron lariat. Nature. 2014;514:193–197. doi: 10.1038/nature13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen H-C, Tseng C-K, Tsai R-T, Chung C-S, Cheng S-C. Link of NTR-mediated spliceosome disassembly with DEAH-box ATPases Prp2, Prp16, and Prp22. Mol Cell Biol. 2013;33:514–525. doi: 10.1128/MCB.01093-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warkocki Z, et al. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nature Struct Mol Biol. 2009;16:1237–1243. doi: 10.1038/nsmb.1729. [DOI] [PubMed] [Google Scholar]
- 40.Chiu Y-F, et al. Cwc25 is a novel splicing factor required after Prp2 and Yju2 to facilitate the first catalytic reaction. Mol Cell Biol. 2009;29:5671–5678. doi: 10.1128/MCB.00773-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krishnan R, et al. Biased Brownian ratcheting leads to pre-mRNA remodeling and capture prior to first-step splicing. Nature Struct Mol Biol. 2013;20:1450–1457. doi: 10.1038/nsmb.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nature Struct Mol Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Semlow DR, Blanco MR, Walter NG, Staley JP. Spliceosomal DEAH-Box ATPases remodel pre-mRNA to activate alternative splice sites. Cell. 2016;164:985–998. doi: 10.1016/j.cell.2016.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tseng C-K, Liu H-L, Cheng S-C. DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA. 2011;17:145–154. doi: 10.1261/rna.2459611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwer B, Guthrie C. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J. 1992;11:5033–5039. doi: 10.1002/j.1460-2075.1992.tb05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villa T, Guthrie C. The Isy1p component of the NineTeen complex interacts with the ATPase Prp16p to regulate the fidelity of pre-mRNA splicing. Genes Dev. 2005;19:1894–1904. doi: 10.1101/gad.1336305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Umen JG, Guthrie C. Prp16p, Slu7p, and Prp8p interact with the 3' splice site in two distinct stages during the second catalytic step of pre-mRNA splicing. RNA. 1995;1:584–597. [PMC free article] [PubMed] [Google Scholar]
- 48.Chen W, et al. Endogenous U2·U5·U6 snRNA complexes in S. pombe are intron lariat spliceosomes. RNA. 2014;20:308–320. doi: 10.1261/rna.040980.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen THD, et al. CryoEM structures of two spliceosomal complexes: starter and dessert at the spliceosome feast. Current Opinion in Structural Biology. 2016;36:48–57. doi: 10.1016/j.sbi.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garrey SM, et al. A homolog of lariat-debranching enzyme modulates turnover of branched RNA. RNA. 2014;20:1337–1348. doi: 10.1261/rna.044602.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schreieck A, et al. RNA polymerase II termination involves C-terminal-domain tyrosine dephosphorylation by CPF subunit Glc7. Nature Struct Mol Biol. 2014;21:175–179. doi: 10.1038/nsmb.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin RJ, Newman AJ, Cheng SC, Abelson J. Yeast mRNA splicing in vitro. J Biol Chem. 1985;260:14780–14792. [PubMed] [Google Scholar]
- 53.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 54.Hardin JW, Warnasooriya C, Kondo Y, Nagai K, Rueda D. Assembly and dynamics of the U4/U6 di-snRNP by single-molecule FRET. Nucleic Acids Res. 2015;43:10963–10974. doi: 10.1093/nar/gkv1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguyen THD, et al. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature. 2015;523:47–52. doi: 10.1038/nature14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scheres SHW. Semi-automated selection of cryo-EM particles in RELION-1.3. J Struct Biol. 2015;189:114–122. doi: 10.1016/j.jsb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bai X-C, Rajendra E, Yang G, Shi Y, Scheres SHW. Sampling the conformational space of the catalytic subunit of human γ-secretase. eLife. 2015;4:1485. doi: 10.7554/eLife.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scheres SHW, Chen S. Prevention of overfitting in cryo-EM structure determination. Nature Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen S, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nature Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu P, et al. Structure of the mRNA splicing complex component Cwc2: insights into RNA recognition. Biochem J. 2012;441:591–597. doi: 10.1042/BJ20111385. [DOI] [PubMed] [Google Scholar]
- 65.van Roon A-MM, et al. 113Cd NMR experiments reveal an unusual metal cluster in the solution structure of the yeast splicing protein Bud31p. Angew Chem Int Ed Engl. 2015;54:4861–4864. doi: 10.1002/anie.201412210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Biasini M, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–8. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kurowski MA, Bujnicki JM. GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 2003;31:3305–3307. doi: 10.1093/nar/gkg557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Price SR, Evans PR, Nagai K. Crystal structure of the spliceosomal U2B"-U2A' protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- 69.Kudla G, Granneman S, Hahn D, Beggs JD, Tollervey D. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc Natl Acad Sci USA. 2011;108:10010–10015. doi: 10.1073/pnas.1017386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang J, et al. The I-TASSER Suite: protein structure and function prediction. Nature Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Walbott H, et al. Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J. 2010;29:2194–2204. doi: 10.1038/emboj.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 73.Nicholls RA, Fischer M, McNicholas S, Murshudov GN. Conformation-independent structural comparison of macromolecules with ProSMART. Acta Crystallogr D. 2014;70:2487–2499. doi: 10.1107/S1399004714016241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brown A, et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D. 2015;71:136–153. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marcia M, Pyle AM. Visualizing Group II Intron Catalysis through the Stages of Splicing. Cell. 2012;151:497–507. doi: 10.1016/j.cell.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Le Hir H, Saulière J, Wang Z. The exon junction complex as a node of post-transcriptional networks. Nature Reviews Molecular Cell Biology. 2016;17:41–54. doi: 10.1038/nrm.2015.7. [DOI] [PubMed] [Google Scholar]
- 77.Bessonov S, Anokhina M, Will CL, Urlaub H, LUhrmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- 78.Alexandrov A, Colognori D, Shu M-D, Steitz JA. Human spliceosomal protein CWC22 plays a role in coupling splicing to exon junction complex deposition and nonsense-mediated decay. Proc Natl Acad Sci USA. 2012;109:21313–21318. doi: 10.1073/pnas.1219725110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Steckelberg A-L, Boehm V, Gromadzka AM, Gehring NH. CWC22 connects pre-mRNA splicing and exon junction complex assembly. Cell Rep. 2012;2:454–461. doi: 10.1016/j.celrep.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 80.Barbosa I, et al. Human CWC22 escorts the helicase eIF4AIII to spliceosomes and promotes exon junction complex assembly. Nature Struct Mol Biol. 2012;19:983–990. doi: 10.1038/nsmb.2380. [DOI] [PubMed] [Google Scholar]
- 81.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 82.Andersen CBF, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 83.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–9. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]