Abstract
Background
Culture is the gold standard for the detection of environmental B. pseudomallei. In general, soil specimens are cultured in enrichment broth for 2 days, and then the culture broth is streaked on an agar plate and incubated further for 7 days. However, identifying B. pseudomallei on the agar plates among other soil microbes requires expertise and experience. Here, we evaluate a lateral flow immunoassay (LFI) developed to detect B. pseudomallei capsular polysaccharide (CPS) in clinical samples as a tool to detect B. pseudomallei in environmental samples.
Methodology/Principal Findings
First, we determined the limit of detection (LOD) of LFI for enrichment broth of the soil specimens. Soil specimens (10 grams/specimen) culture negative for B. pseudomallei were spiked with B. pseudomallei ranging from 10 to 105 CFU, and incubated in 10 ml of enrichment broth in air at 40°C. Then, on day 2, 4 and 7 of incubation, 50 μL of the upper layer of the broth were tested on the LFI, and colony counts to determine quantity of B. pseudomallei in the broth were performed. We found that all five soil specimens inoculated at 10 CFU were negative by LFI on day 2, but four of those five specimens were LFI positive on day 7. The LOD of the LFI was estimated to be roughly 3.8x106 CFU/ml, and culture broth on day 7 was selected as the optimal sample for LFI testing. Second, we evaluated the utility of the LFI by testing 105 soil samples from Northeast Thailand. All samples were also tested by standard culture and quantitative PCR (qPCR) targeting orf2. Of 105 soil samples, 35 (33%) were LFI positive, 25 (24%) were culture positive for B. pseudomallei, and 79 (75%) were qPCR positive. Of 11 LFI positive but standard culture negative specimens, six were confirmed by having the enrichment broth on day 7 culture positive for B. pseudomallei, and an additional three by qPCR. The LFI had 97% (30/31) sensitivity to detect soil specimens culture positive for B. pseudomallei.
Conclusions/Significance
The LFI can be used to detect B. pseudomallei in soil samples, and to select which samples should be sent to reference laboratories or proceed further for bacterial isolation and confirmation. This could considerably decrease laboratory workload and assist the development of a risk map for melioidosis in resource-limited settings.
Author Summary
Burkholderia pseudomallei is an environmental Gram-negative bacillus and the causative agent of melioidosis. Culture and PCR assays are standard diagnostic tools used to detect B. pseudomallei in the environment. However, those tests require experienced microbiologists and are regularly conducted only in a few research laboratories worldwide. In this study, we demonstrated that the prototype lateral flow immunoassay (LFI) developed to detect B. pseudomallei capsular polysaccharide (CPS) in clinical samples could be used to detect B. pseudomallei in environmental samples. We found that the LFI can be used to detect B. pseudomallei in experimentally spiked soil specimens. Next, we evaluated the sensitivity of LFI using 105 soil samples collected in Northeast Thailand. We found that the LFI had high sensitivity to detect B. pseudomallei in the soil. We propose that the LFI could be used to detect environmental B. pseudomallei in resource-limited settings. Soil samples positive for LFI could be sent to reference laboratories for confirmation with culture or molecular methods. The use of LFI could assist in the development of a global risk map for melioidosis.
Introduction
The Gram-negative Burkholderia pseudomallei is a soil-dwelling organism and also the cause of melioidosis, an often fatal infectious disease [1,2]. Melioidosis can be difficult to diagnose due to its diverse clinical manifestations. The diagnostic confirmation relies on microbiological culture, which is often unavailable in resource-restricted regions of the world [3]. Even with such facilities, B. pseudomallei may be dismissed as a culture contaminant [4], or be misidentified by standard identification methods including API 20NE and automated bacterial identification systems [5,6]. The disease occurs as a result of skin inoculation, inhalation and ingestion of environmental B. pseudomallei [7]. The organism is intrinsically resistant to a wide range of antimicrobials, and treatment with ineffective antimicrobials may result in case fatality rates (CFRs) exceeding 70% [8,9]. A recent spatial modeling study estimated there to be 165,000 human melioidosis cases per year worldwide, of which 89,000 die [10]. The study also estimated that melioidosis is severely underreported in the 45 countries in which it is known to be endemic and that B. pseudomallei is likely present in a further 34 countries in which melioidosis has never been reported [10].
Defining the distribution of B. pseudomallei in countries where B. pseudomallei is likely present but melioidosis has never been reported is important, since this will provide policy makers with evidence for raising awareness of this disease among healthcare workers and microbiology laboratories in these areas [11]. Environmental sampling can be used to identify areas where people are at risk even before cases are recognized. For example, the first environmental survey around Vientiane City (Lao PDR) in 1998 demonstrated the presence of B. pseudomallei prior to the recognition of human disease [12]. This environmental finding resulted in an effort to identify B. pseudomallei from clinical specimens, with the first case of melioidosis being identified in 1999 [13]. Since then, more than 920 culture-positive melioidosis patients have been identified in Lao PDR [14]. Environmental sampling can also be used to confirm the endemicity of melioidosis after identifying melioidosis cases in new areas. Recent findings include the detection of environmental B. pseudomallei after case reports in Gabon [15] and Bangladesh [16].
Culture and PCR assays are commonly used to detect B. pseudomallei in the environment, but both tests require experienced microbiologists and are only available in a few research laboratories worldwide. For the culture method, soil specimens are initially cultured in selective broth for 2 days, and then the upper layer of the broth is streaked on an agar plate and incubated for a further 7 days [11]. Identifying B. pseudomallei on agar plates among other soil microbes is time-consuming and requires expertise and experience. For effective molecular detection, soil specimens are also first enriched in selective broth for 2 days prior to nucleic acid amplification [17,18]. Despite the higher sensitivity than culture, PCR assays also require experienced personnel and positive controls, and are relatively costly for resource-limited settings.
Recently, a lateral flow immunoassay (LFI) was developed to detect B. pseudomallei capsular polysaccharide (CPS) in clinical samples [19]. In a pilot study, the LFI was shown to have 100% sensitivity for all urine samples containing B. pseudomallei greater than 1.2×104 CFU/ml [19]. Here, we evaluate the LFI as a tool to detect B. pseudomallei in the enrichment broth of the environmental samples. We found that the LFI had high sensitivity in our setting.
Methods
Determining the limit of detection (LOD) of LFI to detect B. pseudomallei in enrichment media on experimental soil samples
B. pseudomallei wild-type strain E08 and 300 grams of soil culture negative for B. pseudomallei collected from Nakorn Ratchasima, Northeast Thailand, were used. The strain E08 was selected as the strain has been frequently used as a representative strain of B. pseudomallei from the environment [20–22]. Soil specimens (10 gram/specimen) were spiked with B. pseudomallei at an inoculum of 10 to 105 colony forming units (CFU) per 10 grams of soil, and incubated in 10 ml of enrichment broth (threonine-basal salt solution plus colistin at 50 mg/liter [TBSS-C50]) in air at 40°C. Unspiked soil samples and soil samples sterilised by autoclaving were included as negative controls. Soil samples sterilised by autoclaving, TBSS-C50 and sterile distilled water, all spiked with B. pseudomallei, were also included as positive controls. After incubation for 2, 4 and 7 days, the upper layer of enrichment broth were tested by LFI, and evaluated by culture for quantity of B. pseudomallei. A total of nine soil specimens were used to cover the range of inoculum from 10 to 105 CFU per 10 grams of soil, while five specimens was used for the lowest inoculum of 10 CFU per 10 grams of soil (Table 1).
Table 1. LFI results and quantitative B. pseudomallei count of enrichment broth of the experimental soil specimens.
| Tube No. | Specimens | B. pseudomallei Inoculation | Enrichment broth on day | LFI test | Quantitative B. pseudomallei count (CFU/ml) | Notes |
|---|---|---|---|---|---|---|
| 1 | soil 10 g a | 105 CFU | 0 | ND | 3.8 x 103 | Expected to have 104 CFU/ml |
| 2 | Positive | 4.9 x 107 | ||||
| 4 | Positive | 1.3 x 108 | Fungi 1+ | |||
| 7 | Positive | 4.5 x 107 | Fungi 2+ | |||
| 2 | soil 10 g a | 104 CFU | 0 | ND | 5.7 x 102 | Expected to have 103 CFU/ml |
| 2 | Positive | 2.6 x 107 | ||||
| 4 | Positive | 8.6 x 106 | Fungi 1+ | |||
| 7 | Positive | 1.2 x 105 | Fungus 3+ | |||
| 3 | soil 10 g a | 103 CFU | 0 | ND | 35 | Expected to have 102 CFU/ml |
| 2 | Positive | 9.1 x 106 | ||||
| 4 | Positive | 2.3 x 107 | Fungi 1+ | |||
| 7 | Positive | 6.1 x 104 | Fungus 4+ | |||
| 4 | soil 10 g a | 102 CFU | 0 | ND | 10 | Expected to have 10 CFU/ml |
| 2 | Positive | 3.8 x 106 | ||||
| 4 | Positive | 5.5 x 106 | Fungi 1+ | |||
| 7 | Positive | 6.4 x 106 | Fungi 2+ | |||
| 5 | soil 10 g a | 10 CFU | 0 | ND | 0 | Expected to have 1 CFU/ml |
| 2 | Negative | 8.4 x 105 | ||||
| 4 | Positive | 7.6 x 105 | Fungi 1+ | |||
| 7 | Positive | 9.0 x 103 | Fungi 2+ | |||
| 6 | soil 10 g a | 10 CFU | 0 | ND | 0 | Expected to have 1 CFU/ml |
| 2 | Negative | 1.0 x 104 | ||||
| 4 | Negative | 2.1 x 106 | ||||
| 7 | Positive | 3.0 x 107 | Fungi 1+ | |||
| 7 | soil 10 g a | 10 CFU | 0 | ND | 0 | Expected to have 1 CFU/ml |
| 2 | Negative | 2.0 x 104 | ||||
| 4 | Negative | 1.9 x 106 | ||||
| 7 | Positive | 1.5 x 107 | Fungi 1+ | |||
| 8 | soil 10 g a | 10 CFU | 0 | ND | 0 | Expected to have 1 CFU/ml |
| 2 | Negative | 9.0 x 103 | ||||
| 4 | Negative | 3.0 x 105 | ||||
| 7 | Positive | 1.8 x 106 | Fungi 1+ | |||
| 9 | soil 10 g a | 10 CFU | 0 | ND | 0 | Expected to have 1 CFU/ml |
| 2 | Negative | 3.0 x 103 | ||||
| 4 | Negative | 1.0 x 105 | Fungi 1+ | |||
| 7 | Negative | 2.0 x 104 | Fungi 1+ |
ND = Not done. Notes show expected quantitative B. pseudomallei count of enrichment broth on day 0, and fungi present on agar plates (1+, 2+, 3+ and 4+ represent the amount of fungi).
a Soil culture negative for B. pseudomallei was used. Soil were enriched with 10 ml of TBSS-C50 (threonine-basal salt solution plus colistin 50 mg/L) and incubated in air at 40°C for 7 days.
The Active Melioidosis Detect (AMD) LFI developed by InBios (Seattle, USA) was used [19]. The evaluated LFI uses a monoclonal antibody (mAb) 4C4 targeting the CPS of B. pseudomallei. MAb 4C4 was sprayed onto a nitrocellulose membrane strip for the test line, and goat anti-chicken IgY (Lampire Biological laboratories, Pennsylvania, USA) was sprayed on the same membrane for the control line. The conjugate pad contained dried 40 nm gold particles conjugated with mAb 4C4 as well as a small amount of gold conjugated with chicken IgY (to react with the control line). The conjugate pad was treated with a borate-based buffer containing a small concentration of detergent and dried for later gold conjugate application. The sample application pad was also treated similarly and dried. The LFI was assembled by combining the sprayed membrane, conjugate pad, and sample pad on top of an adhesive plastic backing. The LFI was stored in air at room temperature until use. A total of 50 μl of the upper layer of the broth was mixed with two drops of lysis buffer and two drops of chase buffer in a clean tube, in which the LFI pad was dipped. After 15 minutes at room temperature, the LFI results were read.
The quantitative count of B. pseudomallei in the enrichment broth was performed as described previously with minor modifications [23]. In short, two 100 μl samples and two 10 μl samples of the upper layer of enrichment broth were spread onto four modified Ashdown’s agar plates using a rotary plater so that the individual colonies could be identified. All plates were then incubated in air at 40°C. The plates were inspected daily for 4 days, and B. pseudomallei was identified as previously described [11]. The LOD of LFI was estimated by comparing the quantitative B. pseudomallei results between samples with positive and negative LFI results. The quantitative counts of B. pseudomallei in specimens with high levels of fungus overgrowth were excluded. The optimal duration of enrichment for LFI testing was determined by selecting the duration that provided the highest proportion of LFI positivity.
Evaluating utility of LFI to detect B. pseudomallei in the enrichment media of soil samples from Northeast Thailand
A total of 105 environmental soil samples were collected from 21 rice paddy fields located in 7 provinces (Burirum, Chaiyaphum, Khon Kaen, Loei, Nakhon Ratchasima, Nong Bua Lam Phu and Udon Thani) in Northeast Thailand during March and June 2015. Three villages per each province were randomly selected for study. We calculated that 105 samples were needed to estimate sensitivity of LFI with the 95% confidence interval of 15%. The randomization was performed using Stata version 14.0 (StataCorp LP, College Station, Texas). A rice field in each village was selected, and the first 5 soil samples collected in each rice field were used for the study. In each rice field, the soil samples was collected 2.5 meters apart. The soil sampling and culture was performed according to the consensus guidelines for environmental sampling of B. pseudomallei developed by DEBWorP [11] with slight modifications. In short, 10 grams of soil were collected at a depth of 30 cm. Each 10 grams of soils were incubated in 10 ml of TBSS-C50 enrichment broth (threonine-basal salt solution with 50 mg/L colistin) at 40°C in air for 7 days. After incubation for 2 and 7 days, 10 μl of the upper layer of enrichment broth were directly plated on Ashdown agar and incubated at 40°C in air further for 7 days [24], and B. pseudomallei was then identified as previously described [11]. In addition, 50 μl of the upper layer of enrichment broth on day 7 was tested on the LFI as described above and 1 ml of the upper layer of enrichment broth on day 7 was stored for qPCR assay. Culture of the upper layer of enrichment broth on day 7 is not normally conducted, but was performed in this study in order to evaluate the presence of B. pseudomallei in soil specimens which were negative by standard culture but LFI positive. Three microbiologists performed culture, LFI and qPCR independently, and they were blinded to the results of the other two tests.
Detecting B. pseudomallei in the enrichment media by quantitative real-time PCR (qPCR)
A 1 mL aliquot of broth on day 7 was stored at -20°C until further processing. Prior to nucleic acid extraction, the broth was spun at 12,000g for 10 min, and all but the pellet and 200 μL of broth were removed. The remaining DNA pellet and broth were processed using the Qiagen Mini Kit using the bacterial extraction protocol, according to the manufacturers instructions. The qPCR assay targeting orf2 of B. pseudomallei type III secretion system (TTS1) was performed as previously described [18].
Statistical analysis
Utility of the LFI was determined by comparing the results between culture of broth on day 2, LFI of broth on day 7 and qPCR of broth on day 7. Culture of broth on day 2 was selected for the main comparison because it is the standard method used for environmental detection of B. pseudomallei in soil samples [11]. The degree of agreement between culture and LFI was expressed using the Kappa index and its P value. This describes the level of association, both positive and negative, beyond that caused by chance, as follows: 0.00–0.20, slight; 0.21–0.40, fair; 0.41–0.60, moderate; 0.61–0.80, substantial; 0.81–1.00, high.
Results
Limit of detection (LOD) of LFI to detect B. pseudomallei in the enrichment media
A total of nine soil specimens (10 gram/specimen) were spiked with B. pseudomallei at an inoculum of 10 to 105 CFU per 10 grams of soil, and incubated in 10 ml of enrichment broth in air at 40°C (Table 1). Immediately after inoculation and mixing, we found that the quantitative count of B. pseudomallei in the broth ranged from 0% to 100% of the inoculum. All 0% results were from five specimens inoculated with 10 CFU of B. pseudomallei per 10 gram of soil (Table 1). The 100% result was the specimen inoculated with 100 CFU of B. pseudomallei per 10 gram of soil, with the quantitative count after inoculation similar to the expected concentration (10 CFU/ml; Table 1).
On day 2, the quantitative count of B. pseudomallei in the broth of five soil specimens spiked with 10 CFU ranged from 3x103 to 8.4x105 CFU/ml. Nonetheless, all of those five specimens were LFI negative (Table 1, Tube No. 5 to 9). On day 4, the quantitative count of B. pseudomallei in the broth of those five specimens ranged from 1x105 to 2.1x106 CFU/ml. Only one specimen (with a quantitative count 7.6x105 CFU/ml) was LFI positive, while the remaining four were LFI negative. On day 7, B. pseudomallei count in the broth of those five specimens ranged from 9x103 to 1.3x108 CFU/ml. There was some difficulty obtaining quantitative counts in the two specimens with low counts (9x103 and 2x104 CFU/ml) due to overgrowth with fungi. Nonetheless, four specimens were LFI positive, while the remaining one with the count of 2x104 CFU/ml was LFI negative (Table 1). S1 Table shows the results of all control specimens.
Using the quantitative counts of all the specimens, we found that the highest count that had a negative LFI result was 2.1x106 CFU/ml (Tube 6), and the next higher count that had LFI positive was 3.8x106 CFU/ml (Tube 4, Table 1). Therefore, we estimated that the LOD of the investigated LFI to detect B. pseudomallei in the enrichment broth of soil samples was about 3.8x106 CFU/ml. As 80% of enrichment broth on day 7 of soil specimens spiked with 10 CFU of B. pseudomallei per 10 grams of soil were LFI positive while only 20% of those enrichment broth on day 4 were LFI positive, the enrichment broth on day 7 was selected as the optimal sample for LFI.
Utility of LFI using soil samples from Northeast Thailand
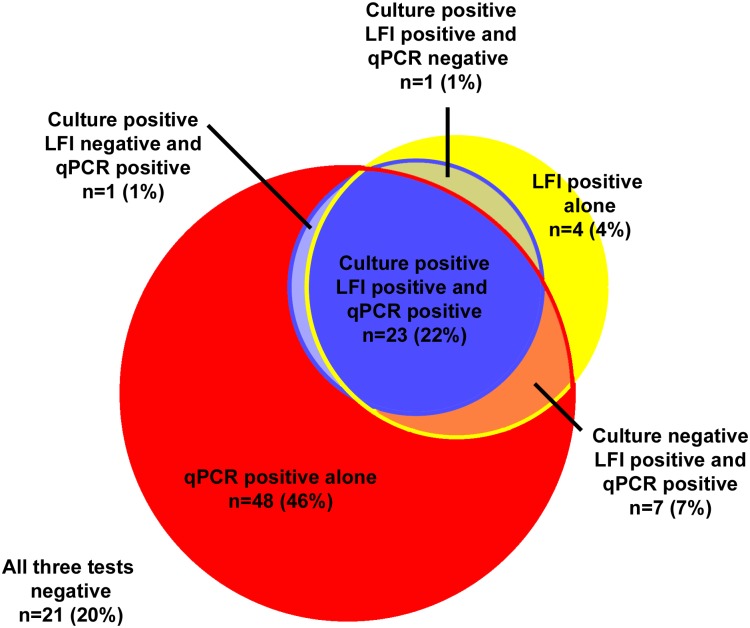
Of 105 soil samples collected in Northeast Thailand, 25 (24%) were culture positive for B. pseudomallei using the standard culture method, 35 (33%) were LFI positive, and 79 (75%) were qPCR positive (Fig 1). Agreement of results between the standard culture and LFI was substantial (Kappa index 0.72, p<0.001). Of 11 specimens LFI positive but the culture of broth on day 2 negative, six had broth on day 7 culture positive for B. pseudomallei, three were qPCR positive but broth on day 7 culture negative, and the other two were both qPCR and culture of broth on day 7 negative. We estimated that the LFI had 97% (30/31; 95% confidence interval 83%-99%) sensitivity for detection of soil specimens culture positive for B. pseudomallei.
Fig 1. Venn diagram showing culture (blue), lateral flow immunoassay (LFI; yellow) and qPCR assay (red) results of 105 soil samples.
Culture was performed according to the consensus guidelines for environmental sampling of B. pseudomallei developed by DEBWorP [11], in which soil specimens were enriched in the enrichment broth for 2 days and the upper layer of enrichment broth on day 2 was streaked on agar plates and observed daily. For the LFI and qPCR assay, soil specimens were enriched in the broth for 7 days and the broth on day 7 was used for the tests.
Discussion
This study demonstrates the utility of LFI as a tool to detect B. pseudomallei in soil samples. The LFI has a high sensitivity (97%) to detect soil specimens culture positive for B. pseudomallei. The test is very simple to use and does not require expertise or new equipment; therefore, LFI could be useful for inexperienced microbiologists who want to conduct an environmental survey in resource-limited settings. One explanation of the high sensitivity of the LFI is that we chose the enriched broth on day 7 as the specimen for LFI. Enrichment of soil specimens increases the concentration of B. pseudomallei in the broth exponentially, and the enrichment step (for 1–2 days) is already necessary for both culture and molecular detection [17,18]. We show that, for the evaluated LFI, enrichment of only 2 days is not sufficient, and 7 days are needed to increase the quantity of B. pseudomallei in the broth from as low as 1 CFU/ml (equivalent to 1 CFU/gram of soil) up to 3.8 x 106 CFU/ml high enough to be detected.
Although culture is the gold standard for the detection of environmental B. pseudomallei, it requires expertise and experience. Colonies of B. pseudomallei on Ashdown agar plates can have seven morphologies [25] and be dismissed as other soil bacteria [4]. In addition, although Ashdown agar contains crystal violet and gentamicin to suppress the growth of other bacteria, overgrowth of other soil bacteria and fungi on agar plates is common [26], particularly after 7 days of incubation. In this study, we report the advantage of LFI for the detection of B. pseudomallei in soil samples as the reading is simple and can be performed by microbiologists otherwise not experienced with B. pseudomallei. Preliminary detection of B. pseudomallei in enrichment media of soil samples by LFI will also reduce the workload of at melioidosis reference laboratories by focusing on the confirmation of identified LFI positives rather than screening large sample numbers. This could enhance the development of a global risk map for melioidosis, particularly in areas without melioidosis reference laboratories [11].
The LOD of LFI in soil enrichment (3.8x106 CFU/ml) is comparatively higher than its previously reported LOD in direct urine samples (1.2 x 104 CFU/ml) [19]. This is probably due to the presence of other organisms and other substances in the soil enrichment.
The long total time required to complete the LFI test (7 days) is not a major problem as the total time required for the culture method is also 9 days. The culture method requires the soil specimen to be in the enrichment broth for 2 days, and then the upper layer of the broth is streaked on plates and incubated for a further 7 days. B. pseudomallei can survive well in soil kept at ambient temperature (24 to 32°C) and away from direct sunlight [11]. Therefore, culture and qPCR assays can be performed later from the soil specimens which are LFI positive. A study reported that the numbers of B. pseudomallei in soil increased moderately over 2 weeks when kept at 20°C [27]. The isolation of environmental B. pseudomallei provides definitive evidence that people in the areas sampled are at risk of melioidosis; therefore, it is important to use the test or a combination of tests with high accuracy rather than a rapid test with low accuracy.
The high proportion of specimens with were culture negative but qPCR positive (Fig 1) could be due to many reasons. First, culture results were based on enrichment of soil samples on day 2, while qPCR was based on that on day 7, resulting in higher number of B. pseudomallei. Second, qPCR has a lower detection limit compared to the culture method [18], and could also detect non-culturable B. pseudomallei [27]. In this study, qPCR confirmed the presence of B. pseudomallei in an additional three specimens that were culture negative but LFI positive. As qPCR is more sensitive than culture and LFI, the molecular methods should be considered if absence of the organism in the environment needs to be confirmed and resources are available [28]. Nonetheless, PCR assays for environmental samples require experienced molecular microbiologists and separate facilities, which are largely not available in areas where melioidosis could be highly endemic.
The LFI may have some limitations. First, the LFI could yield false positive results if a variant of B. thailandensis which produces CPS similar to B. pseudomallei is present in the soil. Second such B. thalandensis variants have been reported in soil from the USA (Texas) [29], Cambodia [30] and Laos [18], but not so far in soil from other areas. Second, the quantitative counts (CFU/ml) in our study could be underestimated due to overgrowth of fungi and other bacteria. Third, the use of LFI may lead to a long total time, as repeated bacterial culture would be needed for the confirmation of B. pseudomallei in the environment (15 days; 7 days of LFI plus 9 days of culture). If transportation of LFI-positive soil samples to reference laboratories is required, it would be preferred within 7-days of completing the LFI tests [27]. Sample site selection and total number of samples per site could be performed according to the consensus guidelines for environmental sampling of B. pseudomallei developed by DEBWorP [11]. Nonetheless, if rapid results are required, molecular methods should be considered [28]. Fourth, the utility of LFI may be different in other regions where the soil physicochemical properties are different, B. pseudomallei numbers lower and/or other organisms are present. Fifth, Our study sample size is not large. Further studies should also investigate the sensitivity and utility of the LFI to detect B. pseudomallei in soil outside Northeast Thailand.
In conclusion, we recommend that the LFI could be used to detect environmental B. pseudomallei in new areas, particularly when the investigation is conducted by microbiologists who have limited experience with the isolation and identification of B. pseudomallei from soil specimens. LFI positive soil specimens could then be sent to reference laboratories for bacterial isolation and confirmation. This could considerably decrease laboratory workload and assist in the development of a global risk map for melioidosis.
Supporting Information
(DOCX)
Acknowledgments
We thank Sittikorn Rongsumlee, Weerawat Wongasa and Malinee Oyuchua for technical assistance.
Data Availability
All relevant data are within the paper.
Funding Statement
This study was funded by the Wellcome trust (106698/B/14/Z) and the National Institute of Allergy and Infectious Diseases (Y1-AI-4906-09). LFI tests were kindly provided by InBios International, Inc., USA. DL is supported by an Intermediate Fellowship awarded by the Wellcome Trust (101103/Z/13/Z). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cheng AC, Currie BJ (2005) Melioidosis: epidemiology, pathophysiology, and management. Clin Microbiol Rev 18: 383–416. 10.1128/CMR.18.2.383-416.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.White NJ (2003) Melioidosis. Lancet 361: 1715–1722. [DOI] [PubMed] [Google Scholar]
- 3.Hoffmaster AR, AuCoin D, Baccam P, Baggett HC, Baird R, et al. (2015) Melioidosis diagnostic workshop, 2013(1). Emerg Infect Dis 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.John TJ, Jesudason MV, Lalitha MK, Ganesh A, Mohandas V, et al. (1996) Melioidosis In India: the tip of the iceberg? Indian J Med Res 103: 62–65. [PubMed] [Google Scholar]
- 5.Weissert C, Dollenmaier G, Rafeiner P, Riehm J, Schultze D (2009) Burkholderia pseudomallei misidentified by automated system. Emerg Infect Dis 15: 1799–1801. 10.3201/eid1511.081719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deepak RN, Crawley B, Phang E (2008) Burkholderia pseudomallei identification: a comparison between the API 20NE and VITEK2GN systems. Trans R Soc Trop Med Hyg 102 Suppl 1: S42–44. [DOI] [PubMed] [Google Scholar]
- 7.Limmathurotsakul D, Peacock SJ (2011) Melioidosis: a clinical overview. Br Med Bull 99: 125–139. 10.1093/bmb/ldr007 [DOI] [PubMed] [Google Scholar]
- 8.White NJ, Dance DA, Chaowagul W, Wattanagoon Y, Wuthiekanun V, et al. (1989) Halving of mortality of severe melioidosis by ceftazidime. Lancet 2: 697–701. [DOI] [PubMed] [Google Scholar]
- 9.Lipsitz R, Garges S, Aurigemma R, Baccam P, Blaney DD, et al. (2012) Workshop on treatment of and postexposure prophylaxis for Burkholderia pseudomallei and B. mallei Infection, 2010. Emerg Infect Dis 18: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Limmathurotsakul D G N, Dance D.A, Messina J. P, Pigott D.M, Moyes C.L, Rolim D.B, Bertherat E, Day N. P, Peacock S. J, Hay I.S (2015) Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nat Microbiology 1: 15008. [DOI] [PubMed] [Google Scholar]
- 11.Limmathurotsakul D, Dance DA, Wuthiekanun V, Kaestli M, Mayo M, et al. (2013) Systematic review and consensus guidelines for environmental sampling of Burkholderia pseudomallei. PLoS Negl Trop Dis 7: e2105 10.1371/journal.pntd.0002105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wuthiekanun V, Mayxay M, Chierakul W, Phetsouvanh R, Cheng AC, et al. (2005) Detection of Burkholderia pseudomallei in soil within the Lao People's Democratic Republic. J Clin Microbiol 43: 923–924. 10.1128/JCM.43.2.923-924.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phetsouvanh R, Phongmany S, Newton P, Mayxay M, Ramsay A, et al. (2001) Melioidosis and Pandora's box in the Lao People's Democratic Republic. Clin Infect Dis 32: 653–654. 10.1086/318713 [DOI] [PubMed] [Google Scholar]
- 14.Rachlin A, Dittrich S, Phommasone K, Douangnouvong A, Phetsouvanh R, et al. (2016) Investigation of Recurrent Melioidosis in Lao People's Democratic Republic by Multilocus Sequence Typing. Am J Trop Med Hyg 94: 1208–1211. 10.4269/ajtmh.15-0909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiersinga WJ, Birnie E, Weehuizen TA, Alabi AS, Huson MA, et al. (2015) Clinical, environmental, and serologic surveillance studies of melioidosis in Gabon, 2012–2013. Emerg Infect Dis 21: 40–47. 10.3201/eid2101.140762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jilani MS, Robayet JA, Mohiuddin M, Hasan MR, Ahsan CR, et al. (2016) Burkholderia pseudomallei: Its Detection in Soil and Seroprevalence in Bangladesh. PLoS Negl Trop Dis 10: e0004301 10.1371/journal.pntd.0004301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaestli M, Mayo M, Harrington G, Watt F, Hill J, et al. (2007) Sensitive and specific molecular detection of Burkholderia pseudomallei, the causative agent of melioidosis, in the soil of tropical northern Australia. Appl Environ Microbiol 73: 6891–6897. 10.1128/AEM.01038-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knappik M, Dance DA, Rattanavong S, Pierret A, Ribolzi O, et al. (2015) Evaluation of Molecular Methods To Improve the Detection of Burkholderia pseudomallei in Soil and Water Samples from Laos. Appl Environ Microbiol 81: 3722–3727. 10.1128/AEM.04204-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houghton RL, Reed DE, Hubbard MA, Dillon MJ, Chen H, et al. (2014) Development of a Prototype Lateral Flow Immunoassay (LFI) for the Rapid Diagnosis of Melioidosis. PLoS Negl Trop Dis 8: e2727 10.1371/journal.pntd.0002727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trakulsomboon S, Dance DA, Smith MD, White NJ, Pitt TL (1997) Ribotype differences between clinical and environmental isolates of Burkholderia pseudomallei. J Med Microbiol 46: 565–570. 10.1099/00222615-46-7-565 [DOI] [PubMed] [Google Scholar]
- 21.Bast A, Krause K, Schmidt IH, Pudla M, Brakopp S, et al. (2014) Caspase-1-dependent and -independent cell death pathways in Burkholderia pseudomallei infection of macrophages. PLoS Pathog 10: e1003986 10.1371/journal.ppat.1003986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gierok P, Kohler C, Steinmetz I, Lalk M (2016) Burkholderia pseudomallei Colony Morphotypes Show a Synchronized Metabolic Pattern after Acute Infection. PLoS Negl Trop Dis 10: e0004483 10.1371/journal.pntd.0004483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith MD, Wuthiekanun V, Walsh AL, White NJ (1995) Quantitative recovery of Burkholderia pseudomallei from soil in Thailand. Trans R Soc Trop Med Hyg 89: 488–490. [DOI] [PubMed] [Google Scholar]
- 24.Limmathurotsakul D, Wuthiekanun V, Amornchai P, Wongsuwan G, Day NP, et al. (2012) Effectiveness of a simplified method for isolation of Burkholderia pseudomallei from soil. Appl Environ Microbiol 78: 876–877. 10.1128/AEM.07039-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chantratita N, Wuthiekanun V, Boonbumrung K, Tiyawisutsri R, Vesaratchavest M, et al. (2007) Biological relevance of colony morphology and phenotypic switching by Burkholderia pseudomallei. J Bacteriol 189: 807–817. 10.1128/JB.01258-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peacock SJ, Chieng G, Cheng AC, Dance DA, Amornchai P, et al. (2005) Comparison of Ashdown's medium, Burkholderia cepacia medium, and Burkholderia pseudomallei selective agar for clinical isolation of Burkholderia pseudomallei. J Clin Microbiol 43: 5359–5361. 10.1128/JCM.43.10.5359-5361.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trung TT, Hetzer A, Gohler A, Topfstedt E, Wuthiekanun V, et al. (2011) Highly sensitive direct detection and quantification of Burkholderia pseudomallei bacteria in environmental soil samples by using real-time PCR. Appl Environ Microbiol 77: 6486–6494. 10.1128/AEM.00735-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall CM, Busch JD, Shippy K, Allender CJ, Kaestli M, et al. (2015) Diverse Burkholderia Species Isolated from Soils in the Southern United States with No Evidence of B. pseudomallei. PLoS One 10: e0143254 10.1371/journal.pone.0143254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duval BD, Elrod MG, Gee JE, Chantratita N, Tandhavanant S, et al. (2014) Evaluation of a latex agglutination assay for the identification of Burkholderia pseudomallei and Burkholderia mallei. Am J Trop Med Hyg 90: 1043–1046. 10.4269/ajtmh.14-0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sim BM, Chantratita N, Ooi WF, Nandi T, Tewhey R, et al. (2010) Genomic acquisition of a capsular polysaccharide virulence cluster by non-pathogenic Burkholderia isolates. Genome Biol 11: R89 10.1186/gb-2010-11-8-r89 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper.