Abstract
Ethylene is crucial in climacteric fruit ripening. The ethylene signal pathway regulates several physiological alterations such as softening, carotenoid accumulation and sugar level reduction, and production of volatile compounds. All these physiological processes are controlled by numerous genes and their expression simultaneously changes at the onset of ripening. Ethylene insensitive 2 (EIN2) is a key component for ethylene signal transduction, and its mutation causes ethylene insensitivity. In tomato, silencing SlEIN2 resulted in a non-ripening phenotype and low ethylene production. RNA sequencing of SlEIN2-silenced and wild type tomato, and differential gene expression analyses, indicated that silencing SlEIN2 caused changes in more than 4,000 genes, including those related to photosynthesis, defense, and secondary metabolism. The relative expression level of 28 genes covering ripening-associated transcription factors, ethylene biosynthesis, ethylene signal pathway, chlorophyll binding proteins, lycopene and aroma biosynthesis, and defense pathway, showed that SlEIN2 influences ripening inhibitor (RIN) in a feedback loop, thus controlling the expression of several other genes. SlEIN2 regulates many aspects of fruit ripening, and is a key factor in the ethylene signal transduction pathway. Silencing SlEIN2 ultimately results in lycopene biosynthesis inhibition, which is the reason why tomato does not turn red, and this gene also affects the expression of several defense-associated genes. Although SlEIN2-silenced and green wild type fruits are similar in appearance, their metabolism is significantly different at the molecular level.
Introduction
The gaseous phytohormone ethylene (C2H4) is essential for developmental and physiological regulation in a variety of higher plants [1] and its effect is more obvious during the ripening process of climacteric fruits [2]. Physiological and molecular genetic analysis have uncovered a pathway for ethylene signal transduction, which is evolutionarily conserved from the receptors of the endoplasmic reticulum membrane to transcription factors in the nucleus [3,4,5,6]. The ethylene receptor (ETR), localized on the endoplasmic reticulum, negatively regulates the ethylene signal transduction occurring upstream [4,7]. Several years later, studies in root hair cells by Dong et al (2010) indicated that receptor ETR1 also localizes at the Golgi network and only partially at the ER [8]. So far seven ETR genes of tomato (Solanum lycopersicum) have been cloned, SlETR1, SlETR2, NR, SlETR4, SlETR5, SLETR6, and SlETR7 [9,10,11,12], present different expression patterns during tomato development [13]. Arabidopsis thaliana constitutive triple response 1 (CTR1), which is a Raf-like protein kinase [5], has multiple homologs in tomato. Some CTR1-like proteins can interact with ETR proteins and negatively regulate signal transduction [14]. In addition, SlCTR1 is sensitive to exo-ethylene treatment and its expression increases during tomato development [15].
In A. thaliana, ethylene insensitive 2 (EIN2), also located in the ER, is a critical positive regulator in the ethylene signal pathway and its C-terminal domain (CEND) can be cleaved and transferred to the nucleus, stabilizing another positive regulation protein, ethylene insensitive 3 (EIN3) [3,16]. ETR1 interacts with EIN2, in which Ser645 is phosphorylated by CTR1 to block the cleavage of CEND and its transfer to the nucleus [6,17,18]. EIN2 is the only protein whose loss-of-function mutation results in complete ethylene insensitivity in the ethylene signal transduction pathway between the nucleus and cytoplasm [16]. Two F-box proteins, ETP1 and ETP2, destroy EIN2 through the ubiquitin pathway [19]. In turn, EIN2’s cleaved CEND can inhibit the expression of the F-box genes EB1 and EBF2, disrupting the accumulation of EIN3 by recognizing their 3′-untranslated regions and transferring them to the P-body. This is accompanied by an exoribonuclease, EIN5, acting in the cytoplasm at translation level [20,21]. Using virus induced gene silence (VIGS), SlEIN2 can be silenced in tomato plants, significantly suppressing fruit ripening [22]. Silencing only one of the functionally redundant SlEILs (coding for EIN3-like proteins) did not produce significantly non-ripening phenotypes [23]. After ethylene insensitive-like (EIL) proteins bind to the promoter regions of ethylene response elements (EREs), the ethylene response factors (ERFs) are able to bind the GCC-box, a conserved sequence of ethylene response genes, and activate the ethylene-induced pathogenesis-related genes [24,25,26]. In banana, which is also a climacteric fruit, the GCC-box motif is homologous to the cis-acting elements of MaEXP1, suggesting that some ERFs might have a role in fruit softening [27].
Fruit ripening is a complicated process, including the accumulation of volatile components, flavonoids formation, pectin degradation, and carotenoid biosynthesis [24,28,29,30]. These diverse processes are regulated by numerous transcription factors and signal transduction pathways [24], among which the ethylene signal pathway is typically found. Unlike the functionally redundant ETRs, CTRs, EILs, and ERFs, the uniqueness of EIN2 might enable it to participate in many ethylene-related metabolic pathways, which probably contributes to the complete ethylene insensitivity resulting from a mutation in the functional domain of SlEIN2. Previous studies have shown that non-ripening tomatoes were obtained by silencing SlEIN2 through VIGS, and recently a new paper from Gao et al (2016) showed that in yellow-fruited tomato 1(yft1) mutant, lower expressed of EIN2 would lead to impaired ethylene biosynthesis and perception, as well as abnormal carotenoid production [22,31]. However, no study has assessed how SlEIN2 regulates fruit ripening at the gene level. Therefore, the present study aims to provide a preliminary analysis of SlEIN2 regulation during fruit ripening using RNA sequencing (RNA-seq). The results showed that silencing SlEIN2 leads to significant changes in the expression of a large number of genes involved in chlorophyll binding proteins, ethylene biosynthesis, lycopene production, defense, etc., as SlEIN2 and the ethylene signal pathway critically upregulate several transcription factors in a feedback loop. The study of SlEIN2 also increases the knowledge on the molecular mechanisms regulating fruit ripening by signal transduction pathways.
Materials and Methods
Plant material and growth conditions
Ailsa Craig tomato seeds preserved in our laboratory were sown in commercially available tomato-cultivation soil and grown in a chamber at 25 ± 2°C, with a relative humidity of 75% and under a light: darkness cycle of 16:8 h, regulated by fluorescent lamps. Tomato plants were watered with a nutrient solution once a week.
Tobacco rattle virus (TRV)-SlEIN2 vector construction
Vectors used VIGS are based on the TRV pTRV1 and pTRV2 (Liu Y et al., 2002). We adopted In-fusion® (Clontech, Nanjing, China), a new cloning technique that does not require T4 DNA ligase and the insertion of a silencing fragment, and has high ligation efficiency. To generate pTRV2-SlEIN2, pTRV2 plasmids were first linearized through digestion with EcoRI and BamHI. As the reverse insertion of the silencing fragment can improve silencing efficiency [22], a 348-bp SlEIN2 fragment was amplified using the forward primer 5′-TAAGGTTACCGAATTCCCTGAATTGGAGCTGTAC-3′, which included a BamHI adaptor (underlined) and the reverse primer 5′- GCTCGGTACCGGATCCTGGAAATGTCCCTGTAGG-3′, which included an EcoRI adaptor (underlined). The resulting product was cloned into pTRV2 using the In-fusion® kit and following the manufacturer’s instructions.
Agrobacterium tumefaciens infiltration
The vectors pTRV1- and pTRV2-SlEIN2, and the control vectors pTRV1 and pTRV2, were transformed into two sets of the A. tumefaciens strain GV3101 and cultured at 200 rpm at 28°C in the Luria—Bertani (LB) medium containing 10 mM 2-N-morpholino ethanesulfonic acid (MES) and 20 mM acetosyringone (AS), with 50 μg/mL of kanamycin, gentamycin, and rifampicin antibiotics. After 16 h, both sets of A. tumefaciens cells were centrifuged and resuspended in the infiltration buffer (10 mM MES, 200 μM AS, 10 mM MgCl2; pH 5.6), until their OD600 ranged between 4 and 7. Bacterial suspensions were set aside for 3–4 h and then combined at a 1:1 ratio, before their infiltration in tomato plants’ using a 1-mL syringe. Ten days after pollination, the carpopodium of tomato plants was perforated and 50–100 μL bacterial solution were infiltrated at the wound site.
RNA isolation and real-time quantitative PCR
Five days after breaker (BK), the orange ripe (OR) pTRV and pTRV-SlEIN2-inoculated, only the green area sampled, fruits were collected, each type including 6 fruits. Mature green (GM) fruits from another control group were also collected to evaluate differences between gene-silenced and authentic unripe fruits. Control and gene-silenced fruits were stored at -80°C before use and their total RNA was isolated using the RNeasy® Mini Kit (Qiagen, Hilden, Germany). Unwanted genomic DNA was digested using DNase I (Tiangen Biotech Co., Beijing, China). The concentration and purity of RNA were measured in a NAS-99 spectrophotometer (ATCGene Inc., New Jersey, United States). The RNA integrity estimated through gel electrophoresis showed a 28S/18S brightness ratio of approximately 2:1. Complementary DNA was then synthesized from 2 μg RNA using the TransScript® II One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (Transgen Biotech Co., LTD., Beijing, China) with oligo(dTs). Virus vectors were detected by PCR using the EasyTaq PCR SuperMix (Transgen Biotech Co., LTD), coat protein, and pTRV-RNA2 specific primers. Amplifications were performed in a Bio-Rad (Hercules, CA, United States) thermocycler under the following conditions: 94°C for 3 min, followed by 30 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 40 s.
Quantitative PCR (qPCR) was performed in the Bio-Rad CFX96 thermocycler using the TransStart Top Green qPCR SuperMix (Transgen Biotech Co., LTD) for 5 min at 95°C, followed by 40 cycles of 15 s at 95°C and 30 s at 60°C. Changes in fluorescence intensity were monitored in each cycle. Three biological replicates, each including two mixed fruits, were included in the PCR and expression levels were determined relatively to that of Actin (ACT1), which was used as the internal control, and analyzed using the 2- ΔCt method [32]. All primers used are listed in S1 Table.
RNA sequencing and assembly of RNA transcripts
Total RNA was isolated from green pTRV-SlEIN2 samples and two groups of pTRV fruits (biological replicates were the same as in qPCR). Total RNA concentration was measured in the NAS-99 spectrophotometer (ATCGene Inc.), and an RNA integrity number ≥ 7.0 was confirmed through gel electrophoresis. Messenger RNA was then enriched using oligo(dTs) coupled with magnetic beads, before being cut into 300-bp fragments (Novogene, Tianjing, China). Complementary DNA libraries were obtained using random hexamers to synthesize the first strand, and adopting DNA polymerase I and dNTPs to generate the second strand. Synthesized cDNA was then purified, its ends were repaired, and adaptors were ligated. After libraries’ preparation, 150-bp pair-end sequencing was performed on an Illumina® Hiseq PE150 (Illumina, Inc., Beijing, China), generating 6 G raw data for each pair-end sequencing.
Raw reads were quality checked and trimmed using cutadapt (version 1.10, https://pypi.python.org/pypi/cutadapt/) and FASTX-Toolkit (version 0.0.13.2 http://hannonlab.cshl.edu/fastx_toolkit/download.html). After removing barcode and adaptor sequences, the resulting clean reads were checked for quality using the Q < 20 threshold. All clean reads were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/) under the accession number SRP076745. Clean reads within each library were aligned with the tomato reference genome (version SL2.50, ftp://ftp.sgn.cornell.edu/tomato_genome) using TopHat (version 2.0.8, http://ccb.jhu.edu/software/tophat/index.shtml). Reads with less than two mismatches were used to construct transcripts using Cufflinks (version 2.0.2, http://cole-trapnell-lab.github.io/cufflinks/). Genes in pTRV-SlEIN2 and pTRV-GM or pTRV-OR fruits were considered as differentially expressed genes (DEGs) if |fold-change| ≥ 2 and Q-value < 0.05.
Gene Ontology (GO) enrichment analysis
GO enrichment analysis was performed using GO-TermFinder (version 0.86, http://search.cpan.org/dist/GO-TermFinder/) based on DEGs, gene identity in the Sol genomics network database to GO terms, gene association, and GO libraries (http://geneontology.org/page/downloads). The threshold of the corrected P-value was 0.05, and genes were classified into the following classes: cellular component, biological process, and molecular function.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis
Fasta format files containing DEGs cDNA or protein sequences were obtained using Perl scripts and KEGG enrichment analysis was then performed in KOBAS (version 2.0, http://kobas.cbi.pku.edu.cn/download.do), based on native blast tools and organism annotation libraries. KEGG pathways with a corrected P-value < 0.05 were analyzed.
Results and Discussion
Silencing SlEIN2 produced non-ripening phenotype and differential expression of several thousand genes
Ethylene regulates several plant physiological activities including development, senescence, flowering, and fruit ripening through signal transduction pathways. As SlEIN2 is an important component of signal transduction, mutations occurring in this gene will effectively block the signal transduction pathway, resulting in plant insensitivity to ethylene. To understand the role of SlEIN2 in the development and ripening of tomato, we obtained SlEIN2-silenced fruits and analyzed their DEGs using RNA-seq.
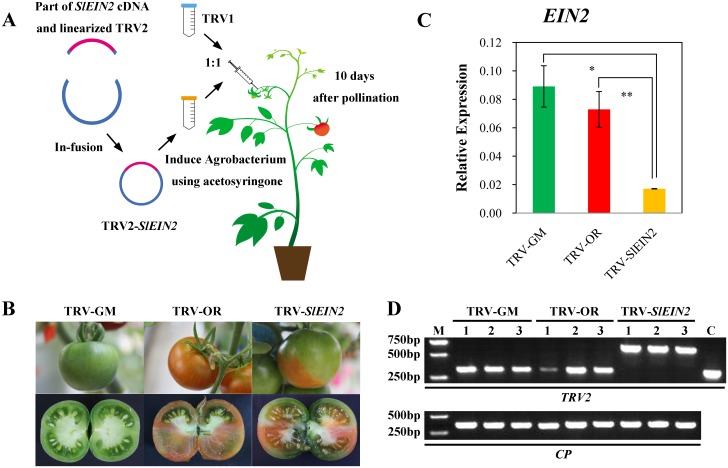
According to the sketch presented in Fig 1A, a mixture of A. tumefaciens GV3101 cultures containing pTRV1- and pTRV2-SlEIN2 or pTRV1 and pTRV2 constructs in a 1:1 ratio, were needle-injected into the carpopodium of wild-type Ailsa Craig tomato fruits 10 days after pollination. The phenotype on the fruit was observed at 5 days after breaker stage. The SlEIN2-silenced fruits were still green whereas the non-SlEIN2-silenced and control fruits were red (Fig 1B). The phenotype presented by SlEIN2-silenced fruits was consistent with our previous study [22]. They remained green, i.e., they were not ripe, and their appearance was very similar to that of pTRV-injected GM fruits.
Fig 1. Acquisition of TRV-SlEIN2 fruits.
(A). Main phases of VIGS. (B). Non-ripening phenotype of attached and dissected TRV-SlEIN2 tomato fruit compared with the two control groups. (C). qPCR analysis of SlEIN2 expression in VIGS and control fruits. The error bar indicates the standard deviation, based on three biological replicates. Asterisks indicate significant differences, according to Student’s t-test (***, P < 0.001). (D). pTRV transmission in fruits. M is the 2 kb marker, and C is the control, using the pTRV2 plasmid as template. 1, 2, and 3 represent the three biological replicates.
To confirm SlEIN2 gene silencing at the molecular level, primers specific to the SlEIN2 genes outside the region targeted for silencing were designed and used in qPCR. Results evidenced a 76% reduction in SlEIN2 transcripts in silenced fruits in relation to pTRV-injected OR fruits. The expression of SlEIN2 in pTRV-injected GM differed from that in pTRV-SlEIN2 fruits, the latter only accounting for 19% of the former (Fig 1C). Consideringan average of ~80% reduction of target endogenous mRNA is normally achived using TRV-VIGS, the samples were feasible for futher studies and analyses [33,34]. As the level of ACT1 transcript was similar in tissues infected with pTRV-SlEIN2 and control vectors, SlEIN2 seems to play an important role in the ethylene signal transduction pathway controlling fruit ripening.
It was reported that SlEIN2 expression reaches its peak at the GM stage, and is reduced after BK, during ripening [35]. The massive accumulation of phosphorylated SlEIN2 in the GM stage is probably related to the upcoming respiration peak and vast changes in fruit substance and color. When fruit development reached the breaker stage, there was few EIN2 in VIGS fruits, and thus interrupted fruits ripening (Fig 2). The correct size of bands shown in Fig 1D evidenced pTRV was well transmitted to the fruits.
Fig 2. Phenotype of SlEIN2-silenced tomato three months after picking, BK+5 phase was initiated.
To understand the molecular mechanism of SIEIN2 regulating tomato fruit ripening, SlEIN2-silenced and control fruit samples (pTRV-GM and pTRV-OR) were analyzed by RNA-seq. All clean reads generated in the sequencing experiment were mapped and aligned with the tomato reference genome (Table 1) Within each file, 79.56 ± 0.62% of the reads were uniquely aligned, suggesting that sequencing results were relatively stable. Discarded multiple-mapped reads (0.60 ± 0.07% of total mapped reads) and the almost uniform 42% GC-contents of sequences are not shown. Selected DEGs (SDEGs) regarding pTRV-GM/pTRV-SlEIN2 and pTRV-OR/pTRV-SlEIN2 fruits are listed in S2 Table.
Table 1. Summary of clean read counts and percentage of unique mapped reads.
| Sample | Clean reads left/right | Left unique mapped | Right Unique mapped | Unique alignment |
|---|---|---|---|---|
| TRV-LeEIN2 1 | 20,795,992 (100%) |
19,032,879 (91.52%) |
17,273,527 (83.06%) |
16,596,614 (79.81%) |
| TRV-LeEIN2 2 | 21,042,397 (100%) |
19,329,124 (91.86%) |
17,422,732 (82.80%) |
16,772,627 (79.71%) |
| TRV-LeEIN2 3 | 22,409,355 (100%) |
20,533,360 (91.63%) |
18,780,469 (83.81%) |
18075341 (80.66%) |
| TRV-GM 1 | 22,011,906 (100%) |
20,122,103 (91.41%) |
18301383 (83.14%) |
17,594,909 (79.93%) |
| TRV-GM 2 | 22,675,897 (100%) |
20,747,156 (91.49%) |
18,765,171 (82.75%) |
18,053,232 (79.61%) |
| TRV-GM 3 | 22,025,764 (100%) |
20,100,048 (91.26%) |
18,052,961 (81.96%) |
17,378,831 (78.90%) |
| TRV-OR 1 | 23,938,433 (100%) |
21,985,222 (91.84%) |
19,547,355 (81.66%) |
18,793,997 (78.51%) |
| TRV-OR 2 | 27,028,002 (100%) |
24,746,795 (91.56%) |
22,246,381 (82.31%) |
21,423,896 (79.27%) |
| TRV-OR 3 | 21,571,779 (100%) |
19,700,720 (91.33%) |
17,838,181 (82.69%) |
17,177,796 (79.63%) |
1, 2, 3, biological replicates.
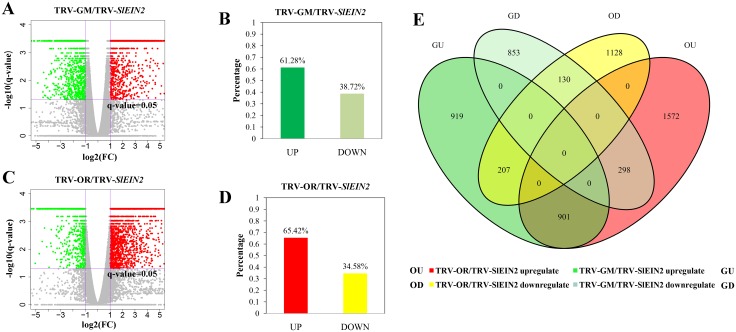
A considerable number of genes changed their expression when there was a deficiency in SlEIN2 (Fig 3A and 3C). In the SlEIN2-silenced fruits, 61.28% of the SDEGs were upregulated and 38.72% were downregulated compared to the GM control group (Fig 3B). Similarly, in OR fruits, 65.42% of the SDEGs were upregulated and 34.58% were downregulated in pTRV-SlEIN2 fruits (Fig 3D). Whereas 901 genes were upregulated in both OU and GU, only 130 genes were downregulated in OD and GD (Fig 3E). These results indicate that silencing SlEIN2 enhances the expression of more genes.
Fig 3. Analyses of differential expressed genes.
(A, C). Volcano diagrams of DEGs. Spots above the threshold line (Q-value = 0.05), indicate that differences are significant. Genes whose expression was less than a half than that displayed in the control group for Q-value < 0.05 are displayed in the green area, while those whose expression was more than two-fold that of the control group are displayed in the red area. Genes in the grey area were neither over- or under-expressed. (B, D). Percentage of up/down regulated SDEGs. SDEGs were screened in the green and red areas. (E) Venn diagram showing the numbers of non-overlapped and overlapped SDEGs in the four conditions tested. OU and OD separately means upregulated and downregulated TRV-OR/TRV-SlEIN2 SDEGs. GU and GD represents upregulated and downregulated TRV-GR/TRV-SlEIN2 SDEGs respectively.
Classifying SDEGs through GO and Pathway enrichments
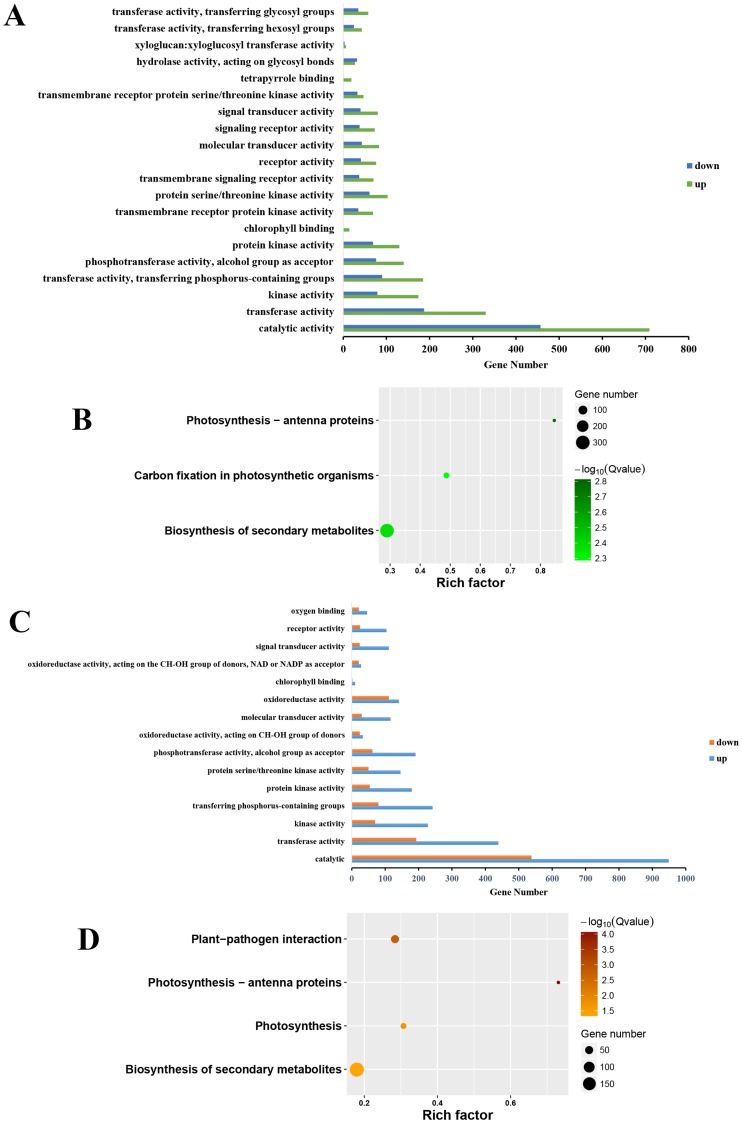
Gene ontology was successfully annotated in molecular function using GO-TermFinder (Fig 4A and 4C), while cell component and biological process not, since SDEGs were not significantly enriched in the two category. A diagram showing molecular functions’ connection is provided in S1 Fig. Two groups were highly enriched: catalytic and transfer activities. The concentration of catalytic proteins such as l-aminocyclopropane-l-carboxylic acid synthase (ACS), which biosynthesizes the precursor of ethylene, and the cell wall decomposition-related enzyme polygalacturonase, drastically changes during fruit ripening [30]. Transfer proteins, such as UDP-glucosyltransferases, which modify anthocyanins and flavonoids by glycosylation increasing their polarity, water solubility [36], and stability, are responsible for fruit ripening, and chlorophyll (Chl) a/b binding proteins, form Chl-protein complexes [37] that take part in photosynthesis.
Fig 4. Molecular function and pathway enrichment analysis of SDEGs.
(A). Molecular function of TRV-GM/TRV-SlEIN2 SDEGs, considering a corrected P-value < 0.05. The X axis indicates the gene number, and Y represents classification. (B). Top 3 pathway enrichment of TRV-GM/TRV-SlEIN2 SDEGs, with a Q-value < 0.05. Rich factor means the number of gene from SDEGs/all gene numbers, in a pathway. (C, D). Molecular function and top 4 pathway enrichment of TRV-OR/TRV-SlEIN2 SDEGs. All the graphic descriptions and parameters are identical to TRV-GM/TRV-SlEIN2.
In TRV-GM/TRV-SlEIN2, SDEGs in the class of Ser/Thr kinase were enriched while in TRV/TRV-SlEIN2 OR fruits they were not, suggesting that most of these genes might not be regulated by SlEIN2. Most SDEGs of TRV-OR/TRV-SlEIN2 exclusively related to redox activities were enriched. NAD/NADP are representative coenzymes in several metabolic activities, better known for tricarboxylic acid cycle and fatty acid oxidation. The genes enriched in NAD/NADP include several unannotated dehydrogenases and respiratory burst oxidases that are NADPH-oxidase homologs and related to plant defense [38], some acyl-CoA reductases and fatty acid oxidases that alter the composition of aroma volatiles, and the electron carrier ferredoxin (S2 Table). In addition, most genes in this class were upregulated in VIGS fruits compared to TRV-OR fruits, suggesting that they might prevent fruit ripening.
Results of the KEGG pathway enrichment analyses showed that SlEIN2 plays a role in regulating the accumulation of chlorophyll binding proteins (Fig 4B and 4D). Light-harvest chlorophyll binding proteins (LHCPs) and chlorophyll are destabilized during fruit senescence through the regulation of staygreen (SGR) [39]. The significant enrichment of the SDEGs in the class of photosynthesis-antenna proteins indicated that LHCPs expression in SlEIN2-silenced fruits differed from both GM and OR fruits. Differences in carbon fixation (dark reaction) between pTRV GM and pTRV-SlEIN2 fruits were found and pTRV OR/pTRV-SlEIN2 SDEGs were enriched on photosynthesis (light reaction can be visualized in the link in S3 Table. Thus, although VIGS fruits were green, the genes they expressed in carbon fixation were similar to those of OR fruits, suggesting that SlEIN2 influenced light reactions but not dark reactions in photosynthesis. In addition, silencing SlEIN2 altered the activities of genes associated with anti-pathogen, such as the respiratory burst oxidase homolog Solyc03g117980, whose fold change was nearly 4.6 in TRV-OR/TRV-SlEIN2 (S2 Table). Although several defense-associated genes were differentially expressed between VIGS and GM fruits, these differences were not significant according to KEGG pathways.
Ripening-associated transcript factors are influenced by SlEIN2 silencing
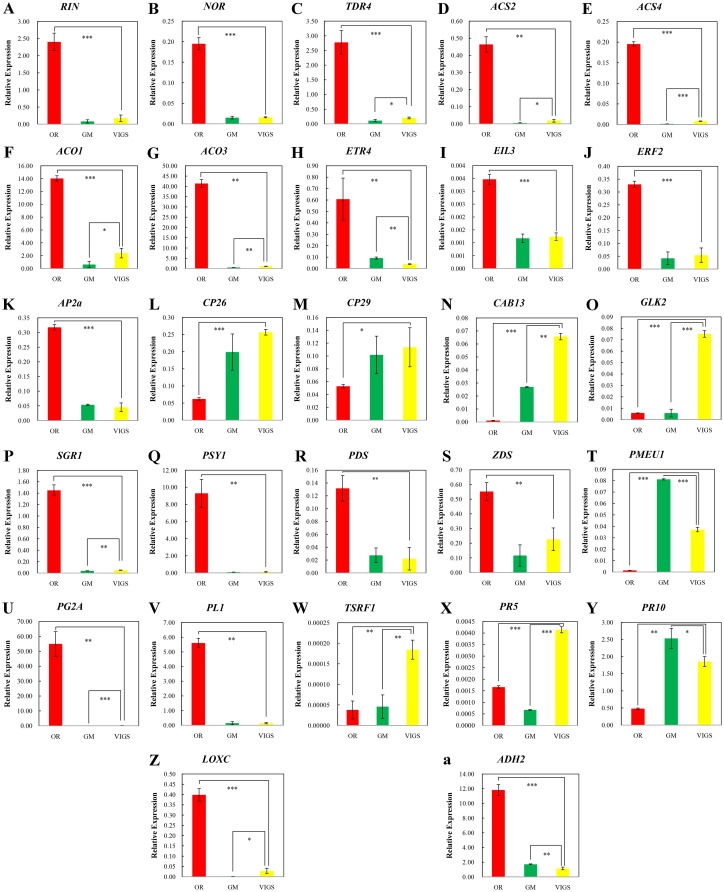
In order to verify the consistency of the RNA-seq results and the gene expression pattern in tomato fruit, 28 differentially expressed and ripening-associated genes were selected from RNA-seq results, and verified by qPCR using RNA-seq materials as template. Gene selection was based on SDEGs statistics (S2 Table) and genes identification and description are listed in Table 2.
Table 2. SGN ID and discription of genes detected in qPCR.
| Gene | SGN ID | Discription |
|---|---|---|
| EIN2 | Solyc09g007870 | Ethylene insensitive 2 |
| RIN | Solyc05g012020 | MADS-box transcription factor MADS-RIN |
| TDR4 | Solyc06g069430 | FRUITFULL-like MADS-box 1 |
| NOR | Solyc10g006880 | NAC domain protein |
| ACS2 | Solyc01g095080 | 1-aminocyclopropane-1-carboxylic acid synthase-2 |
| ACS4 | Solyc05g050010 | 1-aminocyclopropane-1-carboxylic acid synthase-4 |
| ACO1 | Solyc07g049530 | 1-aminocyclopropane-1-carboxylate oxidase 1 |
| ACO3 | Solyc09g089580 | 1-aminocyclopropane-1-carboxylate oxidase 3 |
| ETR4 | Solyc06g053710 | ethylene receptor homolog |
| EIL3 | Solyc01g096810 | Ethylene insensitive 3 class transcription factor |
| ERF2 | Solyc09g075420 | Ethylene responsive transcription factor 2b |
| AP2a | Solyc03g044300 | AP2-like ethylene-responsive transcription factor |
| CP26 | Solyc06g063370 | Chlorophyll a-b binding protein 1A |
| CP29 | Solyc09g014520 | Chlorophyll a-b binding protein 6A |
| CAB13 | Solyc07g063600 | Chlorophyll a-b binding protein 13 |
| GLK2 | Solyc10g008160 | Transcription factor (Fragment) |
| SGR1 | Solyc08g080090 | Senescence-inducible chloroplast staygreen protein 2 |
| PSY1 | Solyc03g031860 | Phytoene synthase 1 |
| PDS | Solyc03g123760 | Phytoene desaturase |
| ZDS | Solyc01g097810 | Zeta-carotene desaturase |
| PMEU1 | Solyc03g123630 | Pectinesterase |
| PG2A | Solyc10g080210 | Polygalacturonase A |
| PL1 | Solyc03g111690 | Pectate lyase |
| TSRF1 | Solyc09g089930 | Ethylene responsive transcription factor 1a |
| PR5 | Solyc08g080670 | Osmotin-like protein |
| PR10 | Solyc09g090990 | Major allergen Mal d 1 |
| LoxC | Solyc01g006540 | Lipoxygenase |
| ADH2 | Solyc06g059740 | Alcohol dehydrogenase 2 |
Ripening inhibitor (RIN), a member of MADS (named by four transcription factors: MCM1, AG, DEFA and SRF) family, is an indispensible and well-known regulator of tomato fruit ripening which positively regulates gene expression by directly binding to its promoter resulting in other transcription factors expression, ethylene production, cell wall decomposition, aroma variation, and RIN expression [40]. The self-regulated transcription factors seems have no upstream regulator, but the ethylene signal pathway is essential for promoting the expression of RIN [41], suggesting that the production of ethylene is probably auto-regulated through RIN, and that the initial ripening is induced by a developmental factor [42]. When SlEIN2 was repressed, the expression of RIN decreased drastically (Fig 5A–5C), along with its target genes, Non ripening (NOR) and Fruitful 1 (TDR4) [40]. Rin expression was at least two-fold lower in TRV-SlEIN2 than in OR fruits, but not significantly differs from it in GM fruits. The NOR is a ripening regulator of the NAC (named by three transcription factors: NAM, ATAF and CUC) family, whose mutation causes a green phenotype compared to the wild type at red ripe stage [43] whereas TDR4 is another transcription factor of the MADS family. It interacts with RIN to regulate the accumulation of lycopene and lipid metabolism during ripening. After silencing TDR4, ripe fruits remained orange whereas wild type were red [44]. Still, pTRV-SlEIN2 was green for more than three months, unlike rin- and nor-silenced fruits, which are yellowish and yellowish-orange [45]. In summary, SlEIN2 affects fruit ripening mainly by affecting RIN expression.
Fig 5. Relative expression of 28 genes covering several aspects associated with ripening.
The error bar indicates the standard deviation, based on three biological replicates. Asterisks indicate significant differences, according to Student’s t-test (*, P<0.05, **, P<0.01, ***, P< 0.001).
SlEIN2 silencing blocked the self-promotion of ethylene biosynthesis and gave an effect to other components in ethylene signal pathway
Several genes involved in ethylene biosynthesis are direct RIN targets, including ACS2, ACS4, and E8 [41]. The expression of ACS2 and ACS4 was four times lower in pTRV-SlEIN2 than in OR fruits (Fig 5D and 5E). In addition, ACO1 and ACO3 gene expressions were also significantly reduced when SlEIN2 was silenced (Fig 5F and 5G). Studies have confirmed that Rin indirectly regulates the expression of ACO1 [19,46], which is a direct target of HB-1; HB-1 is directly regulated by RIN [41,47]. The promoter of ACO3 is the target of ERF2 [48], an ethylene responsive component at the end of the signal pathway. Thus, the abrupt drop in the expressions of these genes might have been sufficient to reduce ethylene production in gene-silenced fruits.
ETR is a negative regulation component upstream of SlEIN2. Tomato has seven ETRs, some of which are relatively stable while others differ from leaf to reproductive tissues [13]. The levels of SlETR4 and SlETR5 increase significantly as fruits mature and ripen, but only SlETR4 changes in response to ethylene treatment [13,49]. Ethylene can enhance the mRNA level of SlETR4, while the protein level was on opposite. [49]. It appears that mRNA of SlETR4 could inhibite its translation. Our study showed that silencing SlEIN2 reduced SlETR4 expression, as a result of inhibiting ethylene biosynthesis (Fig 5H), and this probably would promote the accumulation of ETR4 receptors When EIN2 decreased, SlEIL3 was downregulated (Fig 5I), and the level of EIL3 in SlEIN2-silenced was very closed to it in GM fruits. As ERF2 was also subjected to the effect of EIN2 (Fig 5J), it might have led to the low ACO3 level in VIGS fruits. APETALA2a (AP2a) is another direct target of RIN and positively regulated by this transcription factor [41]. AP2a negatively regulates ethylene biosynthesis and signal [50], indicating that the accumulation of ethylene can be self-promoted and self-limited through ethylene signal pathway. The level of this gene decreased more than five-fold (Fig 5K) as RIN reduced.
Silencing SlEIN2 has an effect on chlorophyll binding proteins
Pathway enrichment unraveled the influence of EIN2 silencing in LHCPs (see the link in S3 Table). The important Photosystem II (PSII) LHCP named CP29, which is located in the core antenna of PSII, has the highest Chl-a/b binding ratio [37]. The expression of CP29 increased nearly two-fold in pTRV-SlEIN2 tomatoes compared to OR fruits, but no significant difference between VIGS and GM fruits (Fig 5M). Two minor antenna proteins, CP24 and CP26, affect the interactions between PSII subunits [51]. The mRNA level of CP26 were similar to those of CP29 (Fig 5L). Tomato tolerance to continuous light (CL), provided by CAB13, contributes to the increase in substantial yield, as CL influences carbohydrate metabolism and photosynthesis [52], and SlEIN2 silencing led to a larger increase in the expression of CAB13 (Fig 5N) than in normal green fruits. GOLDEN-LIKE (GLK) and SGR are not LHCPs but also contribute to the photosynthetic capacity, and SGR can directly interact with LHCPII, a family of LHCPs belonging to PSII, separating the assembled LHCPII and leading to Chl degradation and plant degreening [39]. The SGR1, another target of RIN [41], also has physical interaction with phytoene synthase 1 (PSY1) and promotes the biosynthesis of carotenoids in tomato [53]. The expression of SGR1 was significantly reduced in SlEIN2-silenced fruits, indicating its positive regulation by ethylene (Fig 5P). Chloroplast development requires GLKs and the expression of GLK2 is typically higher than that of GLK1, especially in fruit shoulder [54]. The SlEIN2-silenced group presented higher levels of GLK2 than GM and OR fruits (Fig 5O). However, RNA-seq and relative expression analyses indicated that SlEIN2 and ethylene act in Chl degradation and negatively regulate photosynthesis.
Silencing SlEIN2 leads to lycopene reduction and cell wall decomposition
PSY, phytoene desaturase (PDS), and ζ-carotenedesaturase (ZDS) are three of the genes involved in carotenoid metabolism and are closely related to the synthesis of lycopene. Although many PSY genes generate phytoene in tomato fruit, PSY1 has a direct interaction with SGR1 and thus was analyzed here. The results showed that these genes were severely inhibited in SlEIN2-silenced fruits, particularly PSY1, whose expression was lowered by, at least, 90% (Fig 5Q–5S). Repressing PDS is sufficient to cause low lycopene content and prevent tomato from reddening [55]. Pectin methylesterase isoenzyme (PMEU1) is a ubiquitously expressed pectinesterase contributing to harden the cell wall, and the reduction of PMEU1 in fruit enhances softening rate [56]. The SlEIN2-silenced fruits presented an increased expression of PMEU1, although not as high as that of GM fruits (Fig 5T). Polygalacturonase 2A (PG2A) and pectin lyase (PL1) are two enzymes related to pectin degradation. Both were expressed in higher amounts than Actin in ripening fruits, but SlEIN2-silencing caused their extreme reduction (Fig 5U and 5V). Although its relative expression was not tested, α-Expansin 1 (EXP1) (Solyc06g051800), which is also a booster for cell wall degradation, had a -3.38-fold change in TRV-OR/TRV-SlEIN2 according to the RNA-seq data (S2 Table). Previous studies have also reported that RIN was associated with the promoters of PG2a and EXP1, and might positively regulate their expression [41,48]. The results obtained here showed that SlEIN2 regulated fruit softening by affecting a series of softening-associated genes.
Silencing SlEIN2 altered the mRNA level of several genes involved in defense and aroma compounds generation
An in vitro study reported that tomato stress responsive factor 1 (TSRF1) interacts with the GCC-box and activates the expression of pathogenesis-related (PR) genes, to strengthen the resistance of tomato to Ralstonia solanacearum, and the study showed that Solyc09g089930 is upregulated by ethylene [57]; however, according to the ementary file of tomato genome research [58], this upregulation is gradually decreased during ripening. Silencing SlEIN2 increased TSRF1 expression and the expression of the PR genes PR5 and PR10 was also enhanced (Fig 5W and 5X). Although not tested, expression of other defense-related genes, such as respiratory burst oxidase, the defense-related WRKY1 Solyc06g066370, and the RIN4 Solyc09g059430 were also enhanced, as they were detected in KEGG pathways and RNA-seq data. The above results suggested that SlEIN2 is involved in transformation of tomato pathogen-defense.
The genes 13-lipoxygenase (LOXC), hydroperoxide lyase, and alcohol dehydrogenase 2 (ADH2) are known to participate in volatiles’ biosynthesis. Their RNAs increase during ripening [24] and ADH2 is a direct RIN target [41]. Silencing SlEIN2 decreased the levels of both ADH2 and LOXC (Fig 5a and 5Z), thereby suggesting that this gene dowregulates the genes involved in aroma biosynthesis.
Conclusions
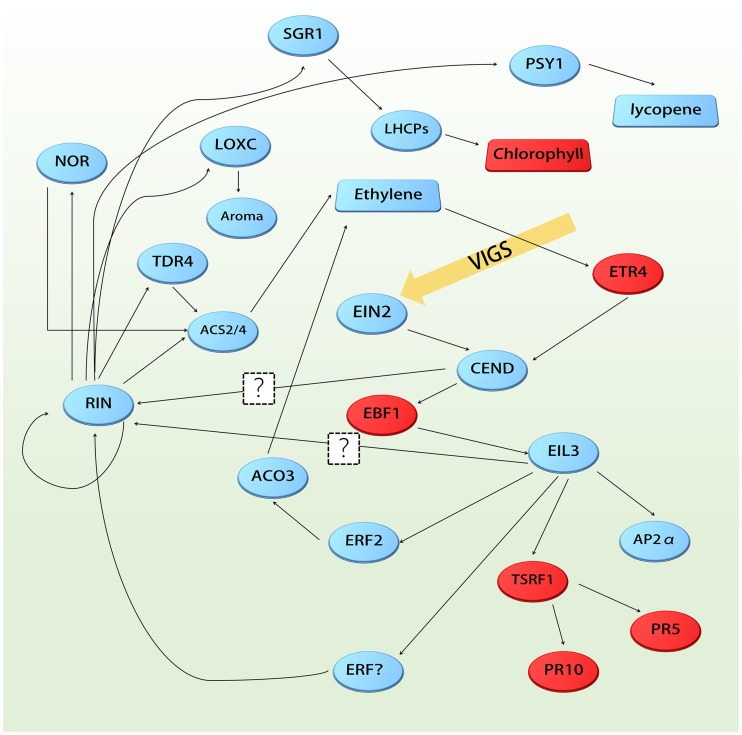
The technique, VIGS, used in the present study is a convenient and powerful tool for targeted gene silencing in tomato, producing the non-ripening phenotype in a short period. In the present study, we assessed SlEIN2 effects on fruit ripening inhibition and this is presented in Fig 6. Silencing SlEIN2 leads to the reduction of RIN in a feedback regulation process, which is generally found upstream of the ethylene signal pathway. In addition, silencing SlEIN2 can decrease the degradation of LHCPs and chlorophyll by reducing RIN expression and that of its target, SGR1. As a result, few SGR1 proteins will interact with PSY1 inhibiting lycopene biosynthesis, which is the reason why tomato does not turn red. Downstream regulation of SlEIN2 is conducted by ERFs and silencing SlEIN2 altered the expression of several defense-associated genes. Although SlEIN2-silenced and GM fruits are similar in appearance, there are significant differences in their secondary metabolites and in antenna-Chl proteins expression.
Fig 6. Regulation route in SlEIN2-silenced fruits.
Red represents upregulated substances and proteins, and blue downregulated substances and proteins.
Supporting Information
(DOCX)
(XLSX)
(XLSX)
(PDF)
Acknowledgments
The authors thank Dr. S.P. Dinesh-Kumar (University of California, Davis) for providing the pTRV1 and pTRV2 vectors.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Nature Science Foundation of China (grants No. 31571898 and No. 31572173) and the Education Foundation of Da Bei Nong Group (No. 1061-2413001). The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Ecker JR (1995) The ethylene signal transduction pathway in plants. Science 268: 667–675. [DOI] [PubMed] [Google Scholar]
- 2.Alexander L, Grierson D (2002) Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J Exp Bot 53: 2039–2055. [DOI] [PubMed] [Google Scholar]
- 3.Wen X, Zhang C, Ji Y, Zhao Q, He W, An F, et al. (2012) Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res 22: 1613–1616. 10.1038/cr.2012.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen YF, Randlett MD, Findell JL, Schaller GE (2002) Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J Biol Chem 277: 19861–19866. 10.1074/jbc.M201286200 [DOI] [PubMed] [Google Scholar]
- 5.Gao Z, Chen YF, Randlett MD, Zhao XC, Findell JL, Kieber JJ, et al. (2003) Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. J Biol Chem 278: 34725–34732. 10.1074/jbc.M305548200 [DOI] [PubMed] [Google Scholar]
- 6.Qiao H, Shen Z, Huang SS, Schmitz RJ, Urich MA, Briggs SP, et al. (2012) Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338: 390–393. 10.1126/science.1225974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hua J, Meyerowitz EM (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94: 261–271. [DOI] [PubMed] [Google Scholar]
- 8.Dong CH, Jang M, Scharein B, Malach A, Rivarola M, Liesch J, et al. (2010) Molecular association of the Arabidopsis ETR1 ethylene receptor and a regulator of ethylene signaling, RTE1. J Biol Chem 285: 40706–40713. 10.1074/jbc.M110.146605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tieman DM, Klee HJ (1999) Differential expression of two novel members of the tomato ethylene-receptor family. Plant Physiol 120: 165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lashbrook CC, Tieman DM, Klee HJ (1998) Differential regulation of the tomato ETR gene family throughout plant development. Plant J 15: 243–252. [DOI] [PubMed] [Google Scholar]
- 11.Liu M, Pirrello J, Chervin C, Roustan JP, Bouzayen M (2015) Ethylene Control of Fruit Ripening: Revisiting the Complex Network of Transcriptional Regulation. Plant Physiol 169: 2380–2390. 10.1104/pp.15.01361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kevany BM, Tieman DM, Taylor MG, Cin VD, Klee HJ (2007) Ethylene receptor degradation controls the timing of ripening in tomato fruit. Plant J 51: 458–467. 10.1111/j.1365-313X.2007.03170.x [DOI] [PubMed] [Google Scholar]
- 13.Klee HJ (2002) Control of ethylene-mediated processes in tomato at the level of receptors. J Exp Bot 53: 2057–2063. [DOI] [PubMed] [Google Scholar]
- 14.Zhong S, Lin Z, Grierson D (2008) Tomato ethylene receptor-CTR interactions: visualization of NEVER-RIPE interactions with multiple CTRs at the endoplasmic reticulum. J Exp Bot 59: 965–972. 10.1093/jxb/ern021 [DOI] [PubMed] [Google Scholar]
- 15.Adams-Phillips L, Barry C, Kannan P, Leclercq J, Bouzayen M, Giovannoni J (2004) Evidence that CTR1-mediated ethylene signal transduction in tomato is encoded by a multigene family whose members display distinct regulatory features. Plant Mol Biol 54: 387–404. 10.1023/B:PLAN.0000036371.30528.26 [DOI] [PubMed] [Google Scholar]
- 16.Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152. [DOI] [PubMed] [Google Scholar]
- 17.Ju C, Yoon GM, Shemansky JM, Lin DY, Ying ZI, Chang J, et al. (2012) CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci U S A 109: 19486–19491. 10.1073/pnas.1214848109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bisson MM, Bleckmann A, Allekotte S, Groth G (2009) EIN2, the central regulator of ethylene signalling, is localized at the ER membrane where it interacts with the ethylene receptor ETR1. Biochem J 424: 1–6. 10.1042/BJ20091102 [DOI] [PubMed] [Google Scholar]
- 19.Qiao H, Chang KN, Yazaki J, Ecker JR (2009) Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev 23: 512–521. 10.1101/gad.1765709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W, Ma M, Feng Y, Li H, Wang Y, Ma Y, et al. (2015) EIN2-directed translational regulation of ethylene signaling in Arabidopsis. Cell 163: 670–683. 10.1016/j.cell.2015.09.037 [DOI] [PubMed] [Google Scholar]
- 21.Merchante C, Brumos J, Yun J, Hu Q, Spencer KR, Enriquez P, et al. (2015) Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell 163: 684–697. 10.1016/j.cell.2015.09.036 [DOI] [PubMed] [Google Scholar]
- 22.Fu DQ, Zhu BZ, Zhu HL, Jiang WB, Luo YB (2005) Virus-induced gene silencing in tomato fruit. Plant J 43: 299–308. 10.1111/j.1365-313X.2005.02441.x [DOI] [PubMed] [Google Scholar]
- 23.Tieman DM, Ciardi JA, Taylor MG, Klee HJ (2001) Members of the tomato LeEIL (EIN3-like) gene family are functionally redundant and regulate ethylene responses throughout plant development. Plant J 26: 47–58. [DOI] [PubMed] [Google Scholar]
- 24.Klee HJ, Giovannoni JJ (2011) Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet 45: 41–59. 10.1146/annurev-genet-110410-132507 [DOI] [PubMed] [Google Scholar]
- 25.Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohme-Takagi M, Shinshi H (1995) Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7: 173–182. 10.1105/tpc.7.2.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trivedi PK, Nath P (2004) MaExp1, an ethylene-induced expansin from ripening banana fruit. Plant Science 167: 1351–1358. [Google Scholar]
- 28.Awad MA (2002) Formation of flavonoids, especially anthocyanin and chlorogenic acid in ‘Jonagold’apple skin: influences of growth regulators and fruit maturity. Sci Hortic-Amsterd: 257–266. [Google Scholar]
- 29.Bramley PM (2002) Regulation of carotenoid formation during tomato fruit ripening and development. J Exp Bot 53: 2107–2113. [DOI] [PubMed] [Google Scholar]
- 30.Brady C (1987) Fruit ripening. Annu Rev Plant Physiol: 155–178. [Google Scholar]
- 31.Gao L, Zhao W, Qu H, Wang Q, Zhao L (2016) The yellow-fruited tomato 1 (yft1) mutant has altered fruit carotenoid accumulation and reduced ethylene production as a result of a genetic lesion in ETHYLENE INSENSITIVE2. Theor Appl Genet 129: 717–728. 10.1007/s00122-015-2660-4 [DOI] [PubMed] [Google Scholar]
- 32.Giorio G, Stigliani AL, D'Ambrosio C (2008) Phytoene synthase genes in tomato (Solanumlycopersicum L.)—new data on the structures, the deduced amino acid sequences and the expression patterns. FEBS J 275: 527–535. 10.1111/j.1742-4658.2007.06219.x [DOI] [PubMed] [Google Scholar]
- 33.Senthil-Kumar M, Mysore KS (2014) Tobacco rattle virus-based virus-induced gene silencing in Nicotiana benthamiana. Nat Protoc 9: 1549–1562. 10.1038/nprot.2014.092 [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Schiff M, Dinesh-Kumar SP (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786. [DOI] [PubMed] [Google Scholar]
- 35.Wang J, Chen G, Hu Z, Chen X (2007) Cloning and characterization of the EIN2-homology gene LeEIN2 from tomato: Full Length Research Article. DNA sequence: 33–38. 10.1080/10425170600986738 [DOI] [PubMed] [Google Scholar]
- 36.Jones PR, Moller BL, Hoj PB (1999) The UDP-glucose:p-hydroxymandelonitrile-O-glucosyltransferase that catalyzes the last step in synthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor. Isolation, cloning, heterologous expression, and substrate specificity. J Biol Chem 274: 35483–35491. [DOI] [PubMed] [Google Scholar]
- 37.Green BR, Pichersky E, Kloppstech K (1991) Chlorophyll a/b-binding proteins: an extended family. Trends Biochem Sci 16: 181–186. [DOI] [PubMed] [Google Scholar]
- 38.Sagi M, Fluhr R (2006) Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol 141: 336–340. 10.1104/pp.106.078089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park SY, Yu JW, Park JS, Li J, Yoo SC, Lee NY, et al. (2007) The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 19: 1649–1664. 10.1105/tpc.106.044891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujisawa M, Shima Y, Higuchi N, Nakano T, Koyama Y, Kasumi T, et al. (2012) Direct targets of the tomato-ripening regulator RIN identified by transcriptome and chromatin immunoprecipitation analyses. Planta 235: 1107–1122. 10.1007/s00425-011-1561-2 [DOI] [PubMed] [Google Scholar]
- 41.Fujisawa M, Nakano T, Shima Y, Ito Y (2013) A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 25: 371–386. 10.1105/tpc.112.108118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yokotani N, Nakano R, Imanishi S, Nagata M, Inaba A, Kubo Y (2009) Ripening-associated ethylene biosynthesis in tomato fruit is autocatalytically and developmentally regulated. J Exp Bot 60: 3433–3442. 10.1093/jxb/erp185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cantu D, Blanco-Ulate B, Yang L, Labavitch JM, Bennett AB, Powell AL (2009) Ripening-regulated susceptibility of tomato fruit to Botrytis cinerea requires NOR but not RIN or ethylene. Plant Physiol: 1434–1449. 10.1104/pp.109.138701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bemer M, Karlova R, Ballester AR, Tikunov YM, Bovy AG, Wolters-Arts M, et al. (2012) The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 24: 4437–4451. 10.1105/tpc.112.103283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bewley JD, Banik M, Bourgault R, Feurtado JA, Toorop P, Hilhorst HW (2000) Endo-beta-mannanase activity increases in the skin and outer pericarp of tomato fruits during ripening. J Exp Bot 51: 529–538. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y, Zhang S (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399. 10.1105/tpc.104.026609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martel C, Vrebalov J, Tafelmeyer P, Giovannoni JJ (2011) The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol 157: 1568–1579. 10.1104/pp.111.181107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Z, Zhang H, Quan R, Wang XC, Huang R (2009) Transcriptional regulation of the ethylene response factor LeERF2 in the expression of ethylene biosynthesis genes controls ethylene production in tomato and tobacco. Plant Physiol 150: 365–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kevany BM, Tieman DM, Taylor MG, Cin VD, Klee HJ (2007) Ethylene receptor degradation controls the timing of ripening in tomato fruit. Plant J 51: 458–467. 10.1111/j.1365-313X.2007.03170.x [DOI] [PubMed] [Google Scholar]
- 50.Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, et al. (2011) Transcriptome and Metabolite Profiling Show That APETALA2a Is a Major Regulator of Tomato Fruit Ripening. The Plant Cell 23: 923–941. 10.1105/tpc.110.081273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Bianchi S, Dall'Osto L, Tognon G, Morosinotto T, Bassi R (2008) Minor antenna proteins CP24 and CP26 affect the interactions between photosystem II subunits and the electron transport rate in grana membranes of Arabidopsis. Plant Cell 20: 1012–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Velez-Ramirez AI, van Ieperen W, Vreugdenhil D, van Poppel PM, Heuvelink E, Millenaar FF (2014) A single locus confers tolerance to continuous light and allows substantial yield increase in tomato. Nat Commun 5: 4549 10.1038/ncomms5549 [DOI] [PubMed] [Google Scholar]
- 53.Sakuraba Y, Park SY, Paek NC (2015) The Divergent Roles of STAYGREEN (SGR) Homologs in Chlorophyll Degradation. Mol Cells 38: 390–395. 10.14348/molcells.2015.0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nadakuduti SS, Holdsworth WL, Klein CL, Barry CS (2014) KNOX genes influence a gradient of fruit chloroplast development through regulation of GOLDEN2-LIKE expression in tomato. Plant J 78: 1022–1033. 10.1111/tpj.12529 [DOI] [PubMed] [Google Scholar]
- 55.Fu DQ, Zhu BZ, Zhu HL, Zhang HX, Xie YH, Jiang WB, et al. (2006) Enhancement of virus-induced gene silencing in tomato by low temperature and low humidity. Mol Cells 21: 153–160. [PubMed] [Google Scholar]
- 56.Phan TD, Bo W, West G, Lycett GW, Tucker GA (2007) Silencing of the major salt-dependent isoform of pectinesterase in tomato alters fruit softening. Plant Physiol 144: 1960–1967. 10.1104/pp.107.096347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang H, Zhang D, Chen J, Yang Y, Huang Z, Huang D, et al. (2004) Tomato stress-responsive factor TSRF1 interacts with ethylene responsive element GCC box and regulates pathogen resistance to Ralstonia solanacearum. Plant Mol Biol 55: 825–834. 10.1007/s11103-004-2140-8 [DOI] [PubMed] [Google Scholar]
- 58.(2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. 10.1038/nature11119 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(XLSX)
(XLSX)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.