Abstract
Identification of non-coding RNAs (ncRNAs) has been significantly improved over the past decade. On the other hand, semantic annotation of ncRNA data is facing critical challenges due to the lack of a comprehensive ontology to serve as common data elements and data exchange standards in the field. We developed the Non-Coding RNA Ontology (NCRO) to handle this situation. By providing a formally defined ncRNA controlled vocabulary, the NCRO aims to fill a specific and highly needed niche in semantic annotation of large amounts of ncRNA biological and clinical data.
Keywords: non-coding RNA, bio-ontologies, domain ontology, reference ontology, ontology development, semantic data annotation
1 Introduction
Non-coding RNAs (ncRNAs), including but not limited to transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), long ncRNAs (lncRNAs), and microRNAs (miRNAs), are special functional RNA molecules that are not translated into protein. Research interest in ncRNA biology has significantly grown, and a large amount of information has been continuously obtained thanks to rapidly developed sequencing technologies in recent years. Unfortunately, semantic annotation and integration of data about ncRNAs lag behind identification of ncRNAs; therefore, effective methodologies are needed to bring together discovery made by the ncRNA research community.
Emerging semantic technologies have been successfully applied to promote more precise communication among scientists in biological, biomedical, and clinical domains (Bard, 2003; Blake, 2004; Blake and Bult, 2006; Huang et al., 2010; Huang et al., 2012; http://neurocommons.org). In particular, the Open Biological and Biomedical Ontologies (OBO) Library (see http://www.obo.sourceforge.net/) has served as an umbrella for different bio-ontologies shared across various domains. However, the OBO Library does not include comprehensive ontologies targeted for the ncRNA domain. Likewise, no such ncRNA ontologies are found in the National Center for Biomedical Ontology (NCBO) BioPortal (see https://bioportal.bioontology.org/) either.
The Non-Coding RNA Ontology (NCRO), to be described in this paper, is the very first comprehensive domain ontology specifically designed for the ncRNA field. The controlled vocabulary that is precisely defined in the NCRO can be utilised as a resource to annotate and integrate ncRNA data generated by relevant communities. In the sense of semantic data annotation and integration, the NCRO is meant to fill a specific and highly needed niche in comprehensive unification of ncRNA biology.
2 Related work
2.1 Research in ncRNA biology
Abnormal expression of ncRNAs is involved in many human diseases (Mattick, 2001; Mattick, 2015). When differentially expressed ncRNAs play regulatory roles in altering target gene expression, further phenotypic effects can be realised. Differential expression of ncRNAs in malignant tissues compared with normal tissues can be exploited as potential therapeutic targets for cancer therapy or as biomarkers used for diagnosis, prediction of patient outcome, or monitoring the effectiveness of cancer therapeutics (Mattick 2015). In recent years serious attempts have been made to effectively deliver ncRNA into tumours in animal models (Babar et al., 2012; Daige et al., 2014; Cheng et al., 2015).
2.2 Research in bio-ontologies
RNA Ontology (RNAO) (Hoehndorf et al., 2011): RNAO is an OBO foundry reference ontology to catalogue the molecular entities composing primary, secondary, and tertiary components of RNA. The goal of the RNAO project is to enable integration and analysis of diverse RNA datasets.
Gene Ontology (GO) (Ashburner et al., 2000): GO is by far the most successful and widely used bio-ontology, consisting of three independent sub-ontologies: biological processes, molecular functions, and cellular components. The GO has been utilised to annotate gene products of model organisms including Homo sapiens.
Ontology for MIcroRNA Target (OMIT): OMIT (Huang et al., 2011; Huang et al., 2014; Huang et al., 2015; Huang et al., 2016) is a miRNA domain ontology being developed as part of the NIH OmniSearch project. The purpose is to establish standard metadata in miRNA domain for more effective identification of miRNAs’ roles in various human diseases.
Sequence Ontology (SO) (Eilbeck et al., 2015): SO is an ontology to capture genomic features and the relationships that obtain between them. This ontology contains the features necessary to annotate a genome with structural features such as gene models and also the terms necessary for the annotation of genomic variants.
PRotein Ontology (PRO) (Natale et al., 2011): Proteins are functional entities in many processes eventually impacted by the regulatory effect of ncRNAs (e.g., miRNA bindings). The PRO, with a particular focus on human proteins and disease-related variants thereof, provides an ontological representation of proteins.
SNOMED CT: Owned and maintained by the International Health Terminology Standard Development Organization, SNOMED CT (see http://www.ihtsdo.org/snomed-ct) is the most comprehensive clinically oriented medical terminology system to promote the use of certified electronic health record (EHR) technology (see http://www.healthit.gov/providers-professionals/meaningful-use-definition-objectives).
NCI Thesaurus (NCIt): NCIt (De Coronado et al., 2009) is a reference biomedical ontology published by the National Cancer Institute (NCI). NCIt terminology includes clinical care, translational & basic research, and public information and administrative activities.
3 Scope and development of the NCRO
The NCRO represents all known subtypes of ncRNA molecules including those created in living organisms as well as those engineered or adapted for some purposes; the structure in each ncRNA type, including sequence and conformation; functions, dispositions, and roles of ncRNAs, as well as processes that are realised in either functions or dispositions; and clinical phenotypes associated with expression of normal and abnormal ncRNAs.
In the development pipeline for the NCRO, we have observed a set of practices proposed by the OBO Foundry Initiative (Smith et al., 2007; see http://www.obofoundry.org/crit.shtml). For example, the ontology should be: freely available; expressed in a standard language; documented for successive versions; orthogonal to existing ontologies; including natural language specifications; developed collaboratively; and used by multiple researchers.
All NCRO terms descend from terms defined in the Basic Formal Ontology (BFO) (see http://www.ifomis.org/bfo/). The BFO is a small, upper-level ontology that is designed for use in supporting information retrieval, analysis, and integration in scientific and other domains. Because the BFO is a well-established upper ontology adopted by all OBO ontologies, our strategy to make the NCRO a BFO-compliant ontology will set the stage for interoperability between the NCRO and other extant OBO ontologies.
The NCRO development is mainly a top-down procedure, where we have utilised the ncRNA domain knowledge provided by cellular biologists and clinical investigators in the project team. The ontology development has also been complemented by a bottom-up approach, in the means that important terms and relations were appended based upon a deep analysis of representative, ncRNA-related databases (Table 1). Additionally, an iterative procedure, which includes a series of interviews, exchanges of documents, refinements, and related documentations, has been followed to make the NCRO a dynamic ontology. Besides a designated project website (see http://omnisearch.soc.southalabama.edu/ontologyfile.php), we have also utilised GitHub (see https://github.com/omnisearch/ncro) to further assist the management and version control of the ontology design and implementation. In addition, being an open-source ontology and following OBO Foundry principles, a GitHub tracker (see https://github.com/omnisearch/ncro-ontology-files/issues) was established to facilitate discussion by an open group of investigators.
Table 1.
List of ncRNA-related databases
| Database name | Brief introduction |
|---|---|
| Ensembl ncRNA | A database of ncRNA annotations. |
| GENCODE | A database for annotation of gene features. |
| lncRNAdb | A reference database for functional lncRNAs. |
| lncRNAtor | A Web portal encompassing lncRNA data. |
| miRBase | A database of miRNA sequences and annotation. |
| NDB | A database of experimentally determined nucleic acids. |
| NONCODE | A database of ncRNAs except for tRNAs and rRNAs. |
| NRED | An ncRNA expression database. |
| Rfam | A database of a collection of RNA families. |
| NONCODE | A database of non-coding RNAs (except for tRNAs and rRNAs). |
| GENCODE | A database for integrated annotation of gene features. |
| miRBase | A database of published miRNA sequences and annotation. |
| TarBase | A database of biologically validated miRNA targets. |
| miRDB | A database for miRNA target prediction and functional annotations. |
| TargetScan | A database about predictions of biological targets of miRNAs. |
| miRGator | A Web portal encompassing miRNA diversity and expression profiles. |
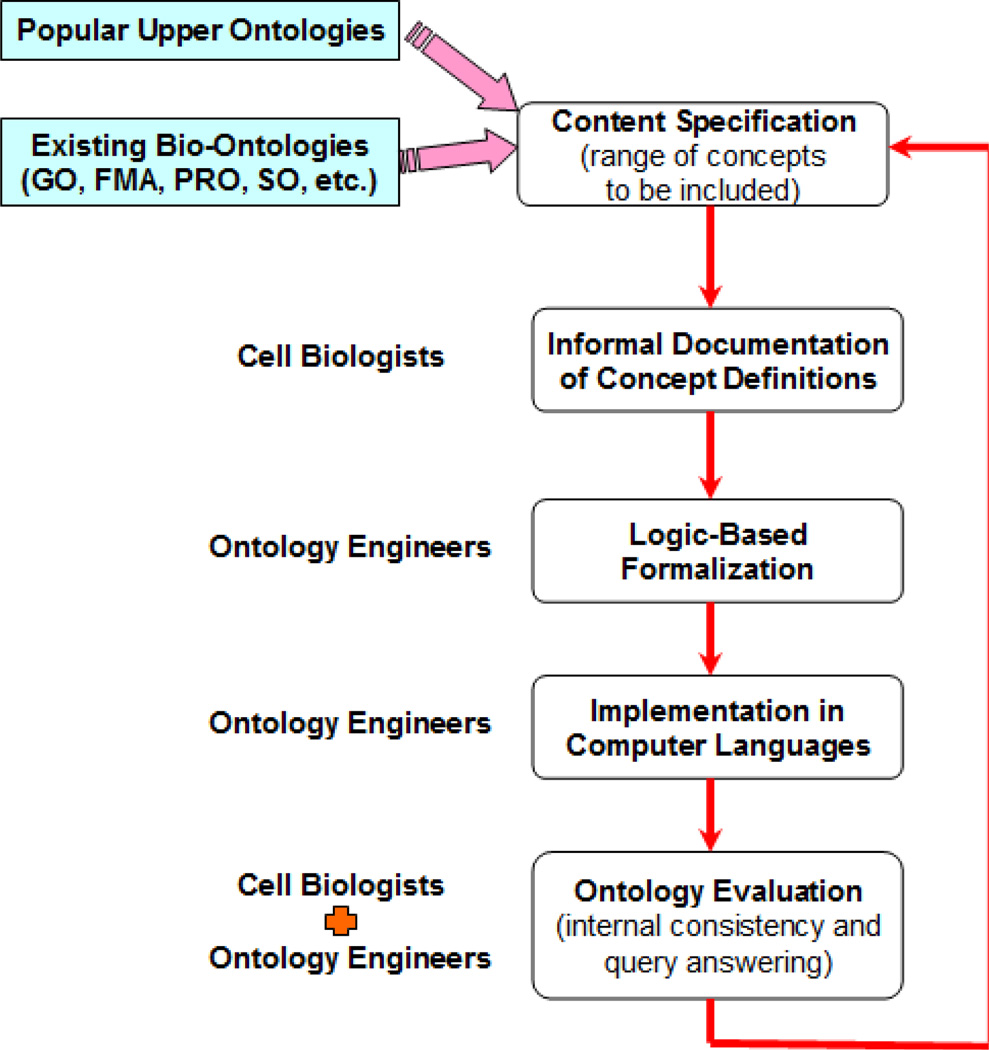
There were five different stages during the ontology development: to specify the range of concepts, an informal documentation of concept definitions, a logic-based formalisation of concepts, to implement in a computer language, and the evaluation. Figure 1 exhibits the flowchart of our iterative ontology development.
Figure 1.
The NCRO ontology construction procedure (see online version for colours)
There exist different formats/languages for describing ontologies, all of which are popular and based on different logics: Web Ontology Language (OWL) (see http://www.w3.org/2004/owl/), OBO, Knowledge Interchange Format (KIF) (see http://www-ksl.stanford.edu/knowledge-sharing/kif/), and Open Knowledge Base Connectivity (OKBC) (see http://www.ai.sri.com/okbc/spec/okbc2/okbc2.html). We have chosen both the OBO and Web Ontology Language (OWL) formats: the former is widely accepted in OBO Foundry community and the latter is recommended by the World Wide Web Consortium (W3C). As for the development tools, we used OBO-Edit (see http://oboedit.org/) to generate an OBO version of the ontology file in the first place; we then utilised the obo-release-manager (OORT) tool (see https://code.google.com/p/owltools/) to convert the ontology file into an equivalent OWL version; finally we verified the converted ontology in Protégé (see http://protege.stanford.edu/).
4 Details of the NCRO ontology
4.1 NCRO terms and relations
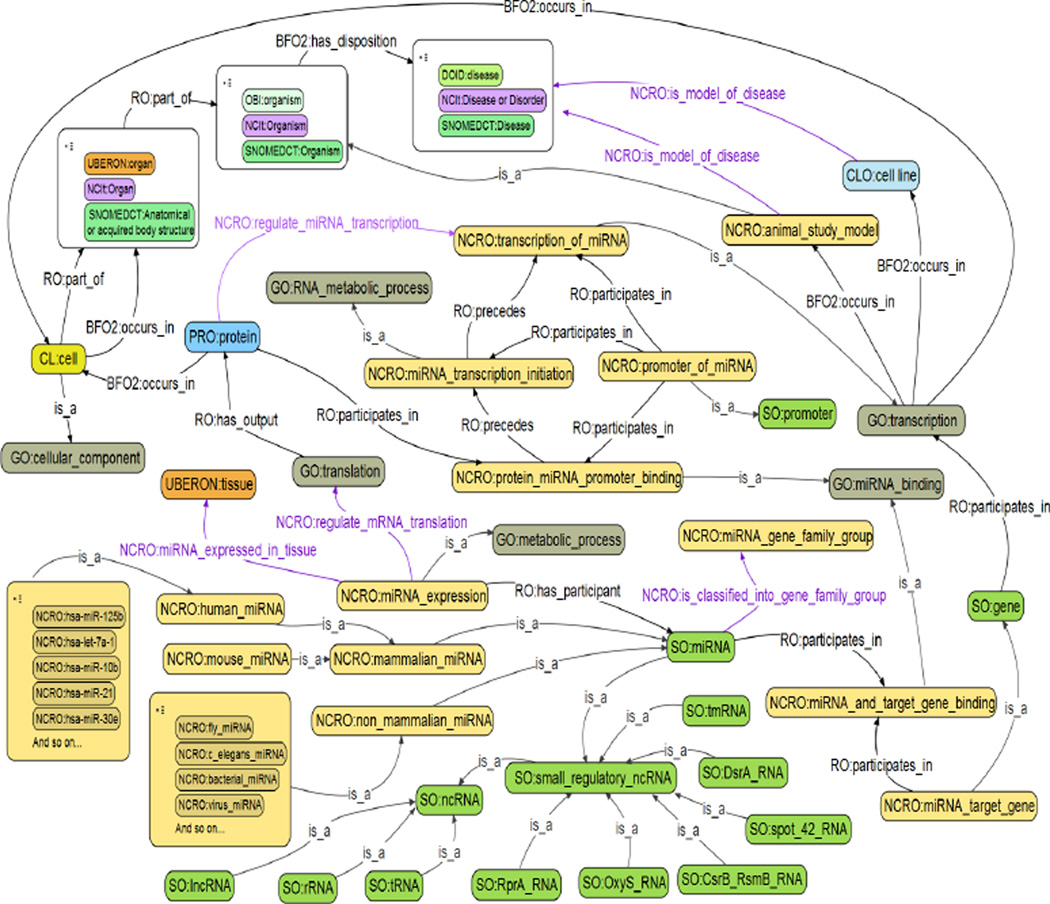
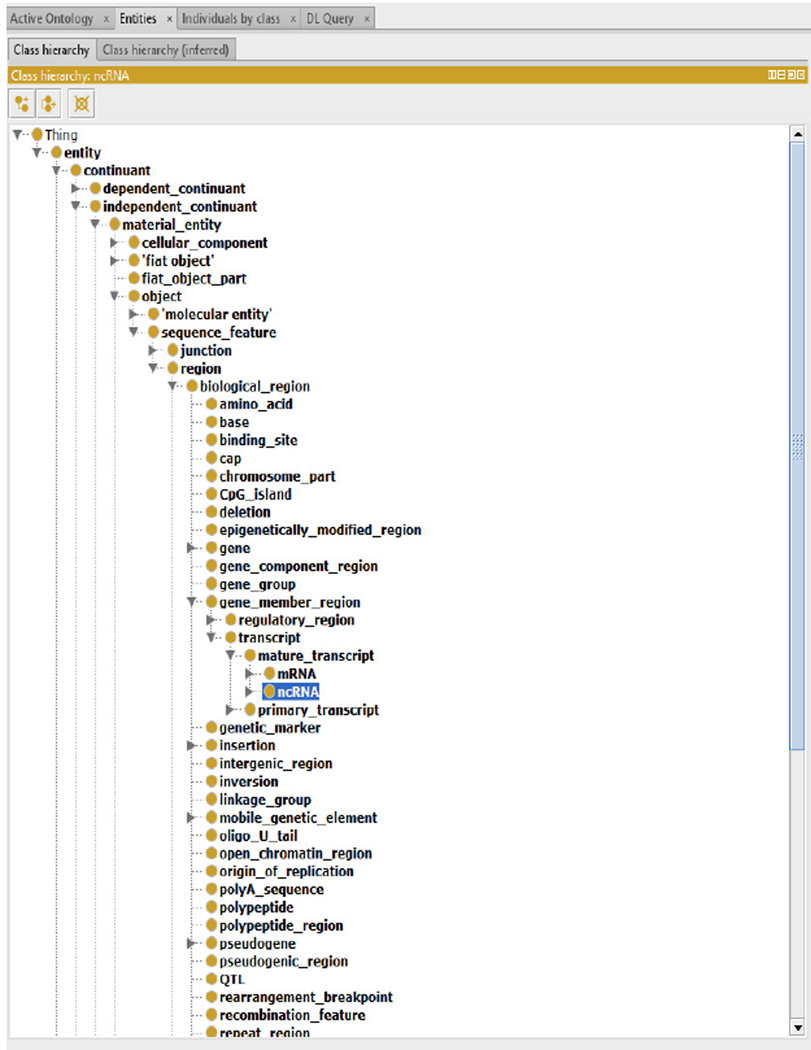
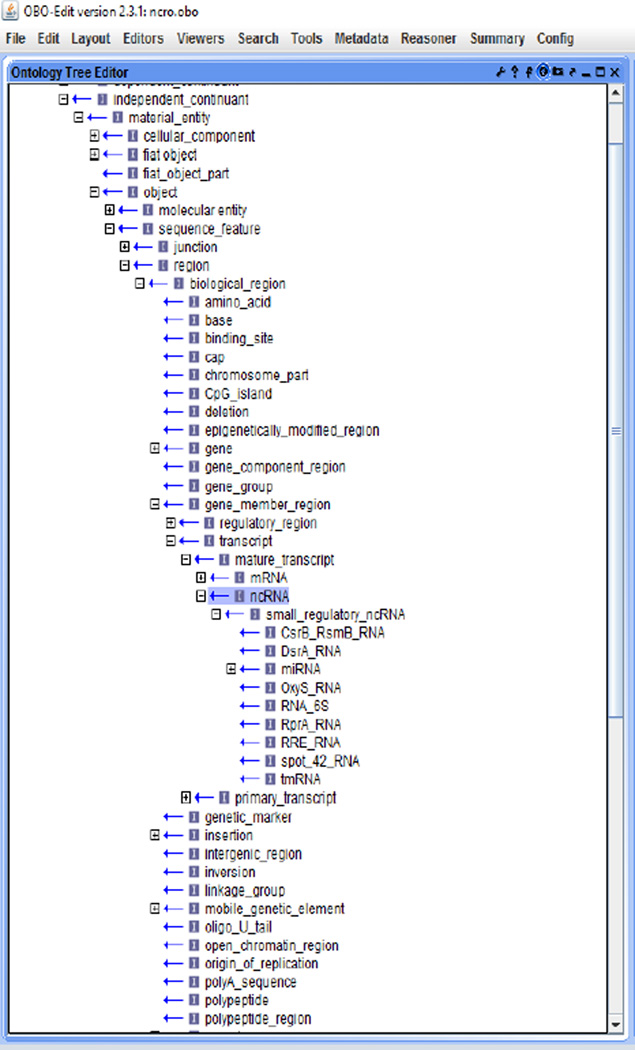
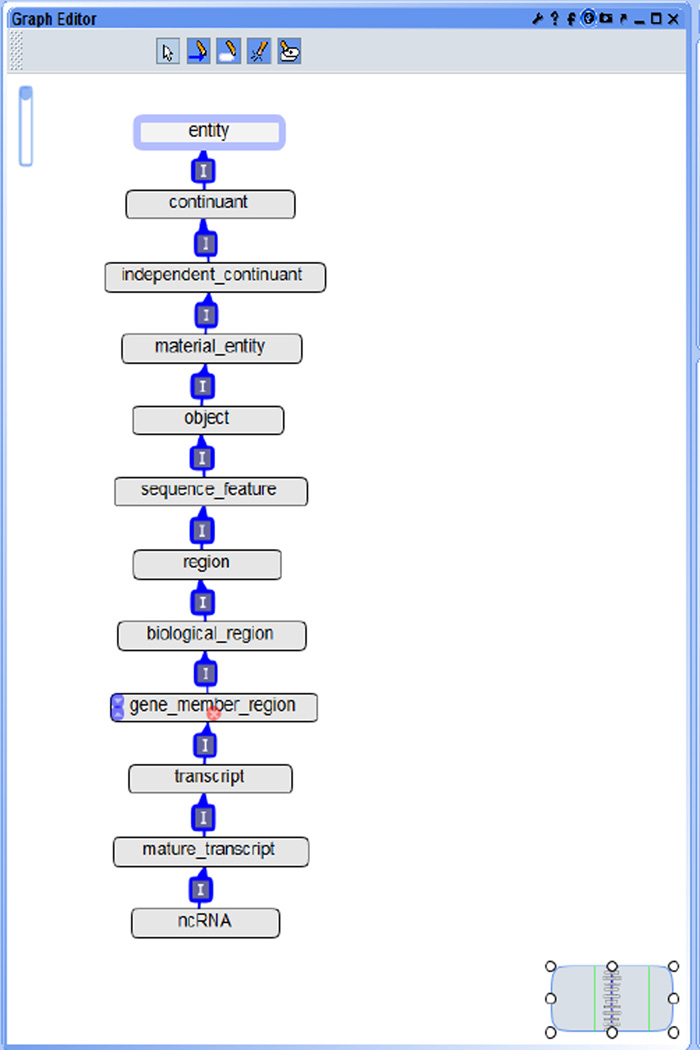
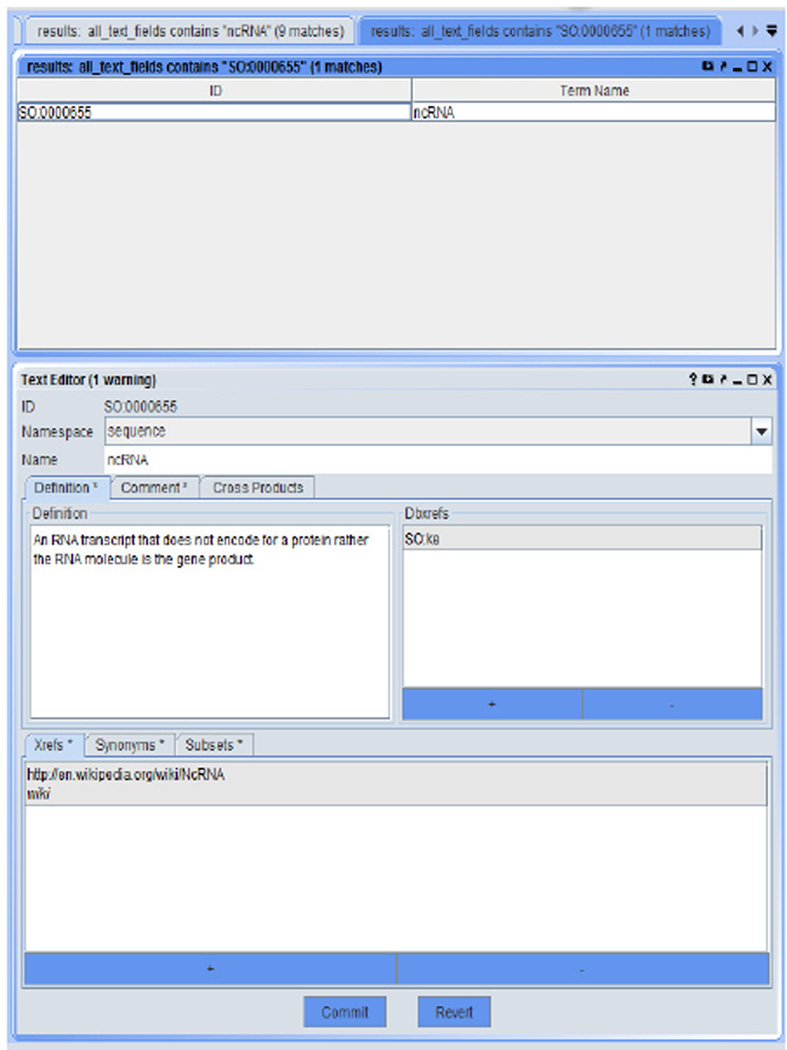
The current version contains a total of 3,060 terms and 45 relations (besides is_a). Figure 2 shows a complete view of the core portion designed in the NCRO, where we use the format of ‘PREFIX:label’ to describe each term or relation. Most of terms were curated with human-readable explanation (Table 2 presents some examples). Figure 3 is a screenshot from the Protégé graphic user interface (GUI), and Figures 4, 5, and 6 are screenshots from the OBO-Edit GUI.
Figure 2.
Core terms and relations in the NCRO ontology (see online version for colours)
Table 2.
Example terms in the NCRO
| Term | Human-readable explanation |
|---|---|
| lncRNA | An non-protein coding transcript (longer than 200 nt). |
| miRNA | A small (22nt) RNA molecule that is the endogenous transcript of a miRNA gene. |
| riboswitch | Part of mRNA, acting as a direct sensor of small molecules to control their own expression. |
| ribozyme | An RNA molecule with catalytic activity. |
| rRNA | Part of ribosome, providing structural scaffolding and catalytic activity. |
| snRNA | A small nuclear RNA that is involved in pre-mRNA splicing and processing. |
| sRNA | A small ncRNA molecule of 50–250 nt. |
| IRES | A sequence element that recruits a ribosomal subunit to internal mRNA for translation initiation. |
| mammalian_miRNA | All miRNAs found in mammalians. |
| human_miRNA | All miRNAs found in Homo sapiens. |
| miRNA_and_target_gene_binding | Binding between miRNA and its targets. |
| protein_miRNA_promoter_binding | Binding between protein and miRNA promoter. |
| miRNA_expression | The expression profile of a miRNA. |
| miRNA_transcription_initiation | The initiation of miRNA transcription process. |
| transcription_of_miRNA | The transcription process of a miRNA. |
| promoter_of_miRNA | The promoter to start the miRNA transcription. |
Figure 3.
The NCRO ontology in the Protégé GUI (see online version for colours)
Figure 4.
The NCRO ontology in the OBO-Edit GUI (1) (see online version for colours)
Figure 5.
The NCRO ontology in the OBO-Edit GUI (2) (see online version for colours)
Figure 6.
The NCRO ontology in the OBO-Edit GUI (3) (see online version for colours)
Note that 83.10% of all terms were defined in the NCRO, and the rest were imported from various extant ontologies. The statistics of all terms is exhibited in Table 3. As for relations, 48.90% were imported from RO, and the rest (51.03%) were defined in the NCRO.
Table 3.
Statistics of terms in the NCRO
| Source Ontology | Percentage (%) |
|---|---|
| Non-Coding RNA Ontology (NCRO) | 83.10 |
| Basic Formal Ontology (BFO) | 1.27 |
| Gene Ontology (GO) | 8.63 |
| Sequence Ontology (SO) | 6.55 |
| PRotein Ontology (PRO) | 0.07 |
| Uberon Multi-Species Anatomy Ontology (UBERON) | 0.07 |
| Chemical Entities of Biological Interest (CHEBI) | 0.07 |
| Cell Ontology (CL) | 0.03 |
| Human Disease Ontology (DOID) | 0.03 |
| Ontology for Biomedical Investigations (OBI) | 0.10 |
4.2 Ontology reasoning to facilitate data annotation and semantic query
The NCRO ontology provides a standardised, well-structured, and formally defined set of terms, along with various relations among these terms, thereby to (1) enable more precise description of ncRNA annotations to identify and integrate like annotations; (2) help establish mappings among diverse sources through cross-references defined in the ontology; and (3) provide necessary software substrates for automated ontology reasoning.
The logical reasoning enabled by the NCRO is able to significantly enhance the query capacity. In other words, more complicated search queries are now enabled. Conventionally, term features and relations among terms need to be hard-coded in software applications in order for them to make logical inferences or connect pieces of data with each other to discover hidden clues that are not explicitly contained in original data. When new terms and relations are added, or when modifications are applied to existing terms and relations, all these details have to be integrated into software applications by revising respective source code. Such a requirement leads to inefficient software development and maintenance. The NCRO can mitigate this challenge because the knowledge about terms and relations is now contained in the ontology rather than in software applications.
5 Conclusions
Important roles are performed by ncRNAs in various molecular functions and different biological and pathological processes; therefore, interest in ncRNA biology has grown throughout biomedicine, biomedical informatics, and clinical sciences. However, the annotation and integration of ncRNA data significantly lag behind their identification because there were no standardised ncRNA nomenclatures. Following this observation, we developed the NCRO ontology, which aims to provide a systematically structured, formally defined ncRNA controlled vocabulary.
The NCRO development is an on-going research effort, and we will continue our investigation along this line of research. All ontology files and design documentations are publicly available on a designated project website (see http://omnisearch.soc.southalabama.edu/ontologyfile.php) and the GitHub project site (see https://github.com/omnisearch/ncro), as well as in both the OBO Library (see http://www.obofoundry.org/cgibin/detail.cgi?id=ncro) and the NCBO BioPortal (see http://bioportal.bioontology.org/ontologies/ncro).
Acknowledgments
Funding for J. Huang was provided in part by the National Cancer Institute (NCI) at the National Institutes of Health (NIH), under the Award Number U01CA180982. Funding for G.M. Borchert was provided in part by Natural Science Foundation (NSF) CAREER grant 1350064 (GMB) awarded by Division of Molecular and Cellular Biosciences (with co-funding provided by the NSF EPSCoR program) and in part by the Abraham A. Mitchell Cancer Research Fund. The views contained in this paper are solely the responsibility of the authors and do not represent the official views, either expressed or implied, of the NIH, NSF, the US Government, or the Abraham A. Mitchell Cancer Research Fund.
Biographies
Jingshan Huang is an Associate Professor in the Biomedical Informatics Group housed in the School of Computing at the University of South Alabama. His research areas are bioinformatics and biomedical informatics; data semantics and web data; and artificial intelligence and big data, with a unifying theme of the semantics and analysis of data as well as their innovative applications on human genomics and transcriptomics. The ultimate research goal of Huang Group is to investigate effective computational methods to help fully integrate large-scale genomics into the clinic, thereby to understand the genetic basis of disease and discover relevant genomic hallmarks, and eventually, to help people live longer, healthier lives. His research has been supported by NIH, NSF, and DOE.
Karen Eilbeck is an Associate Professor in the Department of Biomedical Informatics at the University of Utah. Her expertise is in understanding and accessing biological data to understand diseases better. Her research involves the annotation of biological sequence, in particular genome sequence. Her NIH funding has included the development of the Sequence Ontology and methods for clinical reporting of variation. She is also leading the Laboratory Submission Working group for the ClinVar database.
Barry Smith is an academic working especially in the fields of ontology and biomedical informatics. From 1976 to 1994, he held appointments in Sheffield, Manchester, and Liechtenstein, and in 1994 he moved to the University at Buffalo, SUNY, where he is currently Julian Park Distinguished Professor of Philosophy and Adjunct Professor of Biomedical Informatics, Computer Science, and Neurology. He is also one of the founders of the Open Biological and Biomedical Ontologies (OBO) Foundry and the founding Director of the National Center for Ontological Research (NCOR).
Judith A. Blake is Professor at Jackson Laboratory. Her research focuses on functional and comparative genome informatics, working on the development of systems to integrate and analyse genetic, genomic, and phenotypic information. She is one of the principal investigators of the Gene Ontology (GO) Consortium, an international effort to provide controlled structured vocabularies for molecular biology that serve as terminologies, classifications and ontologies to further data integration, analysis and reasoning.
Dejing Dou is an Associate Professor in the Computer and Information Science Department at the University of Oregon and leads the Advanced Integration and Mining (AIM) Lab. His general research areas include ontology, data mining, data integration, information extraction, and biomedical and health informatics. In particular, he focuses on three critical challenges in processing data and knowledge: heterogeneity, reusability, and scalability. Besides his other federal funded projects, he was the PI of NIH Neural ElectroMagnetic Ontologies (NEMO) project and is currently the PI of NIH Semantic Mining of Activity, Social, and Health data (SMASH) project.
Weili Huang is the Principle Investigator at the Miracle Query, Inc. Her research areas include clinical pharmacology, healthcare informatics, regulatory affairs, and pharmaceutics. Before moving to Miracle Query, she was a pharmacologist reviewer for the Center for Drug Evaluation and Research (CDER) at the US Food and Drug Administration (FDA).
Darren A. Natale is the Team Lead - Protein Science in Protein Information Resource, and a Research Assistant Professor in the Department of Biochemistry and Molecular & Cellular Biology in the Georgetown University Medical Center. He leads the NIH PRotein Ontology (PRO) project. In addition, he is also a curator for the UniProt Knowledgebase.
Alan Ruttenberg is the Director of Clinical and Translational Data Exchange in the School of Dental Medicine at the University at Buffalo, SUNY. He is responsible for building systems, technological and social, that enable wider sharing of translationally relevant information. He also trains graduate students in the cross-disciplinary skills required for building ontologies using semantic technologies. He has developed ontologies for a variety of disciplines, including imaging and atlasing, neuroscience, cell biology, information artefacts, immunology, molecular pathways, anatomy, disease, and cell biology.
Jun Huan is a Professor in the Department of Electrical Engineering and Computer Science at the University of Kansas. He directs the Data Science and Computational Life Sciences Laboratory in the University Information and Telecommunication Technology Center. He works on data science, machine learning, data mining, big data, and interdisciplinary topics including bioinformatics. His research is recognised internationally, including being a recipient of the NSF CAREER Award (2009) among other honours.
Michael T. Zimmermann is an Assistant Professor of Biomedical Informatics in the Division of Biomedical Statistics and Informatics, College of Medicine at Mayo Clinic. His research spans network biology of vaccine response, the pharmacogenomics of chemotherapy response, and applications of bioinformatics and structural biology to the interpretation of Variants of Unknown Significance in the context of an Individualised Medicine clinic.
Guoqian Jiang is an Associate Professor and Associate Consultant in the Division of Biomedical Statistics and Informatics, College of Medicine at Mayo Clinic. He conducts research focused mainly on semantic model building and quality auditing of large-scale biomedical terminologies/ ontologies and data standards, collaborative authoring and annotation of biomedical terminologies/ontologies, and clinical phenotyping and their integration with clinical data standards and Semantic Web technologies.
Yu Lin is a consultant ontologist for BD2K-LINCS Data Coordination and Integration Center at the University of Miami. Her research expertise areas are ontology development and ontology engineering.
Bin Wu is the Director of Endocrinology Department at the First Affiliated Hospital of Kunming Medical University. He is also an Associate Professor at Kunming Medical University. His expertise areas are Diabetes, Obesity, and microRNAs.
Harrison J. Strachan is a student in the School of Computing at the University of South Alabama, advised by Dr. Jingshan Huang.
Nisansa de Silva is a PhD student in the Computer and Information Science Department at the University of Oregon, advised by Dr. Dejing Dou and co-advised by Dr. Jingshan Huang.
Mohan Vamsi Kasukurthi is a student in the School of Computing at the University of South Alabama, advised by Dr. Jingshan Huang.
Vikash Kumar Jha is a student in the School of Computing at the University of South Alabama, advised by Dr. Jingshan Huang.
Yongqun He is an Associate Professor of Lab Animal Medicine, Microbiology, Immunology, and Bioinformatics at the University of Michigan, Ann Arbor. His Lab includes both dry-lab (bioinformatics lab) and wet-lab (microbiology and immunology lab). The former covers four different topics: Ontology Development, Ontology Tool Development, Literature Mining, and Bayesian Network Modelling. The latter focuses on the analysis of caspase-2-mediated cell death mechanism and its role in Brucella pathogenesis and immunity.
Shaojie Zhang is an Associate Professor of Computer Science at the University of Central Florida. With general research interest being Computational Biology and Bioinformatics, he is particularly focusing on ncRNA gene discovery, RNA 3D structure analysis, and computational genomics.
Xiaowei Wang is an Assistant Professor of Cancer Biology at the Washington University in St. Louis, where he leads the RNAi research lab. His research is focused on microRNAs. By combining computational data modelling with high-throughput experimental data, his Lab is developing novel computational algorithms to more accurately predict the genes targeted by microRNAs. Furthermore, they are also carrying out large-scale profiling experiments to identify microRNAs that are involved in cancer development.
Zixing Liu is a Research Scientist in Mitchell Cancer Institute at the University of South Alabama. His research focuses on breast diseases and microRNAs.
Glen M. Borchert is an Assistant Professor in the Biology Department (College of Arts and Sciences), as well as an Assistant Professor in the Pharmacology Department (College of Medicine), at the University of South Alabama. His main research areas are microRNAs, Transposable Elements, and Genomics. He was a recipient of the NSF CAREER Award (2014).
Ming Tan is the Chief of Cell Death and Metabolism Research Center, Vincent F. Kilborn, Jr. Cancer Research Scholar, and an Associate Professor of Oncologic Sciences in Mitchell Cancer Institute at the University of South Alabama. His research areas are metabolism and cancer, microRNA and cancer, cancer metastasis, therapeutic resistance, and targeted cancer therapy. His research is supported by NIH and Vincent F. Kilborn, Jr. Cancer Research foundation. He serves on several editorial boards for scientific journals, including PLOS ONE and Scientific Reports (Nature Publishing Group).
Footnotes
This paper is a revised and expanded version of a paper entitled ‘A domain ontology for the non-coding RNA field’ presented at the ‘2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM-15)’, Washington D.C., November 2015.
Contributor Information
Jingshan Huang, School of Computing, University of South Alabama, Shelby Hall, Room 1123, 150 Jaguar Drive Mobile, AL 36688, USA, huang@southalabama.edu.
Karen Eilbeck, Department of Biomedical Informatics, University of Utah School of Medicine, Salt Lake City, Utah, USA, keilbeck@genetics.utah.edu.
Barry Smith, University at Buffalo – SUNY, Buffalo, New York 14260, USA, phismith@buffalo.edu.
Judith A. Blake, Jackson Laboratory, Bar Harbor, Connecticut 04609, USA, Judith.Blake@jax.org
Dejing Dou, Computer and Information Science Department, University of Oregon, Eugene, Oregon 97403, USA, dou@cs.uoregon.edu.
Weili Huang, Miracle Query, Inc., Eugene, Oregon 97405, USA, weili.huang@miraclequery.com.
Darren A. Natale, Georgetown University Medical Center, Washington DC 20007, USA, dan5@georgetown.edu
Alan Ruttenberg, University at Buffalo – SUNY, Buffalo, New York 14260, USA, alanruttenberg@gmail.com.
Jun Huan, Department of Electrical Engineering and Computer Science, University of Kansas, Lawrence, Kansas 66045, USA, jhuan@ittc.ku.edu.
Michael T. Zimmermann, Division of Biomedical Statistics and Informatics, College of Medicine at Mayo Clinic, Rochester, Minnesota 55905, USA, Zimmermann.Michael@mayo.edu
Guoqian Jiang, Division of Biomedical Statistics and Informatics, College of Medicine at Mayo Clinic, Rochester, Minnesota 55905, USA, Jiang.Guoqian@mayo.edu.
Yu Lin, Data Coordination and Integration Center, University of Miami, Miami, Florida 33146, USA, linikujp@gmail.com.
Bin Wu, Endocrinology Department, Kunming Medical University, Kunming, Yunnan, 650032 China, wu.bin.kmu@qq.com.
Harrison J. Strachan, School of Computing, University of South Alabama, Mobile, Alabama 36688, USA, hjs1221@jagmail.southalabama.edu
Nisansa de Silva, Computer and Information Science, University of Oregon, Eugene, Oregon 97403, USA, nisansa@cs.uoregon.edu.
Mohan Vamsi Kasukurthi, School of Computing, University of South Alabama, Mobile, Alabama 36688, USA, mk1530@jagmail.southalabama.edu.
Vikash Kumar Jha, School of Computing, University of South Alabama, Mobile, Alabama 36688, USA, vj1521@jagmail.southalabama.edu.
Yongqun He, Lab Animal Medicine, Microbiology, Immunology and Bioinformatics, University of Michigan, Ann Arbor, Michigan 48109, USA, yongqunh@med.umich.edu.
Shaojie Zhang, Department of Computer Science, University of Central Florida, Orlando, Florida 32816, USA, shzhang@cs.ucf.edu.
Xiaowei Wang, Cancer Biology, Washington University in St. Louis, St. Louis, Missouri 63130, USA, xwang@radonc.wustl.edu.
Zixing Liu, Mitchell Cancer Institute, University of South Alabama, Mobile, Alabama 36604, USA, zixingliu@health.southalabama.edu.
Glen M. Borchert, Department of Biology, University of South Alabama, Mobile, Alabama 36688, USA, borchert@southalabama.edu
Ming Tan, Mitchell Cancer Institute, University of South Alabama, Mobile, Alabama 36604, USA, mtan@health.southalabama.edu.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babar IA, Cheng CJ, Booth CJ, Liang X, Weidhaas JB, Saltzman WM, Slack FJ. Nanoparticle-based therapy in an in vivo microrna-155 (mir-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci USA. 2012;109(26):E1695–E1704. doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard J. Ontologies: formalising biological knowledge for bioinformatics. Bioessays. 2003;25(5):501–506. doi: 10.1002/bies.10260. [DOI] [PubMed] [Google Scholar]
- Blake JA. Bio-ontologies-fast and furious. Nat Biotechnol. 2004;22(6):773–774. doi: 10.1038/nbt0604-773. [DOI] [PubMed] [Google Scholar]
- Blake JA, Bult CJ. Beyond the data deluge: data integration and bio-ontologies. J. Biomed Inform. 2006;39(3):314–320. doi: 10.1016/j.jbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Cheng CJ, Bahal R, Babar IA, Pincus Z, Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM, Saltzman WM, Slack FJ. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518(7537):107–110. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daige CL, Wiggins JF, Priddy L, Nelligan-Davis T, Zhao J, Brown D. Systemic delivery of a mir34a mimic as a potential therapeutic for liver cancer. Mol Cancer Ther. 2014;13(10):2352–2360. doi: 10.1158/1535-7163.MCT-14-0209. [DOI] [PubMed] [Google Scholar]
- De Coronado S, Wright LW, Fragoso G, Haber MW, Hahn-Dantona EA, Hartel FW, Quan SL, Safran T, Thomas N, Whiteman L. The NCI Thesaurus quality assurance life cycle. J Biomed Inform. 2009;42(3):530–539. doi: 10.1016/j.jbi.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Eilbeck K, Lewis SE, Mungall CJ, Yandell M, Stein L, Durbin R, Ashburner M. The sequence ontology: a tool for the unification of genome annotations. Genome Biol. 2005 Apr;6(5) doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatima R, Akhade VS, Pal D, Rao SM. Long noncoding RNAs in development and cancer: potential biomarkers and therapeutic targets. Mol Cell Ther. 2015 Jun;3 doi: 10.1186/s40591-015-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehndorf R, Batchelor C, Bittner T, Dumontier M, Eilbeck K, Knight R, Mungall CJ, Richardson JS, Stombaugh J, Westhof E, Zirbel CL, Leontis NB. The RNA Ontology (RNAO): an ontology for integrating RNA sequence and structure data. Applied Ontology. 2011;6(1):53–89. [Google Scholar]
- Huang J, Dang J, Borchert GM, Eilbeck K, Zhang H, Xiong M, Jiang W, Wu H, Blake JA, Natale DA, Tan M. OMIT: dynamic, semi-automated ontology development for the microRNA domain. PLOS ONE. 2014 Jul;9(7) doi: 10.1371/journal.pone.0100855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Dou D, Dang J, Pardue JH, Qin X, Huan J, Gerthoffer WT, Tan M. Knowledge acquisition, semantic text mining, and security risks in health and biomedical informatics. World J Biol Chem. 2012;3(2):27–33. doi: 10.4331/wjbc.v3.i2.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Dou D, He L, Dang J, Hayes PJ. Ontology-based knowledge discovery and sharing in bioinformatics and medical informatics: a brief survey; Proc. 7th International Conference on Fuzzy Systems and Knowledge Discovery, FSKD-2010; 2010. Aug, [Google Scholar]
- Huang J, Gutierrez F, Dou D, Blake JA, Eilbeck K, Natale DA, Smith B, Lin Y, Wang X, Liu Z, Tan M, Ruttenberg A. A semantic approach for knowledge capture of microRNA-target gene interactions. Proc. BHI Workshop at 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM-2015) 2015 Nov;7(25) [Google Scholar]
- Huang J, Gutierrez F, Strachan HJ, Dou D, Huang W, Smith B, Blake JA, Eilbeck K, Natale DA, Lin Y, Wu B, de Silva N, Wang X, Liu Z, Borchert GM, Tan M, Ruttenberg A. OmniSearch: a semantic search system based on the Ontology for MIcroRNA Target (OMIT) for microRNA-target gene interaction data. J Biomed Semantics. 2016 May;7(25) doi: 10.1186/s13326-016-0064-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Townsend C, Dou D, Liu H, Tan M. OMIT: a domain-specific knowledge base for MicroRNA target prediction. Pharm Res. 2011;28(12):3101–3104. doi: 10.1007/s11095-011-0573-8. [DOI] [PubMed] [Google Scholar]
- Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2(11):986–991. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays. 2003;25(10):930–939. doi: 10.1002/bies.10332. [DOI] [PubMed] [Google Scholar]
- Natale DA, Arighi CN, Barker WC, Blake JA, Bult CJ, Caudy M, Drabkin HJ, D’Eustachio P, Evsikov AV, Huang H, Nchoutmboube J, Roberts NV, Smith B, Zhang J, Wu C. The protein ontology: a structured representation of protein forms and complexes. Nucleic Acids Res. 2011;39:D539–D545. doi: 10.1093/nar/gkq907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, Leontis N, Rocca-Serra P, Ruttenberg A, Sansone SA, Scheuermann RH, Shah N, Whetzel PL, Lewis S. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25(11):1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]