Torsins are essential, disease-relevant ATPases, but their function is unknown. Monitoring of nuclear envelope morphology after deletion of multiple Torsins or their cofactors reveals a robust inner nuclear membrane–blebbing phenotype in HeLa cells. Nucleoporins and ubiquitin are defining molecular components of these omega-shaped blebs.
Abstract
The human genome encodes four Torsin ATPases, the functions of which are poorly understood. In this study, we use CRISPR/Cas9 engineering to delete all four Torsin ATPases individually and in combination. Using nuclear envelope (NE) blebbing as a phenotypic measure, we establish a direct correlation between the number of inactivated Torsin alleles and the occurrence of omega-shaped herniations within the lumen of the NE. A similar, although not identical, redundancy is observed for LAP1 and LULL1, which serve as regulatory cofactors for a subset of Torsin ATPases. Unexpectedly, deletion of Tor2A in a TorA/B/3A-deficient background results in a stark increase of bleb formation, even though Tor2A does not respond to LAP1/LULL1 stimulation. The robustness of the observed phenotype in Torsin-deficient cells enables a structural analysis via electron microscopy tomography and a compositional analysis via immunogold labeling. Ubiquitin and nucleoporins were identified as distinctively localizing components of the omega-shaped bleb structure. These findings suggest a functional link between the Torsin/cofactor system and NE/nuclear pore complex biogenesis or homeostasis and establish a Torsin-deficient cell line as a valuable experimental platform with which to decipher Torsin function.
INTRODUCTION
Torsins belong to the superfamily of ATPases associated with a variety of cellular activities (AAA+) proteins, which typically use the energy from ATP hydrolysis to perform mechanical work on substrate proteins (Hanson and Whiteheart, 2005). Four Torsins are encoded in the human genome: TorsinA (TorA), TorsinB (TorB), Torsin2A (Tor2A), and Torsin3A (Tor3A; Rose et al., 2015; Laudermilch and Schlieker, 2016). Torsins are the only members of the AAA+ family that reside in the lumen of the endoplasmic reticulum (ER) and contiguous perinuclear space (PNS; Goodchild and Dauer, 2004, 2005; Vander Heyden et al., 2009). TorA and TorB are membrane associated through a short N-terminal hydrophobic sequence, whereas Tor3A and Tor2A are soluble (Rose et al., 2015). The most-studied member is TorA because an in-frame deletion of one of two consecutive glutamate residues near the C-terminus of TorA (TorA ∆E) causes the autosomal-dominant, severe movement disorder DYT1 dystonia (Ozelius et al., 1997). Although the precise functions of Torsins remain undefined, numerous lines of evidence point to a functional role for TorA at the nuclear envelope (NE; Goodchild et al., 2005; Nery et al., 2008; Vander Heyden et al., 2009; Jokhi et al., 2013; Liang et al., 2014; Weisheit and Dauer, 2015).
The hallmark phenotype observed upon TorA deletion or manipulation is the “blebbing” or herniation of the inner nuclear membrane (INM) into the PNS (Goodchild et al., 2005; Jokhi et al., 2013; Liang et al., 2014; Rose et al., 2014; Weisheit and Dauer, 2015). This phenotype was first reported in a knockout (KO) mouse model of TorA (Goodchild et al., 2005), although blebbing was restricted to neural tissue, in which TorA is highly expressed relative to other Torsins (Jungwirth et al., 2010). More recently, several mouse models bearing TorA KO or point mutations in neural tissues displayed dystonic symptoms and NE blebbing (Liang et al., 2014; Pappas et al., 2015; Weisheit and Dauer, 2015). When apparently normal TorA KO murine embryonic fibroblasts (MEFs) were treated with short hairpin RNA against TorB, NE blebbing was observed in these cells, suggesting functional redundancy between the two Torsins (Kim et al., 2010). Consistent with this idea, NE blebbing in TorA KO mice is confined to a specific neurodevelopmental window, and the disappearance of blebs correlates with increased expression of TorB (Tanabe et al., 2016). Similar NE abnormalities have also been reported in Drosophila melanogaster (Jokhi et al., 2013) and Caenorhabditis elegans (VanGompel et al., 2015) upon manipulation of the respective Torsin variants in these organisms, suggesting that Torsin function at the NE is conserved.
The lamina-associated polypeptide 1 (LAP1) and luminal domain–like LAP1 (LULL1) are cofactors that activate the ATPase activity of either TorA or TorB (Zhao et al., 2013). Only LULL1 can additionally activate Tor3A, whereas neither cofactor can activate Tor2A’s putative ATPase activity (Zhao et al., 2013). LAP1 and LULL1 are type II integral membrane proteins; LAP1 binds to the nuclear lamina and resides in the INM (Foisner and Gerace, 1993), whereas LULL1 localizes throughout the ER (Goodchild and Dauer, 2005; Goodchild et al., 2015; Vander Heyden et al., 2009). Both proteins activate Torsin by adopting AAA-like folds in their luminal domains and forming a closely apposed interface with Torsin to complement the Torsin active site (Brown et al., 2014; Sosa et al., 2014; Demircioglu et al., 2016).
We postulated that LAP1 or LULL1 can form a composite, membrane-spanning machine with Torsin in either the NE or ER, respectively, the stoichiometry of which has yet to be established (Brown et al., 2014; Rose et al., 2015; Laudermilch and Schlieker, 2016), but it is unknown whether LAP1 and LULL1 participate with Torsin in completely independent pathways or share some overlap in their functions. NE blebbing was observed in LAP1 KO mice in several tissues, and the blebs were similar to those observed in TorA KO mice (Kim et al., 2010). No NE blebbing was observed in wild-type MEFs upon LULL1 knockdown (Kim et al., 2010), and a LULL1 KO in HeLa cells reduces herpes simplex virus-1 growth without overtly affecting NE membrane dynamics (Turner et al., 2015).
Because Torsin expression varies widely among tissues in mice (Goodchild et al., 2005; Jungwirth et al., 2010) and Torsins are subject to distinct regulation by ATPase-activating cofactors (Zhao et al., 2013), it is unclear to what degree the four members of Torsin family overlap in their function. Another key question relates to the composition of the NE-resident blebs seen upon Torsin or LAP1 manipulation. We previously noted that there exists a distinct electron density at the bleb neck, the identity of which is likely key toward deciphering Torsin function (Laudermilch and Schlieker, 2016).
In this study, we use the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system (Ran et al., 2013) to engineer HeLa cell lines of individual or combined KOs of Torsins or LAP1 and LULL1 and demonstrate a functional redundancy between all four Torsins and synthetic genetics effects between the cofactors. The robustness of the blebbing phenotype enabled us to conduct a structural and compositional analysis of the blebs, revealing nucleoporins (nups) and ubiquitin (Ub) as constituents of the neck and lumen of the blebs, respectively. These results establish an altogether Torsin-deficient cell line as a robust experimental platform with which to study Torsin function.
RESULTS AND DISCUSSION
Generation of Torsin- and cofactor-knockout cell lines
To systematically address whether functional redundancy exists on the level of Torsins and cofactors, we created genetic deletions of Torsins or their cofactors alone or in combination in HeLa cells (Figure 1). Several of these have been reported (Turner et al., 2015), including individual and double KOs of LAP1B, LULL1, TorA, and TorB. It has recently been reported that there is a second LAP1 isoform in human cells, designated LAP1C, which is generated by an alternative transcription start site and ultimately results in a truncation of the N-terminus of the canonical LAP1B isoform (Santos et al., 2014; Figure 1A). We designed a CRISPR guide RNA to target only the LAP1B isoform and generated a specific KO of this isoform (Figure 1B; Turner et al., 2015). Attempts to delete both LAP1 isoforms with the CRISPR/Cas9 system by targeting the guide RNA to regions shared by LAP1B and LAP1C failed, presumably because LAP1 is essential for mitosis (Neumann et al., 2010). However, we were able to achieve efficient knockdown of LAP1C in LAP1B/LULL1 KO cells using small interfering RNAs (siRNAs) targeting both LAP1 isoforms, as judged by a significant reduction of the LAP1C level in immunoblots (Figure 1B). This finding suggests that a more careful interpretation of LAP1 data based on knockdowns or KOs is warranted, since the desired perturbation might not always affect both isoforms.
FIGURE 1:
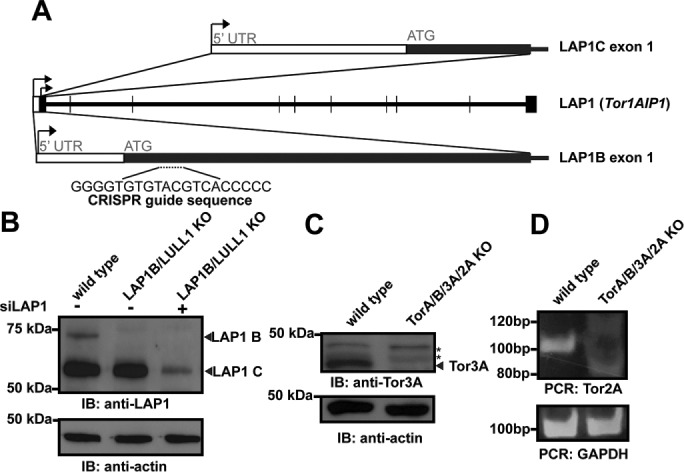
Deletion of Torsins and cofactors from HeLa cells. (A) Diagram of the Tor1AIP1 locus. According to Santos et al. (2014), the first LAP1 exon is different between LAP1B and LAP1C, depending on the transcription start site (arrows), whereas the remaining exons are shared between the two isoforms. The CRISPR guide used to target LAP1B is shown. (B) Immunoblot using an anti-LAP1 antibody in wild-type, LAP1B KO, or siLAP1-treated cells. Loading control: actin. (C) Immunoblot using an anti-Tor3A antibody in wild-type or TorA/B/3A KO cells. Loading control: actin. Asterisks: nonspecific background bands. (D) PCR of the Tor2A gene with primers flanking the CRISPR guide site in wild-type or TorA/B/3A/2A KO cells. Loading control: GAPDH.
We also generated cell lines containing a triple KO of TorA/B/3A and a quadruple KO of TorA/B/3A/2A (Figure 1, C and D). We validated these KOs with an anti-Tor3A antibody (Figure 1C). Lacking suitable Tor2A antibodies, we used PCR-based genotyping to validate the presence of insertion/deletion mutations in the Tor2A gene (Figure 1D).
Deleting multiple Torsins or cofactors results in NE blebbing
To determine whether multiple deletions of Torsins have synthetic effects on NE architecture, we conducted a comparative electron microscopic (EM) ultrastructural analysis of the cell lines we generated (Figure 2). Individual deletion of Torsins, LAP1B, or LULL1 led to minimal perturbations of the NE (Figure 2D; unpublished data). The blebbing phenotype was significantly increased in a double KO of LAP1B and LULL1, with an average of 1.0 bleb/30 μm of NE (Figure 2, B and D). We found a moderate but not statistically significant increase in blebbing upon siRNA treatment against LAP1C in LAP1B/LULL1 double KO cells (Figures 1B and 2D). It is likely that this mild synthetic genetic effect would be more pronounced if the LAP1C knockdown efficacy were higher (note that the LAP1C levels are reduced to ∼20% of wild type; Figure 1B). In any event, the observation that LAP1 and LULL1 have a synthetic effect in NE blebbing is surprising, since these proteins have distinct localization patterns (Goodchild and Dauer, 2005; Goodchild et al., 2015; Vander Heyden et al., 2009). Because the extraluminal domain of LULL1 is within the size limit to passively diffuse into the INM (Boni et al., 2015; Ungricht et al., 2015), it is possible that LULL1 can access the INM but would not be retained there without binding to the nuclear lamins. Alternatively, it has been reported that LULL1 is needed to route Torsin to the NE (Vander Heyden et al., 2009; Goodchild et al., 2015), whereas LAP1 would function with Torsin at the NE. Another speculative possibility is that LAP1 and LULL1 set up an intraluminal, antiparallel gradient responsible for spatiotemporal control of ATPase activity and that any additive perturbation of this gradient would be increasingly detrimental for Torsin localization and function.
FIGURE 2:
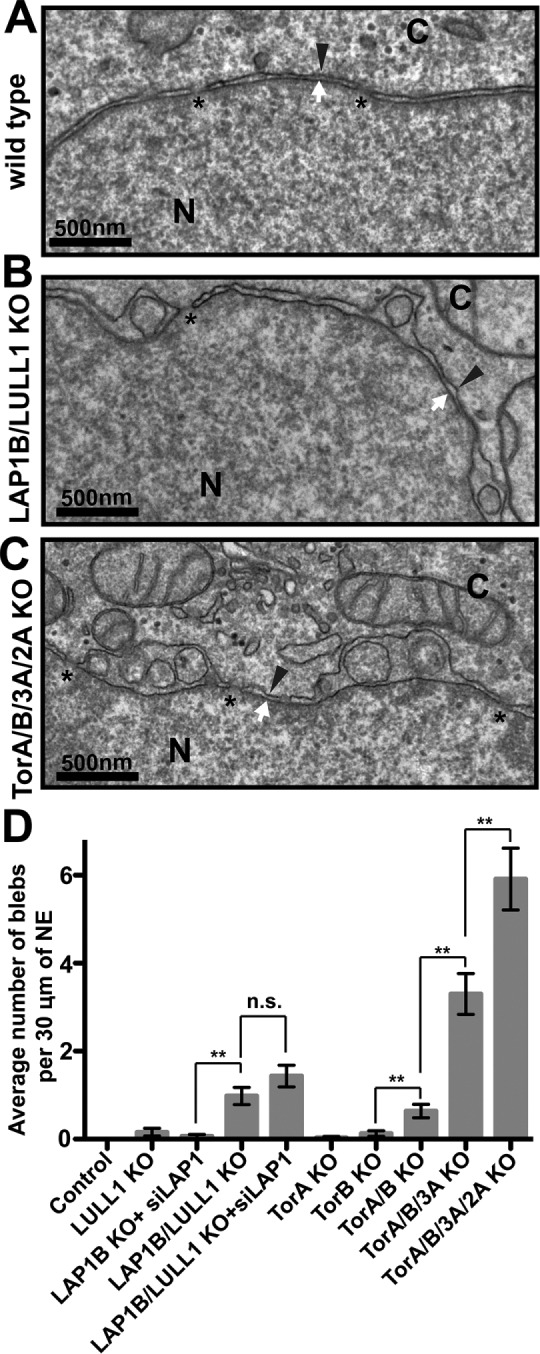
Torsin or cofactor deletion results in blebbing of the NE. (A–C) EM of (A) a wild-type HeLa cell, (B) a LAP1B/LULL1 KO cell, and (C) a TorA/B/3A/2A KO cell. Asterisk, nuclear pore; black arrowhead, ONM; white arrow, INM. N, nucleus; C, cytoplasm. (D) Quantification of the average number of blebs per 30 μm of NE in different genetic backgrounds. Thirty EM sections per cell line were counted. Error bars, SEM. ns, not significant. **p < 0.05.
We found a significant increase in the penetrance of the blebbing phenotype in a double KO of TorA and TorB, but this did not match the effect of knocking out both the LAP1 and LULL1 cofactors (Figure 2D), as there was an average of 0.6 bleb/30 μm of NE in TorA/B KO cells. Because Tor3A’s ATPase activity is activated by LULL1 (Zhao et al., 2013), we engineered a cell line harboring a triple KO of TorA, TorB, and Tor3A. We found a significant increase in the penetrance of the phenotype in this mutant, as >95% of EM sections exhibited blebs with an average of 3.3 blebs/30 μm of NE. Finally, we engineered a cell line containing a KO of all four Torsins and found the most severe blebbing phenotype under these conditions, with nearly every EM section containing at least one bleb and an average of 5.9 blebs/30 μm of NE (Figure 2, C and D). This was unexpected, since both LAP1 and LULL1 can activate the ATPase activity of TorA and TorB, only LULL1 can activate Tor3A, and neither protein can activate Tor2A (Zhao et al., 2013). Together these data suggest that, although a subset of Torsins and LAP1 or LULL1 can form composite machines (Brown et al., 2014; Sosa et al., 2014; Rose et al., 2015; Laudermilch and Schlieker, 2016), Torsins may additionally have cofactor-independent functions. Alternatively, the possibility remains that unidentified Torsin cofactor(s) have not been accounted for in this study.
The striking redundancy between Torsins in a human cell line also helps to explain why it has been extremely challenging to determine Torsins’ precise functions, since our data show that knocking down or deleting a single human Torsin is unlikely to result in an overt cellular phenotype. However, individual Torsins may also operate in independent pathways or have tissue-specific functions, although they clearly share functional overlap in the context of NE integrity despite their distinct regulation and differences in membrane association.
EM tomography reveals uniform morphological features of NE blebs
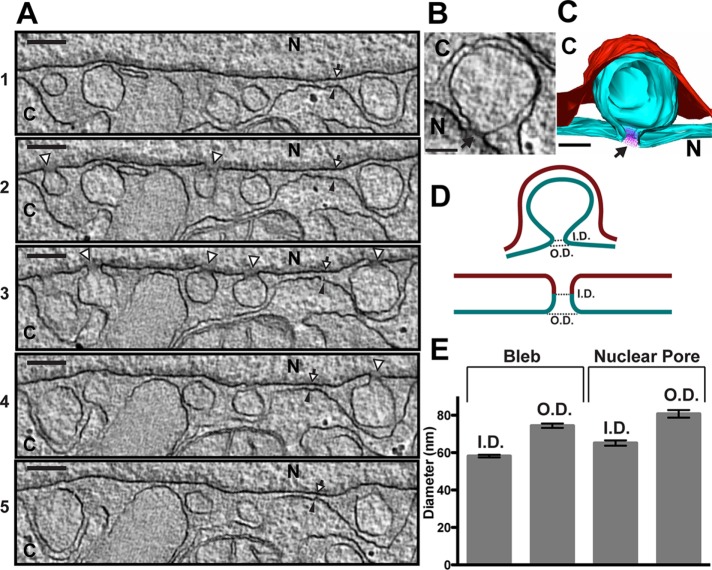
Having established that the NE blebbing phenotype in HeLa cells is most penetrant upon multiple deletions of Torsins or cofactors, we asked whether we could better characterize the structure and molecular composition of the blebs. We used EM tomography to determine whether the blebs observed in TorA/B/3A/2A KO cells represented a mixed population of free and attached vesicles or are all attached to the INM (Figure 3A). At certain sections in the tomogram, the blebs appear to be free vesicles trapped between the INM and outer nuclear membrane (ONM; Figure 3A, 1 and 5, and Supplemental Movie Figure 3.mov). However, collecting three-dimensional information from a tilt series of a 640-nm-thick section (see Figure 3C for a model) allowed us to determine that each bleb is still attached by a small “neck” to the INM. For example, the two blebs in the center of each image in Figure 3A appear to be free in the PNS in 1, 4, and 5, but 3 clearly shows that the both blebs are attached to the INM. This membrane herniation represents an energetically unfavorable state, and proteinaceous components are likely required to stabilize the positive curvature at the neck from the nuclear side or the negative curvature at the luminal side. Indeed, we observe an amorphous electron density specifically at the necks of the blebs from the nuclear side (Figure 3, B and C, arrow). We also note that the curvature and shape of the membrane at the neck are reminiscent of the curvature and shape of nuclear pores (Alber et al., 2007; Knockenhauer and Schwartz, 2016; Kosinski et al., 2016; Lin et al., 2016). We therefore measured the dimensions (Figure 3D) of both blebs and nuclear pores and found the overall dimensions to be very similar (Figure 3E), although the bleb neck (59-nm inner diameter) is slightly smaller than the nuclear pore complex (NPC; 65-nm inner diameter). We previously suggested that Torsin may be involved in mediating NPC assembly or disassembly (Laudermilch and Schlieker, 2016), and a recent report showed that mutation of the C. elegans Torsin variant, OOC-5, resulted in blebbing and mislocalized, extranuclear NPC components (VanGompel et al., 2015). Together these observations raise the key question of whether the blebs observed upon Torsin deletion in HeLa cells contain nups.
FIGURE 3:
EM tomography of TorA/B/3A/2A KO cells. (A) Individual sections from an EM tomogram constructed from a tilt series of a 640-nm-thick section of TorA/B/3A/2A KO cells. The arrowheads with white fill and black outline in 2–4 point to the necks of the blebs. White arrow with black outline, INM; black arrowhead, ONM. Scale bar, 200 nm. (B) Enlarged image of a bleb from a tomogram. Scale bar, 100 nm. (C) Model produced from a tomogram. The INM and bleb are cyan, the ONM is red, and the electron density at the neck is modeled in purple (also black arrow in B). Scale bar, 100 nm. (D) Depiction of the method used to measure the neck of the blebs (left) and nuclear pore (right). The INM is cyan and the ONM is red. ID, inner diameter; OD, outer diameter. (E) Measurements of ID and OD of blebs and nuclear pores in the tomogram from A. Thirty blebs and 15 nuclear pores were measured. Error bars, SEM.
Nucleoporins and ubiquitin localize to distinct regions of NE blebs
Owing to the relatively small size of the blebs, which is at or below the resolution limit of light microscopy, we used immunogold labeling in concert with EM to enable a compositional analysis. Using an antibody (MAb414) that recognizes four phenylalanine-glycine (FG) nups of the NPC central channel (Figure 4A; Davis and Blobel, 1986; Sukegawa and Blobel, 1993), we found that at least a subset of these nups localize specifically to the neck of the blebs in TorA/B/3A/2A KO cells (Figure 4B). Therefore we propose that the electron density seen at the curvature of the bleb necks (Figure 3, B and C) is composed, at least in part, of nups, consistent with the idea that nups form a protocoatomer and stabilize membrane curvature (Devos et al., 2004; Brohawn et al., 2008; Kelley et al., 2015; Stuwe et al., 2015). Although we have been unable to find antibodies for proteins belonging to other nuclear pore subcomplexes that work efficiently in immunogold labeling, it is difficult to imagine that central channel FG nups could be present at the neck without at least some of the scaffolding nups present. It is possible that the blebs result from perturbed NPC formation, either during interphase or mitosis. For example, the blebs may represent either stalled NPC assembly intermediates or defective NPCs. This would be consistent with work in budding yeast, in which endosomal sorting complexes required for transport and the AAA+ ATPase Vps4 act in a quality control pathway that prevents NPC misassembly, perhaps through the removal of defective NPC assembly intermediates (Webster et al., 2014, 2016; Webster and Lusk, 2016). Moreover, the concept that defective NPCs might be covered by membrane seals was suggested based on the presence of similar NE herniations in budding yeast strains lacking Nup116, a central channel nup (Wente and Blobel, 1993). Finally, similar NE herniations have been observed in yeast upon deletion of Apq12, which is believed to participate in NPC biogenesis by influencing membrane fluidity (Scarcelli et al., 2007). Although there is no direct Torsin orthologue in yeast, it is tempting to speculate that Torsin may be an essential metazoan component of analogous quality control–type processes.
FIGURE 4:
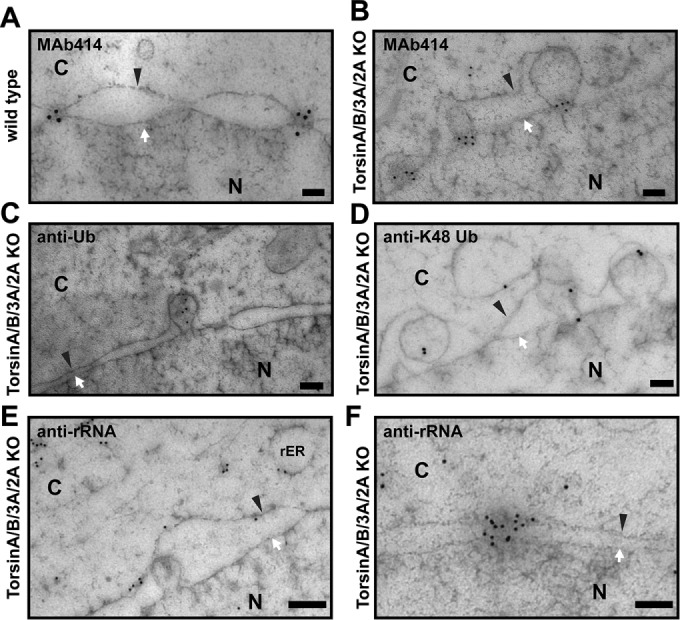
Nucleoporins and ubiquitin localize to distinct structural elements of the blebs. (A, B) EM images of immunogold labeling of (A) wild-type cells and (B) TorA/B/3A/2A KO cells with an anti-FG nup antibody (MAb414). (C, D) EM images of immunogold labeling of TorA/B/3A/2A KO cells with (C) an anti-Ub antibody and (D) an anti–K48 Ub antibody. (E, F) EM images of immunogold labeling of TorA/B/3A/2A KO cells with an anti-rRNA antibody (Y10B). White arrow, INM; black arrowhead, ONM. N, nucleus; C, cytoplasm. Scale bar, 100 nm.
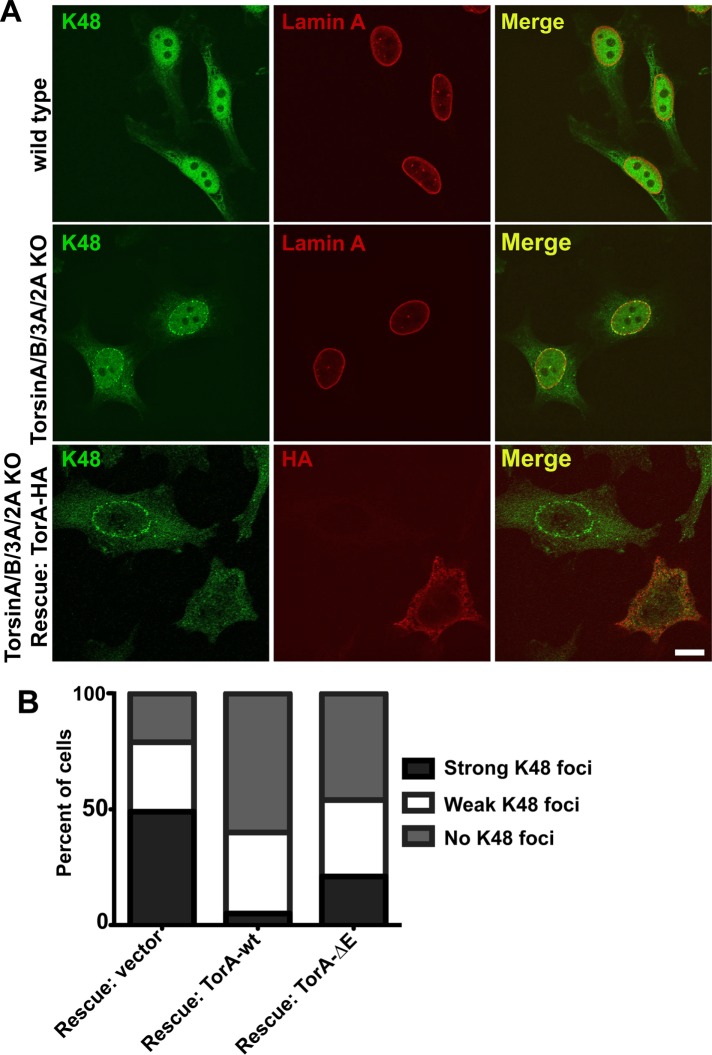
We also found that Ub localizes to the lumen of the blebs in TorA/B/3A/2A KO cells (Figure 4C), consistent with previous evidence (Liang et al., 2014), and that K48-linked Ub localizes to the blebs (Figure 4D). Consistently, we observed the accumulation of K48-Ub foci around the NE in TorA/B/3A/2A KO cells with confocal microscopy (Figure 5A). As expected, this phenotype was potently although not completely rescued by transient transfection of wild-type TorA-HA (Figure 5A, bottom, left cell, and B), whereas the potency of TorA ∆E-HA in rescuing this phenotype was significantly reduced (Figure 5B). Although ubiquitylation has emerged as a regulatory event for various cellular processes, K48-linked Ub is most commonly associated with targeting proteins for degradation by the proteasome (Komander and Rape, 2012). Several previous studies suggested a role for Torsin in the ER-associated degradation pathway (ERAD; Chen et al., 2010; Nery et al., 2011), so it is plausible to suggest a link between the blebs and protein quality control. However, we found no difference in the expression levels of several ER quality control components or the induction of the unfolded protein response (UPR) in TorA/B/3A/2A KO cells relative to WT cells (Supplemental Figure S1). We also determined that ERAD components do not colocalize with K48 foci in TorA/B/3A/2A KO cells (Supplemental Figure S2). Together these results suggest that Torsin participates in a pathway distinct from the canonical ERAD pathway.
FIGURE 5:
K48-linked ubiquitin conjugates accumulate in the nuclear periphery in TorA/B/3A/2A KO cells. (A) Confocal microscopy of wild-type cells (top) or TorA/B/3A/2A KO cells (middle and bottom) stained with an anti–K48 Ub antibody. Top and middle, lamin A was used as a counterstain. Bottom, cells were transfected with a TorA-HA rescue construct and stained with an anti-HA antibody. Scale bar, 10 μm. (B) Percentage of TorA/B/3A/2A KO cells showing K48 Ub foci around the NE with different rescue constructs. At least 100 cells per rescue construct were counted. p < 0.05 for TorA-HA compared with either vector or TorA ∆E-HA and for TorA ∆E-HA compared with vector.
A recent study identified an essential role for Torsin in cellular lipid metabolism and reported an increase in the levels of phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), and phosphatidylinositol (PI) in U2OS cells in a Torsin/LULL1 overexpression-based system (Grillet et al., 2016). Given that NE abnormalities were previously observed in response to lipid perturbation (Schneiter et al., 1996; Surma et al., 2013), we wanted to exclude the formal possibility that the observed structures merely result from perturbations in the total levels of those four lipids. Of note, we did not observe material differences in the levels of PC, PE, PI, and PS in TorA/B/3A/2A KO and LULL1 KO cells compared with wild-type cells (Supplemental Figure S3).
It remains possible that the Ub accumulation is due to general transport through NPCs at the bleb neck. In an effort to distinguish between these possibilities, we used an antibody that recognizes rRNA (Lerner et al., 1981), since ribosomal subunits are frequently trafficked through NPCs. As expected, this antibody efficiently labeled nucleoli (unpublished data), rough ER, and the ONM (Figure 4E), which we attribute to sites of rRNA synthesis in the nucleoli and sites of membrane or luminal protein synthesis on the ER and ONM. However, despite searching extensively, we found no evidence for an accumulation of rRNA in the blebs in TorA/B/3A/2A KO cells (Figure 4E), even though we found rRNA being trafficked through normal NPCs (Figure 4F). Together these data suggest that the NPC elements found in the blebs do not constitute a fully functional NPC. Instead, it appears that the observed Ub accumulation in the bleb lumen is a specific feature of all blebs. Whether such spatially confined ubiquitylated species result indirectly from Torsin dysfunction—for example, resulting from perturbed trafficking or assembly of NPC components—or represents a failure to remove misassembled or unfolded proteins from the NE remains to be established. Another possibility is that the omega-shaped structure is a “frozen” NPC assembly intermediate similar to the recently observed structures occurring during interphase NPC assembly (Otsuka et al., 2016), the completion of which would require the activity of Torsin ATPases.
In conclusion, we established a pronounced redundancy on the level of both Torsin ATPases and their regulatory cofactors. Furthermore, we established that the blebs are continuous with the INM and that nups and Ub localize to defined subregions of the blebs. This cell line and the identification of nups as a defining component of the blebs will enable directed approaches toward defining the precise function of Torsin ATPases and their cofactors in NE/NPC homeostasis.
MATERIALS AND METHODS
Cell culture and generation of knockout cell lines
HeLa cells were purchased from the American Type Culture Collection (Manassas, VA; CCL-2) and were regularly determined to be mycoplasma negative by the absence of extranuclear 4′,6-diamidino-2-phenylindole staining. Cells were cultured in 10% (vol/vol) fetal bovine serum (Sigma-Aldrich, St. Louis, MO) in DMEM. The CRISPR/Cas9 system (Ran et al., 2013) was used to generate knockout HeLa cell lines as described previously (Turner et al., 2015). As guide RNA sequences, we used Tor2a, 5′-CGCGCGTCGCAGCCGCCATC-3′, and Tor3a, 5′-GCGCCACGGACCGCGAAGCA-3′. Genotyping PCR was performed as described previously (Tsai et al., 2016) with the following primers: Tor2a forward, 5′-GAGACCAGCCCGAGCAGCC-3′; Tor2a reverse, 5′-GTTGACTGCCCCACAACGAG-3′; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward, 5′-CGACCGGAGTCAACGGATTTGGTCG-3′; and GAPDH reverse, 5′-GGCAACAATATCCACTTTACCAGA-3′.
Immunoblotting
Immunoblotting was performed as described previously (Rose et al., 2014). The following antibodies were used: rabbit polyclonal anti-LAP1 (Covance custom antiserum; Denver, PA) at 1:5000; rabbit polyclonal anti-Tor3A (Covance custom antiserum) at 1:3000; mouse monoclonal anti–β-actin (AB_306371; Abcam, Cambridge, MA) at 1:2000; anti-p97 (AB_298039, Abcam) at 1:2000; anti-PDI (AB_303304; Abcam) at 1:2000; anti-Bip (AB_303300; Abcam) at 1:2000; anti–proteasome 20S α4 subunit (AB_10541440; Enzo Life Sciences, Uniondale, NY); anti-calnexin (AB_1310022; Abcam) at 1:2000; anti-Hrd1 (a gift from Malaiyalam Mariappan, Yale School of Medicine) at 1:1000; and anti–α-tubulin (AB_477583; Sigma-Aldrich) at 1:4000.
siRNA knockdown
siRNA knockdown was performed as described previously (Rose et al., 2014), except that cells were harvested for immunoblotting 48 h posttransfection. The following LAP1 siRNA pairs were used in a pool: 5′-CCUUAGUCCUGACUGUCUU[dT][dT]-3′ and 5′-AAGACAGUCAGGACUAAGG[dT][dT]-3′; 5′-GAAAUGAAGACGCGAAGGA[dT][dT]-3′ and 5′-UCCUUCGCGUCUUCAUUUC[dT][dT]-3′; and 5′GAAUAAGUACCAAGGUCAA[dT][dT]-3′ and 5′-UUGACCUUGGUACUUAUUC[dT][dT]-3′.
Electron microscopy
Electron microscopy was performed by the Center for Cellular and Molecular Imaging, Yale School of Medicine, with the following procedures: to prepare samples for morphology and membrane processing, cells in culture dishes were fixed in 2.5% (vol/vol) glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4, for 1 h. Buffer rinsed cells were scraped in 1% (wt/vol) gelatin and sedimented in 2% (wt/vol) agar. Chilled blocks were trimmed and processed following the protocol using reduced osmium and thiocarbohydrazide-osmium liganding as described by Ellisman and colleagues (University of California, San Diego; Deerinck et al., 2010). The samples were embedded in Durcupan ACM resin (Electron Microscopy Science [EMS], Hatfield, PA) and polymerized overnight at 60°C. Hardened blocks were cut using a Leica UltraCut UC7. Sections of 60 and 250 nm were collected on Formvar/carbon-coated grids and stained using 2% (wt/vol) uranyl acetate and lead citrate. The 60-nm sections were viewed in an FEI (Hillsboro, OR) Tencai Biotwin TEM at 80 kV. Images were taken using Morada CCD and iTEM (Olympus, Center Valley, PA) software. For electron tomography, 250-nm sections were used with tilt angles from 60° to −60° and imaged using FEI Tecnai TF20 at 200 kV. Data was collected using an FEI Eagle 4K × 4K digital camera. The volume reconstruction and modeling were done using Imod and segmentation done using 3Dmod (Boulder Laboratory for 3-Dimensional Electron Microscopy of Cells, Department of MCD Biology, University of Colorado, Boulder, CO).
Samples for immunolabeling were prepared with high-pressure freezing and freeze substitution. Unfixed samples were gently scraped and high-pressure frozen using a Leica (Wetzlar, Germany) EM HPM100 at 2000 psi. The frozen samples were freeze-substituted using a Leica AFS at −90°C using 0.1% (wt/vol) uranyl acetate/acetone for 50 h, rinsed in acetone, and infiltrated for 10 h at −45°C with Lowicryl HM20 resin (EMS). Samples were placed in gelatin capsules and ultraviolet hardened at −45°C. Hardened blocks were sectioned using a Leica UltraCut UC7, and 60-nm ultrathin sections were collected on Formvar/carbon-coated nickel grids for immunolabeling viewing. For immunolabeling of sections, grids were placed section side down on drops of 0.1 M ammonium chloride to quench untreated aldehyde groups and then blocked for nonspecific binding on 1% (wt/vol) fish-skin gelatin in phosphate-buffered saline (PBS). Single-labeled grids were incubated with primary antibody rabbit anti-ubiquitin at 1:50 dilution (AB_2315524; Dako, Chicago, IL), mouse anti-MAb414 at 1:50 dilution (AB_448181; Abcam), rabbit anti–K48-Ub (AB_11213655; Millipore, Billerica, MA) at 1:200 dilution or mouse anti-Y10B at 1:200 dilution (a gift from Joan Steitz, Yale University, Howard Hughes Medical Institute), which required a rabbit anti-mouse bridge (AB_234003; Jackson ImmunoResearch, West Grove, PA), and labeled with 10 nm Protein A-gold particles (Utrecht Medical Center, Utrecht, Netherlands). All grids were rinsed in PBS, fixed using 1% (vol/vol) glutaraldehyde for 5 min, rinsed, dried, and contrast stained using 2% (wt/vol) uranyl acetate and lead citrate.
Statistics and single-cell analysis
For EM experiments, 30 EM cross-sections per cell line were imaged, and the number of blebs per cross-section was determined. NE circumferences per cross-section were measured with ImageJ. The average NE circumference per cross-section was ∼33 μm, so the bleb counts were reported as the number of blebs per 30 μm of NE to account for different sizes of the cross-sections imaged. Statistical analysis was performed in GraphPad (La Jolla, CA) Prism using an unpaired t test.
For indirect immunofluorescence experiments, single-cell analysis was performed as described previously (Rose et al., 2014). Samples were blinded, and cells were binned into four groups by average fluorescence intensity and assessed as strongly exhibiting K48 foci, weakly exhibiting K48 foci, or not exhibiting K48 foci. Cells from the two bins expressing the lowest levels of TorA-HA or TorA ∆E-HA were included in Figure 5B. Statistical analysis was performed in Excel (Microsoft, Redmond, WA) using a chi-squared analysis.
Immunofluorescence microscopy
Indirect immunofluorescence and confocal microscopy were performed as previously described (Rose et al., 2014; Tsai et al., 2016). The following antibodies were used, all at 1:500 dilution: anti–K48 Ub (AB_11213655; Millipore); anti–lamin A (AB_306909, Abcam); anti-p97 (AB_298039, Abcam); anti-PDI (AB_303304, Abcam); anti–proteasome 20S α4 subunit (AB_10541440; Enzo Life Sciences); anti-Hrd1 (a gift from Malaiyalam Mariappan); and anti-calnexin (AB_1310022; Abcam).
XBP-1 splicing
Induction of the UPR by tunicamycin and detection of XBP-1 splicing were performed as described previously (Zhao et al., 2016).
Lipidomics
Lipid extraction, mass spectrometry, and data analysis were performed by Lipotype GmbH (Dresden, Germany) according to established methods (Herzog et al., 2011; Sampaio et al., 2011).
Supplementary Material
Acknowledgments
We thank Patrick Lusk, Mark Hochstrasser, and members of the Schlieker laboratory for critically reading the manuscript, Joan Steitz for sharing the Y10B antibody, and Malaiyalam Mariappan for sharing the Hrd1 antibody. This work was supported by National Institutes of Health Grants DP2OD008624-01 and 1R01GM114401-01A1 to C.S. and CMB TG T32GM007223 to E.L.
Abbreviations used:
- AAA+
ATPases associated with a variety of cellular activities
- NE
nuclear envelope
- NPC
nuclear pore complex
- nup
nucleoporin
- PNS
perinuclear space
- Tor
Torsin.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-07-0511) on October 26, 2016.
REFERENCES
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Boni A, Politi AZ, Strnad P, Xiang W, Hossain MJ, Ellenberg J. Live imaging and modeling of inner nuclear membrane targeting reveals its molecular requirements in mammalian cells. J Cell Biol. 2015;209:705–720. doi: 10.1083/jcb.201409133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 2008;322:1369–1373. doi: 10.1126/science.1165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RS, Zhao C, Chase AR, Wang J, Schlieker C. The mechanism of Torsin ATPase activation. Proc Natl Acad Sci USA. 2014;111:E4822–E4831. doi: 10.1073/pnas.1415271111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Burdette AJ, Porter JC, Ricketts JC, Fox SA, Nery FC, Hewett JW, Berkowitz LA, Breakefield XO, Caldwell KA, et al. The early-onset torsion dystonia-associated protein, torsinA, is a homeostatic regulator of endoplasmic reticulum stress response. Hum Mol Genet. 2010;19:3502–3515. doi: 10.1093/hmg/ddq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LI, Blobel G. Identification and characterization of a nuclear pore complex protein. Cell. 1986;45:699–709. doi: 10.1016/0092-8674(86)90784-1. [DOI] [PubMed] [Google Scholar]
- Deerinck TJ, Bushong EA, Thor A, Ellisman MH. NCMIR methods for 3D EM: A new protocol for preparation of biological specimens for serial block face scanning electron microscopy. 2010. Available at https://ncmir.ucsd.edu/sbem-protocol (accessed 1 July 2016)
- Demircioglu FE, Sosa BA, Ingram J, Ploegh HL, Schwartz TU. Structures of TorsinA and its disease-mutant complexed with an activator reveal the molecular basis for primary dystonia. Elife. 2016;5:e17983. doi: 10.7554/eLife.17983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foisner R, Gerace L. Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell. 1993;73:1267–1279. doi: 10.1016/0092-8674(93)90355-t. [DOI] [PubMed] [Google Scholar]
- Goodchild RE, Buchwalter AL, Naismith TV, Holbrook K, Billion K, Dauer WT, Liang CC, Dear ML, Hanson PI. Access of torsinA to the inner nuclear membrane is activity dependent and regulated in the endoplasmic reticulum. J Cell Sci. 2015;128:2854–2865. doi: 10.1242/jcs.167452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild RE, Dauer WT. Mislocalization to the nuclear envelope: an effect of the dystonia-causing torsinA mutation. Proc Natl Acad Sci USA. 2004;101:847–852. doi: 10.1073/pnas.0304375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild RE, Dauer WT. The AAA+ protein torsinA interacts with a conserved domain present in LAP1 and a novel ER protein. J Cell Biol. 2005;168:855–862. doi: 10.1083/jcb.200411026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild RE, Kim CE, Dauer WT. Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron. 2005;48:923–932. doi: 10.1016/j.neuron.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Grillet M, Dominguez Gonzalez B, Sicart A, Pottler M, Cascalho A, Billion K, Hernandez Diaz S, Swerts J, Naismith TV, Gounko NV, et al. Torsins are essential regulators of cellular lipid metabolism. Dev Cell. 2016;38:235–247. doi: 10.1016/j.devcel.2016.06.017. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- Herzog R, Schwudke D, Schuhmann K, Sampaio JL, Bornstein SR, Schroeder M, Shevchenko A. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 2011;12:R8. doi: 10.1186/gb-2011-12-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokhi V, Ashley J, Nunnari J, Noma A, Ito N, Wakabayashi-Ito N, Moore MJ, Budnik V. Torsin mediates primary envelopment of large ribonucleoprotein granules at the nuclear envelope. Cell Rep. 2013;3:988–995. doi: 10.1016/j.celrep.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungwirth M, Dear ML, Brown P, Holbrook K, Goodchild R. Relative tissue expression of homologous torsinB correlates with the neuronal specific importance of DYT1 dystonia-associated torsinA. Hum Mol Genet. 2010;19:888–900. doi: 10.1093/hmg/ddp557. [DOI] [PubMed] [Google Scholar]
- Kelley K, Knockenhauer KE, Kabachinski G, Schwartz TU. Atomic structure of the Y complex of the nuclear pore. Nat Struct Mol Biol. 2015;22:425–431. doi: 10.1038/nsmb.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CE, Perez A, Perkins G, Ellisman MH, Dauer WT. A molecular mechanism underlying the neural-specific defect in torsinA mutant mice. Proc Natl Acad Sci USA. 2010;107:9861–9866. doi: 10.1073/pnas.0912877107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knockenhauer KE, Schwartz TU. The nuclear pore complex as a flexible and dynamic gate. Cell. 2016;164:1162–1171. doi: 10.1016/j.cell.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Kosinski J, Mosalaganti S, von Appen A, Teimer R, DiGuilio AL, Wan W, Bui KH, Hagen WJ, Briggs JA, Glavy JS, et al. Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science. 2016;352:363–365. doi: 10.1126/science.aaf0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudermilch E, Schlieker C. Torsin ATPases: structural insights and functional perspectives. Curr Opin Cell Biol. 2016;40:1–7. doi: 10.1016/j.ceb.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA, Jr, Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang CC, Tanabe LM, Jou S, Chi F, Dauer WT. TorsinA hypofunction causes abnormal twisting movements and sensorimotor circuit neurodegeneration. J Clin Invest. 2014;124:3080–3092. doi: 10.1172/JCI72830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DH, Stuwe T, Schilbach S, Rundlet EJ, Perriches T, Mobbs G, Fan Y, Thierbach K, Huber FM, Collins LN, et al. Architecture of the symmetric core of the nuclear pore. Science. 2016;352:aaf1015. doi: 10.1126/science.aaf1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery FC, Armata IA, Farley JE, Cho JA, Yaqub U, Chen P, da Hora CC, Wang Q, Tagaya M, Klein C, et al. TorsinA participates in endoplasmic reticulum-associated degradation. Nat Commun. 2011;2:393. doi: 10.1038/ncomms1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery FC, Zeng J, Niland BP, Hewett J, Farley J, Irimia D, Li Y, Wiche G, Sonnenberg A, Breakefield XO. TorsinA binds the KASH domain of nesprins and participates in linkage between nuclear envelope and cytoskeleton. J Cell Sci. 2008;121:3476–3486. doi: 10.1242/jcs.029454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka S, Bui KH, Schorb M, Hossain MJ, Politi AZ, Koch B, Eltsov M, Beck M, Ellenberg J. Nuclear pore assembly proceeds by an inside-out extrusion of the nuclear envelope. Elife. 2016;5:e19071. doi: 10.7554/eLife.19071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozelius LJ, Hewett JW, Page CE, Bressman SB, Kramer PL, Shalish C, de Leon D, Brin MF, Raymond D, Corey DP, et al. The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat Genet. 1997;17:40–48. doi: 10.1038/ng0997-40. [DOI] [PubMed] [Google Scholar]
- Pappas SS, Darr K, Holley SM, Cepeda C, Mabrouk OS, Wong JM, LeWitt TM, Paudel R, Houlden H, Kennedy RT, et al. Forebrain deletion of the dystonia protein torsinA causes dystonic-like movements and loss of striatal cholinergic neurons. Elife. 2015;4:e08352. doi: 10.7554/eLife.08352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AE, Brown RSH, Schlieker C. Torsins: not your typical AAA+ ATPases. Crit Rev Biochem Mol Biol. 2015;50:532–549. doi: 10.3109/10409238.2015.1091804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AE, Zhao C, Turner EM, Steyer AM, Schlieker C. Arresting a Torsin ATPase reshapes the endoplasmic reticulum. J Biol Chem. 2014;289:552–564. doi: 10.1074/jbc.M113.515791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaio JL, Gerl MJ, Klose C, Ejsing CS, Beug H, Simons K, Shevchenko A. Membrane lipidome of an epithelial cell line. Proc Natl Acad Sci USA. 2011;108:1903–1907. doi: 10.1073/pnas.1019267108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos M, Domingues SC, Costa P, Muller T, Galozzi S, Marcus K, da Cruz e Silva EF, da Cruz e Silva OA, Rebelo S. Identification of a novel human LAP1 isoform that is regulated by protein phosphorylation. PLoS One. 2014;9:e113732. doi: 10.1371/journal.pone.0113732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarcelli JJ, Hodge CA, Cole CN. The yeast integral membrane protein Apq12 potentially links membrane dynamics to assembly of nuclear pore complexes. J Cell Biol. 2007;178:799–812. doi: 10.1083/jcb.200702120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R, Hitomi M, Ivessa AS, Fasch EV, Kohlwein SD, Tartakoff AM. A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol Cell Biol. 1996;16:7161–7172. doi: 10.1128/mcb.16.12.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa BA, Demircioglu FE, Chen JZ, Ingram J, Ploegh HL, Schwartz TU. How lamina-associated polypeptide 1 (LAP1) activates Torsin. Elife. 2014;3:e03239. doi: 10.7554/eLife.03239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuwe T, Correia AR, Lin DH, Paduch M, Lu VT, Kossiakoff AA, Hoelz A. Nuclear pores. Architecture of the nuclear pore complex coat. Science. 2015;347:1148–1152. doi: 10.1126/science.aaa4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukegawa J, Blobel G. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell. 1993;72:29–38. doi: 10.1016/0092-8674(93)90047-t. [DOI] [PubMed] [Google Scholar]
- Surma MA, Klose C, Peng D, Shales M, Mrejen C, Stefanko A, Braberg H, Gordon DE, Vorkel D, Ejsing CS, et al. A lipid E-MAP identifies Ubx2 as a critical regulator of lipid saturation and lipid bilayer stress. Mol Cell. 2013;51:519–530. doi: 10.1016/j.molcel.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe LM, Liang CC, Dauer WT. Neuronal nuclear membrane budding occurs during a developmental window modulated by torsin paralogs. Cell Rep. 2016;16:3322–3333. doi: 10.1016/j.celrep.2016.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PL, Zhao C, Turner E, Schlieker CD. The Lamin B receptor is essential for cholesterol synthesis and perturbed by disease-causing mutations. Elife. 2016;5:e16011. doi: 10.7554/eLife.16011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner EM, Brown RS, Laudermilch E, Tsai PL, Schlieker C. The Torsin activator LULL1 is required for efficient growth of herpes simplex virus 1. J Virol. 2015;89:8444–8452. doi: 10.1128/JVI.01143-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungricht R, Klann M, Horvath P, Kutay U. Diffusion and retention are major determinants of protein targeting to the inner nuclear membrane. J Cell Biol. 2015;209:687–703. doi: 10.1083/jcb.201409127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heyden AB, Naismith TV, Snapp EL, Hodzic D, Hanson PI. LULL1 retargets TorsinA to the nuclear envelope revealing an activity that is impaired by the DYT1 dystonia mutation. Mol Biol Cell. 2009;20:2661–2672. doi: 10.1091/mbc.E09-01-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanGompel MJ, Nguyen KC, Hall DH, Dauer WT, Rose LS. A novel function for the Caenorhabditis elegans torsin OOC-5 in nucleoporin localization and nuclear import. Mol Biol Cell. 2015;26:1752–1763. doi: 10.1091/mbc.E14-07-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster BM, Colombi P, Jager J, Lusk CP. Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell. 2014;159:388–401. doi: 10.1016/j.cell.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster BM, Lusk CP. Border safety: quality control at the nuclear envelope. Trends Cell Biol. 2016;26:29–39. doi: 10.1016/j.tcb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster BM, Thaller DJ, Jager J, Ochmann SE, Borah S, Lusk CP. Chm7 and Heh1 collaborate to link nuclear pore complex quality control with nuclear envelope sealing. EMBO J. 2016;35:2447–2467. doi: 10.15252/embj.201694574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisheit CE, Dauer WT. A novel conditional knock-in approach defines molecular and circuit effects of the DYT1 dystonia mutation. Hum Mol Genet. 2015;24:6459–6472. doi: 10.1093/hmg/ddv355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Blobel G. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J Cell Biol. 1993;123:275–284. doi: 10.1083/jcb.123.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Brown RS, Chase AR, Eisele MR, Schlieker C. Regulation of Torsin ATPases by LAP1 and LULL1. Proc Natl Acad Sci USA. 2013;110:E1545–E1554. doi: 10.1073/pnas.1300676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Brown RS, Tang CA, Hu CA, Schlieker C. Site-specific proteolysis mobilizes TorsinA from the membrane of the endoplasmic reticulum in response to ER stress and B cell stimulation. J Biol Chem. 2016;291:9469–9481. doi: 10.1074/jbc.M115.709337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.