The PI4KIIs play important roles in membrane trafficking through the production of PI(4)P. This study reveals a novel function of PI4KIIβ in regulating invadopodia formation and illustrates the importance of maintaining specific pools of trans-Golgi network–endosomal PI(4)P.
Abstract
The type II phosphatidylinositol 4-kinase (PI4KII) enzymes synthesize the lipid phosphatidylinositol 4-phosphate (PI(4)P), which has been detected at the Golgi complex and endosomal compartments and recruits clathrin adaptors. Despite common mechanistic similarities between the isoforms, the extent of their redundancy is unclear. We found that depletion of PI4KIIα and PI4KIIβ using small interfering RNA led to actin remodeling. Depletion of PI4KIIβ also induced the formation of invadopodia containing membrane type I matrix metalloproteinase (MT1-MMP). Depletion of PI4KII isoforms also differentially affected trans-Golgi network (TGN) pools of PI(4)P and post-TGN traffic. PI4KIIβ depletion caused increased MT1-MMP trafficking to invasive structures at the plasma membrane and was accompanied by reduced colocalization of MT1-MMP with membranes containing the endosomal markers Rab5 and Rab7 but increased localization with the exocytic Rab8. Depletion of PI4KIIβ was sufficient to confer an aggressive invasive phenotype on minimally invasive HeLa and MCF-7 cell lines. Mining oncogenomic databases revealed that loss of the PI4K2B allele and underexpression of PI4KIIβ mRNA are associated with human cancers. This finding supports the cell data and suggests that PI4KIIβ may be a clinically significant suppressor of invasion. We propose that PI4KIIβ synthesizes a pool of PI(4)P that maintains MT1-MMP traffic in the degradative pathway and suppresses the formation of invadopodia.
INTRODUCTION
The two human type II phosphatidylinositol 4-kinase (PI4KII) enzymes display 68% sequence identity across their conserved catalytic domains and exist in multiple subcellular membrane compartments (Balla and Balla, 2006; Minogue and Waugh, 2012). Since the molecular characterization of the PI4KIIs, a general model of PI4KII function in post–trans-Golgi network (TGN) traffic has gradually emerged in which the kinases generate phosphatidylinositol 4-phosphate (PI(4)P), which is required for the recruitment of clathrin adaptors needed in TGN-to-endosomal traffic. PI4KIIs have been most extensively described in endosomes (Balla et al., 2002), the TGN (Wang et al., 2003), and at the plasma membrane (Minogue et al., 2006), as has PI(4)P (Hammond et al., 2014). The PI4KIIα isoform generates PI(4)P, which interacts with AP-1 (Wang et al., 2003), GGAs (Wang et al., 2007), and AP-3 (Salazar et al., 2005; Craige et al., 2008; Mossinger et al., 2012) and therefore has roles in TGN-to-endosome and TGN-to–late endosomal trafficking known to be important in transport to the lysosome. More recently, a mechanism has been described involving the redirection of PI from phosphatidylinositol 3-phosphate (PI(3)P) into PI(4)P synthesis, which is required for exocytic traffic from endosomes. This pathway requires PI4KIIα and is significant because it represents an important step in the recycling of plasma membrane cargoes such as β1-integrin and the transferrin receptor (Ketel et al., 2016). PI4KIIα has also been implicated in human cancer (Li et al., 2010, 2014). PI4KIIβ functions in TGN-to-endosomal traffic by directly interacting with AP-1, playing a role in developmental signaling by controlling the endosomal traffic of Frizzled, a receptor for Wnt (Wieffer et al., 2013). The PI4KII isoforms therefore produce lipids needed for different steps in post-TGN trafficking.
Protease-dependent matrix remodeling performs key physiological functions during embryo development and in the pathogenesis of metastatic disease. In metastatic cells, proteolytic degradation of extracellular matrix (ECM) is mediated by extracellular proteases such as the membrane type I metalloproteinase (MT1-MMP), whose increased expression is associated with a poor prognosis in a wide variety of human tumors (Egeblad and Werb, 2002). ECM invasion by tumor-derived cells is mediated in part by specialized actin- and phosphotyrosine-rich protrusions termed invadopodia, which are capable of degrading extracellular matrix through the directed membrane traffic of MMPs (Poincloux et al., 2009). The overexpression of components of invadopodia such as cortactin in breast and squamous carcinomas and ovarian, bladder, and lung cancers is associated with a more aggressive phenotype and poorer patient prognosis (Paz et al., 2014). MT1-MMP is a key component of mature invadopodia and has been implicated in breaching of basement membranes and invasion through interstitial collagen (Sabeh et al., 2004; Hotary et al., 2006). Trafficking to the plasma membrane from endosomal compartments is a recognized pathway for the delivery of MT1-MMP to invadopodia (Steffen et al., 2008; Yu et al., 2012). However, the molecular machinery controlling the complex trafficking of this key protease is not fully understood.
The PI4KII enzymes have been associated with both the development of breast cancer (Li et al., 2014) and metastasis of hepatocellular carcinoma (HCC; Mazzocca et al., 2008), but links between the PI4KIIs and cellular processes such as actin reorganization during oncogenesis are unclear. We therefore investigated the role of PI4KIIα and PI4KIIβ in actin remodeling and found that depletion of the enzymes had starkly different effects on the actin cytoskeleton. We also found that PI4KIIβ depletion specifically induced the formation of invadopodia and was sufficient to convert HeLa and MCF-7 cells into an invasive phenotype. Loss of PI4KIIα and PI4KIIβ affected different PI(4)P TGN pools and post-TGN traffic. The steady-state distribution of MT1-MMP was also shifted from an endosomal to an exocytic trafficking route. Finally, database analyses indicated that PI4KIIβ was lost in human cancers, indicating that PI4KIIβ may be a clinically significant tumor suppressor.
RESULTS
Depletion of PI4KII isoforms differentially affects the actin cytoskeleton
We used small interfering RNA (siRNA) to induce loss of PI4KII function. Gene silencing was confirmed by immunoblotting 72 h after transfection of HeLa cells with isoform-specific siRNA SMARTpools. We repeatedly achieved at least 85% reduction in levels of PI4KIIα and PI4KIIβ compared with nontargeted controls (Figure 1, A and B, and Supplemental Figure S1, A and B). When we specifically depleted PI4KII isoforms in this way, we noted that monolayers of cells treated with PI4KIIβ siRNA were more dispersed than with PI4KIIα and controls (unpublished results), as described previously (Mazzocca et al., 2008). To investigate this further, we stained the actin cytoskeleton with fluorescently labeled phalloidin and found that, in comparison to controls, loss of PI4KIIα led to increased actin stress fiber formation at leading edges (Figure 1C). Cells also showed some clear focal colocalization with phosphotyrosine (Figure 1C). Loss of PI4KIIβ, on the other hand, resulted in complete loss of actin stress fibers and increased the intensity of cortical actin staining (Figure 1C), as well as of the formation of scattered, actin-rich punctae staining positive for phosphotyrosine (Figure 1C), which were absent in PI4KIIα and control siRNAs. Actin remodeling was confirmed by analyzing the ratio of cortical actin:stress fiber signals (Figure 1D), and normal actin phenotypes were restored by reexpression of siRNA-resistant PI4KIIβ (Figure 1E and Supplemental Figure S1, C–F). The same rescue experiment with overexpressed GFP-PI4KIIα led to increased levels of cortical actin (Figure 1E), a likely consequence of increasing PI(4)P levels (Henmi et al., 2016), which would be expected to alter the actin cytoskeleton. These data indicate that loss of PI4KIIα and PI4KIIβ has markedly different effects on the actin cytoskeleton.
FIGURE 1:
PI4KIIα and PI4KIIβ exert different effects on the actin cytoskeleton. (A, B) Total cell lysates of PI4KIIα, PI4KIIβ, and control siRNA-transfected HeLa cells were analyzed for protein expression by Western blotting; α-tubulin served as a control for protein levels. (C) HeLa cells transfected with the siRNA SMARTpool as indicated for 36 h were reseeded onto collagen matrix and then costained for tyrosine phosphorylated proteins (red) and the actin cytoskeleton (green). Confocal sections were acquired at 0.25-μm intervals and rendered into orthogonal views. Single XY-sections of the basal surface. Single pY and F-actin channels are shown below. Scale bars, 10 μm. (D) Ratio of cortical actin to stress fiber intensity from three independent experiments (n = 20 cells). (E) Cells were also transfected with single siRNA oligos and rescued by reexpression of siRNA-resistant PI4KIIα and PI4KIIβ. Data are presented as mean ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001; ns, not significant.
Loss of PI4KIIβ induces the formation of invadopodia
We sought to confirm the identity of the scattered actin-rich punctae in Figure 1C formed as a result of loss of PI4KIIβ. Initially, we costained F-actin along with the early endosomal marker EEA1; however, these showed minimal colocalization (Supplemental Figure S2A), indicating that these punctae were distinct from actin comets localized to endosomal membranes (Taunton et al., 2000). Furthermore, these structures typically had average diameters of ≤0.25 μm, mostly clustered in regions under the nucleus, and were thus suggestive of invadopodia (Garcia et al., 2014). To confirm the identity of these punctae, we immunostained cells for actin and cortactin, the coincidence of which is diagnostic of invadopodia and related structures (Garcia et al., 2014).
We found that loss of PI4KIIβ, but not of PI4KIIα, induced the formation of punctae staining positive for both markers in HeLa cells (Figure 2A). When we acquired confocal z-series data from PI4KIIβ-depleted cells, we observed the colocalization of actin and cortactin at the ventral surface of gelatin-coated coverslips (Figure 2B), thereby confirming that these structures were indeed invadopodia. As further confirmation, we performed a positive control by treating cells with the phorbol ester phorbol-12-myristate-13-acetate (PMA; Tatin et al., 2006) and found that the actin- and cortactin-rich structures were indistinguishable from those obtained by depletion of PI4KIIβ (Figure 2B). Key constituents of invadopodia are the matrix metalloproteinase MT1-MMP (Poincloux et al., 2009) and β1-integrin (Destaing et al., 2010). To determine whether the ventral structures contained known components of mature invadopodia, we stained siRNA-treated cells plated on coverslips coated with a three-dimensional (3D) layer of collagen for MT1-MMP and β1-integrin or cortactin. Confocal microscopy demonstrated the colocalization of these markers with MT1-MMP (Figure 2, C and D), particularly on the ventral surface of PI4KIIβ-depleted cells (Figure 2, E and F). Furthermore, these structures were seen to penetrate the 3D ECM when we performed Z-series confocal microscopy (Figure 2, E and F).
FIGURE 2:
Depletion of PI4KIIβ induces invadopodia formation. (A) Confocal images showing immunostaining of cortactin (green) and F-actin (red). (B) Confocal images showing comparisons between HeLa cells transfected with PI4KIIβ siRNA and cells treated with 50 nM PMA. (C–F) Confocal images showing cells grown on thick layers of collagen and coimmunostained for MT1-MMP (magenta), (C) cortactin (green), or (D) β1-integrin (green). (E, F) XZ rendering of 20 confocal sections acquired at 0.25-μm intervals, showing localization of MT1-MMP to ventral structures containing cortactin (E) or β1-Integrin (F). Horizontal lines indicate the position of the ECM surface. Scale bars, 20 μm.
Loss of PI4KIIβ causes increased matrix degradation and migration through collagen gel
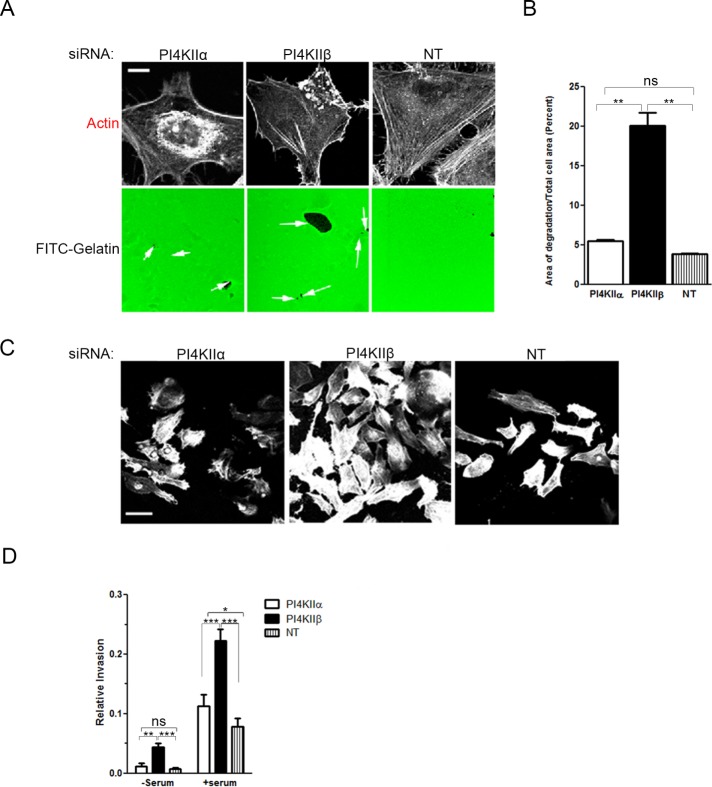
MT1-MMP requires posttranslational processing to become active (Williams and Coppolino, 2011). We therefore determined whether the structures formed upon the loss of PI4KIIβ expression were capable of ECM degradation. We plated target and control siRNA-transfected HeLa cells on an ECM composed of cross-linked fluorescein isothiocyanate (FITC)–conjugated gelatin. In comparison with PI4KIIα and control siRNA–transfected cells, loss of PI4KIIβ resulted in an approximately fourfold increase in fluorescent matrix degradation in comparison with PI4KIIα (Figure 3, A and B). Areas of degradation also corresponded to clusters of actin-rich punctae (Figure 3A), demonstrating that they were the source of the gelatinolytic activity.
FIGURE 3:
PI4KIIβ knockdown promotes ECM degradation and cell migration. (A) HeLa cells were reseeded onto FITC-gelatin–coated coverslips at 58 h posttransfection and then labeled for F-actin (red). Areas of proteolytic degradation are marked with arrows. (B) Percentage areas of degradation per total cell area in a 4 × 104 μm2 field. Data are mean ± SEM (n = 30; experiment performed in triplicate). (C, D) HeLa cells were reseeded onto Transwell inserts coated with type I collagen gel (100 μl of 2 mg/ml) 58 h posttransfection and then placed in a chemoattractant (10% FCS) for 12 h. (C) Representative fields of cells that traversed the collagen-coated membrane of a Transwell chamber in response to chemoattractant. (D) Quantitation of invasion expressed as the ratio of cells that migrated toward chemoattractant relative to the total number of cells seeded onto the serum-free upper chamber. Data are presented as mean ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001; ns, not significant.
We also performed Transwell invasion assays to measure the ability of PI4KII-depleted cells to invade a 3D collagen matrix in response to chemoattractant (serum). Invasion in the absence of serum was relatively low but significantly higher in PI4KIIβ siRNA cells. The introduction of serum to the lower chamber increased the number of migrating cells, with PI4KIIβ cells invading at approximately three times control values and two times that recorded for PI4KIIα-depleted cells (Figure 3, C and D, and Supplemental Figure S2B).
PI4KIIα and PI4KIIβ isoforms synthesize separate pools of TGN PI(4)P involved in TGN-to-endosome traffic
PI4KII isoforms are not known to directly influence the actin cytoskeleton; instead, they control trafficking from the TGN to endosomes through the recruitment of clathrin adaptors. To address the question of how PI4KIIβ knockdown leads to increased matrix degradation, we asked whether the PI4KII isoforms synthesized separate pools of PI(4)P and whether loss of this PI(4)P affected post-TGN membrane traffic. We visualized the PI(4)P generated by each PI4KII isoform by inhibiting PI4KIII activities with wortmannin in control and siRNA-treated cells. When we stained siRNA-treated cells with the PI(4)P reporter glutathione S-transferase (GST)–FAPP-PH, we noted that the intensity of staining in the Golgi region was affected in both PI4KIIα- and PI4KIIβ-knockdown cells but included signal from PI(4)P synthesized by the Golgi enzyme PI4KIIIβ (Supplemental Figure S3A). We therefore treated the same cells with wortmannin to inhibit PI4KIIIβ and fixed and stained the cells with recombinant GST-P4C, which is an unbiased PI(4)P reporter (Luo et al., 2015). The residual GST-P4C signal allowed us to report the PI(4)P derived from the activity of PI4KIIα (PI(4)PPI4KIIα) and PI4KIIβ (PI(4)PPI4KIIβ; Figure 4, A–C, and Supplemental Figure S4D), which in both cases localized to membranes in the Golgi region and on cytoplasmic vesicles (Figure 4, A–C). Cells in which PI4KIIα was depleted showed residual PI(4)PPI4KIIβ staining that colocalized with TGN46 (Figure 4, A and D). In contrast, PI(4)PPI4KIIα predominantly colocalized with the TGN marker syntaxin 6 (Figure 4, B and D). Colocalization of PI(4)P with syntaxin 6 and TGN46 was restored upon expression of plasmids containing silent mutations rendering them resistant to siRNA (Figure 4, E and F, and Supplemental Figure S3, E and F). This demonstrates that the wortmannin-insensitive PI(4)PPI4KIIα and PI(4)PPI4KIIβ pools generated by the PI4KIIs exist in distinct subdomains of TGN/endosomal membranes and that knockdown of each isoform affects a metabolically separate PI(4)P compartment.
FIGURE 4:
PI4KIIα and PI4KIIβ siRNA affects separate pools of PI(4)P at the TGN and perturbs TGN-endosomal traffic. (A–C) Recombinant GST-tagged protein containing the P4C domain from the Legionella pneumoniae SidC gene (GST-P4C) was used to indirectly stain membrane pools of PI(4)P after wortmannin treatment. Samples were costained with (A) TGN46 or (B) syntaxin 6 or (C) transfected with mCherry-tagged (CI-M6PR). (D) Pearson’s r calculated for colocalization between PI(4)P and different TGN markers (20 cells, three independent experiments). (E, F) Pearson’s r for colocalization between PI(4)P and (E) TGN46 or (F) syntaxin 6 after siRNA-mediated silencing with or without subsequent transfection with siRNA-resistant constructs. (G) Confocal images showing immunostaining for GFP-tagged CI-M6PR and adaptor AP-1. Scale bars, 10 μm. (H) Pearson’s r for colocalized pixel intensities between the green (GFP) and magenta (AP-1) channels (20 cells, three independent experiments). (I) HeLa cells were transfected with the indicated siRNA oligos and then serum starved and stimulated with 100 ng/ml EGF for indicated times before analysis of EGFR levels by Western blotting. (J) Signal intensities were quantified by densitometry. Data are mean ± SEM (n = 3; three independent experiments). *p < 0.05, **p < 0.01, ***p < 0.001; ns, not significant.
PI(4)P at the TGN is known to control the recruitment of the clathrin adaptor AP-1 in TGN-to-endosomal traffic (Wang et al., 2003; Wieffer et al., 2013), and we therefore reasoned that loss of TGN PI(4)P would affect this trafficking route. We investigated how loss of TGN PI(4)P altered the steady-state distribution of the cation-independent mannose-6-phosphate receptor (CI-M6PR), finding that PI4KIIα and PI4KIIβ led to a significant loss in colocalization between PI(4)P and this AP-1 cargo (Figure 4, C and D). This effect was particularly marked in PI4KIIβ cells, which also displayed a more compact staining pattern in the Golgi region and an absence of vesicular M6PR (Figure 4C). We also stained cells transfected with GFP-M6PR for AP-1 and found that PI4KIIβ siRNA cells showed reduced colocalization between them (Figure 4, G and H), indicating defective TGN-to-endosomal traffic. We tested the consequences on endolysosomal traffic by performing an epidermal growth factor receptor (EGFR) degradation assay and found that depletion of both PI4KIIα and PI4KIIβ significantly impaired ligand-induced degradation of the EGFR (Figure 4, I and J). Given that loss of either PI4KII isoform impairs EGFR degradation, we coimmunostained MT1-MMP with the lysosomal marker CD63 and found that both PI4KIIα and PI4KIIβ siRNA resulted in reduced colocalization (Supplemental Figure S4, A and B). Collectively these results show that loss of either PI(4)P pool affects TGN-to-endosome traffic and ultimately the degradation of cargo destined for lysosomes.
Loss of PI4KIIβ leads to increased exocytic trafficking of MT1-MMP
MT1-MMP is delivered to invadopodia from endosomal compartments (Steffen et al., 2008; Wiesner et al., 2013). We therefore considered that altered intracellular traffic might play a role in the increased localization of MT1-MMP to invadopodia. We analyzed cell surface MT1-MMP staining versus total cellular MT1-MMP using fluorescence-activated cell sorting (FACS) and found that loss of either PI4KII isoform led to an increase in total cellular MT1-MMP levels compared with controls (Supplemental Figure S4C); however, loss of PI4KIIβ led to an approximately threefold increase in MT1-MMP at the cell surface compared with control and PI4KIIα siRNA-transfected cells (Figure 5A).
FIGURE 5:
PI4KIIβ depletion alters trafficking of MT1-MMP. (A) Surface-labeled and total MT1-MMP were analyzed using flow cytometry, and the relative percentage of MT1-MMP present on the cell surface versus total cellular MT1-MMP was calculated. (B) Confocal immunofluorescence images showing MT1-MMP (magenta) and the indicated Rab GTPases (green). (C) Pearson’s r for colocalization of MT1-MMP and the indicated Rab GTPases. Data are presented as means ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001; ns, not significant.
Increased surface MT1-MMP could result from a decreased rate of endocytosis or an increased rate of exocytic trafficking. We therefore analyzed changes to the complex trafficking itinerary of MT1-MMP by coimmunostaining MT1-MMP and a selective panel of Rab GTPases known to control traffic of cargo in endosomal and Golgi membranes (Wiesner et al., 2013). On loss of PI4KIIα and PI4KIIβ, MT1-MMP displayed decreased colocalization with early endosomal Rab5 and the late endosomal Rab7 (Figure 5, B and C). A significant increase in colocalization was observed with the post-Golgi Rab8, but meaningful differences were not detected in the colocalization of MT1-MMP with late endosomal Rab9 or the recycling endosomal Rab11 (Figure 5, B and C). This analysis demonstrates that MT1-MMP trafficking is redirected from an early endosomal (EE)–late endosomal (LE) route to the plasma membrane via a Rab8 exocytic route.
Loss of PI4KIIβ confers invasiveness on MCF-7 cells
We asked whether PI4KIIβ depletion was sufficient to convert the minimally invasive MCF-7 epithelial cell line into an invasive migratory phenotype. To test this, we evaluated the ability of PI4KIIβ-depleted MCF-7 cells to degrade FITC-gelatin matrix and invade a collagen layer in the Transwell chamber assay. PI4KIIβ-depleted MCF-7 cells degraded FITC-gelatin to an extent comparable to the highly invasive MDA-MDB-231 line (Figure 6, A and B). Furthermore, they displayed increased invasiveness and the ability to invade collagen ECM and cross the membrane toward a chemoattractant (Figure 6C). These findings indicate that PI4KIIβ suppresses cell invasion in vitro.
FIGURE 6:
PI4KIIβ depletion is sufficient to confer a migratory and invasive phenotype. (A) siRNA-treated MCF-7 and untreated MDA-MB-231 cells were reseeded onto FITC-gelatin–coated coverslips (green) at 58 h posttransfection and then labeled for F-actin (red). (B) Percentage areas of proteolytic degradation per total cell in a 4 × 104 μm2 field. Data are mean ± SEM (n = 30; experiments done in triplicate). (C) HeLa cells were reseeded onto collagen-coated Transwell inserts at 58 h posttransfection and placed in a chemoattractant (10% FCS) for 12 h. Histogram shows index of cell migration through an artificial barrier in response to chemoattractant. Data are mean ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001; ns, not significant.
Loss of PI4KIIβ is associated with human cancers
A region of human chromosome 4 encompassing the gene for PI4KIIβ (PI4K2B) contains tumor suppressor genes associated with sporadic colorectal carcinoma (Zheng et al., 2008). We therefore interrogated oncogenomic and transcriptomic data in the CBioPortal and Oncomine databases and found an association between heterozygous loss of PI4K2B and lung (squamous cell and adenocarcinoma), esophageal, pancreatic, prostate, breast, liver, and various other epithelial cancers (Figure 7A and Supplemental Figure S5). In pancreatic tumors, homozygous loss of PI4K2B was detected in 2.75% of samples, heterozygous loss in 43.1%, and gain in 12.8%. Copy number was normal in 39.44% (n = 109 samples; Figure 7B). In HCC, both PI4K2B alleles were deleted in 0.52%, 23.68% displayed heterozygous deletion, 64.73% had normal copy numbers, and 10.52% showed gain of PI4K2B (n = 190; Figure 7C). Significant reductions in PI4KIIβ mRNA expression were detected in invasive breast carcinoma (n = 59; Figure 7D). We also analyzed survival data in the small number of cases available (Ciriello et al., 2015) and found that loss of PI4KIIβ expression correlated with poor survival in patients diagnosed with carcinoma of the breast (Figure 7E). Together these data support the hypothesis that loss of PI4KIIβ is a risk factor in cancers.
FIGURE 7:
Database analysis shows that loss of PI4KIIβ expression is associated with cancers. (A) Cross-cancer copy number alteration (CNA) summary of 18 selected cancer studies (including lung, esophageal, breast, prostate, and pancreatic tumors) for heterozygous loss of the PI4K2B allele. Threshold: minimum 5% altered samples, cBioPortal analysis (Cerami et al., 2012; Gao et al., 2013). adeno, adenocarcinoma; NEPC, neuroendocrine prostate cancer; Squ, squamous cell carcinoma. Sources of the data are given in parentheses: TCGA, The Cancer Genome Atlas; BCRC, British Columbia Cancer Research Center; UTSW, University of Texas Southwestern; FHCRC, Fred Hutchinson Cancer Research Center; SU2C, Stand Up To Cancer/Prostate Cancer Foundation (PCF); MICH, University of Michigan. (B) PI4K2B putative copy number alterations for pancreatic cancers (deletion or amplification) was analyzed using cBioPortal. (C) Overall change in PI4K2B copy number (deletion or amplification) in HCC was analyzed using cBioPortal. (D) Analysis of PI4KIIβ mRNA expression (Oncomine) in recurrent breast carcinoma (Finak breast, reporter: A_23_P18598). (E) Overall Kaplan–Meier survival graph of breast carcinoma case analysis (cBioPortal) with PI4KIIβ expression alteration (Ciriello et al., 2015).
DISCUSSION
Here we identify a post-TGN trafficking pathway that negatively regulates endolysosomal traffic and the formation of invadopodia. Loss of the PI4KII isoforms had dramatically different and unexpected effects on the actin cytoskeleton. An additional and striking feature of PI4KIIβ-depleted cells was the appearance of mature and functional invadopodia on the ventral surface.
PI4KIIβ plays a role in the development of a migratory phenotype in HCC (Mazzocca et al., 2008), but there is limited evidence for the direct involvement of mammalian PI4KII isozymes in actin regulation. In contrast, several studies have described PI4KII functions in post-TGN traffic through PI(4)P biosynthesis and direct binding to clathrin adaptors AP-1 and AP-3 (Wang et al., 2003, 2007; Salazar et al., 2005; Craige et al., 2008), both pathways that contribute to endolysosomal traffic. PI4KIIα controls LE traffic of the EGFR (Minogue et al., 2006), endolysosomal traffic mediated by AP-3 (Craige et al., 2008), and that of lysosomal membrane proteins and hydrolases such as the enzyme β-glucocerebosidase (Jovic et al., 2012). This explains the similar effects of PI4KIIα and PI4KIIβ depletion on EGFR degradation and LE traffic (Figure 4, I and J). PI4KIIβ localizes to the TGN and recruits AP-1, where it controls the endosomal sorting of Frizzled by traffic to degradative compartments (Wieffer et al., 2013). There is therefore a substantial amount of data from our study and in published work indicating that each PI4KII contributes to TGN-EE/LE traffic by acting at different points in a pathway leading to lysosomal degradation. The other principal way in which the PI4KII isoforms differ is in the regulation of their PI4KII activity. The reported low activity of PI4KIIβ (Balla et al., 2002) and the need to recruit this isoform to membranes (Jung et al., 2011) illustrate the importance of regulating its activity and suggest that this isoform is subject to tight regulation.
Few studies have investigated the role of PI4KIIs in TGN PI(4)P synthesis. Our data demonstrate for the first time that the PI4KII enzymes synthesize separate TGN pools of PI(4)P that are defined by the markers syntaxin 6 and TGN46. This is consistent with our previous description of a TGN-endosomal compartment enriched in syntaxin 6 containing highly active PI4KIIα (Simonsen et al., 1999; Waugh et al., 2003). The structurally unrelated, wortmannin-sensitive PI4KIIIβ also contributes to PI(4)P synthesis in the Golgi of MDCK cells (Weixel et al., 2005), indicating that the Golgi complex contains at least three metabolically separate PI(4)P pools.
Cells such as HeLa and MCF-7 normally display minimal invasive potential and typically have low levels of surface MT1-MMP, reflecting the importance of controlling surface metalloproteinase activity (Poincloux et al., 2009). Loss of either PI4KII isoform led to increased levels of the metalloproteinase, consistent with a general impairment of endolysosomal degradative traffic; however, only depletion of PI4KIIβ caused increased levels of surface MT1-MMP, indicating that intracellular traffic of MT1-MMP was affected in an isoform-specific manner.
The reduced colocalization of MT1-MMP with Rab7, which acts at the EE-LE boundary, in both PI4KII-depleted cells demonstrates that the proteinase is not sorted to the LE. This is consistent with the elevated levels of MT1-MMP observed in both PI4KII knockdowns (Supplemental Figure S4C), the impaired degradation of the EGFR (Figure 4, I and J), and the failure of MT1-MMP to traffic to CD63-containing lysosomes (Supplemental Figure 4, A and B). The most dramatic change in colocalization was a PI4KIIβ-specific increase in association with Rab8 membranes. Rab8 operates in a post-Golgi exocytic pathway that determines directional traffic to protrusions of the plasma membrane (Hattula et al., 2006) and is known to mediate the trafficking of MT1-MMP (Bravo-Cordero et al., 2007). A mechanistic investigation of Rab8 was outside the scope of our study, but it is interesting to note that this GTPase was recently shown to regulate the cortical actin cytoskeleton (Bravo-Cordero et al., 2016), suggesting that the mistrafficking of Rab8 may contribute to the actin phenotype.
We were unable to detect increased trafficking of MT1-MMP-GFP to invasive structures in live cell experiments; however, our attempts were fraught with technical difficulties arising from the substantial MT1-MMP signal present in multiple subcellular compartments that obscured invadopodia dynamics. The trafficking of MT1-MMP is known to be complex, and surface levels are controlled by endocytosis and exocytosis from several compartments (Poincloux et al., 2009). The increased localization of MT1-MMP to the plasma membrane, increased association with invasive protrusions, loss of colocalization in Rab5- and Rab7-positive compartments, and increased colocalization with exocytic Rab8 membranes indicate that loss of PI4KIIβ and the associated pool of TGN PI(4)P causes disequilibrium between the endolysosomal and exocytic trafficking routes controlling plasma membrane levels of MT1-MMP. Our data support a model in which MT1-MMP traffic is directed away from the default endolysosomal pathway and forced into an exocytic pathway. One possible consequence of reduced MT1-MMP traffic into the endolysosomal pathway is increased recycling. Although we did not directly address this possibility, it remains likely because we also detected increased invadopodial localization of β1-integrin, a molecule known to be recycled through a pathway involving Rab8 and cotrafficked with MT1-MMP (Macpherson et al., 2014). Of interest, the recent finding that PI4KIIα directs exocytic traffic of β1-integrin from endosomes to the plasma membrane when endosomal PI(3)P is lost (Ketel et al., 2016) suggests that endosomal PI(4)PPI4KIIα may play a role in β1-integrin delivery. Further investigation is required to determine whether loss of PI(4)PPI4KIIβ results in an increase in PI(4)PPI4KIIα and whether this is accompanied by increased β1-integrin recycling. Integrin traffic to the plasma membrane plays important roles in tissue organization and is known to be dependent on PI(3)P (Ribeiro et al., 2011). Cross-talk between this PI monophosphate and the PI(4)P produced by the PI4KIIs is an unanswered question.
The conversion of the minimally invasive HeLa and MCF-7 cell lines to an invasive phenotype and the fact that tumor suppressor genes map closely to the chromosomal location of the PI4K2B allele (Zheng et al., 2008) led us to perform database searches. These showed that loss of heterozygosity of the PI4K2B allele and PI4KIIβ underexpression were associated with numerous cancers of epithelial origin. PI4KIIβ underexpression was also associated with poorer patient survival; however, the low statistical power of the studies led to a relatively low probability value (p = 0.262). Although we are cautious in drawing conclusions, we are aware that this finding may be highly significant and therefore speculate that PI4KIIβ acts as a metastasis suppressor by maintaining PI(4)P-dependent post-TGN traffic into the endolysosomal pathway. The association of PI4KIIβ with relatively common human cancers (lung, breast, liver, pancreas, and others) warrants further investigation, particularly because of the possibility that this putative suppressor pathway could be rescued by inhibition of PI(4)P phosphatases acting at the TGN.
It is notable that a developmental role for PI4KIIβ in zebrafish has been described (Wieffer et al., 2013). The pathological process of tumor cell invasion has many similarities with important processes during embryogenesis along with neurite outgrowth and angiogenesis. It is therefore possible that PI4KIIβ activity is subject to regulation controlling physiological invasion. This hypothesis points to a PI(4)P-regulated switch that determines the decision between lysosomal degradation of cargoes and their traffic to the plasma membrane that may be fundamentally important in the intracellular traffic of key molecules such as integrins and matrix metalloproteinases that determine tissue remodeling during development.
MATERIALS AND METHODS
Cell lines, culture, and transfection
HeLa and MCF7 cell lines were cultured in DMEM (Life Technologies, Paisley, UK) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich, Dorset, UK) and maintained at 37°C and 5% CO2. MDA-MB-231 cells were cultured in Leibowitz’s L-15 medium (Life Technologies) supplemented with 15% FBS and 2 mM l-glutamine (Life Technologies) and maintained at 37°C, 0.1% CO2. All media were supplemented with antibiotics (1000 U/ml penicillin and 0.1 mg/ml streptomycin; Sigma-Aldrich). DharmaFECT (GE Healthcare Dharmacon, Lafayette, CO) and Lipofectamine (Life Technologies) were used to transfect siRNAs and plasmids, respectively.
Antibodies and reagents
All primary antibodies with corresponding dilutions are listed in Supplemental Table S1. Highly cross-adsorbed Alexa Fluor–conjugated secondary antibodies and Alexa Fluor–conjugated phalloidin (1:250 dilution) were purchased from Molecular Probes (Life Technologies). Hoechst 33342 trihydrochloride was also from Molecular Probes and was used at 1:5000 for nuclear staining. Human recombinant EGF was purchased from Sigma-Aldrich. Unless otherwise stated, other reagents were from Sigma Aldrich.
Plasmids
siRNA-resistant GFP-tagged PI4KIIα and hemagglutinin-tagged PI4KIIβ were generous gifts from Volker Haucke (Leibniz Institut für Molekulare Pharmakologie, Berlin, Germany). mCherry and GFP-tagged CI-M6PR were kindly provided by Mihaela Anitei (Technische Universität, Dresden, Germany).
Small interfering RNAs
We silenced PI4KIIα and PI4KIIβ using ON-TARGETplus SMARTpools siRNA oligonucleotides (GE Healthcare Dharmacon). Each comprised a mix of four siRNA duplexes targeting specific sequences within the named genes. We also used an ON-TARGETplus nontargeting pool (D-001810-10-05) as control. Gene silencing was confirmed by Western blotting of whole-cell lysates. PI4KII-knockdown phenotypes were rescued using single siRNA duplexes and the ectopic expression of siRNA resistant plasmids as described previously (Mossinger et al., 2012; Wieffer et al., 2013). Further details are given in the Supplemental Materials and Supplemental Table S2.
Western immunoblotting
Cells were lysed in RIPA buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 0.5% sodium deoxycholate, 1% Triton X-100, 10% glycerol, 0.5 mM ethylene diamine tetraacetic acid [EDTA], 0.5 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N’,N’-tetraacetic acid [EGTA], 10 mM sodium fluoride, 10 mM sodium pyrophosphate, 0.2 mM Na3VO4) and 1× cOmplete Protease Inhibitor (Roche, Mannheim, Germany). Protein content was determined using the Lowry method (Bio-Rad, Hertfordshire, UK), and lysates were solubilized in 2× Laemmli sample buffer, separated using SDS–PAGE, blotted, and probed with respective antibodies. Blots were imaged on a FluorChem M Multifluor imaging system (ProteinSimple, San Jose, CA) and analyzed by densitometry using its gel analysis function.
Immunofluorescence microscopy
Cells were grown on glass coverslips, fixed, and probed with antibodies of interest as previously (Minogue et al., 2006). Unless otherwise stated, images were acquired using the 63×/1.4 numerical aperture lens on a Zeiss LSM 510 confocal microscope. Randomly chosen representative fields were selected and 12-bit images acquired using identical detector gain and offset settings. Images were quantitatively analyzed for fluorescence intensities, pixel correlations, area measurements, and cell densities using ImageJ software (National Institutes of Health, Bethesda, MD). Full details are available in the Supplemental Materials.
Fluorescent matrix degradation
The ability of cells to degrade the extracellular matrix was analyzed using fluorescent gelatin matrix–coated culture dishes prepared according to Bowden et al. (1999). Targeted or control siRNA-transfected cells were seeded onto FITC-gelatin–coated coverslips and cultured for 12–18 h. Matrix degradation was calculated as the total area of matrix degradation (black holes) per total cell area expressed as a percentage (full details are given in the Supplemental Materials).
MT1-MMP trafficking
We used an established method (Kean et al., 2009). Briefly, MCF-7 cells were reseeded onto coverslips thinly coated with collagen (5 μg/cm2) at 36 h posttransfection and grown in complete DMEM for 20 h at 37°C. Cells were then serum starved for 4 h to facilitate internalization of MT1-MMP. For trafficking of endogenous MT1-MMP, cells were pulsed with anti–MT1-MMP antibody (Abcam, Cambridge, UK) for 1 h at 4°C. Cells were subsequently fixed with 3.7% formaldehyde, permeabilized, and immunostained.
For flow cytometry experiments, we induced internalization of MT1-MMP by serum starvation and stimulated surface expression as described. Cells were lifted using FACS buffer (5 mM EDTA in phosphate-buffered saline [PBS], pH 7.4) and subsequent steps carried out at 4°C. Cells were probed with rabbit anti–MT1-MMP (Abcam) for 1 h, washed three times with PBS, and then labeled with anti–rabbit Alexa Fluor antibody. Cells were analyzed using a FACS LSRII cell sorter (BD Biosciences, Oxford, UK), and ∼10,000 cells were counted per sample. Postacquisition analysis was performed using Flowjo software, version V10 (FlowJo LLC, Ashland, OR).
Transwell migration assay
This was performed in 8-μm-pore, collagen-coated Transwell chambers (Corning, Tewksbury, MA). Cells were transfected with target or control siRNA in six-well plates and, after 48 h, resuspended in serum-free medium and seeded at 5 × 104 into the upper chamber of the Transwell. Cells were allowed to migrate toward 10% FBS plus DMEM for 12 h. Migratory cells at the lower chamber were fixed and stained with Alexa 488–conjugated phalloidin, imaged using similar optical settings (20× Objective, Zeiss LSM 510 confocal microscope), and counted in five random fields using ImageJ. Histograms represent relative invasion of the Transwell chamber, calculated as the number of cells counted at the lower chamber relative to the total number of cells seeded onto the upper chamber (details are give in the Supplemental Materials).
Data analysis
Prism 5 (GraphPad, La Jolla, CA) was used for statistical analysis. Data are presented as mean ± SEM from at least three independent experiments. Statistical comparisons were performed using one-way analysis of variance followed by Tukey’s posttest at a 95% confidence interval.
Oncogenomic database analysis
The cBIO Cancer Genomics Portal (http://cbioportal.org/; Cerami et al., 2012; Gao et al., 2013) and Oncomine (Rhodes et al., 2004) are open access resources for cancer genomics data sets containing data from 69 cancer genomics studies with 17,177 samples (cBIO) and 715 data sets with 86,733 samples (Oncomine). Heterozygous loss of the PI4K2B allele was analyzed in all cancers, as were copy number alterations in pancreatic cancers and hepatocellular carcinoma. In addition, correlations of PI4KIIβ mRNA expression, recurrence, metastatic events, or overall survival in breast cancer cases were investigated.
Supplementary Material
Acknowledgments
We are grateful to Volker Haucke (Leibniz Institut für Molekulare Pharmakologie, Berlin, Germany) for reagents. GFP-CI-M6PR was provided by Mihaela Anitei (Technische Universität, Dresden, Germany). We acknowledge Justin Hsuan, in whose laboratory the studies were initiated. We are grateful to Robin Ketteler (Laboratory for Molecular Cell Biology, University College London) for help and advice. This work was supported by a National Universities Commission (NUC) of Nigeria PRESSID Scholarship (G.O.A. and S.M.) and Medical Research Council core funding to J.K. (ref. MC_EX_G0800785).
Abbreviations used:
- CI-M6PR
cation-independent mannose 6-phosphate receptor
- CNA
copy number alteration
- ECM
extracellular matrix
- EE
early endosome
- EEA1
early endosomal autoantigen 1
- EGFR
epidermal growth factor receptor
- GFP
green fluorescent protein
- GST
glutathione-S-transferase
- LE
late endosome
- MT1-MMP
membrane type I matrix metalloproteinase
- NT
nontargeting siRNA
- P4C
PI(4)P-binding domain of SidC
- PI4KIIα
phosphatidylinositol 4-kinase type II alpha
- PI4KIIβ
phosphatidylinositol 4-kinase type II beta
- PI(3)P
phosphatidylinositol 3-phosphate
- PI(4)P
phosphatidylinositol 4-phosphate.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-08-0564) on October 26, 2016.
REFERENCES
- Balla A, Balla T. Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends Cell Biol. 2006;16:351–361. doi: 10.1016/j.tcb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Balla A, Tuymetova G, Barshishat M, Geiszt M, Balla T. Characterization of type II phosphatidylinositol 4-kinase isoforms reveals association of the enzymes with endosomal vesicular compartments. J Biol Chem. 2002;277:20041–20050. doi: 10.1074/jbc.M111807200. [DOI] [PubMed] [Google Scholar]
- Bowden ET, Barth M, Thomas D, Glazer RI, Mueller SC. An invasion-related complex of cortactin, paxillin and PKCmu associates with invadopodia at sites of extracellular matrix degradation. Oncogene. 1999;18:4440–4449. doi: 10.1038/sj.onc.1202827. [DOI] [PubMed] [Google Scholar]
- Bravo-Cordero JJ, Cordani M, Soriano SF, Diez B, Munoz-Agudo C, Casanova-Acebes M, Boullosa C, Guadamillas MC, Ezkurdia I, Gonzalez-Pisano D, et al. A novel high-content analysis tool reveals Rab8-driven cytoskeletal reorganization through Rho GTPases, calpain and MT1-MMP. J Cell Sci. 2016;129:1734–1749. doi: 10.1242/jcs.174920. [DOI] [PubMed] [Google Scholar]
- Bravo-Cordero JJ, Marrero-Diaz R, Megias D, Genis L, Garcia-Grande A, Garcia MA, Arroyo AG, Montoya MC. MT1-MMP proinvasive activity is regulated by a novel Rab8-dependent exocytic pathway. EMBO J. 2007;26:1499–1510. doi: 10.1038/sj.emboj.7601606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craige B, Salazar G, Faundez V. Phosphatidylinositol-4-kinase type II alpha contains an AP-3-sorting motif and a kinase domain that are both required for endosome traffic. Mol Biol Cell. 2008;19:1415–1426. doi: 10.1091/mbc.E07-12-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destaing O, Planus E, Bouvard D, Oddou C, Badowski C, Bossy V, Raducanu A, Fourcade B, Albiges-Rizo C, Block MR. beta1A integrin is a master regulator of invadosome organization and function. Mol Biol Cell. 2010;21:4108–4119. doi: 10.1091/mbc.E10-07-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia E, Machesky LM, Jones GE, Anton IM. WIP is necessary for matrix invasion by breast cancer cells. Eur J Cell Biol. 2014;93:413–423. doi: 10.1016/j.ejcb.2014.07.008. [DOI] [PubMed] [Google Scholar]
- Hammond GR, Machner MP, Balla T. A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J Cell Biol. 2014;205:113–126. doi: 10.1083/jcb.201312072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattula K, Furuhjelm J, Tikkanen J, Tanhuanpaa K, Laakkonen P, Peranen J. Characterization of the Rab8-specific membrane traffic route linked to protrusion formation. J Cell Sci. 2006;119:4866–4877. doi: 10.1242/jcs.03275. [DOI] [PubMed] [Google Scholar]
- Henmi Y, Morikawa Y, Oe N, Ikeda N, Fujita A, Takei K, Minogue S, Tanabe K. PtdIns4KIIalpha generates endosomal PtdIns(4)P and is required for receptor sorting at early endosomes. Mol Biol Cell. 2016;27:990–1001. doi: 10.1091/mbc.E15-08-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotary K, Li XY, Allen E, Stevens SL, Weiss SJ. A cancer cell metalloprotease triad regulates the basement membrane transmigration program. Genes Dev. 2006;20:2673–2686. doi: 10.1101/gad.1451806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovic M, Kean MJ, Szentpetery Z, Polevoy G, Gingras AC, Brill JA, Balla T. Two phosphatidylinositol 4-kinases control lysosomal delivery of the Gaucher disease enzyme, beta-glucocerebrosidase. Mol Biol Cell. 2012;23:1533–1545. doi: 10.1091/mbc.E11-06-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Barylko B, Lu D, Shu H, Yin H, Albanesi JP. Stabilization of phosphatidylinositol 4-kinase type IIbeta by interaction with Hsp90. J Biol Chem. 2011;286:12775–12784. doi: 10.1074/jbc.M110.178616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kean MJ, Williams KC, Skalski M, Myers D, Burtnik A, Foster D, Coppolino MG. VAMP3, syntaxin-13 and SNAP23 are involved in secretion of matrix metalloproteinases, degradation of the extracellular matrix and cell invasion. J Cell Sci. 2009;122:4089–4098. doi: 10.1242/jcs.052761. [DOI] [PubMed] [Google Scholar]
- Ketel K, Krauss M, Nicot AS, Puchkov D, Wieffer M, Muller R, Subramanian D, Schultz C, Laporte J, Haucke V. A phosphoinositide conversion mechanism for exit from endosomes. Nature. 2016;529:408–412. doi: 10.1038/nature16516. [DOI] [PubMed] [Google Scholar]
- Li J, Lu Y, Zhang J, Kang H, Qin Z, Chen C. PI4KIIalpha is a novel regulator of tumor growth by its action on angiogenesis and HIF-1alpha regulation. Oncogene. 2010;29:2550–2559. doi: 10.1038/onc.2010.14. [DOI] [PubMed] [Google Scholar]
- Li J, Zhang L, Gao Z, Kang H, Rong G, Zhang X, Chen C. Dual inhibition of EGFR at protein and activity level via combinatorial blocking of PI4KIIalpha as anti-tumor strategy. Protein Cell. 2014;5:457–468. doi: 10.1007/s13238-014-0055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Wasilko DJ, Liu Y, Sun J, Wu X, Luo ZQ, Mao Y. Structure of the Legionella virulence factor, SidC reveals a unique PI(4)P-specific binding domain essential for its targeting to the bacterial phagosome. PLoS Pathog. 2015;11:e1004965. doi: 10.1371/journal.ppat.1004965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson IR, Rainero E, Mitchell LE, van den Berghe PV, Speirs C, Dozynkiewicz MA, Chaudhary S, Kalna G, Edwards J, Timpson P, Norman JC. CLIC3 controls recycling of late endosomal MT1-MMP and dictates invasion and metastasis in breast cancer. J Cell Sci. 2014;127:3893–3901. doi: 10.1242/jcs.135947. [DOI] [PubMed] [Google Scholar]
- Mazzocca A, Liotta F, Carloni V. Tetraspanin CD81-regulated cell motility plays a critical role in intrahepatic metastasis of hepatocellular carcinoma. Gastroenterology. 2008;135:244–256.e241. doi: 10.1053/j.gastro.2008.03.024. [DOI] [PubMed] [Google Scholar]
- Minogue S, Waugh MG. The phosphatidylinositol 4-kinases: don’t call it a comeback. Subcell Biochem. 2012;58:1–24. doi: 10.1007/978-94-007-3012-0_1. [DOI] [PubMed] [Google Scholar]
- Minogue S, Waugh MG, De Matteis MA, Stephens DJ, Berditchevski F, Hsuan JJ. Phosphatidylinositol 4-kinase is required for endosomal trafficking and degradation of the EGF receptor. J Cell Sci. 2006;119:571–581. doi: 10.1242/jcs.02752. [DOI] [PubMed] [Google Scholar]
- Mossinger J, Wieffer M, Krause E, Freund C, Gerth F, Krauss M, Haucke V. Phosphatidylinositol 4-kinase IIalpha function at endosomes is regulated by the ubiquitin ligase Itch. EMBO Rep. 2012;13:1087–1094. doi: 10.1038/embor.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz H, Pathak N, Yang J. Invading one step at a time: the role of invadopodia in tumor metastasis. Oncogene. 2014;33:4193–4202. doi: 10.1038/onc.2013.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poincloux R, Lizarraga F, Chavrier P. Matrix invasion by tumour cells: a focus on MT1-MMP trafficking to invadopodia. J Cell Science. 2009;122:3015–3024. doi: 10.1242/jcs.034561. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro I, Yuan L, Tanentzapf G, Dowling JJ, Kiger A. Phosphoinositide regulation of integrin trafficking required for muscle attachment and maintenance. PLoS Genet. 2011;7:e1001295. doi: 10.1371/journal.pgen.1001295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeh F, Ota I, Holmbeck K, Birkedal-Hansen H, Soloway P, Balbin M, Lopez-Otin C, Shapiro S, Inada M, Krane S, et al. Tumor cell traffic through the extracellular matrix is controlled by the membrane-anchored collagenase MT1-MMP. J Cell Biol. 2004;167:769–781. doi: 10.1083/jcb.200408028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar G, Craige B, Wainer BH, Guo J, De Camilli P, Faundez V. Phosphatidylinositol-4-kinase type II alpha is a component of adaptor protein-3-derived vesicles. Mol Biol Cell. 2005;16:3692–3704. doi: 10.1091/mbc.E05-01-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Gaullier JM, D’Arrigo A, Stenmark H. The Rab5 effector EEA1 interacts directly with syntaxin-6. J Biol Chem. 1999;274:28857–28860. doi: 10.1074/jbc.274.41.28857. [DOI] [PubMed] [Google Scholar]
- Steffen A, Le Dez G, Poincloux R, Recchi C, Nassoy P, Rottner K, Galli T, Chavrier P. MT1-MMP-dependent invasion is regulated by TI-VAMP/VAMP7. Curr Biol. 2008;18:926–931. doi: 10.1016/j.cub.2008.05.044. [DOI] [PubMed] [Google Scholar]
- Tatin F, Varon C, Genot E, Moreau V. A signalling cascade involving PKC, Src and Cdc42 regulates podosome assembly in cultured endothelial cells in response to phorbol ester. J Cell Sci. 2006;119:769–781. doi: 10.1242/jcs.02787. [DOI] [PubMed] [Google Scholar]
- Taunton J, Rowning BA, Coughlin ML, Wu M, Moon RT, Mitchison TJ, Larabell CA. Actin-dependent propulsion of endosomes and lysosomes by recruitment of N-WASP. J Cell Biol. 2000;148:519–530. doi: 10.1083/jcb.148.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sun HQ, Macia E, Kirchhausen T, Watson H, Bonifacino JS, Yin HL. PI4P promotes the recruitment of the GGA adaptor proteins to the trans-Golgi network and regulates their recognition of the ubiquitin sorting signal. Mol Biol Cell. 2007;18:2646–2655. doi: 10.1091/mbc.E06-10-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YJ, Wang J, Sun HQ, Martinez M, Sun YX, Macia E, Kirchhausen T, Albanesi JP, Roth MG, Yin HL. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell. 2003;114:299–310. doi: 10.1016/s0092-8674(03)00603-2. [DOI] [PubMed] [Google Scholar]
- Waugh MG, Minogue S, Anderson JS, Balinger A, Blumenkrantz D, Calnan DP, Cramer R, Hsuan JJ. Localization of a highly active pool of type II phosphatidylinositol 4-kinase in a p97/valosin-containing-protein-rich fraction of the endoplasmic reticulum. Biochem J. 2003;373:57–63. doi: 10.1042/BJ20030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixel KM, Blumental-Perry A, Watkins SC, Aridor M, Weisz OA. Distinct Golgi populations of phosphatidylinositol 4-phosphate regulated by phosphatidylinositol 4-kinases. J Biol Chem. 2005;280:10501–10508. doi: 10.1074/jbc.M414304200. [DOI] [PubMed] [Google Scholar]
- Wieffer M, Cibrian Uhalte E, Posor Y, Otten C, Branz K, Schutz I, Mossinger J, Schu P, Abdelilah-Seyfried S, Krauss M, Haucke V. PI4K2beta/AP-1-based TGN-endosomal sorting regulates Wnt signaling. Curr Biol. 2013;23:2185–2190. doi: 10.1016/j.cub.2013.09.017. [DOI] [PubMed] [Google Scholar]
- Wiesner C, El Azzouzi K, Linder S. A specific subset of RabGTPases controls cell surface exposure of MT1-MMP, extracellular matrix degradation and three-dimensional invasion of macrophages. J Cell Sci. 2013;126:2820–2833. doi: 10.1242/jcs.122358. [DOI] [PubMed] [Google Scholar]
- Williams KC, Coppolino MG. Phosphorylation of membrane type 1-matrix metalloproteinase (MT1-MMP) and its vesicle-associated membrane protein 7 (VAMP7)-dependent trafficking facilitate cell invasion and migration. J Biol Chem. 2011;286:43405–43416. doi: 10.1074/jbc.M111.297069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Zech T, McDonald L, Gonzalez EG, Li A, Macpherson I, Schwarz JP, Spence H, Futo K, Timpson P, et al. N-WASP coordinates the delivery and F-actin-mediated capture of MT1-MMP at invasive pseudopods. J Cell Biol. 2012;199:527–544. doi: 10.1083/jcb.201203025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng HT, Jiang LX, Lv ZC, Li DP, Zhou CZ, Gao JJ, He L, Peng ZH. Are there tumor suppressor genes on chromosome 4p in sporadic colorectal carcinoma. World J Gastroenterol. 2008;14:90–94. doi: 10.3748/wjg.14.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.