Abstract
Expression of pituitary and placental members of the human GH and chorionic somatomammotropin (CS) gene family is directed by an upstream remote locus control region (LCR). Pituitary-specific expression of GH requires direct binding of Pit-1 (listed as POU1F1 in the HUGO database) to sequences marked by a hypersensitive site (HS) region (HS I/II) 14·6 kb upstream of the GH-N gene (listed as GH1 in the HUGO database). We used human embryonic kidney 293 (HEK293) cells overexpressing wild-type and mutant Pit-1 proteins as a model system to gain insight into the mechanism by which Pit-1 gains access to the GH LCR. Addition of Pit-1 to these cells increased DNA accessibility at HS III, located 28 kb upstream of the human GH-N gene, in a POU homeodomain-dependent manner, as reflected by effects on histone hyperacetylation and RNA polymerase II activity. Direct binding of Pit-1 to HS III sequences is not supported. However, the potential for binding of ETS family members to this region has been demonstrated, and Pit-1 association with this ETS element in HS III sequences requires the POU homeodomain. Also, both ETS1 and ELK1 co-precipitate from human pituitary extracts using two independent sources of Pit-1 antibodies. Finally, overexpression of ELK1 or Pit-1 expression in HEK293 cells increased GH-N RNA levels. However, while ELK1 overexpression also stimulated placental CS RNA levels, the effect of Pit-1 appeared to correlate with ETS factor levels and target GH-N preferentially. These data are consistent with recruitment and an early role for Pit-1 in remodeling of the GH LCR at the constitutively open HS III through protein–protein interaction.
Introduction
The five human GH/chorionic somatomammotropin (CS) gene family members include pituitary GH (GH-N, listed as GH1 in the HUGO database), placental GH variant (GH-V, GH2), and the placenta-expressed CS genes (CS-A (CSH1), CS-B (CSH2), and CS-L (CSHL1)), contained within a single 47 kb locus on chromosome 17 (Chen et al. 1989). The five GH/CS genes are aligned in the same transcriptional orientation in the order 5′-GH-N/CS-L/CS-A/GH-V/CS-B-3′, share 90–99% nucleotide sequence similarity, and are believed to have evolved by gene duplication (Chen et al. 1989). The separate expression of GH family members in pituitary and placenta distinguishes the GH/CS locus from the extensively studied β-globin locus, where expression is restricted to a single tissue type (erythroid tissues) with varied expression during developmental stages (Bulger et al. 1999). Thus, the GH/CS locus offers a useful model to investigate the molecular mechanisms involved in tissue-specific gene regulation.
The expression of the GH/CS genes is largely under the control of a remote locus control region (LCR), located 14–32 kb upstream of the GH-N promoter and encompassing a total of five hypersensitive sites (HS I–V; Jones et al. 1995, Jin et al. 2009). The appearance of HS I/II is specific to pituitary somatotrophs, while HS IV is unique to placental tissue. HS III and HS V appear in both tissues, and more recently, it was reported that HS III is in fact ubiquitous (Kimura et al. 2007). Even though HS I/II are required for pituitary expression of GH-N ( Jin et al. 1999, Shewchuk et al. 1999), the inclusion of the entire, intact LCR encompassing HS I–V was required to recapitulate appropriate levels of pituitary-specific human GH-N expression in transgenic mice ( Jones et al. 1995, Bennani-Baiti et al. 1998). Distinct patterns of histone acetylation and methylation were associated with pituitary and placental chromatin (Ho et al. 2004, Kimura et al. 2004). In the pituitary, these epigenetic modifications are dependent on i) the presence of HS I (Ho et al. 2002) and ii) the extension of LCR transcription (measured by RNA polymerase (pol) II activity) from HS I/II in the direction of the GH-N gene (Ho et al. 2008). Chromatin conformation capture (3C) analysis has revealed the inclusion of the GH-N promoter, specific to pituitary chromatin, in a common complex with both the HS I/II and the HS III, V regions (Ho et al. 2008). This physical association of the GH-N promoter with the LCR complex is also dependent on both the presence of HS I and transcriptional activity downstream from the LCR. Interestingly, in the absence of GH-N promoter juxtaposition (and, consequently, GH-N expression), the regions containing HS I/II and HS III, V remain in close approximation (Ho et al. 2008).
HS I/II contain multiple Pit-1(POU1F1)-binding sites that are sufficient to confer pituitary-specific expression in vivo (Jin et al. 1999, Shewchuk et al. 1999). The binding of Pit-1 in this region is known to be an important step in the development of pituitary GH-N expression (Shewchuk et al. 2006), which defines the somatotroph. However, Pit-1 protein is expressed in three of the five hormone-producing cell types of the anterior pituitary including prolactin-producing lactotrophs and thyroid-stimulating hormone-producing thyrotrophs (Li et al. 1990, Radovick et al. 1992). If the presence of Pit-1 alone were sufficient for gaining access to HS I/II and/or activation of the GH locus, then GH-N RNA expression might be expected in lactotrophs and thyrotrophs as well as in somatotrophs. Nonetheless, the ability to identify and dissect the events resulting from the appearance of Pit-1 in terms of GH-N expression has been hampered by both the inherent difficulty in obtaining human embryonic cells of the pre-somatotroph lineage and the differences between the GH(CS) locus in primates and non-primates.
Here, we have pursued the expression of Pit-1 in a human embryonic kidney 293 (HEK293), and hence non-pituitary/placenta, cell line as a model system to examine potential early-stage interactions between Pit-1 and the human GH gene locus. Effects on chromatin associated with increased DNA accessibility were detected at HS III, in spite of the presence of high-affinity Pit-1 DNA elements at HS I/II and their absence at HS III. Evidence for an effect mediated through protein–protein interaction and independent of direct DNA binding using Pit-1 mutants is discussed.
Materials and methods
Cell culture, plasmid construction, and gene transfer
HEK293 cells were maintained at 37 °C in a monolayer culture in DMEM (pH 7·4) supplemented with 5% fetal bovine serum and antibiotics in a humidified air/CO2 (19:1) atmosphere. The c-myc/Pit-1 expression vector was obtained by inserting the human Pit-1 cDNA downstream of the pCMV-myc expression vector (Clontech Laboratories, Inc). The cDNA for wild-type (wt) and mutant Pit-1 proteins, including complete deletion of the POU homeodomain (ΔPOUHD), and POU-specific domain (ΔPOUS), as well as partial deletion of the N-terminal trans-activation domain(ΔN8–128), were kindly provided by Dr H Ingraham (Ingraham et al. 1990b), and were re-inserted into the expression vector pcDNA3.1. The construct containing a hybrid thymidine kinase (TK) promoter/firefly luciferase (luc) gene with a 1·6 kb BglII fragment containing HS I/II from the human GH LCR inserted upstream (HS I/IITKp.luc) is described elsewhere as 1.6GTKp.luc ( Jin et al. 1999). Transient gene transfer was performed using a high-transfection efficiency method ‘Trans-IT293 reagent’ (Mirus Bio Corp., Madison, WI, USA) according to the manufacturer’s directions. Cells were harvested 48 h after transfection, and nuclei and RNA were isolated. Luc activity was measured after cell lysis in Tris-Triton buffer (100 mM Tris–HCl (pH 7·8) and 0·1% Triton X-100) using a photon-counting luminometer (Lumat LB9507; Berthold Technologies, Oakville, TN, USA). Values were normalized with protein concentration, which was assessed using the Bradford protein assay and BSA as a standard.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assay (EMSA) was performed essentially as described previously (Norquay et al. 2006). Competitor oligonucleotide was pre-incubated with cellular nuclear extracts and poly(dI-dC) in a reaction buffer (10 mM Tris–HCl (pH 7·5); 50 mM NaCl; 1 mM dithiothreitol; 1 mM EDTA; and 5% (v/v) glycerol) for 10 min at room temperature. Radiolabeled oligonucleotide probes were added, and the reaction mixtures were incubated for a further 10 min at room temperature. Antibodies to c-myc (Clontech #631604), Pit-1 (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA, X-7, sc-442), ETS1 (Santa Cruz Biotechnology Inc., sc-111X), and ELK1 (Santa Cruz Biotechnology Inc., sc-355) were added to the binding reaction mixtures, and incubated for 10 min further at room temperature. Recombinant Pit-1 protein (Santa Cruz Biotechnology Inc., sc-4014) was also used. The DNA–protein complexes were resolved in non-denaturing 5% (w/v) polyacrylamide gels and visualized by autoradiography. The sequences of the oligonucleotide probes can be found in Supplementary Table 1, see section on supplementary data given at the end of this article.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed using a modified human tissue procedure as described previously (Norquay et al. 2003). In brief, cells were harvested 48 h after transfection in cold PBS buffer, and crosslinked with formaldehyde before lysis and sonication of chromatin. Insoluble material was removed by centrifugation, and the DNA content was measured by spectrophotometry. Chromatin was pre-cleared for 3 h, immunoprecipitated overnight at 4 °C, pelleted by the addition of protein A/G sepharose, and reverse crosslinked at 68 °C. DNA was isolated using QIAquick columns (Qiagen), and was further analyzed by PCR. PCR was performed using 10 ng of input DNA or 5 μl of eluted (bound) DNA per PCR (Taq DNA pol; Qiagen) at an annealing temperature of 55 °C for 27 cycles. Primers used for PCRs can be found in Supplementary Table 2, see section on supplementary data given at the end of this article. A region of the fibroblast growth factor-16 (FGF16) gene, which is expressed predominantly in embryonic brown adipose tissue and in postnatal heart, was used as a control for non-specific sequences present in our bound sample as described previously for the assessment of pituitary and placental chromatin (Norquay et al. 2003, 2006, Jin et al. 2004).
RNA preparation and non-coding transcription analysis
Total RNA was extracted from transfected HEK293 cells using QIA shredder and RNeasy Plus Mini Kit (Qiagen). A series of sense and anti-sense strand-specific reverse transcriptase (RT) primers were generated to detect bi-directional transcripts in approximately every 1 kb beginning upstream of HS V and downstream across HS I/II and to the GH-N promoter (Supplementary Table 3, see section on supplementary data given at the end of this article). For RT-PCR analysis, 1 μg of RNA was reverse transcribed with the addition of 5 picomolar sequence-specific RT primer first, and then with MMLV transcriptase (Invitrogen Life Technologies). Minus RT reactions were also set up to confirm the absence of genomic DNA contamination. Ten percent of the RT reaction mixture was used for PCR. PCR primers (Supplementary Table 4, see section on supplementary data given at the end of this article) were designed downstream of each sequence-specific RT primer in the sense direction. PCR was performed at an annealing temperature of 55 °C for 30 cycles using Taq DNA pol (Qiagen).
Protein blotting and immunoprecipitation
For the detection of wt and mutant Pit-1 proteins in transfected HEK293 cells, 20-μg whole cell protein from transfected cells was analyzed by protein blotting as described previously (Norquay et al. 2006). Polyclonal goat antibodies to the amino terminus of human Pit-1 were used to detect wt and mutant Pit-1 proteins (N-20, #sc-16288, Santa Cruz Biotechnology Inc.). For immunoprecipitation (IP) experiments, all the steps were carried out at 4 °C. Initially, 200-μg nuclear extract from transfected HEK293 cells was pre-cleared for 1 h with 20-μl protein A/G plus agarose. Pre-cleared extracts were then equally divided into control and experimental groups for IP with 2-μg normal mouse serum (Santa Cruz Biotechnology Inc., sc-45051) or c-myc monoclonal antibody (Clontech Laboratories, Inc.) respectively. IP was carried out overnight, and on the following day, extracts were incubated with protein A/G plus agarose for 3 h. Washes were done for 2 min using the following: supplemented NET buffer (50 mM Tris, pH 7·5; 500 mM NaCl; 0·1% NP-40; and 1 mM EDTA), followed by NET buffer (50 mM Tris, pH 7·5; 150 mM NaCl; 0·1% NP-40; and 1 mM EDTA), and a final wash buffer (10 mM Tris and 0·1% NP-40). The immunoprecipitates were then extracted with 0·4, 0·6, and 1·0 M KCl. Protein was recovered using 10K Nanosep centrifugal devices (PALL Life Sciences) according to the manufacturer’s instructions. The protein fraction was then assessed by EMSA with different radiolabeled oligonucleotides.
IP using the ‘Universal Magnetic Co-IP’ kit were performed according to the manufacture’s instructions. In brief, 2-g post-mortem human pituitary tissue samples (obtained from the Human Pituitary Repository, Protein and Polypeptide Laboratory, University of Manitoba) were used to harvest nuclear proteins according to the kit instruction. Three hundred-microgram nuclear proteins were used in each IP reaction. The antibodies against ETS1 (N-276, sc-111, Santa Cruz Biotechnology Inc.) and ELK1 (I-20, sc-355, Santa Cruz Biotechnology Inc.), as well as non-immune normal rabbit serum, were used to precipitate the associated proteins. Purified proteins were then resolved by SDS-PAGE, immunoblotted, and detected with two separate Pit-1 antibodies (N-20, #sc-16288, and affinity purified rabbit antibodies to a polyhistidine fusion protein including full-length rat Pit-1, X-7, #sc-442, Santa Cruz Biotechnology Inc).
DNA affinity purification
Forty picomoles of double-stranded biotinylated oligonucleotides (bioETS/HS III) were coupled to 0·2 mg of streptavidin magnetic beads (Promega Corp.), and affinity purification of nuclear extract from transfected HEK293 cells was done at room temperature as described previously (Norquay et al. 2006). Beads were washed three times, and then, bound protein complexes were removed from the beads by boiling for 5 min in Laemmli sample buffer, followed by immunoblotting.
Quantitative real-time PCR
Quantitative RT-PCR (qRT-PCR) analyses were performed in an iCycler (Bio-Rad) with specific primers designed for GH-N, CS-A/B, and GH-V respectively (Supplementary Table 2). Reaction volumes of 30 μl consisted of the following components: 2·5 mM MgCl2, 0·025% DMSO, 0·6 μl (1:1000 dilution) of SYBR green (Sigma, S9430), 0·3 μl (1:1000 dilution) of fluorescein calibration dye (Bio-Rad, Cat# 170-8780), and 0·75 units of platinum Taq DNA pol (Invitrogen). Thermal cycling was initiated with a 4-min denaturation at 95 °C followed by 40 cycles of 95 °C for 30 s, and annealing at 60 °C for 15 s and 72 °C for 30 s. Standard curves were generated using plasmids containing the amplicon sequences for GH-N, CS-A/B, GH-V, and GAPDH. Minus RT control reactions were performed using the same PCR primers and thermal cycle conditions to detect evidence of genomic DNA contamination. Specific amplifications were identified by a single peak in the melting curve and a single band in the final PCR product visualized on an agarose gel. The gene expression level in each sample (absolute quantification) was calculated from the standard curve and normalized to human GAPDH expression as appropriate. Tests were run in duplicate on independent samples. All amplicon sequences were confirmed by sequencing.
Statistical analysis
Statistical analysis was performed using GraphPad Instat software (GraphPad, La Jolla, CA, USA). For single comparisons, paired t-tests were applied, and for multiple comparisons, one-way ANOVA was used with the Tukey–Kramer or Dunnet’s post-test as appropriate. A value of P<0·05 is considered statistically significant (*P<0·05; **P<0·01; ***P<0·001).
Results
Evidence for increased DNA accessibility at HS III through the association with an intact Pit-1 POU homeodomain
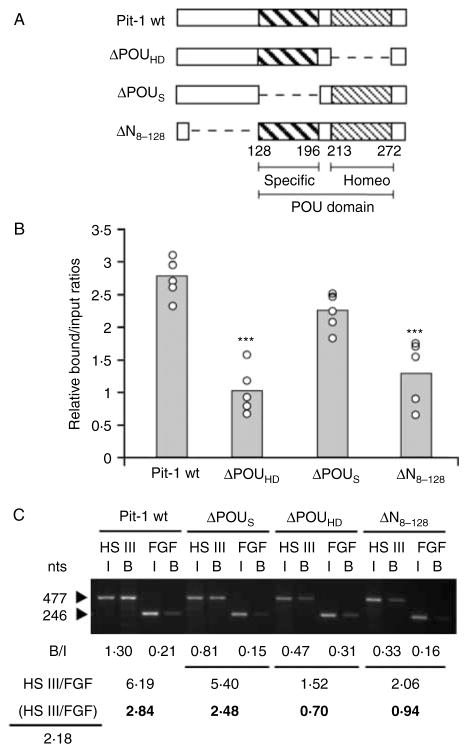
In the absence of human pituitary pre-somatotrophs, wt and mutated Pit-1 proteins were compared for their effects on chromatin remodeling and DNA accessibility in the (inactive) GH LCR in HEK293 cells. Expression vectors for wt Pit-1 and for previously reported mutants lacking three significant regions of the Pit-1 protein were generated: these include complete ΔPOUHD, complete ΔPOUS, and partial ΔN8–128 (Ingraham et al. 1990b). For acetylation, antibodies to hyperacetylated histone H4 were used in combination with PCR to detect the HS III region by ChIP assay in cells 48 h after gene transfer. The bound/input ratios were expressed relative to FGF16 as described for the assessment of transfected c-myc/Pit-1 expression (Cattini et al. 2006). In a series of five independent experiments, expression of wt Pit-1 was associated with increased histone H4 hyperacetylation at HS III (Fig. 1), as observed previously with c-myc/Pit-1 expression (Cattini et al. 2006). However, this effect was lost with the ΔPOUHD or the N-terminal trans-activation domain (ΔN8–128). In contrast, disruption of the POU-specific domain (ΔPOUS) did not result in a loss of hyperacetylation.
Figure 1.
Effect of Pit-1 deletions on histone H4 hyperacetylation at HS III. (A) HEK293 cells were transfected with wild-type and mutant Pit-1 expression vectors. (B) Nuclei from transfected HEK293 cells were analyzed by ChIP for hyperacetylated histone H4 at HS III. Bound/input (B/I) ratios for wild-type (wt) Pit-1 and each mutant were determined by gel electrophoresis and densitometry analysis, and were normalized to values obtained with unrelated FGF16. Corrected values are expressed as a fold increase relative to non-transfected HEK293 cells. The values for five independent experiments are shown. Mean values are indicated by gray-filled bars. Statistical analysis was performed by one-way ANOVA with the Tukey–Kramer post-hoc test for multiple comparisons. ***P<0·001. ΔPOUHD, POU homeodomain deletion; ΔPOUS, POU-specific domain deletion; ΔN8–128, N-terminal domain deletion. (C) Representative PCR results for the HS III and FGF16 (FGF) regions in input, I, and bound, B, fractions from HEK293 cell ChIP assays. For cells transfected with wild-type and mutant Pit-1, values for B/I ratio for HS III and FGF are shown, as well as HS III/FGF16 values. The HS III/FGF16 value for non-transfected cells was 2·18, and was used to generate relative values (bold) for fold effect.
The effect of Pit-1 on DNA accessibility to RNA pol II was assessed through the detection of relative levels of bi-directional (sense and anti-sense) non-coding RNA transcripts. RNA was isolated from cells 48 h after gene transfer and assessed using specific RT and PCR primers. Mean values for relative (to GAPDH) transcript levels at sites within the GH LCR were obtained (Fig. 2A). The highest and most significant levels of transcripts were observed associated with and immediately downstream of HS III.
Figure 2.
Fold effect of Pit-1 deletions on the level of non-coding transcripts along the GH/CS locus in transfected HEK293 cells. The Pit-1 expression vectors were each introduced into HEK293 cells by transient transfection and assessed after 48 h. Random transcripts (anti-sense and/or sense) were detected using sequence-specific reverse transcription primers followed by PCR. All values are normalized to GAPDH, which is presumed to be constitutively active, in each group individually, and then corrected relative to the non-transfected HEK293 group. (A) The mean relative RNA level at each region along the GH/CS locus is shown for c-myc/Pit-1 (Cattini et al. 2006) in the sense and anti-sense directions. Bars represent S.E.M. Statistical analysis was done by one-way ANOVA followed by Dunnet’s post-test for multiple comparisons with a single control (n=6), *P<0·05, **P<0·01. (B) The mean relative fold effects at each region along the GH/CS locus are shown for wild-type Pit-1 (wt) and each of the Pit-1 mutants (ΔPOUHD, ΔPOUS, and ΔN8–128) in the sense direction (n=3–5). The relative position of each amplicon along the GH/CS locus is also shown. GHp, GH promoter; GH-N, GH gene. Bars represent S.E.M.
To further characterize Pit-1 action, the fold effect of mutated versus wt Pit-1 proteins on DNA accessibility to RNA pol II was assessed through the detection of relative levels of (sense) non-coding RNA transcripts. RNA was again isolated from cells 48 h after gene transfer, and assessed using specific RT and PCR primers reflecting transcript levels in the GH LCR as indicated (Fig. 2B). The results obtained with wt and mutated Pit-1 are normalized to GAPDH RNA levels to allow relative effects to be compared. As such, values are expressed as a fold effect of Pit-1 (wt or mutant) expression. Only disruption of the POU homeodomain resulted in a decrease in relative transcript levels and, thus, presumably RNA pol II activity; in contrast, both the N-terminal and ΔPOUS mutants were still associated with increased transcript levels and, thus, appeared to retain the ability to stimulate RNA pol II activity (Fig. 2B).
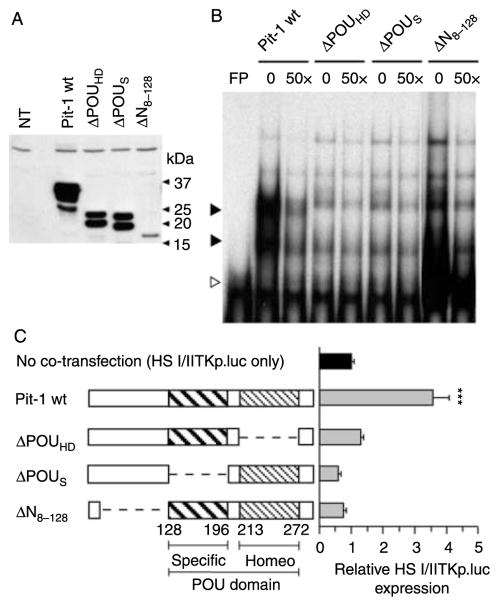
Based on the literature, disruption of both the POU homeodomain and POU-specific domain is expected to interfere with DNA binding (Ingraham et al. 1990b). Given the differential effects of these expression vectors on DNA accessibility (Figs 1 and 2), binding and trans-activation properties of these mutants were re-assessed. Expression of wt and mutant Pit-1 proteins in transfected HEK293 cells was assessed by electrophoresis and immunoblotting using antibodies raised against total Pit-1 protein (Fig. 3A). Major bands of the expected sizes were detected as appropriate. To assess binding of the different Pit-1 products to DNA, 1 μg of nuclear extract from transfected HEK293 cells was used with GH promoter (GHp) Pit-1 oligonucleotide probe in an EMSA (Fig. 3B). The GHp Pit-1 oligonucleotide has been shown to direct pituitary-specific expression of reporter gene, to possess pituitary-specific binding activity, and to specifically bind to recombinant Pit-1 protein (Lefevre et al. 1987, Nickel et al. 1991, Nachtigal et al. 1993, Jin et al. 1999). Major binding complexes were detected with wt Pit-1 (solid arrowhead) and the N-terminal mutant ΔN8–128 (open arrowhead), and both could be competed with an unlabeled probe. Major complexes were not suggested with nuclear extracts containing Pit-1 mutants with deletion of either POU domain.
Figure 3.
Characterization of mutant Pit-1 proteins in transiently transfected HEK293 cells. (A) HEK293 cells were transiently transfected with expression vectors for wild-type and mutant Pit-1 proteins. Expression was analyzed by protein immunoblotting with anti-Pit-1 antibodies. NT, non-transfected; Pit-1wt, wild-type Pit-1; ΔPOUHD, Pit-1 POU homeodomain deletion; ΔPOUS, Pit-1 POU-specific domain deletion; ΔN8–128, Pit-1 N-terminal trans-activation domain deletion. Protein sizes are indicated in kilodaltons. (B) Nuclear extract (1 μg) from cells transfected with the indicated forms of Pit-1 was assessed by EMSA using a radiolabeled Pit-1-binding site from the human GH-N promoter (GH-N–Pit-1). Both wild-type (solid arrowhead) and N-terminal-deleted Pit-1 (open arrowhead), but not the Pit-1 POU deletion mutants, bind to the Pit-1 DNA element. An excess of unlabeled Pit-1 element (50×) was added as a specific competitor. Note: no specific complexes were detected when non-transfected HEK293 nuclear extract was used. (C) Schematic showing linear structure of Pit-1 wild type and mutants that were co-transfected with the HS I/IITKp.luc reporter gene. Firefly luciferase activity was normalized to protein and expressed as per μg protein lysate. Corrected values are expressed as a percentage of HS I/IITKp.luc activity, which is arbitrarily set to 1·0 (black-filled bar). Statistical analysis was performed by one-way ANOVA followed by the Tukey–Kramer post-test for multiple comparisons (n=6). The mean value for HS I/IITKp.luc was 9·12±0·51 (relative light units ×104). ***P<0·001. Bars represent S.E.M.
The ability of Pit-1 mutant proteins to trans-activate the HS I/IITKp.luc reporter gene was assessed in HEK293 cells. Cells were transfected with HS I/IITKp.luc and co-transfected with wt, ΔN8–128, ΔPOUS, or ΔPOUHD Pit-1. The activity in cell lysate was assessed after 48 h, and was corrected for DNA uptake through co-transfection with Renilla luciferase. Results are expressed as fold increase relative to HS I/IITKp.luc activity with no co-transfection (Fig. 3C). Expression of wt Pit-1 significantly increased TK promoter activity. In contrast, there was no significant stimulation with mutations of Pit-1 in the DNA binding (ΔPOUHD and ΔPOUS) or trans-activation (ΔN8–128) domains. Thus, in spite of the differential effects of ΔPOUHD and ΔPOUS Pit-1 on DNA accessibility at HS III, this does not appear to correlate with DNA binding.
Pit-1 does not bind to Pit-1/Oct-1-like DNA elements in HS III-related sequences corresponding to nucleotides −28 225/−27 652 of the GH-N gene
The possible lack of a requirement for DNA binding was pursued. Initial inspection of HS I/II sequences in the GH LCR did not reveal Pit-1 DNA elements based on the identity with the known consensus (Pit-1) element available at that time (Bennani-Baiti et al. 1998). However, given the pituitary-specific nature of HS I/II, we pursued the presence of Pit-1-like elements in this region and reported Pit-1 binding ( Jin et al. 1999). This was confirmed by others (Shewchuk et al. 1999), and was shown subsequently in elegant studies to be essential for remodeling of the GH locus and GH-N expression in transgenic mice in vivo (Ho et al. 2002). As such, the potential for direct binding of Pit-1 to HS III-related DNA was first pursued by sequence analysis using the MatInspector 2.2 binding site detection program based on the TRANSFAC 4.0 database (Quandt et al. 1995, Wingender et al. 2000). A 574 bp fragment of the HS III-related sequences, corresponding to nucleotides −28 225/−27 652 and possessing both binding and functional activities (Jin et al. 2004), was analyzed using a consensus Pit-1 motif 5′-TWTAW-TAATWCAT-3′. There was no match for Pit-1; however, four Oct-1(POU2F1)-like elements (OLEa–d) were identified (Fig. 4A). Both Pit-1 and Oct-1 are members of the POU-domain family of transcriptional activators, which can bind to DNA as heterodimers or associate with each other in the absence of DNA, via their POU domains (Voss et al. 1991).
Figure 4.
Pit-1 does not associate directly with Oct-1-like elements located in the HS III-related sequences. (A) The sequence (5′ –3′ ) of the HS III fragment (−28 225/−27 652 in relation to the GH-N gene) is shown. Four Oct-1-like elements (OLEa–d) according to the MatInspector search are highlighted. The corresponding subfragments used in further EMSA studies are underlined. The ETS site according to MatInspector search is also indicated. (B) EMSA was done using a radiolabeled oligonucleotide containing the consensus Oct-1 DNA element as a specific probe and nuclear extract from non-transfected HEK293 cells. Unlabeled Oct-1 and mutant Oct-1 (Oct-1m) elements, as well as four Oct-1-like elements (OLEa, b, c, and d), were used as competitor oligonucleotides at 25- and 50-fold mass excess of probe. The mobility of a specific complex is indicated with a solid arrowhead. (C) EMSA was performed using the consensus Oct-1 DNA-binding element as a specific probe with nuclear proteins from HEK293 cells expressing Pit-1. Unlabeled Oct-1 and Oct-1m oligonucleotides as well as an oligonucleotide containing the Pit-1 element from the GH-N promoter (GHp Pit-1) were used as specific competitors at 25- and 50-fold mass excess of probe. Specific Pit-1 antibodies (Santa Cruz Biotechnology Inc., X-7, sc-442) were also used. Specific complexes are indicated with solid arrowheads. The ‘supershift’ is indicated by an open arrowhead. (D) EMSA was done using radiolabeled GHp Pit-1 oligonucleotide as a specific probe with recombinant Pit-1 protein (Santa Cruz Biotechnology Inc., sc-4014). The mobility of the specific complex is indicated with a solid arrowhead. (E) The recombinant Pit-1 protein was also incubated with the radiolabeled Oct-1 consensus element. Competitor oligonucleotides, including OLEa–d, were used at 25- and/or 50-fold mass excess of probe. The mobility of a specific complex is indicated by a solid arrowhead. FP, free probe; NE, nuclear extract; NRS, normal rabbit serum.
As a first approximation for ‘direct’ or ‘indirect’ association of Pit-1 with HS III sequences via an Oct-1 DNA element, nuclear proteins were isolated from HEK293 cells (Fig. 4B) and HEK293 cells transfected with Pit-1 (HEK293/Pit-1; Fig. 4C), and assessed by EMSA using a consensus Oct-1 motif (5′-ACAGCTTACGTTTAGTGATCAA-3′) as a DNA probe. A low-mobility complex was competed effectively with a 25× molar excess of Oct-1 oligonucleotide, but not with a mutated Oct-1 oligonucleotide (solid arrowhead I, Fig. 4B); this is consistent with the presence of Oct-1-binding activity in HEK293 nuclear protein. In contrast, no competition of this complex was observed with 25× molar excess of OLEa, b, c, or d, although some competition and thus suggestion of binding were observed with a 50× molar excess of OLEc (Fig. 4B). Similarly, when HEK293/Pit-1 nuclear protein was used together with an Oct-1 consensus element as a probe, high-mobility complexes consistent with direct Pit-1 binding (e.g. solid arrowhead II) were observed in addition to the low-mobility complex (solid arrowhead I, Fig. 4C). Again, these complexes were all competed effectively with a consensus Oct-1 oligonucleotide. The participation of Pit-1 in the high-affinity/mobility complexes was confirmed by specific competition with Pit-1 oligonucleotide, and ‘supershift’ with Pit-1 antibodies (open arrowhead, Fig. 4C).
The ability of the four Oct-1-like sites (OLEa–d) to compete with recombinant Pit-1 protein was assessed by EMSA. As a positive control, the ability of Pit-1 protein to bind to the proximal Pit-1 GH-1 element of the GH-N promoter (GHp Pit-1) was confirmed, and a specific high-mobility complex was observed (solid arrowhead, Fig. 4D) as described previously ( Jin et al. 1999). The EMSA was repeated using a consensus Oct-1 element (5′-ACAGCTTACGTTTAGTGATCAA-3′) as the free probe. Again, a specific complex of similar mobility was observed with Pit-1 protein (solid arrowhead, Fig. 4E). This complex was competed effectively with 25× mass excess of unlabeled Oct-1 oligonucleotide. In contrast, there was no comparable competition with 50× excess of OLEa–d. The four OLEs (a–d) were also tested as specific probes for HEK293 versus HEK293/Pit-1 binding by EMSA. No specific binding related to Pit-1 expression was observed with OLEa–d as suggested by the lack of novel and specific complexes observed with the nuclear extract from HEK293/Pit-1 versus HEK293 cells (data not shown; Yang 2010).
Pit-1 and ETS family members participate in a common complex in human pituitary samples
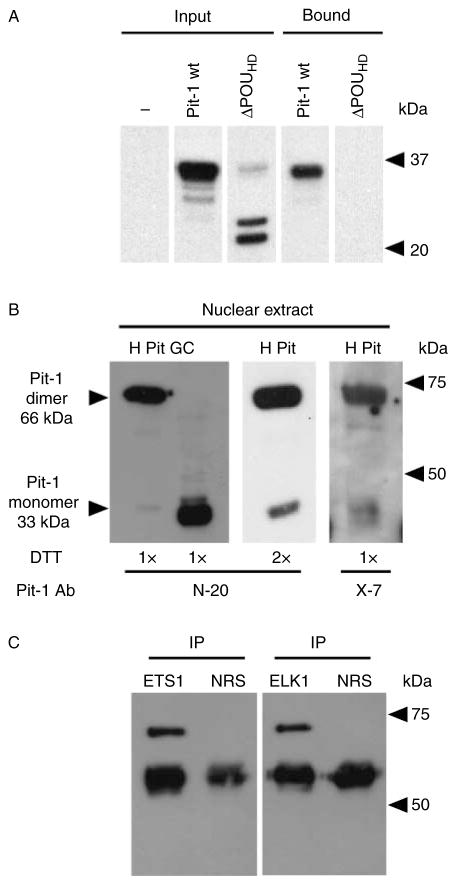
Previously, we showed that ETS factors can associate with the HS III-related sequences shown in Fig. 4A (Jin et al. 2004). Furthermore, a POU homeodomain-dependent interaction between Pit-1 and an ETS family member (ETS1) was shown to mediate prolactin expression (Bradford et al. 1997, 2000). To further examine the potential interaction between Pit-1 and ETS factors in the HEK293 cell system, and particularly in association with the HS III-related sequences, a 41 bp HS III oligonucleotide ( Jin et al. 2004) containing the OLEd sequences and thus ETS DNA element (Fig. 4A and Supplementary Table 1) was conjugated with magnetic beads and used to extract nuclear proteins from HEK293 cells transfected with wt or mutant Pit-1 lacking the POU homeodomain (ΔPOUHD). The input and bound fractions were analyzed by SDS-PAGE and immunoblotting (Fig. 5A). When probed with Pit-1 antibodies, both wt and mutant Pit-1 appeared in the input fraction, but only wt Pit-1 was detected after bead extraction.
Figure 5.
Pit-1 participates in a common complex with ETS family members derived from human pituitary tissue. (A) The ETS/HS III oligonucleotide was conjugated to magnetic beads, and mixed with nuclear proteins from non-transfected (NT) HEK293 cells and from HEK293 cells transfected with wild-type (wt) as well as POU homeodomain-deleted (ΔPOUHD) Pit-1 cDNAs. After mixing with the conjugated oligonucleotides, the bound fractions as well as input fractions were analyzed by electrophoresis and protein immunoblotting with Pit-1 antibodies (N-20). (B) Twenty micrograms of nuclear protein from human pituitary tissue samples (harvested using the Universal Magnetic Co-IP kit) as well as rat pituitary GC cells were resolved by SDS-PAGE, immunoblotted, and detected with Pit-1 antibodies (N-20). Additional human pituitary tissue samples were treated with either 2× dithiothreitol before SDS-PAGE and immunoblotting using N-20 Pit-1 antibodies, or SDS-PAGE and immunoblotting using X-7 Pit-1 antibodies. The bands that are consistent with the expected size for monomer and homodimer forms of Pit-1 at ~33 and 66 kDa respectively are indicated by arrowheads. (C) Immunoprecipitation (IP) was performed using nuclear extracts from human pituitary tissue samples to detect the Pit-1/ELK1 interaction using the Universal Magnetic Co-IP kit according to the manufacturer’s instructions; note: the first lane of (B) represents the ‘input’ human pituitary sample for this IP. Antibodies against ETS1 (N-276) and ELK1 (I-20), as well as non-immune normal rabbit serum (NRS), were used to precipitate the associated proteins. Purified proteins were resolved by SDS-PAGE, immunoblotted, and detected with Pit-1 antibodies (N-20). The mobilities of molecular weight markers (in kDa) are shown.
A potential interaction between ETS1 and ELK1 with Pit-1 was pursued in human pituitary tissue. Samples from human anterior pituitary and rat anterior pituitary tumor GC cells were first assessed after immunoblotting alone as positive sources of Pit-1 protein (controls). The major specific band detected in the control human pituitary sample with a goat antibody to an amino-terminal peptide of human Pit-1 (N-20) had an apparent molecular weight of 66 kDa, consistent with a Pit-1 homodimer. A band of ~33 kDa (or monomer) was also detected in the human sample, but it was the major band detected in GC cell nuclear protein (Fig. 5B). It is unclear why a human Pit-1 dimer exists or is preferred under the assay conditions. However, when the pituitary sample was treated/reduced with twice the concentration of dithiothreitol, a 33 kDa band was more readily detectable, although the 66 kDa species was still dominant (Fig. 5B). Also, the 66 kDa band was the major species detected when a second Pit-1 antibody (rabbit polyclonal to rat Pit-1, X-7) was used to assess the isolated (input) human pituitary protein. This again indicates that the 66 kDa band has Pit-1 immunoreactivity and thus structure.
To assess the possible ETS family/Pit-1 interaction, nuclear protein was isolated from human anterior pituitary samples and immunoprecipitated using ETS1 and ELK1 antibodies, followed by immunoblotting using specific mouse monoclonal antibodies to Pit-1. Pit-1 immunoreactivity was detected after IP of human pituitary tissue with both ETS1 and ELK1 antibodies (Fig. 5C).
To further assess a possible interaction between Pit-1 and ETS1 or ELK1 in the context of the effect observed in HEK293 cells after gene transfer, nuclear protein was isolated 48 h after transfection of HEK293 cells with c-myc/Pit-1 or c-myc tag alone. The nuclear protein was immunoprecipitated with antibody against c-myc or with non-immune mouse serum (NMS), and analyzed by SDS-PAGE and immunoblotting (Fig. 6A). ETS1 was not detected after IP (with c-myc antibodies) and immunodetection with ETS1 antibodies (upper panel, Fig. 6A). By contrast, a minor band of the appropriate size was observed with ELK1 antibodies, although detection was made difficult because of co-localization of other bands and, presumably, the low levels reflected in the extract before precipitation (lower panel, Fig. 6A).
Figure 6.
Pit-1 expressed in HEK293 cells associates with ELK1 at HS III. (A) HEK293 cells were transfected with c-myc-tagged Pit-1, and nuclear protein (input) was immunoprecipitated (bound) with c-myc antibody or normal mouse serum (NMS). The resulting fractions were analyzed by protein blotting and probed with antibodies for the presence of ETS1 or ELK1. Migration positions of molecular weight markers (in kDa) are shown at right. (B) EMSA using a radiolabeled 41 bp HS III oligonucleotide containing an ETS domain-binding site from HS III was performed (Jin et al. 2004). Nuclear protein from c-myc/Pit-1-transfected HEK293 cells was immunoprecipitated with c-myc antibody and eluted at increasing salt concentrations. Closed arrowhead shows a shifted product that appears specifically with c-myc precipitation. (C) The 0·4 M KCl fraction from (B) was used in a further mobility shift assay with HS III as the probe in combination with antibodies (Ab) against ELK1 and ETS1. The band specific to c-myc antibody precipitation is competed with ELK1 antibody but not with ETS1 antibody. (D) The radiolabeled (41 bp HS III m) probe contains a 7 bp mutation previously shown to abrogate ELK1 binding (Jin et al. 2004). The bound product from the 0·4 M KCl fraction of the immunoprecipitate also failed to bind.
To clarify this result, the immunoprecipitates generated from HEK293 cells transfected with c-myc/Pit-1, using either c-myc antibodies or NMS, were extracted with 0·4, 0·6, and 1·0 M KCl, and assessed for the ability to bind to the previously identified (41 bp) ETS DNA element in HS III ( Jin et al. 2004) by EMSA (Fig. 6B). A single shifted product was observed in the 0·4 M KCl fraction following IP with c-myc antibodies, but not in the other elution fractions or after precipitation with NMS (arrowhead, Fig. 6B). The specificity of this interaction was assessed with a subsequent EMSA using the 0·4 M KCl c-myc fraction in the presence of ELK1 or ETS1 antibodies. Competition of the shifted band (arrowhead) was detected with the addition of ELK1 antibodies but not with that of ETS1 antibodies (Fig. 6C). The EMSA was also repeated using the 41 bp HS III oligonucleotide, in which a 7 bp mutation of the ETS DNA element was introduced. The band associated with the 0·4 M KCl fraction and wt 41 bp HS III oligonucleotide was lost (Fig. 6D).
Pit-1 but not ELK1 specifically increases endogenous GH-N versus CS gene expression in transfected HEK293 cells
The levels of endogenous GH/CS RNA were assessed in HEK293 cells transfected with human Pit-1 or ELK1 cDNA by real-time RT-PCR using primer sets designed for GH-N, CS-A/B, and GH-V transcripts respectively (Fig. 7). Expression of GH-N RNA was increased significantly about sixfold in the presence of wt Pit-1. In contrast, neither CS-A/B nor GH-V RNA levels were increased significantly. Human GH/CS RNA levels were also assessed in HEK293 cells overexpressing the ELK1 cDNA. A 13-fold increase in GH-N RNA levels was observed; however, significant 3- to 4-fold increases in CS RNA levels were also detected. When Pit-1 and ELK1 were co-expressed, GH-N levels were increased 23-fold relative to non-transfected cells. Perhaps more importantly, Pit-1 again was associated with a significant and specific (10-fold) increase in GH-N RNA levels but not with (placental) CS or GH-V RNA levels relative to ELK1 overexpression alone (Fig. 7).
Figure 7.
Expression of Pit-1 specifically stimulates endogenous GH-N RNA expression in HEK293 cells. Human GH/CS RNA expression was assessed by quantitative real-time PCR (qRT-PCR) after expression of Pit-1 and ELK1 alone or in combination in HEK293 cells. The RNA level in each sample was calculated from the standard curve and normalized to human GAPDH expression as appropriate. RNA levels after transfection are presented as fold increase above basal levels in non-transfected HEK293 cells, which are arbitrarily set to 1·0. Statistical analysis was performed by one-way ANOVA with the Tukey–Kramer post-hoc test for multiple comparisons (n=3–5). **P<0·01; *** and ###P<0·001.
Discussion
The POU homeodomain transcription factor Pit-1 plays an essential role in the activation of the GH locus in the pituitary through binding to HS I/II and proximal promoter (Jones et al. 1995, Jin et al. 1999, Shewchuk et al. 1999). However, a role for Pit-1 alone is not supported by the absence of GH-N expression in pituitary lactotrophs and thyrotrophs, both of which express Pit-1 protein (Li et al. 1990, Radovick et al. 1992). Studies using the mouse pre-somatotroph progenitor GHFT1 cell line demonstrated that HS III was the only HS within the GH LCR that showed a high level of histone H3 and H4 acetylation after stable gene transfer, suggesting the enrichment of HS III in the acetylated chromatin fraction in the Pit-1-containing pituitary-derived cell line (Elefant et al. 2000). Thus, it is reasonable to propose that HS III serves as an initial ‘entry’ point for recruitment of tissue-specific factors such as Pit-1, which can then trigger chromatin remodeling and subsequent assembly of additional activator proteins (Cattini et al. 2006). In the current study, introduction of exogenous Pit-1 into non-pituitary HEK293 increased the accessibility of HS III-related DNA sequences to binding proteins through evidence of increased hyperacetylation and RNA pol II activity (Figs 1 and 2). Thus, this supports the notion that HS III represents an early point of interaction between Pit-1 and the GH/CS LCR.
The ability of Pit-1 to increase RNA pol II activity (as an indirect indication of DNA accessibility to transcription factors) appears to require an intact Pit-1 POU homeodomain. Functionally, this differs from the observed Pit-1-induced hyperacetylation of histone H4, which appears to require both the POU homeodomain and the N-terminal trans-activation domain. This interpretation, of course, assumes similar levels of Pit-1 expression. It was shown in a transgenic mouse model that the insertion of a TerF termination element downstream of HS I/II repressed RNA pol II tracking, but had no effect on histone hyperacetylation in this region (Ho et al. 2006). The results presented here appear to extend this idea by i) showing that HS III may act as a (ubiquitously available) entry point for changes to HS I/II that lead to GH expression in pituitary cells, and ii) linking the previously demonstrated mechanistic difference (Ho et al. 2006) to Pit-1 function. The POU homeodomain is responsible for low-affinity, non-specific DNA binding (Ingraham et al. 1990b), and for protein–protein interactions (Bradford et al. 2000). The N-terminal trans-activation domain has been shown previously to be responsible for transcriptional activation (Ingraham et al. 1990a), and it is thus intriguing that this region might not be required for RNA pol II activity in the GH LCR, but is required for chromatin modifications. Thus, in addition to transcriptional activation, the N-terminal domain may contribute to the recruitment of factors, such as histone acetyltransferases, which modify chromatin.
Our data also indicate that Pit-1 does not associate directly with DNA sequences in the HS III region. Potential candidate binding sites were identified based on similarity to related Oct-1-like DNA elements, but no Oct-1 or Pit-1 (direct or indirect) interaction was detected (Fig. 4B and E). However, HS III-related sequences do contain an ETS DNA element that lies upstream and in close proximity to OLEd (Fig. 4A), and this was shown previously to bind to ETS factors by EMSA, as well as to associate with ELK1 in human pituitary chromatin in situ ( Jin et al. 2004). Our results obtained with human GH-N-related sequences (Fig. 5) as well as those of others in the context of rat prolactin gene regulation (Bradford et al. 1997, 2000, Gutierrez-Hartmann et al. 2007) suggest that Pit-1 and ETS factors, including ELK1 and ETS1, can participate in a common human pituitary protein complex. However, it should be noted that no additional band was detected using the OLEd as a probe and comparing nuclear extracts from HEK293 cells without or with Pit-1 expression by EMSA (Yang 2010). This might reflect a low level of endogenous ELK1 and thus Pit-1/ETS/factor complex and/or instability of the complex under the EMSA conditions employed.
Pit-1 was shown previously to associate with ETS1 via the Pit-1 POU homeodomain, and act synergistically to activate the rat prolactin promoter (Bradford et al. 1997, 2000). An ETS-binding motif in the Pit-1 POU homeodomain was both necessary and sufficient for direct binding of Pit-1 to ETS1 in a DNA-independent manner. Thus, the POU homeodomain has been linked to protein–protein interactions and function (Gutierrez-Hartmann et al. 2007) as well as to low-affinity DNA binding (Ingraham et al. 1990b). Our data are consistent with these observations and a role for the Pit-1 POU homeodomain in the context of the human GH-N locus. Effects on hyperacetylation and DNA accessibility at HS III were reduced significantly with the expression of the ΔPOUS mutant versus wt Pit-1 (Figs 1 and 2). These effects do not appear to require direct binding of Pit-1 to the DNA, as the POU-specific domain mutant, which also affects DNA binding (Fig. 3B), was not significantly different from wt Pit-1. Finally, wt Pit-1 but not the POU homeodomain mutant Pit-1 was able to associate, presumably indirectly, with an ETS element-containing HS III oligonucleotide (Fig. 5A), raising the possibility that Pit-1 may interact with the HS III region via this ETS element. To our knowledge, this is the first report to suggest co-operation between the ETS family and Pit-1 in the regulation of pituitary GH.
In addition to providing evidence that Pit-1 can induce further chromatin remodeling, and at HS III, Pit-1 was also able to preferentially stimulate endogenous (human) GH-N RNA expression in HEK293 cells (Fig. 7). In contrast, overexpression of the ETS factor ELK1 resulted in increased (placental) CS and GH-V as well as GH-N transcripts. ETS factors are known to have enhancer-like activity in pituitary, placental, and non-pituitary/placental cells and, as indicated previously, ETS factor binding to DNA sequences in the HS III region of the GH LCR was demonstrated ( Jin et al. 2004). Interestingly, the relative response to Pit-1 expression appears to be linked to ETS factor and/or ELK1 levels, as GH-N RNA levels increased from 6- to 23-fold in the presence of basal ETS family levels versus overexpressed ELK1 (Fig. 7). These data also support a functional role for Pit-1 and ETS family members in the control of human GH gene expression.
It must also be noted that the specific increase in GH-N versus CS/GH-V levels observed with Pit-1 expression occurs regardless of ELK1 overexpression (Fig. 7). This suggests that in addition to an important role in GH-N gene activation, Pit-1 may help to restrict the expression of the other (placental) members of the GH family in a pituitary context. Previous studies have implicated Pit-1 in the pituitary repression of the placental GH/CS family members (Nachtigal et al. 1993, Cattini et al. 2006, Norquay et al. 2006). Finally, the observation that overexpression of ELK1 in HEK293 cells resulted in a significant increase in endogenous CS as well as GH transcripts raises the possibility of a role for ETS factors in the activation of the GH/CS LCR in both the pituitary and placenta, or reflects an apparent ‘general’ stimulation of transcriptional activity (Gutierrez-Hartmann et al. 2007). Thus, this effect might be independent of Pit-1 and HS III-related sequences.
Expression of exogenous Pit-1 in HEK293 cells resulted in significant chromatin remodeling at the endogenous HS III region (Fig. 1). HS III was originally reported to be present in both pituitary and placental chromatin, but it is now described as ‘constitutive’ (Jones et al. 1995, Kimura et al. 2004). The ‘poised’ nature of the chromatin at HS III and the results presented here raise and are consistent with the possibility that Pit-1 associates with a member (or members) of the ETS family via its POU homeodomain, and thereby gains access to the GH LCR through HS III. What is less clear is whether the ability of Pit-1 (and/or ETS factor) to stimulate RNA pol II activity, as indicated by effects on DNA accessibility and endogenous GH-N RNA, has any further role in facilitating Pit-1 and HS I/II interaction. The ability of ELK1 to associate with the HS III region in human pituitary tissue in situ and participate in a common complex with Pit-1, as suggested through co-IP and gel shift/competition assays (Figs 5 and 6), raises the possibility that the resulting stimulation of RNA pol II activity and increased DNA accessibility observed at HS III extend out from this point; this effect might be more pronounced in progenitor pituitary cells. In this scenario, Pit-1 is recruited by ELK1 (or possibly by other ETS family members), to the already ‘poised’ HS III site, resulting in further chromatin remodeling and recruitment of additional proteins. The ‘cascade’ of chromatin remodeling along the locus may then open up HS I/II and result in direct protein–DNA association of Pit-1 to its binding sites within HS I/II, an essential step in establishing a functional GH-N promoter in the pituitary (Ho et al. 2002).
Alternatively, an ETS factor/Pit-1 interaction might play a role in maintaining a chromatin configuration that supports appropriate GH-N promoter activity in the pituitary. Recent studies using the chromatin conformation capture (3C) assay suggest that HS III, V and HS I/II regions interact in pituitary chromatin, with looping out of intervening sequences (Ho et al. 2008). The presence of this ‘specificity’ loop can be linked with appropriate (site of integration independent and pituitary specific) expression of GH-N in transgenic mice (Ho et al. 2002). A second loop between HS I/II and the GH-N promoter region is associated with efficient promoter activity (Ho et al. 2002, 2008). It is therefore noteworthy that ELK1 and Pit-1 have been shown to associate with HS III and HS I/II regions respectively in situ ( Jin et al. 1999, 2004, Shewchuk et al. 1999). This raises the possibility that ELK1 (and/or other ETS factors) and Pit-1 participate in a common complex that supports the generation and/or maintenance of the ‘specificity’ loop involving HS III, V and HS I/II regions of chromatin. The roles for ETS factor/Pit-1 interaction in recruitment of Pit-1 or formation and/or maintenance of a looped chromatin configuration in the GH LCR are, of course, not mutually exclusive.
In summary, a specific role for HS III in the GH/CS locus is suggested by its presence and open configuration, as well as by the predicted molecular interactions that are probably part of the LCR complex. These interactions are suggested by the requirement of HS I–III, V for appropriate human GH-N gene expression (in transgenic mice), and the looping of chromatin to bring the HS III, V region in close proximity with HS I/II and the GH-N promoter in vivo (Cattini et al. 2006, Ho et al. 2008). Consistent with this idea, introduction of Pit-1 into (human) HEK293 cells resulted in further chromatin remodeling and increased DNA accessibility extending from HS III. These effects of Pit-1 appear independent of DNA binding but rather via protein–protein interaction involving the POU homeodomain. Clearly, Pit-1 alone is necessary but not sufficient to limit appropriate GH-N gene activation in pituitary somatotrophs, given the presence of Pit-1 protein in both lactotrophs and thyrotrophs. The potential interaction of Pit-1 and ETS factors in human pituitary samples and the presence of an ETS site at HS III raise the possibility that members of the ETS family may act as co-factors with Pit-1 that contribute to differential recruitment and/or activation and expression of specific genes, such as human GH-N and prolactin, thereby helping to define a particular cell type.
Supplementary Material
Acknowledgments
Funding
This work was supported by a grant from the Canadian Institutes of Health Research (MT-10853). X Yang is the former recipient of a National Sciences and Engineering Research Council Doctoral Postgraduate Scholarship and a Manitoba Health Research Council Studentship.
Footnotes
This is linked to the online version of the paper at http://dx.doi.org/10.1677/JME-10-0017.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- Bennani-Baiti IM, Asa SL, Song D, Iratni R, Liebhaber SA, Cooke NE. DNase I-hypersensitive sites I and II of the human growth hormone locus control region are a major developmental activator of somatotrope gene expression. PNAS. 1998;95:10655–10660. doi: 10.1073/pnas.95.18.10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford AP, Wasylyk C, Wasylyk B, Gutierrez-Hartmann A. Interaction of Ets-1 and the POU-homeodomain protein GHF-1/Pit-1 reconstitutes pituitary-specific gene expression. Molecular and Cellular Biology. 1997;17:1065–1074. doi: 10.1128/mcb.17.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford AP, Brodsky KS, Diamond SE, Kuhn LC, Liu Y, Gutierrez-Hartmann A. The Pit-1 homeodomain and beta-domain interact with Ets-1 and modulate synergistic activation of the rat prolactin promoter. Journal of Biological Chemistry. 2000;275:3100–3106. doi: 10.1074/jbc.275.5.3100. [DOI] [PubMed] [Google Scholar]
- Bulger M, van Doorninck JH, Saitoh N, Telling A, Farrell C, Bender MA, Felsenfeld G, Axel R, Groudine M. Conservation of sequence and structure flanking the mouse and human beta-globin loci: the beta-globin genes are embedded within an array of odorant receptor genes. PNAS. 1999;96:5129–5134. doi: 10.1073/pnas.96.9.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattini PA, Yang X, Jin Y, Detillieux KA. Regulation of the human growth hormone gene family: possible role for Pit-1 in early stages of pituitary-specific expression and repression. Neuroendocrinology. 2006;83:145–153. doi: 10.1159/000095522. [DOI] [PubMed] [Google Scholar]
- Chen EY, Liao YC, Smith DH, Barrera-Saldana HA, Gelinas RE, Seeburg PH. The human growth hormone locus: nucleotide sequence, biology, and evolution. Genomics. 1989;4:479–497. doi: 10.1016/0888-7543(89)90271-1. [DOI] [PubMed] [Google Scholar]
- Elefant F, Su Y, Liebhaber SA, Cooke NE. Patterns of histone acetylation suggest dual pathways for gene activation by a bifunctional locus control region. EMBO Journal. 2000;19:6814–6822. doi: 10.1093/emboj/19.24.6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Hartmann A, Duval DL, Bradford AP. ETS transcription factors in endocrine systems. Trends in Endocrinology and Metabolism. 2007;18:150–158. doi: 10.1016/j.tem.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Ho Y, Elefant F, Cooke NE, Liebhaber SA. A defined locus control region determinant links chromatin domain acetylation with long-range gene activation. Molecular Cell. 2002;9:291–302. doi: 10.1016/s1097-2765(02)00447-1. [DOI] [PubMed] [Google Scholar]
- Ho Y, Liebhaber SA, Cooke NE. Activation of the human GH gene cluster: roles for targeted chromatin modification. Trends in Endocrinology and Metabolism. 2004;15:40–45. doi: 10.1016/j.tem.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Ho Y, Elefant F, Liebhaber SA, Cooke NE. Locus control region transcription plays an active role in long-range gene activation. Molecular Cell. 2006;23:365–375. doi: 10.1016/j.molcel.2006.05.041. [DOI] [PubMed] [Google Scholar]
- Ho Y, Tadevosyan A, Liebhaber SA, Cooke NE. The juxtaposition of a promoter with a locus control region transcriptional domain activates gene expression. EMBO Reports. 2008;9:891–898. doi: 10.1038/embor.2008.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingraham HA, Albert VR, Chen RP, Crenshaw EB, III, Elsholtz HP, He X, Kapiloff MS, Mangalam HJ, Swanson LW, Treacy MN, et al. A family of POU-domain and Pit-1 tissue-specific transcription factors in pituitary and neuroendocrine development. Annual Review of Physiology. 1990a;52:773–791. doi: 10.1146/annurev.ph.52.030190.004013. [DOI] [PubMed] [Google Scholar]
- Ingraham HA, Flynn SE, Voss JW, Albert VR, Kapiloff MS, Wilson L, Rosenfeld MG. The POU-specific domain of Pit-1 is essential for sequence-specific, high affinity DNA binding and DNA-dependent Pit-1–Pit-1 interactions. Cell. 1990b;61:1021–1033. doi: 10.1016/0092-8674(90)90067-o. [DOI] [PubMed] [Google Scholar]
- Jin Y, Surabhi RM, Fresnoza A, Lytras A, Cattini PA. A role for A/T-rich sequences and Pit-1/GHF-1 in a distal enhancer located in the human growth hormone locus control region with preferential pituitary activity in culture and transgenic mice. Molecular Endocrinology. 1999;13:1249–1266. doi: 10.1210/mend.13.8.0332. [DOI] [PubMed] [Google Scholar]
- Jin Y, Norquay LD, Yang X, Gregoire S, Cattini PA. Binding of AP-2 and ETS-domain family members is associated with enhancer activity in the hypersensitive site III region of the human growth hormone/chorionic somatomammotropin locus. Molecular Endocrinology. 2004;18:574–587. doi: 10.1210/me.2003-0405. [DOI] [PubMed] [Google Scholar]
- Jin Y, Lu SY, Fresnoza A, Detillieux KA, Duckworth ML, Cattini PA. Differential placental hormone gene expression during pregnancy in a transgenic mouse containing the human growth hormone/chorionic somatomammotropin locus. Placenta. 2009;30:226–235. doi: 10.1016/j.placenta.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BK, Monks BR, Liebhaber SA, Cooke NE. The human growth hormone gene is regulated by a multicomponent locus control region. Molecular and Cellular Biology. 1995;15:7010–7021. doi: 10.1128/mcb.15.12.7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura AP, Liebhaber SA, Cooke NE. Epigenetic modifications at the human growth hormone locus predict distinct roles for histone acetylation and methylation in placental gene activation. Molecular Endocrinology. 2004;18:1018–1032. doi: 10.1210/me.2003-0468. [DOI] [PubMed] [Google Scholar]
- Kimura AP, Sizova D, Handwerger S, Cooke NE, Liebhaber SA. Epigenetic activation of the human growth hormone gene cluster during placental cytotrophoblast differentiation. Molecular and Cellular Biology. 2007;27:6555–6568. doi: 10.1128/MCB.00273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre C, Imagawa M, Dana S, Grindlay J, Bodner M, Karin M. Tissue-specific expression of the human growth hormone gene is conferred in part by the binding of a specific trans-acting factor. EMBO Journal. 1987;6:971–981. doi: 10.1002/j.1460-2075.1987.tb04847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Crenshaw EB, III, Rawson EJ, Simmons DM, Swanson LW, Rosenfeld MG. Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU-domain gene pit-1. Nature. 1990;347:528–533. doi: 10.1038/347528a0. [DOI] [PubMed] [Google Scholar]
- Nachtigal MW, Nickel BE, Cattini PA. Pituitary-specific repression of placental members of the human growth hormone gene family. A possible mechanism for locus regulation. Journal of Biological Chemistry. 1993;268:8473–8479. [PubMed] [Google Scholar]
- Nickel BE, Nachtigal MW, Bock ME, Cattini PA. Differential binding of rat pituitary-specific nuclear factors to the 5′-flanking region of pituitary and placental members of the human growth hormone gene family. Molecular and Cellular Biochemistry. 1991;106:181–187. doi: 10.1007/BF00230184. [DOI] [PubMed] [Google Scholar]
- Norquay LD, Yang X, Sheppard P, Gregoire S, Dodd JG, Reith W, Cattini PA. RFX1 and NF-1 associate with P sequences of the human growth hormone locus in pituitary chromatin. Molecular Endocrinology. 2003;17:1027–1038. doi: 10.1210/me.2003-0025. [DOI] [PubMed] [Google Scholar]
- Norquay LD, Yang X, Jin Y, Detillieux KA, Cattini PA. Hepatocyte nuclear factor-3alpha binding at P sequences of the human growth hormone locus is associated with pituitary repressor function. Molecular Endocrinology. 2006;20:598–607. doi: 10.1210/me.2005-0221. [DOI] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Research. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radovick S, Nations M, Du Y, Berg LA, Weintraub BD, Wondisford FE. A mutation in the POU-homeodomain of Pit-1 responsible for combined pituitary hormone deficiency. Science. 1992;257:1115–1118. doi: 10.1126/science.257.5073.1115. [DOI] [PubMed] [Google Scholar]
- Shewchuk BM, Asa SL, Cooke NE, Liebhaber SA. Pit-1 binding sites at the somatotrope-specific DNase I hypersensitive sites I, II of the human growth hormone locus control region are essential for in vivo GH-N gene activation. Journal of Biological Chemistry. 1999;274:35725–35733. doi: 10.1074/jbc.274.50.35725. [DOI] [PubMed] [Google Scholar]
- Shewchuk BM, Ho Y, Liebhaber SA, Cooke NE. A single base difference between Pit-1 binding sites at the GH promoter and locus control region specifies distinct Pit-1 conformations and functions. Molecular and Cellular Biology. 2006;26:6535–6546. doi: 10.1128/MCB.00267-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss JW, Wilson L, Rosenfeld MG. POU-domain proteins Pit-1 and Oct-1 interact to form a heteromeric complex and can cooperate to induce expression of the prolactin promoter. Genes and Development. 1991;5:1309–1320. doi: 10.1101/gad.5.7.1309. [DOI] [PubMed] [Google Scholar]
- Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Pruss M, Reuter I, Schacherer F. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Research. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. PhD Thesis. University of Manitoba; Winnipeg, Canada: 2010. A study of the role of transcription factors and chromatin remodeling in the activation/repression of the human growth hormone/chorionic somatomammotropin genes. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.