Abstract
Few gene targets of Visual System Homeobox 2 (VSX2) have been identified despite its broad and critical role in the maintenance of neural retina (NR) fate during early retinogenesis. We performed VSX2 ChIP-seq and ChIP-PCR assays on early stage optic vesicle-like structures (OVs) derived from human iPS cells (hiPSCs), which highlighted WNT pathway genes as direct regulatory targets of VSX2. Examination of early NR patterning in hiPSC-OVs from a patient with a functional null mutation in VSX2 revealed mis-expression and upregulation of WNT pathway components and retinal pigmented epithelium (RPE) markers in comparison to control hiPSC-OVs. Furthermore, pharmacological inhibition of WNT signaling rescued the early mutant phenotype, whereas augmentation of WNT signaling in control hiPSC-OVs phenocopied the mutant. These findings reveal an important role for VSX2 as a regulator of WNT signaling and suggest that VSX2 may act to maintain NR identity at the expense of RPE in part by direct repression of WNT pathway constituents.
Keywords: Human induced pluripotent stem cells (hiPSCs), VSX2, WNT, optic vesicles
Graphical abstract
Visual systems homeobox 2 (VSX2) is a key transcription factor involved in neural retinal development. However, surprisingly few gene regulatory targets are known for VSX2 beyond Microphthalmia induced transcription factor (MITF), a protein involved in the development of retinal pigmented epithelium (RPE). We used patient-specific human pluripotent stem cells carrying a mutation in VSX2 that abolishes DNA binding activity (R200Q), combined with pharmacological manipulations and chromatin immunoprecipitation, to demonstrate a novel interaction between VSX2 and WNT signaling genes. In VSX2R200Q cultures, VSX2 binding to WNT signaling genes is abolished and optic vesicle cells aberrantly adopt an RPE fate in lieu of neural retina. This effect that can be partially overcome by inhibition of WNT signaling, or induced in wild type cells through promotion of WNT signaling. This study also presents the first unbiased list of VSX2 targets, thus providing a resource for identification of other genes and mechanisms involved in this biological process.
INTRODUCTION
The vertebrate eye develops from the anterior neural plate during late gastrulation as a result of intrinsic programming and signals from surrounding tissues, which in turn leads to the expression of a group of transcription factors that specify the eye field1, 2. Following eye field specification, the bilateral optic pits evaginate to form unpatterned optic vesicles (OVs). OVs then undergo invagination, resulting in the creation of the bilayered optic cup, the inner and outer aspects of which become the neural retina (NR) and retinal pigmented epithelium (RPE), respectively.
OV patterning, optic cup formation, and subsequent retinal maturation are influenced by inductive signaling molecules, which are thought to include Fgfs3–5, Tgfβ superfamily members, Hedgehog,6, 7 and/or Wnts8–12. In addition to extrinsic instructional cues, there are multiple transcription factors that play crucial roles in early NR and RPE development. Two such factors are Microphthalmia-associated transcription factor (Mitf) and Visual system homeobox 2 (Vsx2). Mitf is initially expressed uniformly throughout the early mammalian OV, while at later stages its expression is restricted to the RPE, where it promotes differentiation, proliferation, and pigmentation13, 14. The initiation of this restriction coincides with the onset of Vsx2 expression in the distal OV, which also serves to demarcate the future NR. Vsx2 is involved in maintenance and proliferation of the neural retinal progenitor cell (NRPC) pool, timing of photoreceptor production, and differentiation of one type of retinal interneuron, the bipolar cell15–18. In addition, Vsx2 has been shown to inhibit Mitf expression directly by binding and repressing Mitf promoter sites19, 20, as well as through protein-protein interactions21. Disruption of Vsx2 in animal models causes retinal defects, microphthalmia, and RPE layer duplication15, 18, 21; similarly, in humans VSX2 mutations result in very small, nonfunctional eyes with malformed retinas22–24. However, the mechanism by which VSX2 influences these diverse processes remains the subject of investigation.
VSX2 expression has also been used in vitro to identify multipotent NRPCs derived from human pluripotent stem cells (hPSCs)25–29. In order to study the role of VSX2 in human retinogenesis and hPSC differentiation, we previously generated human induced PSCs (hiPSCs) from a microphthalmic individual bearing a homozygous R200Q mutation in VSX2 (VSX2R200Q) and an unaffected sibling. The R200Q mutation eliminates the ability of VSX2 to bind DNA21, 24, 30, thus rendering it unable to directly regulate gene expression. Populations of 3-dimensional OV-like structures were derived from the hiPSC lines (hiPSC-OVs) and differentiated into retinal cell types in a manner analogous to human retinogenesis in vivo28, 31. VSX2R200Q hiPSC-OVs demonstrated proliferation defects, enhanced RPE differentiation at the expense of NR, and absence of bipolar cells, which closely approximates the vertebrate disease phenotype31. Interestingly, RNA-seq performed on early VSX2R200Q hiPSC-OVs revealed upregulation of a striking number of genes belonging to the WNT pathway, which led us to further examine the association between VSX2 and WNT signaling during early retinal differentiation.
In the present study, we show that major components of the WNT pathway are gene targets for VSX2 binding during retinal differentiation in hiPSCs, and that pharmocological manipulation of the WNT pathway alters the expected phenotypes of both wild-type control (VSX2WT) and VSX2R200Q hiPSC-OVs. Our findings indicate an important role for VSX2 as a direct regulator of WNT signaling and suggest that VSX2 may maintain NR identity in early hiPSC-OVs in part by antagonizing expression of WNT pathway constituents.
MATERIALS AND METHODS
Cell culture
hiPSCs derived from a microphthalmic patient24 with homozygous R200Q mutations in VSX2 and an unaffected sibling were maintained on irradiated mouse embryonic fibroblasts and differentiated toward retina as previously described31. Briefly, on day 0 (d0), embryoid bodies (EBs) are lifted with 2 mg/ml dispase in the absence of FGF2 to initiate retinal differentiation. On d7, EBs are plated on laminin-coated wells, and by d10, 90% of cells express markers of anterior neuroectoderm26. By d12, MITF expression is detected in a subset of the cells, followed on d14 by expression of VSX2, which marks the production of NRPCs20. By d18–20, NRPC and RPE progenitors are established and OV structures consisting predominantly of NRPCs are manually separated and cultured en masse. These hiPSC-OVs subsequently generate all retinal neuron types in a sequence and time frame approximating normal human retinogenesis29, 31.
Immunocytochemistry
EBs were plated on laminin-coated coverslips on d7 of retinal differentiation and fixed with 4% paraformaldehyde after an additional 7–11 days of differentiation. D35 and d50 hiPSC-OVs were fixed for 1 hour at RT in 4% paraformaldehyde, cryopreserved in 15–30% sucrose, and cut into 11 μm cryosections. Samples were blocked in 10% normal donkey serum, 0.5% Triton X100, 1% fish gelatin, and 5% bovine serum albumin for 45 min at room temperature and incubated with primary antibody overnight at 4°C in a humidified chamber (primary antibody sources and dilutions are listed in Table S1). AF546-, AF488-, and AF633- conjugated secondary antibodies (Thermo Fisher) were diluted 1:500 in blocking buffer and incubated at room temperature for 30 min. Samples were mounted in Prolong Gold antifade + DAPI (Thermo Fisher) and images were taken on a Nikon A1R-Si laser scanning confocal microscope (Nikon). Cell counts were performed with Nikon Elements module D and plotted with Graph Pad Prism 6.
WNT agonist and antagonist treatments
EBs were plated at d7 on laminin-coated plastic wells or laminin-coated glass coverslips and treated with the WNT antagonist IWP2 (5 μM) (Tocris) from d12 to d20 or with the WNT agonist CHIR99021 (3 μM) (Tocris) from d14 to d20. DMSO served as the vehicle control for all experiments. Treated coverslips were fixed at d18 and immunostained as described above. Treated wells were collected after 30 days of differentiation and total RNA was extracted with RNAeasy spin columns (Qiagen) and reverse transcribed with iScript cDNA kit (BioRad) according to manufacturers' instructions. Quantitative PCR was performed with SSO Advanced SybrGreen master mix (BioRad) on a Step One Plus Real Time PCR system (Thermo Fisher). Relative expression was normalized to the geometric mean of two reference genes and the average 2−ΔΔCq ± SEM of three replicates was plotted using Graph Pad Prism 6. Statistical significance (p<0.05) was calculated with an unpaired two-tailed Student's t-test. Primer sequences are listed in Table S2.
Chromatin Immunoprecipitation
Human iPSC-OVs were manually selected and differentiated to d30, fixed in 1% formaldehyde for 10 min at room temperature, washed, and lysed in Pierce IP lysis buffer (Thermo Fisher) supplemented with 40 μl/ml protease cocktail inhibitor P8340 (Sigma Aldrich). Cleared lysate was sheared in a Q700 ultrasonic processor (Qsonica) equipped with a cup horn, and shearing was monitored using 1% agarose gel electrophoresis. 10% volume was reserved for input and the remainder was incubated with 2 μg sheep anti-VSX2 antibody (Exalpha) overnight at 4°C with rocking. Immunoprecipitates were collected on protein-G conjugated Dynabeads (Invitrogen), washed 5x with sterile PBS, and eluted in 10 mM Tris/1 mM EDTA pH 8 + 1% SDS. Three volumes of 1% SDS, 0.1M NaHCO3, and 200 mM NaCl were added to input and IP samples and crosslinks were reversed by incubation at 65°C for 4 hours. DNA was extracted with phenol:chloroform:isoamyl alcohol, ethanol precipitated, and quantified with the Qubit high sensitivity double stranded DNA kit (Thermo Fisher). 8–10 ng of DNA were prepared for deep sequencing with either the Illumina ChIP-Seq DNA or the TruSeq ChIP Sample Preparation Kit (Illumina) and quantified with a Qubit fluorometer. All samples were loaded at a final concentration of 8 pM and sequenced on the Illumina HiSeq 2500.
ChIP-seq analysis
ChIP-seq reads were aligned to the hg19 Homo sapiens assembly using Bowtie32. Duplicate reads were removed within each replicate. Transcription factor binding sites were first called after combining reads from both replicates using the R software package SPP33 with the following settings: detection window halfsize = 300, False Discovery Rate (FDR) = 0.05. After subtracting the normalized input read counts34, the number of ChIP binding reads were computed for every binding site within each replicate, and only sites having at least 10 ChIP binding reads within both replicates were considered high confidence sites. Transcription factor binding motifs were analyzed in an unbiased fashion using HOMER version 4.635.
ChIP-PCR
Chromatin from both VSX2WT and VSX2R200Q hiPSC-OVs were immunoprecipitated as described above and purified DNA was diluted 1:10 for PCR. Genomic coordinates for selected WNT pathway peaks were used to generate primers, and sites in the MITF-H promoter known either to be bound or not to be bound by VSX220 were used as positive and negative controls for these experiments, respectively. Primer sequences are listed in Table S2 and PCR analysis was performed with 2X PCR master mix (Promega) (35 cycles and Tm = 58°C) followed by visualization on a 2% agarose gel.
RESULTS
VSX2 binds a subset of WNT pathway genes
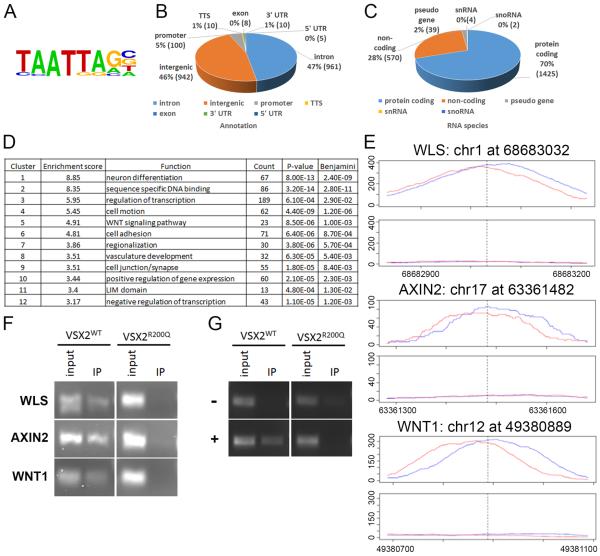
In a previous study, transcriptome comparison of VSX2WT and VSX2R200Q hiPSC-OVs suggested a regulatory role for VSX2 in WNT signaling, which in turn might contribute to the overproduction of RPE at the expense of NR seen in mutant OVs31. To further investigate this possibility, we performed unbiased searches for VSX2 DNA binding sites in two independent samples of d30 VSX2WT hiPSC-OVs using chromatin immunoprecipitation followed by massively parallel DNA sequencing (ChIP-Seq). Western blot analysis of cell lysates and immunoprecipitates demonstrated VSX2 antibody specificity (Fig. S1A), although differences in binding affinities between lots of VSX2 polyclonal antibodies resulted in the second ChIP-seq displaying weaker signal strength. We then compared merged ChIP and input samples for peak calling and further stipulated that peaks be present in both replicates, which resulted in a list of 2038 high confidence VSX2 binding sites (Table S3). Examination of the DNA regions occupied by VSX2 in these analyses revealed a consensus binding motif identical to that previously found for VSX2 in various mammalian systems (Fig. 1A)21, 24, 30. Furthermore, 236 of these sites were located within 2.5 kb of a transcription start site. The majority of peaks were split between intronic and intergenic sequences (Fig. 1B), characteristic of enhancer targets, and more than half of the identified loci (1425 peaks) were associated with protein-coding genes (Fig. 1C), indicative of VSX2's role as a transcriptional regulator. Notably, DAVID annotation clustering36, 37 not only identified numerous functions consistent with the known roles of VSX2 in early OV development, but also highlighted the WNT signaling pathway as a functional target of VSX2 (Fig. 1D). Target sites were identified in multiple WNT pathway genes, including WLS, AXIN2, and WNT1 (Table 1).
Figure 1. VSX2 ChIP-seq and ChIP-PCR analyses demonstrating binding of VSX2 to WNT pathway genes in d30 VSX2WT but not VSX2R200Q hiPSC-OVs.
Confirmation of the VSX2 consensus binding motif in VSX2WT hiPSC-OV ChIP-seq target sequences (A). Distribution of high confidence VSX2 DNA binding targets in VSX2WT hiPSC-OVs as categorized by genomic location (B) or RNA species (C). List of GO terms with greater than 3-fold enrichment generated via DAVID functional analysis of high confidence ChIP-seq peaks (D). Peak localization and genomic coverage maps (± 200 bp toward the 5' or 3' end) for WLS, AXIN2, and WNT1 (red and blue lines represent forward and reverse DNA strand reads, respectively). The top panel designates ChIP coverage and the lower panel shows the input control for each gene (E). VSX2 ChIP-PCR from VSX2WT or VSX2R200Q d30 hiPSC-OVs confirming direct binding of VSX2WT, but not VSX2R200Q, to targets identified proximal to the WNT signaling pathway genes WLS, AXIN2, and WNT1 (F). Regions in the MITF-H promoter previously shown by ChIP-PCR to be bound (+) or not bound (−) by VSX220 served as positive and negative controls, respectively (amplified from the same chromatin preparation presented in panel F) (G). See also Fig. S1.
Table 1.
List of WNT-related Genes Identified by VSX2 ChIP-SEQ
| ID | Gene Name |
|---|---|
| CXXC4 | CXXC finger 4 |
| DIXDC1 | DIX domain containing 1 |
| FBXW4 | F-box and WD repeat domain containing 4 |
| RSPO2 | R-spondin 2 homolog (Xenopus laevis) |
| WWOX | WW domain containing oxidoreductase |
| AXIN2 | axin 2 |
| CSNK1A1 | casein kinase 1, alpha 1 |
| CTNNB1 | catenin (cadherin-associated protein), beta 1, 88kDa |
| DKK2 | dickkopf homolog 2 (Xenopus laevis) |
| FRAT2 | frequently rearranged in advanced T-cell lymphomas 2 |
| FZD1 | frizzled homolog 1 (Drosophila) |
| FZD5 | frizzled homolog 5 (Drosophila) |
| FRZB | frizzled-related protein |
| NXN | nucleoredoxin |
| SFRP1 | secreted frizzled-related protein 1 |
| SFRP2 | secreted frizzled-related protein 2 |
| TLE1 | similar to transducin-like enhancer of split 1 |
| TCF7L2 | transcription factor 7-like 2 (T-cell specific, HMG-box) |
| TLE4 | transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| WLS | WNT ligand secretion mediator |
| WNT1 | wingless-type MMTV integration site family, member 1 |
| WNT4 | wingless-type MMTV integration site family, member 4 |
| WNT7B | wingless-type MMTV integration site family, member 7B |
To verify the potential for the canonical WNT pathway to serve as a direct target for VSX2 binding and regulation during early hiPSC-OV differentiation, we performed confirmatory ChIP-PCR analysis focusing on the three highest scoring WNT pathway genes: WLS, WNT1, and AXIN2 (Fig. 1F; peak coverage for each is shown in Fig. 1E). Consensus binding sites from the MITF-H promoter that were previously shown to be either bound or unbound by VSX2 were also included as positive and negative controls, respectively (Fig. 1G)20. Two additional WNT pathway genes, SMAD3 and LEF1, with strong peaks in the first ChIP-seq analysis (peak coverage shown in Fig. S1B) but not in the second (and thus not included in the high confidence list) were also subjected to ChIP-PCR (Fig. S1C). All five WNT targets were amplified from immunoprecipitates of d30 VSX2WT hiPSC-OVs, but not VSX2R200Q hiPSC-OVs (Fig. 1F and Fig. S1C). Thus, our findings confirm that these genes are direct targets for VSX2 binding during retinal development.
Misexpression of an essential canonical WNT pathway component in neural retinal progenitors in VSX2R200Q hiPSC-OVs
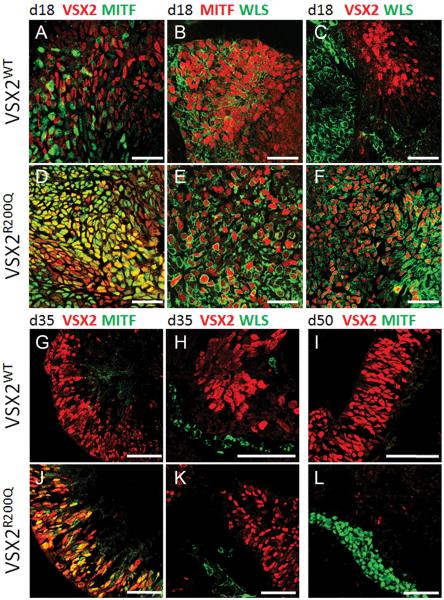
To further investigate effects on the WNT pathway stemming from the loss of VSX2 DNA binding ability, VSX2WT and VSX2R200Q hiPSCs underwent retinal differentiation for 18 days, a time point at which the expression of MITF and VSX2 (indicative of RPE progenitors and NRPCs, respectively) becomes mutually exclusive20. D18 VSX2WT and VSX2R200Q hiPSC-OVs were then immunostained for VSX2, MITF, and the critical canonical WNT pathway protein WLS (also known as GPR177), which is necessary for WNT secretion and activation (Fig. 2A–F, Fig. S2)38. Expression of MITF was excluded from VSX2+ cells in VSX2WT hiPSC-OVs at d18, consistent with VSX2's known role as a repressor of MITF (Fig. 2A)16, 18. Similarly, WLS expression was observed in MITF+ (Fig 2B), but not VSX2+ (Fig 2C), cells at d18. In stark contrast, VSX2+ cells in d18 VSX2R200Q hiPSC-OVs co-labeled with both MITF and WLS (Fig. 2D–F).
Figure 2. Comparison of VSX2, MITF, and WLS localization in VSX2WT and VSX2R200Q hiPSC-OVs.
D18 OVs from VSX2WT (A–C) or VSX2R200Q (D–F) hiPSCs were immunostained for VSX2 (red) and MITF (green) (A, D), MITF (red) and WLS (green) (B, E), or VSX2 (red) and WLS (green) (C, F). D35 and d50 VSX2WT hiPSC-OVs (G–I) were examined for VSX2 and MITF (G,I) or VSX2 and WLS (H) expression and compared to d35 (J,K) and d50 (L) VSX2R200Q hiPSC-OVs. Identical images with DAPI-labeled nuclei are shown in Fig. S2. Scale bars = 50 μm.
We next examined hiPSC-OV cultures differentiated to d35, a time point at which the RPE and NR domains are well-established, and to d50, when RPE maturation and retinal neurogenesis are fully underway28, 29. An abundant population of VSX2+ NRPCs was present in d35 VSX2WT hiPSC-OVs, which did not co-label with MITF or WLS (Fig. 2G and H; also see Fig. S2G and H). By d50, VSX2WT hiPSC-OVs consisted primarily of VSX2+ NRPCs with rare MITF+ RPE cells (Fig. 2I). However, in d35 VSX2R200Q hiPSC-OVs, a subpopulation of VSX2+ cells continued to co-express MITF, although co-labeling between VSX2 and WLS was no longer observed (Fig. 2J and K; also see Fig. S2J and K). By d50, VSX2R200Q hiPSC-OVs contained MITF+ RPE with no detectable VSX2+ cells (Fig. 2L). Together, these data demonstrate that, in the absence of functional VSX2 DNA binding activity, both MITF and WLS exhibit abnormal and prolonged expression in VSX2+ cells. However, other factors must be involved in the regulation of WLS expression in hiPSC-OVs, since it is eventually turned off even in mutant NRPCs.
The canonical WNT pathway is active in early VSX2R200Q hiPSC-OVs
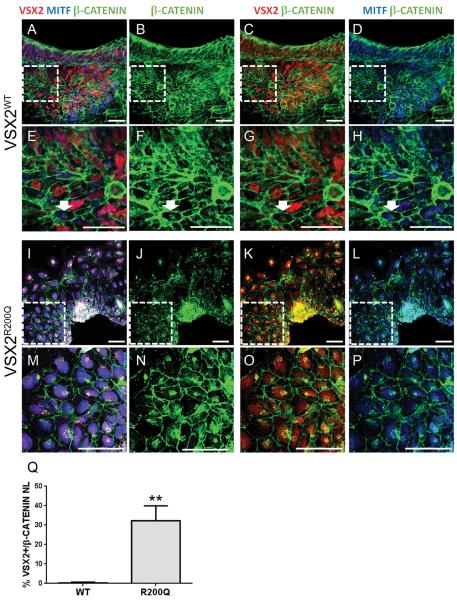
We next asked whether there were differences in WNT signaling activity between VSX2WT and VSX2R200Q hiPSC-OVs by examining βCATENIN nuclear localization (NL), a hallmark of canonical WNT pathway activation. For these experiments, we focused on d14 cultures as they contain a transient mixed population of MITF+/VSX2−, MITF+/VSX2+, and MITF−/VSX2+ cells as uncommitted hiPSC-OV cells transition to either NRPCs or early RPE precursors20. In VSX2WT hiPSC-OVs, βCATENIN NL was detected in MITF+/VSX2− cells and rare MITF+/VSX2+ cells but not in MITF−/VSX2+ cells (Fig. 3A–H; also see Fig S3). In d14 VSX2R200Q hiPSC-OVs, only MITF+/VSX2− and MITF+/VSX2+ populations were present, and many cells demonstrated βCATENIN NL (Fig. 3I–P; also see Fig S3). The percentage of VSX2+/βCATENIN NL+ co-labeled cells within the total VSX2+ cell population was quantified, revealing a mean of 0.37 ± 0.90% double-positive cells in VSX2WT cultures and 32.1 ± 19.3% double-positive cells in VSX2R200Q cultures (p=0.01) (Fig. 3Q). These results show that inactivation of canonical WNT signaling coincides with the onset of VSX2 expression in d14 VSX2WT hiPSC-OVs, whereas WNT signaling remains active in cells expressing mutant VSX2R200Q, which lacks DNA binding ability21, 24, 30.
Figure 3. βCATENIN localization in d14 VSX2WT and VSX2R200Q hiPSC-OVs.
D14 VSX2WT (A–H) or VSX2R200Q (I–P) hiPSC-OVs were immunolabeled with βCATENIN (green), VSX2 (red), and MITF (blue) primary antibodies. Examples of βCATENIN nuclear localization are designated by white arrows (E–H and M–P). Panels E–H and M–P are cropped magnifications of the outlined areas in panels A–D and I–J, respectively. Also see Fig. S3 for identical images with βCATENIN and DAPI-labeled nuclei. (Q) Graph of percent of βCATENIN nuclear localization in VSX2+ nuclei for WT and R200Q d14 hiPSC-OVs. **p=0.01. Scale bars = 50 μm.
Pharmacological manipulation of WNT signaling interconverts early VSX2WT and VSX2R200Q hiPSC-OV phenotypes
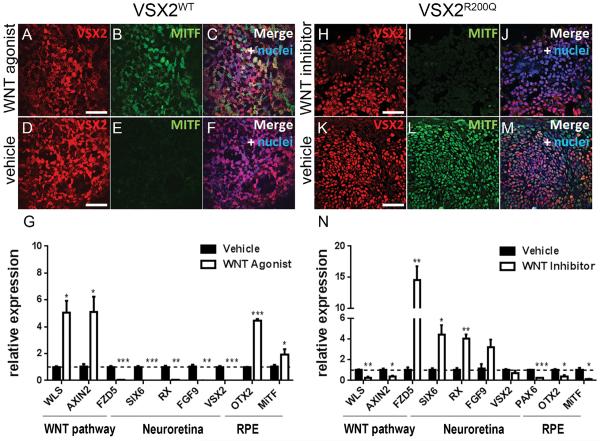
The observation that canonical WNT activity is inversely associated with the presence of functional VSX2 suggested that stimulation or inhibition of WNT signaling might alter the phenotypes of VSX2WT and VSX2R200Q hiPSC-OVs, respectively. Since d12 through d20 represents the critical period for hiPSC-OV formation, we applied pharmacological treatments at various intervals within that time frame, followed by continued growth of cultures without exogenous WNT modulation until d30. When VSX2WT hiPSC-OVs were treated with the WNT agonist CHIR99021 beginning at d12, cultures were pushed toward a nonretinal fate (data not shown). However, when CHIR99021 treatment of VSX2WT hiPSC-OVs was initiated on d14, aberrant MITF and VSX2 co-labeling was detected on d18, analogous to what was observed in VSX2R200Q hiPSC-OVs (Figure 4A–C; compare Fig. 4C to Fig. 2D). Control, vehicle-treated VSX2WT hiPSC-OVs showed the typical segregation of VSX2 and MITF expression at d18 (Fig. 4D–F). Upon removal of CHIR99021 at d20 and further culture to d30, expression of WNT pathway (WLS and AXIN2) and RPE (MITF and OTX2) genes were increased, whereas NR-related genes (VSX2, SIX6, RX, FGF9) were down-regulated (Fig. 4G). By contrast, CHIR99021 treatment of VSX2R200Q hiPSC-OVs over the same time periods had no significant effect (data not shown).
Figure 4. Interconversion of early VSX2WT and VSX2R200Q hiPSC-OV phenotypes by pharmacological manipulation of WNT signaling.
Immunocytochemistry analysis on d18 VSX2WT hiPSC-OVs treated with the WNT agonist CHIR99201 (A–C) or vehicle (D–F) showing VSX2 (red) and MITF (green) coexpression (A–C) or lack thereof (D–F). RT-qPCR analysis of d30 VSX2WT hiPSC-OVs treated from d14–d20 with vehicle or CHIR99201 (G). Immunocytochemistry analysis on d18 VSX2R200Q hiPSC-OVs treated with the WNT inhibitor IWP2 (H–J) or vehicle (K–M) showing lack of coexpression (H–J) or coexpression (K–M) of VSX2 (red) and MITF (green). (N) RT-qPCR analysis of d30 VSX2R200Q hiPSC-OVs treated from d12–d20 with vehicle or inhibitor. Nuclei are shown in blue. *p< 0.01; **p<0.001;***p<0.0001. Scale bars = 50 μm.
Next, VSX2R200Q hiPSC-OVs were treated with the WNT inhibitor IWP2. Unlike WNT agonist treatment of VSX2WT hiPSC-OVs, IWP2 treatment of VSX2R200Q hiPSC-OVs beginning at d12 did not lead to the production of nonretinal lineages. Thus, we applied IWP2 from d12 to d20 for all experiments. When sampled at d18 (Fig. 4H–M), abundant VSX2+ cells were present in IWP2-treated hiPSC-OVs (Fig. 4H), similar to vehicle-treated hiPSC-OVs (Fig. 4K). However, unlike vehicle-treated cultures, MITF expression was no longer detected in mutant VSX2+ cells at d18 in the presence of IWP2 (compare Fig. 4I to Fig. 4L). Likewise, when IWP2 treatment of VSX2R200Q hiPSC-OVs was discontinued at d20, followed by an additional 10 days of culture to promote further differentiation, WNT and RPE gene expression decreased and NR-related gene expression increased relative to d30 vehicle-treated VSX2R200Q hiPSC-OVs (Fig. 4N). Thus, pharmacological blockade of WNT signaling during early NR:RPE patterning partially rescues the VSX2R200Q hiPSC-OV phenotype at d30, while early augmentation of WNT signaling in VSX2WT hiPSC-OVs mimics the effects of the VSX2R200Q mutation.
DISCUSSION
The role of VSX2 as a transcriptional repressor is well-documented, although the number of known gene targets for VSX2 is small considering its pleiotropic effects throughout retinogenesis19, 30, 39–41. One of the earliest and most important functions of VSX2 is in the maintenance of a proliferating pool of NRPCs in the developing OV, which it accomplishes at least in part by repressing MITF expression, as demonstrated in both animal and hPSC model systems16, 18, 20. In this report we provide evidence that VSX2 can regulate expression of WNT pathway genes in early hiPSC-OVs, and that repression of WNT signaling by VSX2 may contribute to the maintenance of NR identity at the expense of RPE. Furthermore, pharmacological manipulation of WNT signaling can partially mimic or rescue the effects of a functional null VSX2 mutation in early hiPSC-OVs.
The Wnt pathway is necessary for axis formation and early embryonic patterning, and also serves as an essential regulator of neural and ocular development in part through the canonical βCatenin/TCF/Lef transcriptional pathway8, 9, 42, 43. With specific regard to ocular development, Wnt signaling is required for proper dorsal-ventral patterning, RPE production, and optic cup morphogenesis in vivo10, 11, 44–47. Conditional knockout (CKO) of βCatenin at an early OV stage in mouse resulted in transdifferentiation of RPE to VSX2+ NRPCs, leading to partial duplication of the NR10, 46. Conversely, ectopic βCatenin activation at later OV stages caused a thickened RPE layer with aberrant Mitf expression in the NR46. Interestingly, Mitf, which acts as a regulator of RPE differentiation and pigmentation, is a direct gene target for the Wnt/βCatenin/Lef transcriptional complex, and Mitf null mutations exhibit a similar RPE to NR phenotype as the βCatenin CKO4, 7.
The WNT pathway has also been exploited in order to enrich for desired cell types in vitro48, 49, including production of NRPCs or RPE cells from hESCs and hiPSCs50–53. Our prior finding that the expression of multiple WNT-related genes was dramatically increased in VSX2R200Q hiPSC-OVs led us to further examine the relationship between VSX2 and the WNT pathway using ChIP and massively parallel DNA sequencing. These analyses showed that VSX2 directly targets a host of genes encoding critical WNT pathway constituents in early hiPSC-OVs. Therefore, results from the present study, combined with prior reports in hiPSCs20 and animal models19, 21 showing direct regulation of MITF expression by VSX2, suggest that VSX2 antagonizes RPE development on multiple levels (Fig. 5). In addition to repressing MITF, VSX2 is capable of downregulating expression of key WNT pathway genes, including WLS, which is required for WNT trafficking and secretion38, 54, and AXIN2, which has a complex role in both βCatenin destabilization and recruitment of key Wnt receptor components to the cell membrane55. These examples, along with others (Tables 1 and S1), point to WNT pathway components as important targets for VSX2-mediated gene repression during early NR development in hiPSCs. However, the observation that WLS is eventually downregulated even in VSX2R200Q mutant NRPCs indicates that gene regulatory mechanisms independent of VSX2 are also in place to control expression of WNT pathway components in hiPSCs.
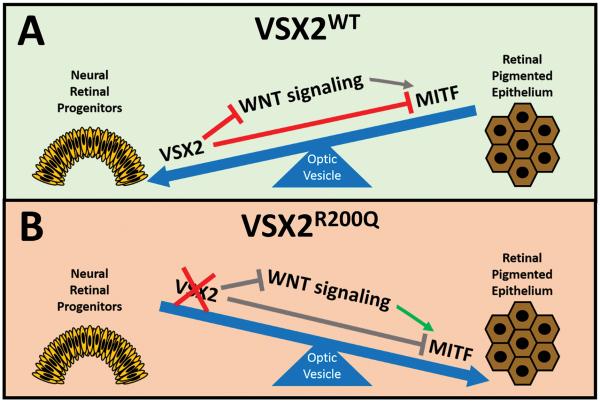
Figure 5. Schematic model depicting the impact of VSX2-mediated regulation of WNT signaling on early retinal patterning in wild-type hiPSC-OVs, and the consequence of the (R200Q)VSX2 mutation.
VSX2WT acts in multiple ways to maintain NR identity. On a gene expression level, VSX2 antagonizes expression of WNT pathway genes and MITF, resulting in the promotion of NR fate at the expense of RPE in hiPSC-OVs (A). In the absence of functional VSX2 (i.e. VSX2R200Q hiPSC-OVs), expression of WNT pathway genes and MITF are unchecked, leading to RPE production over NR (B).
In summary, our findings indicate a heretofore undescribed role for VSX2 as a regulator of WNT signaling in the developing retina, and suggest a mechanism whereby VSX2 maintains NR identity in part by regulation of WNT pathway genes. This study also offers the first unbiased search for VSX2 binding sites in a mammalian developmental model system, which may lead to the discovery of other genes and mechanisms involved in the maintenance of NR identity.
CONCLUSION
This study utilized an hiPSC model of a retinal developmental disorder, combined with pharmacological manipulations, chromatin immunoprecipitation, and massively parallel DNA sequencing to gain insight into a novel mechanism underlying NR vs. RPE fate choice during human retinogenesis. We show that the NRPC transcriptional factor VSX2 is a direct regulator of numerous WNT-related genes and that VSX2-mediated antagonism of WNT signaling plays an important role in the maintenance of NR identity at the expense RPE. Our findings also underscore the utility of hiPSCs in investigating the earliest stages of human ocular development, which are otherwise inaccessible to study.
Supplementary Material
Western blot of lysate from d30 VSX2WT (W) or VSX2R200Q (R) hiPSC-OVs before VSX2 antibody incubation (input) or after incubation with (+Ab) or without (−Ab) VSX2 antibody (ChIP) (M = molecular weight marker). Blue arrowheads point to the expected 47 kD size for VSX2 protein and the white asterisks mark the antibody heavy and light chains. The same VSX2 antibody was used for IP and Western blot detection (A). Peak localization and genomic coverage maps (± 200 bp toward the 5' or 3' end) for LEF1 and SMAD3 (red and blue lines represent forward and reverse DNA strand reads, respectively). The top panel designates ChIP coverage and the lower panel shows the input control for each gene (B). VSX2 ChIP-PCR from d30 VSX2WT or VSX2R200Q hiPSC-OVs confirms direct binding of VSX2WT, but not VSX2R200Q, to target sites proximal to the WNT signaling genes LEF1 and SMAD3 (C).
Identical images shown in Figure 2 with the addition of the nuclear stain DAPI (blue). D18 OVs from VSX2WT (A–C) or VSX2R200Q (D–F) hiPSCs were immunostained for VSX2 (red) and MITF (green) (A, D), MITF (red) and WLS (green) (B, E), or VSX2 (red) and WLS (green) (C,F). D35 and d50 VSX2WT hiPSC-OVs (G–I) were examined for VSX2 and MITF (G, I) or VSX2 andWLS (H) co-labeling and compared to d35 (J, K) and d50 (L) VSX2R200Q hiPSC-OVs. Scale bars = 50 μm.
Day 14 VSX2WT (left panels) or VSX2R200Q (right panels) hiPSC-OVs immunostained for βCATENIN (green) and DAPI (purple) to visualize nuclei. The lower panels are cropped magnifications of the outlined areas in the upper panels. An example of rare βCATENIN nuclear localization in VSX2WT cultures is marked with a white arrow. Scale bars = 5 μm.
High confidence VSX2 target gene list from ChIP-seq analysis.
ACKNOWLEDGEMENTS
We thank Jennifer Bolin and Angela Elwell for technical assistance and Scott Swanson for bioinformatics analysis.
This work was supported by the National Institutes of Health grants R01EY21218 (to DMG), P30HD03352 and Research to Prevent Blindness.
Footnotes
AUTHOR CONTRIBUTIONS Elizabeth E Capowski: conception and design, collection of data, data analysis and interpretation, manuscript writing
Lynda S Wright: conception and design, collection of data, data analysis and interpretation, manuscript writing
Kun Liang: Data anlysis and interpretation
M. Joseph Phillips: conception and design, data analysis and interpretation
Kyle Wallace: collection of data
Anna Petelinsek: collection of data
Anna Hagstrom: collection of data
Isabel Pinilla: collection of data
Katarzyna Borys: collection of data
Jessica Lien: collection of data
Jee Hong Min: collection of data
Sunduz Keles: financial support, data analysis and interpretation
James A Thomson: financial support, data analysis and interpretation
David M Gamm: financial support, conception and design, data analysis and interpretation, manuscript writing, final approval of manuscript
DISCLOSURES The authors indicate no potential conflicts of interest.
References
- 1.Chow RL, Lang RA. Early eye development in vertebrates. Annu Rev Cell Dev Biol. 2001;17:255–296. doi: 10.1146/annurev.cellbio.17.1.255. [DOI] [PubMed] [Google Scholar]
- 2.Zuber ME, Gestri G, Viczian AS, et al. Specification of the vertebrate eye by a network of eye field transcription factors. Development. 2003;130:5155–5167. doi: 10.1242/dev.00723. [DOI] [PubMed] [Google Scholar]
- 3.Hyer J, Mima T, Mikawa T. FGF1 patterns the optic vesicle by directing the placement of the neural retina domain. Development. 1998;125:869–877. doi: 10.1242/dev.125.5.869. [DOI] [PubMed] [Google Scholar]
- 4.Nguyen M, Arnheiter H. Signaling and transcriptional regulation in early mammalian eye development: a link between FGF and MITF. Development. 2000;127:3581–3591. doi: 10.1242/dev.127.16.3581. [DOI] [PubMed] [Google Scholar]
- 5.Vogel-Hopker A, Momose T, Rohrer H, et al. Multiple functions of fibroblast growth factor-8 (FGF-8) in chick eye development. Mech Dev. 2000;94:25–36. doi: 10.1016/s0925-4773(00)00320-8. [DOI] [PubMed] [Google Scholar]
- 6.Fuhrmann S. Eye morphogenesis and patterning of the optic vesicle. Curr Top Dev Biol. 2010;93:61–84. doi: 10.1016/B978-0-12-385044-7.00003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez-Morales JR, Rodrigo I, Bovolenta P. Eye development: a view from the retina pigmented epithelium. Bioessays. 2004;26:766–777. doi: 10.1002/bies.20064. [DOI] [PubMed] [Google Scholar]
- 8.de Iongh RU, Abud HE, Hime GR. WNT/Frizzled signaling in eye development and disease. Front Biosci. 2006;11:2442–2464. doi: 10.2741/1982. [DOI] [PubMed] [Google Scholar]
- 9.Fuhrmann S. Wnt signaling in eye organogenesis. Organogenesis. 2008;4:60–67. doi: 10.4161/org.4.2.5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagglund AC, Berghard A, Carlsson L. Canonical Wnt/beta-catenin signalling is essential for optic cup formation. PLoS One. 2013;8:e81158. doi: 10.1371/journal.pone.0081158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steinfeld J, Steinfeld I, Coronato N, et al. RPE specification in the chick is mediated by surface ectoderm-derived BMP and Wnt signalling. Development. 2013;140:4959–4969. doi: 10.1242/dev.096990. [DOI] [PubMed] [Google Scholar]
- 12.Veien ES, Rosenthal JS, Kruse-Bend RC, et al. Canonical Wnt signaling is required for the maintenance of dorsal retinal identity. Development. 2008;135:4101–4111. doi: 10.1242/dev.027367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bumsted KM, Barnstable CJ. Dorsal retinal pigment epithelium differentiates as neural retina in the microphthalmia (mi/mi) mouse. Invest Ophthalmol Vis Sci. 2000;41:903–908. [PubMed] [Google Scholar]
- 14.Tsukiji N, Nishihara D, Yajima I, et al. Mitf functions as an in ovo regulator for cell differentiation and proliferation during development of the chick RPE. Dev Biol. 2009;326:335–346. doi: 10.1016/j.ydbio.2008.11.029. [DOI] [PubMed] [Google Scholar]
- 15.Burmeister M, Novak J, Liang MY, et al. Ocular retardation mouse caused by Chx10 homeobox null allele: impaired retinal progenitor proliferation and bipolar cell differentiation. Nat Genet. 1996;12:376–384. doi: 10.1038/ng0496-376. [DOI] [PubMed] [Google Scholar]
- 16.Horsford DJ, Nguyen MT, Sellar GC, et al. Chx10 repression of Mitf is required for the maintenance of mammalian neuroretinal identity. Development. 2005;132:177–187. doi: 10.1242/dev.01571. [DOI] [PubMed] [Google Scholar]
- 17.Livne-Bar I, Pacal M, Cheung MC, et al. Chx10 is required to block photoreceptor differentiation but is dispensable for progenitor proliferation in the postnatal retina. Proc Natl Acad Sci U S A. 2006;103:4988–4993. doi: 10.1073/pnas.0600083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowan S, Chen CM, Young TL, et al. Transdifferentiation of the retina into pigmented cells in ocular retardation mice defines a new function of the homeodomain gene Chx10. Development. 2004;131:5139–5152. doi: 10.1242/dev.01300. [DOI] [PubMed] [Google Scholar]
- 19.Bharti K, Liu W, Csermely T, et al. Alternative promoter use in eye development: the complex role and regulation of the transcription factor MITF. Development. 2008;135:1169–1178. doi: 10.1242/dev.014142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capowski EE, Simonett JM, Clark EM, et al. Loss of MITF expression during human embryonic stem cell differentiation disrupts retinal pigment epithelium development and optic vesicle cell proliferation. Hum Mol Genet. 2014;23:6332–6344. doi: 10.1093/hmg/ddu351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zou C, Levine EM. Vsx2 controls eye organogenesis and retinal progenitor identity via homeodomain and non-homeodomain residues required for high affinity DNA binding. PLoS Genet. 2012;8:e1002924. doi: 10.1371/journal.pgen.1002924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bar-Yosef U, Abuelaish I, Harel T, et al. CHX10 mutations cause non-syndromic microphthalmia/ anophthalmia in Arab and Jewish kindreds. Hum Genet. 2004;115:302–309. doi: 10.1007/s00439-004-1154-2. [DOI] [PubMed] [Google Scholar]
- 23.Faiyaz-Ul-Haque M, Zaidi SH, Al-Mureikhi MS, et al. Mutations in the CHX10 gene in non-syndromic microphthalmia/anophthalmia patients from Qatar. Clin Genet. 2007;72:164–166. doi: 10.1111/j.1399-0004.2007.00846.x. [DOI] [PubMed] [Google Scholar]
- 24.Percin EF, Ploder LA, Yu JJ, et al. Human microphthalmia associated with mutations in the retinal homeobox gene CHX10. Nat Genet. 2000;25:397–401. doi: 10.1038/78071. [DOI] [PubMed] [Google Scholar]
- 25.Lamba DA, Karl MO, Ware CB, et al. Efficient generation of retinal progenitor cells from human embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:12769–12774. doi: 10.1073/pnas.0601990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer JS, Shearer RL, Capowski EE, et al. Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2009;106:16698–16703. doi: 10.1073/pnas.0905245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osakada F, Ikeda H, Mandai M, et al. Toward the generation of rod and cone photoreceptors from mouse, monkey and human embryonic stem cells. Nat Biotechnol. 2008;26:215–224. doi: 10.1038/nbt1384. [DOI] [PubMed] [Google Scholar]
- 28.Phillips MJ, Wallace KA, Dickerson SJ, et al. Blood-derived human iPS cells generate optic vesicle-like structures with the capacity to form retinal laminae and develop synapses. Invest Ophthalmol Vis Sci. 2012;53:2007–2019. doi: 10.1167/iovs.11-9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer JS, Howden SE, Wallace KA, et al. Optic vesicle-like structures derived from human pluripotent stem cells facilitate a customized approach to retinal disease treatment. Stem Cells. 2011;29:1206–1218. doi: 10.1002/stem.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark AM, Yun S, Veien ES, et al. Negative regulation of Vsx1 by its paralog Chx10/Vsx2 is conserved in the vertebrate retina. Brain Res. 2008;1192:99–113. doi: 10.1016/j.brainres.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phillips MJ, Perez ET, Martin JM, et al. Modeling human retinal development with patient-specific induced pluripotent stem cells reveals multiple roles for visual system homeobox 2. Stem Cells. 2014;32:1480–1492. doi: 10.1002/stem.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang K, Keles S. Normalization of ChIP-seq data with control. BMC Bioinformatics. 2012;13:199. doi: 10.1186/1471-2105-13-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 38.Das S, Yu S, Sakamori R, et al. Wntless in Wnt secretion: molecular, cellular and genetic aspects. Front Biol (Beijing) 2012;7:587–593. doi: 10.1007/s11515-012-1200-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dorval KM, Bobechko BP, Fujieda H, et al. CHX10 targets a subset of photoreceptor genes. J Biol Chem. 2006;281:744–751. doi: 10.1074/jbc.M509470200. [DOI] [PubMed] [Google Scholar]
- 40.Reichman S, Kalathur RK, Lambard S, et al. The homeobox gene CHX10/VSX2 regulates RdCVF promoter activity in the inner retina. Hum Mol Genet. 2010;19:250–261. doi: 10.1093/hmg/ddp484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowan S, Cepko CL. A POU factor binding site upstream of the Chx10 homeobox gene is required for Chx10 expression in subsets of retinal progenitor cells and bipolar cells. Dev Biol. 2005;281:240–255. doi: 10.1016/j.ydbio.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 42.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 43.Munoz-Descalzo S, Hadjantonakis AK, Arias AM. Wnt/ss-catenin signalling and the dynamics of fate decisions in early mouse embryos and embryonic stem (ES) cells. Semin Cell Dev Biol. 2015;47–48:101–109. doi: 10.1016/j.semcdb.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carpenter AC, Smith AN, Wagner H, et al. Wnt ligands from the embryonic surface ectoderm regulate `bimetallic strip' optic cup morphogenesis in mouse. Development. 2015;142:972–982. doi: 10.1242/dev.120022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cho SH, Cepko CL. Wnt2b/beta-catenin-mediated canonical Wnt signaling determines the peripheral fates of the chick eye. Development. 2006;133:3167–3177. doi: 10.1242/dev.02474. [DOI] [PubMed] [Google Scholar]
- 46.Fujimura N, Taketo MM, Mori M, et al. Spatial and temporal regulation of Wnt/beta-catenin signaling is essential for development of the retinal pigment epithelium. Dev Biol. 2009;334:31–45. doi: 10.1016/j.ydbio.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Westenskow P, Piccolo S, Fuhrmann S. Beta-catenin controls differentiation of the retinal pigment epithelium in the mouse optic cup by regulating Mitf and Otx2 expression. Development. 2009;136:2505–2510. doi: 10.1242/dev.032136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clevers H, Loh KM, Nusse R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346:1248012. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- 49.Van Camp JK, Beckers S, Zegers D, et al. Wnt signaling and the control of human stem cell fate. Stem Cell Rev. 2014;10:207–229. doi: 10.1007/s12015-013-9486-8. [DOI] [PubMed] [Google Scholar]
- 50.Leach LL, Buchholz DE, Nadar VP, et al. Canonical/beta-catenin Wnt pathway activation improves retinal pigmented epithelium derivation from human embryonic stem cells. Invest Ophthalmol Vis Sci. 2015;56:1002–1013. doi: 10.1167/iovs.14-15835. [DOI] [PubMed] [Google Scholar]
- 51.Lupo G, Novorol C, Smith JR, et al. Multiple roles of Activin/Nodal, bone morphogenetic protein, fibroblast growth factor and Wnt/beta-catenin signalling in the anterior neural patterning of adherent human embryonic stem cell cultures. Open Biol. 2013;3:120167. doi: 10.1098/rsob.120167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maruotti J, Sripathi SR, Bharti K, et al. Small-molecule-directed, efficient generation of retinal pigment epithelium from human pluripotent stem cells. Proc Natl Acad Sci U S A. 2015;112:10950–10955. doi: 10.1073/pnas.1422818112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakano T, Ando S, Takata N, et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 54.Najdi R, Proffitt K, Sprowl S, et al. A uniform human Wnt expression library reveals a shared secretory pathway and unique signaling activities. Differentiation. 2012;84:203–213. doi: 10.1016/j.diff.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song X, Wang S, Li L. New insights into the regulation of Axin function in canonical Wnt signaling pathway. Protein Cell. 2014;5:186–193. doi: 10.1007/s13238-014-0019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Western blot of lysate from d30 VSX2WT (W) or VSX2R200Q (R) hiPSC-OVs before VSX2 antibody incubation (input) or after incubation with (+Ab) or without (−Ab) VSX2 antibody (ChIP) (M = molecular weight marker). Blue arrowheads point to the expected 47 kD size for VSX2 protein and the white asterisks mark the antibody heavy and light chains. The same VSX2 antibody was used for IP and Western blot detection (A). Peak localization and genomic coverage maps (± 200 bp toward the 5' or 3' end) for LEF1 and SMAD3 (red and blue lines represent forward and reverse DNA strand reads, respectively). The top panel designates ChIP coverage and the lower panel shows the input control for each gene (B). VSX2 ChIP-PCR from d30 VSX2WT or VSX2R200Q hiPSC-OVs confirms direct binding of VSX2WT, but not VSX2R200Q, to target sites proximal to the WNT signaling genes LEF1 and SMAD3 (C).
Identical images shown in Figure 2 with the addition of the nuclear stain DAPI (blue). D18 OVs from VSX2WT (A–C) or VSX2R200Q (D–F) hiPSCs were immunostained for VSX2 (red) and MITF (green) (A, D), MITF (red) and WLS (green) (B, E), or VSX2 (red) and WLS (green) (C,F). D35 and d50 VSX2WT hiPSC-OVs (G–I) were examined for VSX2 and MITF (G, I) or VSX2 andWLS (H) co-labeling and compared to d35 (J, K) and d50 (L) VSX2R200Q hiPSC-OVs. Scale bars = 50 μm.
Day 14 VSX2WT (left panels) or VSX2R200Q (right panels) hiPSC-OVs immunostained for βCATENIN (green) and DAPI (purple) to visualize nuclei. The lower panels are cropped magnifications of the outlined areas in the upper panels. An example of rare βCATENIN nuclear localization in VSX2WT cultures is marked with a white arrow. Scale bars = 5 μm.
High confidence VSX2 target gene list from ChIP-seq analysis.