Clostridium difficile is transmitted between hosts in the form of a dormant spore, and germination by C. difficile spores is required to initiate infection, because the toxins that are necessary for disease are not deposited on the spore form. Importantly, the C. difficile spore germination pathway represents a novel pathway for bacterial spore germination. Prior work has shown that the order of events during C. difficile spore germination (cortex degradation and DPA release) is flipped compared to the events during B. subtilis spore germination, a model organism. Here, we further characterize the C. difficile spore germination pathway and summarize our findings indicating that DPA release by germinating C. difficile spores occurs through a mechanosensing mechanism in response to the degradation of the spore cortex.
KEYWORDS: Clostridium difficile, DPA, cortex, dipicolinic acid, germination, mechanosensing, osmolytes, spoVAC, spore
ABSTRACT
Classically, dormant endospores are defined by their resistance properties, particularly their resistance to heat. Much of the heat resistance is due to the large amount of dipicolinic acid (DPA) stored within the spore core. During spore germination, DPA is released and allows for rehydration of the otherwise-dehydrated core. In Bacillus subtilis, 7 proteins are encoded by the spoVA operon and are important for DPA release. These proteins receive a signal from the activated germinant receptor and release DPA. This DPA activates the cortex lytic enzyme CwlJ, and cortex degradation begins. In Clostridium difficile, spore germination is initiated in response to certain bile acids and amino acids. These bile acids interact with the CspC germinant receptor, which then transfers the signal to the CspB protease. Activated CspB cleaves the cortex lytic enzyme, pro-SleC, to its active form. Subsequently, DPA is released from the core. C. difficile encodes orthologues of spoVAC, spoVAD, and spoVAE. Of these, the B. subtilis SpoVAC protein was shown to be capable of mechanosensing. Because cortex degradation precedes DPA release during C. difficile spore germination (opposite of what occurs in B. subtilis), we hypothesized that cortex degradation would relieve the osmotic constraints placed on the inner spore membrane and permit DPA release. Here, we assayed germination in the presence of osmolytes, and we found that they can delay DPA release from germinating C. difficile spores while still permitting cortex degradation. Together, our results suggest that DPA release during C. difficile spore germination occurs though a mechanosensing mechanism.
IMPORTANCE Clostridium difficile is transmitted between hosts in the form of a dormant spore, and germination by C. difficile spores is required to initiate infection, because the toxins that are necessary for disease are not deposited on the spore form. Importantly, the C. difficile spore germination pathway represents a novel pathway for bacterial spore germination. Prior work has shown that the order of events during C. difficile spore germination (cortex degradation and DPA release) is flipped compared to the events during B. subtilis spore germination, a model organism. Here, we further characterize the C. difficile spore germination pathway and summarize our findings indicating that DPA release by germinating C. difficile spores occurs through a mechanosensing mechanism in response to the degradation of the spore cortex.
INTRODUCTION
Clostridium difficile is a Gram-positive, spore-forming, strict anaerobe that most commonly infects immunocompromised or antibiotic-treated hosts. Many antibiotics have a broad spectrum of activity that disrupts the normal microbiota, which provides colonization resistance to C. difficile infection (1). This disruption enables C. difficile to colonize and cause disease (2). In a host, C. difficile secretes two toxins (TcdA and TcdB) that damage the colonic epithelium and elicit the primary symptoms of disease (3). Though disease is caused by vegetative cells, it is the spore form that makes possible the transition between the aerobic environment and hosts (4).
Spores are metabolically dormant forms of bacteria that are resistant to many harsh conditions (e.g., heat, desiccation, antibiotics) (5, 6). The spore structure is conserved across most spore-forming, Gram-positive bacteria and is important for maintaining the spore’s resistance properties. Contained within the spore core is genomic DNA bound by small acid-soluble proteins (SASPs) and a large quantity of Ca-dipicolinic acid (DPA) (7, 8). The SASPs protect the DNA from UV damage, and DPA packaging helps protect the core from heat by excluding water (7, 9, 10). Surrounding the core is an inner spore membrane, where many of the Ger-type germinant receptors found in Bacilli, and most Clostridia, are located (6). Surrounding the inner membrane is a thin layer of germ cell wall peptidoglycan and a thick layer of specialized cortex peptidoglycan. In cortex peptidoglycan, many of the N-acetylmuramic acid residues have been converted to muramic-δ-lactam residues and are the targets for cortex-degrading enzymes (8). An outer membrane surrounds the cortex and functions as a scaffold with which to build the coat layer. In some spore-forming bacteria, including C. difficile, an additional exosporium layer surrounds the spore coat (6).
Spores remain metabolically dormant until specific signals, termed germinants, are detected by receptors (germinant receptors) in the spore. Germinant recognition by germinant receptors leads to the irreversible initiation of the germination process. In B. subtilis, a model organism for studying sporulation and germination, the l-alanine germinant is recognized at the inner spore membrane by the GerA germinant receptor (which is composed of GerAA-AB-AC proteins), while l-asparagine, d-glucose, d-fructose, and K+ ions (AGFK) are recognized by GerB and GerK (which are composed of GerBA-BB-BC and GerKA-KB-KC, respectively) (8). The activation of these germinant receptors triggers the release of cations and DPA from the core. The mechanism of DPA release is unclear; however, the proteins encoded by the spoVA operon (in shorthand, SpoVAA-AB-AC-AD-AEa-AEb-AF) play a role (11–16). In B. subtilis, the release of DPA activates the spore cortex lytic enzyme (SCLE) CwlJ, and the actions of CwlJ and SleB lead to cortex degradation (8). This mechanism of spore germination is similar across most spore-forming bacteria studied to date.
C. difficile spore germination is triggered by a combination of certain bile acids and amino acids (5, 17–20). In contrast to the mechanisms of germination observed in Bacilli and most Clostridia, C. difficile does not encode the Ger-type germinant receptors (21). Instead, C. difficile uses the germination-specific, pseudoprotease CspC as the bile acid germinant receptor (22). In Clostridium perfringens, CspA, CspB, and CspC are active proteases with the potential to cleave the SCLE, pro-SleC, to its active form (5, 23–28). Interestingly, C. difficile CspA and CspC are pseudoproteases, as their catalytic triads are not complete. Due to the apparent lack of catalytic activity, we proposed a working model where activated CspC signals CspB to cleave pro-SleC to an active form. SleC activation initiates cortex degradation. Recently, another protein, GerS, was identified to play an important role during C. difficile spore germination (29). Spores lacking GerS fail to degrade cortex but still process SleC into its active form (29). In contrast to the mechanisms of germination observed in B. subtilis, during C. difficile spore germination the DPA contained within the core is released after cortex degradation begins (30). Whereas B. subtilis releases DPA through a pore presumably formed by the proteins encoded by the spoVA operon, C. difficile does not encode the entire operon (21, 31). Instead, C. difficile encodes three homologues: spoVAC, spoVAD, and spoVAE (31). In B. subtilis, the spoVA proteins are required for the completion of sporulation, likely due to defects in DPA packaging (32); SpoVAD is thought to act as a DPA binding protein (11), helping to package DPA in the core during spore formation. Moreover, a recent study identified a mobile genetic element that carries spoVAC, spoVAD, and spoVAE, and the resulting overexpression of the proteins encoded by these genes led to accumulation of up to 50% more DPA in the spores and increased heat resistance (10).
B. subtilis SpoVAC has been shown to have mechanosensing properties (13). If SpoVAC functions as a mechanosensing protein in C. difficile, it could respond to the changes in osmolarity observed at the inner spore membrane due to the removal of constraints placed upon the dormant, dehydrated core by the cortex layer. Cortex degradation may allow pores to open in response to the lower osmotic pressure of the environment relative to that in the DPA-rich core. We hypothesized that a spore germinating in an environment with an osmolyte concentration equal to or higher than that of the core would affect DPA release. Here, we investigated the role high osmolyte concentrations play in cortex degradation and DPA release during C. difficile spore germination. We found that high osmolyte concentrations can block DPA release from the core while permitting cortex degradation. Our data suggest that DPA release during germination by C. difficile spores is due to changes in osmolarity that occur during cortex degradation.
RESULTS
Measuring DPA content of SpoVAC-deficient spores.
In a previous study, we determined that cortex degradation precedes DPA release and we hypothesized that SpoVAC may trigger the SpoVA channel to release DPA in a mechanosensing fashion (30). Recently, a mutation in spoVAC was shown to affect packaging and release of DPA from the C. difficile spore (31). Using the CodA-based allelic exchange system (33), we created a mutant strain in which the spoVAC sequence was truncated, with only the first 30 bp and last 30 bp of spoVAC present. Spores were prepared from wild-type C. difficile R20291, C. difficile MBF02 (ΔspoVAC), and C. difficile MBF02 pMB15 (pspoVAC). The DPA content of these spores was measured by boiling the spores, a condition that artificially releases the stored DPA (22). Similar to the results of a recent study (31), spores derived from the spoVAC mutant strain contained approximately 1% of the DPA content found in the wild-type strain (Fig. 1), and this could be complemented by expressing spoVAC in trans. These results confirmed prior observations indicating that spoVAC is important for packaging DPA during spore formation (31).
FIG 1 .
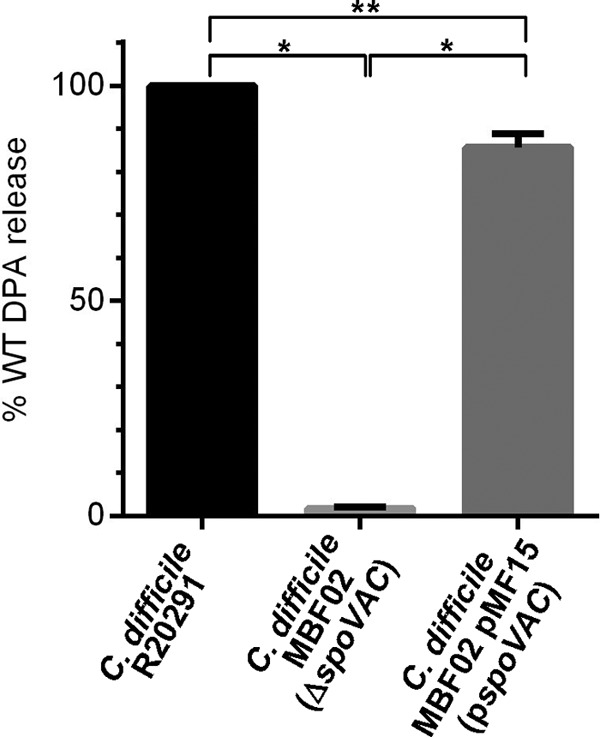
SpoVAC is important for DPA packaging. Equal amounts of spores derived from C. difficile strains R20291 (WT), MBF02 (ΔspoVAC), and MBF02 pMF15 (pspoVAC) were boiled for 15 min, and the amount of DPA was quantified based on Tb3+ fluorescence. The data shown represent the average results from three independent experiments, and the error bars represent the standard deviations from the means. All values are reported as the percentage release relative to that of C. difficile R20291. *, P < 0.001; **, P < 0.05.
High sorbitol concentrations delay the onset of C. difficile spore germination.
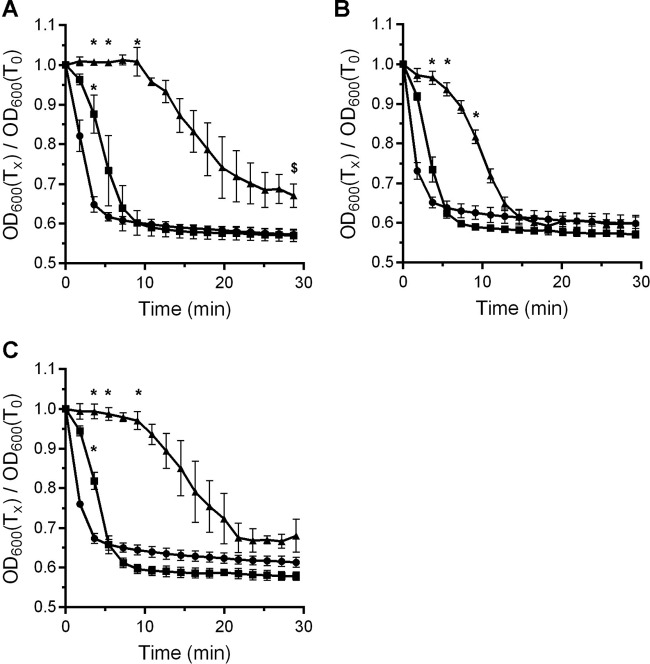
To test the effects of osmolytes on C. difficile spore germination, we measured the change in the optical density at 600 nm (OD600) of spores suspended in either buffer supplemented with taurocholate (TA) and glycine or buffer supplemented with TA, glycine, and an osmolyte (either sorbitol, trehalose, or sucrose). The OD germination assay is a simple method for observing how germination proceeds on the whole (the change from a phase bright spore to a phase dark spore) but does not give detailed information about individual steps (e.g., DPA release, cortex degradation). When C. difficile R20291 spores were suspended in buffer supplemented with both TA and glycine, a rapid decrease in the OD600 of the suspension was observed. However, when C. difficile R20291 spores were suspended in buffer supplemented with TA, glycine, and increasing amounts of sorbitol (0%, 19%, and 38%), the wild-type spores demonstrated a significant delay in the drop in the OD600 (Fig. 2A), suggesting that the increasing osmolyte concentration blocked or slowed C. difficile spore germination.
FIG 2 .
Osmolytes delay germination by C. difficile spores. C. difficile R20291 spores were germinated in buffer supplemented with taurocholate and glycine in the absence of sorbitol (filled circles) or in buffer supplemented with 19% (filled squares) or 38% (filled triangles) osmolyte (sorbitol [A], trehalose [B], or sucrose [C]). For clarity, every fifth data point is plotted, and the data represent the average results from three independent experiments. Error bars represent the standard deviations from the means. *, P < 0.01; $, P < 0.05.
To understand if the above observations were due to sorbitol alone or if other osmolytes could substitute for sorbitol, we measured germination in trehalose (Fig. 2B) or sucrose (Fig. 2C). As observed for germination in the presence of sorbitol, C. difficile spores germinating in the presence of 19% trehalose or sucrose had a short delay in germination. However, C. difficile spore germination was significantly delayed in the presence of 38% trehalose or sucrose (Fig. 2B and C, respectively). These results suggest that C. difficile spore germination can be delayed under increased osmolyte concentrations.
Osmolytes block DPA release during C. difficile spore germination.
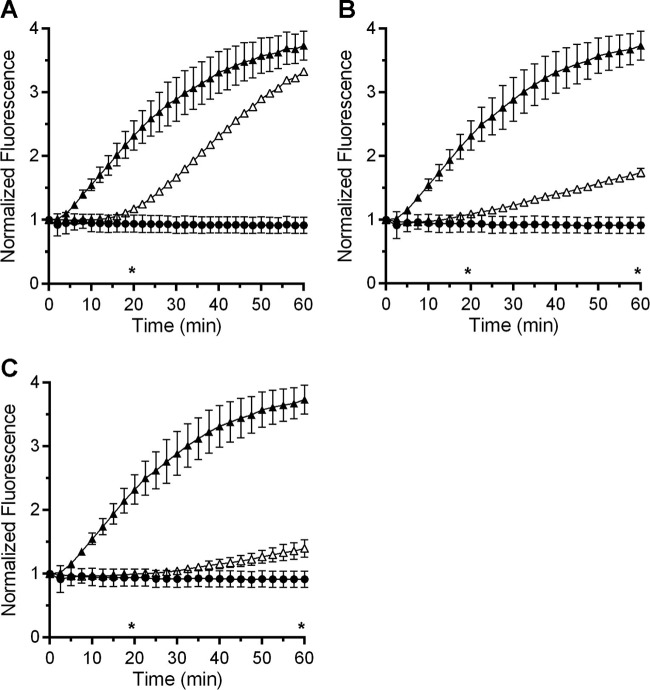
To understand if the osmolyte-mediated delay in germination by C. difficile spores was specific to the OD assay conditions or if we would observe similar changes when measuring DPA release, we monitored DPA release by germinating spores in the presence of TbCl3 (34, 35). During germination, the released DPA complexes with Tb3+, which results in fluorescence of the lanthanide metal. Thus, DPA release by germinating spores is measured in real time by monitoring DPA-dependent Tb3+ fluorescence. C. difficile R20291 spores were added to buffer supplemented with TA and glycine alone or TA, glycine, and sorbitol, and DPA release was monitored as a readout for germination (Fig. 3A). In the absence of sorbitol, spores rapidly released their stored DPA and an increase in Tb3+ fluorescence was observed (Fig. 3A). Interestingly, spores delayed the release of DPA in the presence of sorbitol. We found that 38% sorbitol led to a delay of ~8 min over the course of the experiment (Fig. 3A). To understand if DPA release could be blocked or delayed by other osmolytes, spores were germinated as described above but sorbitol was replaced with trehalose (Fig. 3B) or sucrose (Fig. 3C). Both trehalose and sucrose delayed the DPA release by germinating C. difficile spores.
FIG 3 .
Osmolytes delay DPA release by C. difficile spores. C. difficile R20291 spores were suspended in buffer containing terbium chloride and glycine alone (closed circles) or supplemented with taurocholate (filled triangles) or with taurocholate and 38% osmolyte (open triangles) (sorbitol [A], trehalose [B], or sucrose [C]). For clarity, every fifth data point is plotted, and the data represent the average results from three independent experiments. Error bars represent the standard deviations from the means. *, P ≤ 0.01. Statistical significance for TA plus glycine conditions compared to 38% sorbitol conditions was only tested at the 20- and 60-min time points.
Prior reports have suggested that there is heterogeneity between different C. difficile strains in terms of their response to germinants (36–38). Though the rates and efficiencies with which C. difficile spores germinate in response to bile acids have been reported, the mechanisms underlying the response to germinants are likely conserved among strains (30, 38, 39). To ensure that our observation that high osmolyte concentrations delay the release of DPA from germinating C. difficile spores was not due to a strain-specific phenotype, we determined whether the C. difficile M68 strain yielded similar results. DPA release by C. difficile M68, ribotype 017, spores was delayed by increased sorbitol or trehalose or sucrose concentrations (see Fig. S1 in the supplemental material). These results suggest that the effects of high osmolyte concentrations on C. difficile spore germination are not strain specific and that high osmolyte concentrations delay the release of DPA from the core of germinating C. difficile spores.
Osmolytes delay DPA release by C. difficile M68 spores. C. difficile M68 spores were suspended in buffer containing terbium chloride and glycine alone (filled circles) or supplemented with taurocholate (filled triangles) or taurocholate and 38% osmolyte (open triangles) (sorbitol [A], trehalose [B], or sucrose [C]). For clarity, every fifth data point is plotted, and the data represent the average results from three independent experiments. Error bars represent the standard deviations from the means. Download Figure S1, TIF file, 0.8 MB (819.7KB, tif) .
Copyright © 2016 Francis and Sorg.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Pro-SleC is cleaved to an active form in high osmolyte concentrations.
The delays in OD change and in DPA release could be the result of the high osmolyte concentration retarding the rate at which germinants (TA and glycine) interact with the germinant receptors. To test if the germinants were still activating germination in the presence of osmolyte, we tested if the SCLE pro-SleC was cleaved to its active form (SleC activation is necessary for cortex degradation). C. difficile spores were suspended in buffer with glycine only, as a negative control, or in buffer supplemented with TA and glycine with or without 38% osmolyte. Samples were taken at the indicated times and processed for immunoblotting. In the absence of TA, spores did not activate pro-SleC (Fig. 4). However, spores rapidly cleaved pro-SleC to its active form in response to TA and glycine. Significantly, the presence of osmolyte had no effect on the timing of SleC activation (Fig. 4).
FIG 4 .
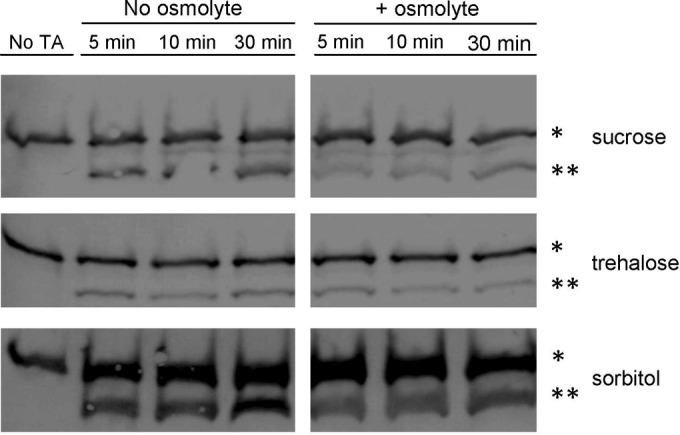
High osmolyte concentrations do not affect SleC activation. Cleavage of pro-SleC (*) to SleC (**) was assayed in buffer supplemented with glycine or buffer supplemented with glycine and taurocholate, with or without 38% osmolyte, at 5, 10, and 30 min.
Detecting cortex degradation in the presence of osmolytes.
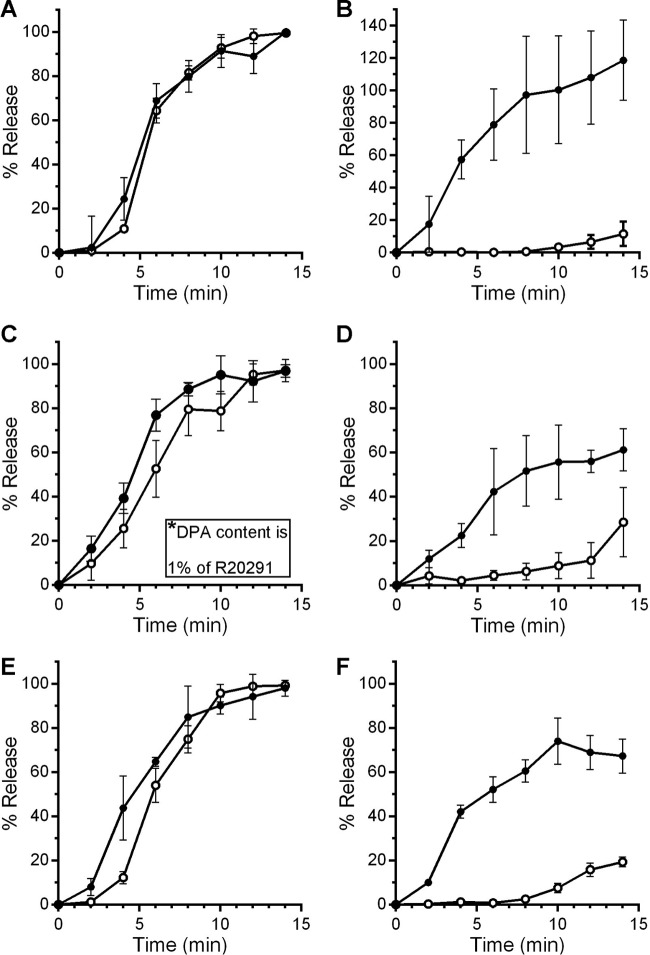
If SleC is activated in the presence of osmolyte, the appearance of cortex fragments in the germinant solution should be readily detected. To detect these fragments, we made use of a colorimetric assay that we, and others, have used to quantify the presence of reducing sugars (27, 28, 30, 40). This assay detects the presence N-acetylglucosamine (NAG) and N-acetylmuramic acid residues (reducing sugars) released during the degradation of cortex peptidoglycan (40). Purified spores were germinated in the presence of TA and glycine and were sampled every 2 min for the presence of both cortex fragments and DPA (Fig. 5). The results for C. difficile R20291 spores were similar to those previously reported for C. difficile strain UK1, as cortex degradation and DPA release seemingly occurred simultaneously (Fig. 5A) for a wild-type strain (using mutant strains, we previously observed that cortex degradation precedes DPA release [30]). When we tested cortex degradation and DPA release in buffer supplemented with 38% sorbitol with TA and glycine (sorbitol was the only osmolyte tested in this assay because it is the only compound among those we tested that does not generate a reducing end), the release of cortex fragments occurred in a similar manner as in the absence of sorbitol, while DPA release was delayed and only began to be evident after 8 min (Fig. 5B).
FIG 5 .
High osmolyte concentrations delay DPA release but not cortex degradation. Purified C. difficile R20291 spores (A and B), C. difficile MBF02 (ΔspoVAC) spores (C and D), or C. difficile MBF02 pMF15 (pspoVAC) spores (E and F) were suspended in buffer supplemented with taurocholate and glycine and no sorbitol (A, C, and E) or the same buffer supplemented with 38% sorbitol (B, D, and F). At the indicated time points, samples were taken for the amount of cortex fragments (filled circles) and DPA (open circles). Values for all graphs were normalized to the average maximum amount of cortex or DPA released by each strain in the absence of sorbitol. The data represent the average results from three independent experiments, and error bars represent the standard deviations from the means.
When germinated in TA with glycine, C. difficile MBF02 (ΔspoVAC) spores released cortex fragments similarly to the wild-type strain (Fig. 5C). DPA release by this strain also occurred; however, the total amount of DPA from the MBF02 strain was only 1% of that found with the R20291 strain (Fig. 1). When the spoVAC mutant spores were germinated in the presence of 38% sorbitol, the release of what little amount of DPA was present was delayed to the very end of the time period, while cortex degradation remained largely unaffected (Fig. 5D). These observations could be complemented by expressing spoVAC in trans from a plasmid (Fig. 5E and F).
Alteration of the osmolyte concentration affects DPA release during germination.
If DPA release is dependent on both cortex degradation and a mechanosensing mechanism, changing the osmolyte concentration during germination should result in a marked change in the rate of DPA release. To test the effects of an osmotic downshift on DPA release by germinating spores, spores were allowed to germinate for 5 min in 1 volume of 38% sorbitol-containing germination buffer (containing TA and glycine). Then, either 2 volumes of germination buffer with sorbitol (leading to no change in osmotic conditions) or 2 volumes of the germination buffer without sorbitol (leading to an osmotic downshift) were then added to the well. Germination was then monitored under these new conditions. In samples that had sorbitol-containing germination buffer added, DPA release was delayed, as expected; the osmotic strength of the solution did not change (Fig. 6). However, diluting the osmolyte to 12.7% (a 3-fold dilution) resulted in a rapid release of DPA from the core (Fig. 6). As controls, samples in which the spores were germinated in germination buffer alone or germination buffer supplemented with 38% sorbitol for the duration of the experiment were included. As expected, in the control samples DPA was released in the no-sorbitol germination buffer, while the sorbitol-containing germination buffer blocked DPA release (Fig. 6). These results suggest that DPA release during germination by C. difficile spores occurs through a mechanosensing mechanism that is dependent on degradation of the spore cortex layer.
FIG 6 .
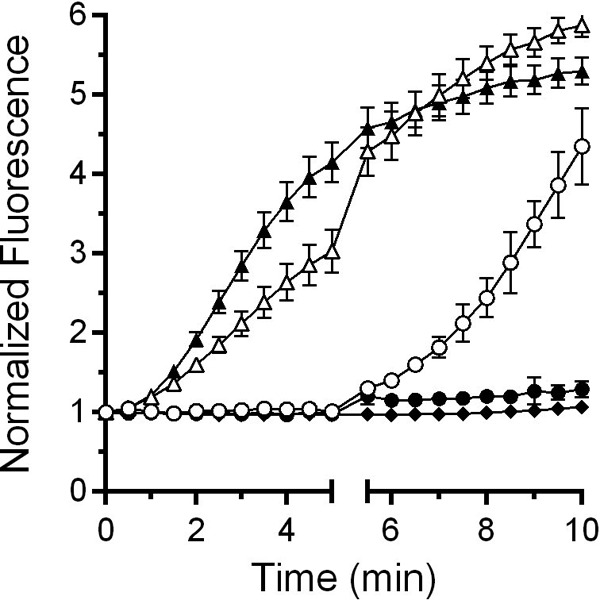
Altering osmotic conditions during germination influences DPA release. Purified C. difficile R20291 spores were suspended in buffer supplemented with taurocholate and glycine alone (filled triangles) or also in the presence of 38% sorbitol (filled diamonds) and allowed to germinate for 10 min. In a separate experiment, spores were germinated for 5 min in buffer supplemented with taurocholate and glycine alone (open triangles) or with 38% sorbitol (open circles). These samples then were diluted with 2 volumes of buffer supplemented with taurocholate and glycine alone. Finally, a separate set of spores were germinated for 5 min in buffer supplemented with taurocholate, glycine, and 38% sorbitol (filled circles), and then 2 volumes of 38% sorbitol-containing germination buffer were added and germination was allowed to continue for another 5 min.
DISCUSSION
The release of DPA from the core of a spore is a crucial step during spore germination. However, the mechanisms of DPA release by germinating spores are poorly understood. Here, we reaffirmed that SpoVAC plays a critical role in DPA import into the core during spore formation (Fig. 1) (31). Unfortunately, our current understanding of the mechanism of DPA release and SpoVAC activity prevents the separation of DPA packaging during C. difficile spore formation from DPA release during germination. Thus, to begin to understand the mechanisms of DPA release by germinating C. difficile spores, we raised the concentration of osmolyte in the germination solution. The increase in osmolyte concentration led to a marked delay in DPA release but not a delay in the activation of SleC, a key step in germination (Fig. 3 and 4). By using an assay that detects the presence of reducing sugars (i.e., cortex fragments) (40), we were able to compare when the cortex was degraded relative to DPA release under conditions where the osmotic strength of the medium was increased. Under these conditions, DPA release was delayed though cortex degradation was unaffected (Fig. 5). Because cortex degradation precedes DPA release during C. difficile spore germination, these results suggest that cortex degradation relieves the constraints put in place by the cortex on the core, leading to production of the mechanosensing protein, SpoVAC, and permitting DPA release.
spoVAC is conserved across most endospore-forming organisms, and the proteins encoded by the spoVA operon play important roles during spore formation and germination (10, 31, 32, 41). In B. subtilis, formation of a dormant, heat-resistant spore is dependent on 5 of the proteins encoded by the spoVA operon: SpoVAA, SpoVAB, SpoVAC, SpoVAD, and SpoVAEb. SpoVAF and SpoVAEa localize to the inner membrane of the spore and have minor roles during spore germination (12). Defects in these proteins reduce germinant-dependent germination but do not affect non-nutrient-mediated spore germination (e.g., exogenous DPA) (12). In B. subtilis, the release of DPA from the core triggers degradation of the spore cortex (8). For DPA to be released from the spore core, a signal first must be transmitted from the germinant receptors to the SpoVA protein. SpoVAF and SpoVAEa may play accessory roles during germination to efficiently receive and transmit the germination signal from GerA or GerB/GerK to the SpoVA complex and permit DPA release. Importantly, C. difficile does not encode ger-type germinant receptors, nor does C. difficile encode spoVAF, but it does encode spoVAE and spoVAD homologues (21). The roles of SpoVAE and SpoVAD during C. difficile spore germination are unknown. However, based on work with B. subtilis, it is likely that SpoVAD is essential for DPA packaging during sporulation (11).
Of the spoVA-encoded proteins, SpoVAC is nearly universally conserved among spore formers (41). Though the number of proteins encoded by the spoVA operon varies between organisms, nearly all encode spoVAC. However, there are a few exceptions. Among the Clostridiales, Bryantella formatexigens only encodes SpoVAA and SpoVAB, and Carboxydibrachium pacificum does not encode any apparent spoVA homologues (41). The nearly universal conservation of spoVAC among spore formers suggests that mechanosensing plays an important role during spore germination.
Mechanosensing during spore germination was previously studied during germination by B. subtilis spores (42–44). Those authors found that a B. subtilis ΔmscL ΔmscS mutant strain germinated similarly to a wild-type strain in response to l-alanine and dodecylamine (43). Thus, the proteins required for osmotic stability during growth play little to no role during germination by B. subtilis spores. However, the mechanosensing SpoVAC protein plays an important role during spore formation and germination (31, 32).
The data supporting SpoVAC as a mechanosensing protein are based partly on the ability of recombinant-expressed SpoVAC-myc-His6 to protect Escherichia coli from an osmotic downshift (13). Those authors found that SpoVAC-myc-His6 protected E. coli to a similar extent as MscL, a well-studied mechanosensing membrane protein (45, 46). The authors further characterized the protein in conductance studies and found that, when recombinant-expressed in E. coli and embedded in lipid vesicles, SpoVAC had a pore size of 4.6 Å (13). This pore size should be large enough to accommodate DPA (a planar molecule with dimensions of 5.2 Å in length and 3.5 Å in width), but the authors mentioned that the vesicles may not recapitulate the lipid content or the hydration state of the inner spore membrane, and this could affect gating of the protein (13). Prior work suggested that the inner spore membrane is in a gel-like state (or in a state that prevents mobility of a protein within the membrane) (47). An interesting hypothesis is that upon the initiation of cortex degradation, local changes at the inner membrane are observed which trigger SpoVAC-mediated DPA release from the core. Subsequently, the change in hydration state of the inner spore membrane at the site of DPA release results in the signal propagating the surrounding SpoVAC proteins. The prediction of such a system would be that high osmolyte concentrations can only block DPA release for so long before a few SpoVAC proteins become activated, randomly. This then would trigger the rest of the SpoVAC proteins to open and would allow DPA to escape the core. In support of this hypothesis, we found that sorbitol, trehalose, or sucrose did not permanently block the release of DPA from germinating spores (Fig. 3; see also Fig. S1 in the supplemental material). These osmolytes only delayed the release of DPA from the germinating spore. In sorbitol-containing germination buffer, the rate with which DPA was released after the initial delay was similar to the rate observed in the absence of sorbitol (Fig. 3A). This suggests that the SpoVAC channel activity is not affected by sorbitol, or water activity, in our assays. In this experiment, sucrose functioned better than trehalose and trehalose functioned better than sorbitol, but DPA release still occurred and the rate of DPA release began to increase at later time points. Importantly though, what little DPA was present in the C. difficile ΔspoVAC strain was still released during spore germination (Fig. 5C), though the value is presented as 100% release, the data were normalized between conditions with and without sorbitol and not between strains. The ΔspoVAC mutant has a low amount of DPA (Fig. 1) (31). This suggests that another protein could function in an accessory role to permit DPA release in response to the change in osmolarity at the inner spore membrane upon cortex degradation, or that the membranes leak DPA in the absence of SpoVAC.
Understanding the process of germination and how DPA release is triggered is likely more straightforward in C. difficile than in other spore-forming organisms. Because only spoVAC, spoVAD, and spoVAE are encoded in C. difficile, the mechanism of DPA release likely does not involve interaction with other factors (e.g., germinant receptors). In C. difficile, the bile acid germinant receptor CspC likely transmits the bile acid signal to CspB, which then cleaves pro-SleC to an active, cortex-degrading form (22, 48, 49). Activated SleC then begins degrading the cortex, allowing expansion and SpoVAC-mediated DPA release, core hydration, and subsequent outgrowth of a vegetative cell. The absence of germinant receptor in the inner spore membrane simplifies the mechanism of DPA release but may result in less specificity and a tendency for an increased amount of spontaneous DPA release. Interestingly, Clostridium perfringens encodes a spoVA system similar to that of C. difficile and also encodes functional germinant receptors (5, 41, 50, 51). How DPA release is signaled in this organism is unknown, but understanding its role would be of important value for understanding how the SpoVAD, SpoVAC, and SpoVAE proteins interact in such a system.
MATERIALS AND METHODS
Bacteria and strains.
Wild-type C. difficile R20291 and C. difficile M68 were routinely grown at 37°C in an anaerobic atmosphere (10% H2, 5% CO2, 85% N2) on brain heart infusion agar supplemented with 5 g/liter yeast extract and 0.1% l-cysteine (BHIS). E. coli DH5α, E. coli HB101(pRK24), and B. subtilis BS49 strains were grown on Luria-Bertani (LB) medium supplemented with antibiotics as needed. Chloramphenicol (20 μg/ml), thiamphenicol (10 μg/ml), kanamycin (50 μg/ml for C. difficile, 20 μg/ml for E. coli), or tetracycline (5 μg/ml for C. difficile, 20 μg/ml for B. subtilis) was added where indicated.
Molecular biology.
Using the codA-dependent allelic exchange strategy (33), we engineered a deletion of spoVAC in C. difficile R20291. To do so, we inserted the Tn916 oriT into pMTL-SC7215, using primers pMTL_SC_7215_tn916_L (CTAGAGTCGACGTCACGCGTCCATGGAGATCTCGAGTAACATCTTCTATTTTTCCCAAATCCTTAC) and pMTL_SC_7215_tn916_R (GGCCAGTGCCAAGCTTGCATGTCTGCAGGCCTCGAGCTAAAGGGAATGTAGATAAATTATTAGGTAATCTGC) to make pMF12, as described previously (22). C. difficile R20291 DNA was used as a template to amplify 1 kb upstream and downstream of the spoVAC deletion using primers spoVAC_ndeI_L (AGCTATGACCGCGGCCGCTGTATCCATATGAGTTCAAAATGGAGATGAAGAGGCAAAAGA), spoVAC_LHF_Rev_II (CTAAAACATCTTAAAAATATAATAAATAATGTCTACATATTTTTTATAATTTTTATCCA), spoVAC_xhoI_L (ATGGATAAAAATTATAAAAAATATGTAGACATTATTTATTATATTTTTAAGATGTTTTAGATGAT), and spoVAC_xhoI_R (TGCCAAGCTTGCATGTCTGCAGGCCTCGAGGTTCTTTAAGGTTAAACATCTCTATACCAC). The resulting 1-kb fragments were stitched together using splicing by overlap extension PCR and subcloned into pMF12 digested with NdeI/XhoI, yielding pMF11. The pMF11 plasmid was introduced into B. subtilis BS49 by using standard techniques. Subsequently, the pMF11 plasmid was introduced into C. difficile R20291 via conjugal transfer from B. subtilis BS49 pMF11, as described previously (22). Tetracycline-sensitive, thiamphenicol-resistant (transposon-negative, plasmid-positive) strains were identified. These isolates were then spread on BHIS supplemented with kanamycin and thiamphenicol to enrich for and identify faster-growing single-crossover integrant clones. The larger colonies were then selected and plated on C. difficile minimal medium supplemented with 50 μg/ml 5-fluorocytosine (FC). The colonies that formed after 48 h were tested for thiamphenicol sensitivity by culturing and by PCR with primers 5′-catP3 (ATGGTATTTGAAAAAATTGATAAAAATAG) and 3′-catP2 (TTAACTATTTATCAATTCCTGCAATTCG) to confirm loss of the plasmid. To confirm the deletion in spoVAC, the colonies were screened by PCR amplification of the spoVAC surrounding region, using spoVAC_ndeI_L and spoVAC_xhoI_R. In order to generate the spoVAC-complementing plasmid, C. difficile spoVAC was amplified using Phusion polymerase with the 5′-spoVAC_Gibson (CATGATTACGAATTCGAGCTCGGTACCCGGGGATCCTAATACTTATGATATGTAGAATAACAAAATATAATAAATATATTACT) and 3′-spoVAC_Gibson (CCAGTGCCAAGCTTGCATGTCTGCAGGCCTCGAGATCTAGTGGTGGTGGTGGTGGTGTAGAGTATTTGCTATCTGTTGAATCGTAT) oligonucleotides. The resulting fragment was cloned via Gibson assembly (52) between the BamHI and XhoI restriction sites of the B. subtilis-C. difficile shuttle vector pJS116 to generate pMF15. The nucleotide sequences were confirmed before use.
Spore formation.
C. difficile strains were plated on reduced BHIS plates and allowed to grow for 4 days, as described previously (17, 19, 30). On day 4, growth was harvested from two plates by scraping the plates with a disposable inoculating loop and suspended into 1.5-ml microcentrifuge tubes containing 1 ml sterile water. Tubes were stored at 4°C overnight. The next day, the contents of each tube were resuspended through pipetting and immediately centrifuged for 1 min at 14,000 × g. The supernatant was removed and the pellet was resuspended in water and centrifuged for 1 min at 14,000 × g; this was repeated for a total of 5 times. After resuspending the pellet again in 1 ml of water, the contents of 2 tubes (2 ml total suspension) were carefully layered over 8 ml of 60% (wt/vol) sucrose. This mixture was centrifuged for 20 min at 4,000 × g. Subsequently, the supernatant was removed and the remaining pellet was suspended in 1 ml of sterile water. As described above, spores were washed 5 times in sterile water. After the final wash, the supernatant was removed and the purified spore preparations were combined in 1 ml of sterile water. After purification, the resulting spore suspension was phase bright and >99.9% of vegetative cells had been removed.
Monitoring initiation of spore germination.
The initiation of spore germination was monitored aerobically at 600 nm (the initiation of C. difficile spore germination is unaffected by the presence of oxygen). C. difficile spore germination was initiated by suspending spores in 50 mM HEPES (pH 7.5), 100 mM NaCl, 100 mM glycine, 10 mM taurocholate, and 19% or 38% (wt/vol) sucrose, trehalose, or sorbitol. Prior to germination, spores were heat shocked for 30 min at 65°C and then placed on ice. Then, 5 μl of spores was diluted into 995 μl of germination buffer and mixed, and the change in the OD600 was measured over time.
Monitoring DPA release.
DPA release was monitored in real time by using terbium fluorescence, as described previously (30, 38). Briefly, an opaque, 96-well plate was prepared with 125-μl aliquots of the germination solutions (see above) supplemented with 800 μM TbCl3. A 5-μl sample of a spore suspension (OD600, 60) was added to each well, and the DPA release was monitored using a SpectraMax M3 fluorescence plate reader (Molecular Devices, Sunnyvale, CA) (excitation, 270 nm; emission, 545 nm; cutoff, 420 nm [appropriate wavelengths for the DPA-Tb3+ complex]).
Protein extraction and Western blotting.
Spores were allowed to germinate in germination buffer (described above). NuPAGE soluble proteins (e.g., SleC) were extracted from 2 × 109/ml purified spores as described previously (38). Proteins were separated by SDS-PAGE and then transferred for 1.5 h at 0.75 A to an Immobilon-P polyvinylidene difluoride 0.45-µm membrane (Millipore). Subsequently, the membrane was blocked for 1 h at room temperature in Tris-buffered saline (TBS) supplemented with 1% (vol/vol) Tween 20 (TBST) and 5% dried skimmed milk. The membrane was then incubated at room temperature for 1 h with rabbit anti-SleC antisera. After incubation with the primary antibody, membranes were washed thrice in TBST for 20 min each. The membranes were then labeled with goat anti-rabbit IgG (Life Technologies, Inc.) for 1 h at room temperature. The membranes were again washed as described above and then exposed to 4-chloro-1-naphthol–diaminobenzidine in peroxide substrate buffer. To stop reactions, membranes were rinsed with water. The membranes were then photographed under white light.
Assay of cortex fragment release by germinating spores.
Cortex fragments were detected according to a method previously reported (30, 40). Briefly, C. difficile spores were heat activated, as described above, and stored on ice until use. A preinoculation 1.0-ml sample was drawn to serve as a blank for cortex fragment detection, and a separate 100-μl sample was taken as a blank for measuring DPA release. A target spore density (OD600) of ~3.0 yielded the best results for detecting cortex fragments, except for MBF02 spores, which required a spore density of ~3.5. A 1.1-ml 0-time point sample was taken immediately after the addition of spores to the germination buffer and centrifuged for 1 min at 14,000 × g. Then, 1.0 ml of this sample was transferred to a fresh tube for cortex fragment analysis and 100 μl was taken to monitor the amount of DPA released. This procedure was repeated at selected time points until the experiment was completed. After all time point samples were collected, the samples were frozen at −80°C and lyophilized.
Lyophilized samples were analyzed for cortex fragments as previously described (40).
Statistical analysis.
Data points represent the means from three independent experiments, and error bars represent standard errors from the means. Statistical analysis between time points, where indicated, was performed using a two-tailed Student’s t test.
ACKNOWLEDGMENTS
We thank members of the Sorg lab and members from Leif Smith’s lab at Texas A&M University for their helpful comments during the preparation of the manuscript.
The project described is supported by awards 5R01AI116895 and 1U01AI124290 from the National Institute of Allergy and Infectious Diseases.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
REFERENCES
- 1.Theriot CM, Young VB. 2015. Interactions between the gastrointestinal microbiome and Clostridium difficile. Annu Rev Microbiol 69:445–461. doi: 10.1146/annurev-micro-091014-104115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Britton RA, Young VB. 2012. Interaction between the intestinal microbiota and host in Clostridium difficile colonization resistance. Trends Microbiol 20:313–319. doi: 10.1016/j.tim.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin-Verstraete I, Peltier J, Dupuy B. 2016. The regulatory networks that control Clostridium difficile toxin synthesis. Toxins (Basel) 8:E153. doi: 10.3390/toxins8050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, Wren BW, Fairweather NF, Dougan G, Lawley TD. 2012. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun 80:2704–2711. doi: 10.1128/IAI.00147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattacharjee D, McAllister KN, Sorg JA. 2016. Germinants and their receptors in Clostridia. J Bacteriol 198:2767–2775. doi: 10.1128/JB.00405-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paredes-Sabja D, Shen A, Sorg JA. 2014. Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol 22:406–416. doi: 10.1016/j.tim.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Setlow P. 2006. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol 101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- 8.Setlow P. 2014. Germination of spores of Bacillus species: what we know and do not know. J Bacteriol 196:1297–1305. doi: 10.1128/JB.01455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Setlow P. 2014. Spore resistance properties. Microbiol Spectr 2. doi: 10.1128/microbiolspec.TBS-0003-2012. [DOI] [PubMed] [Google Scholar]
- 10.Berendsen EM, Boekhorst J, Kuipers OP, Wells-Bennik MH. 2016. A mobile genetic element profoundly increases heat resistance of bacterial spores. ISME J 10:2633–2642. doi: 10.1038/ismej.2016.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Davis A, Korza G, Zhang P, Li YQ, Setlow B, Setlow P, Hao B. 2012. Role of a SpoVA protein in dipicolinic acid uptake into developing spores of Bacillus subtilis. J Bacteriol 194:1875–1884. doi: 10.1128/JB.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perez-Valdespino A, Li Y, Setlow B, Ghosh S, Pan D, Korza G, Feeherry FE, Doona CJ, Li YQ, Hao B, Setlow P. 2014. Function of the SpoVAEa and SpoVAF proteins of Bacillus subtilis spores. J Bacteriol 196:2077–2088. doi: 10.1128/JB.01546-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Velásquez J, Schuurman-Wolters G, Birkner JP, Abee T, Poolman B. 2014. Bacillus subtilis spore protein SpoVAC functions as a mechanosensitive channel. Mol Microbiol 92:813–823. doi: 10.1111/mmi.12591. [DOI] [PubMed] [Google Scholar]
- 14.Vepachedu VR, Setlow P. 2005. Localization of SpoVAD to the inner membrane of spores of Bacillus subtilis. J Bacteriol 187:5677–5682. doi: 10.1128/JB.187.16.5677-5682.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vepachedu VR, Setlow P. 2007. Role of SpoVA proteins in release of dipicolinic acid during germination of Bacillus subtilis spores triggered by dodecylamine or lysozyme. J Bacteriol 189:1565–1572. doi: 10.1128/JB.01613-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang G, Yi X, Li YQ, Setlow P. 2011. Germination of individual Bacillus subtilis spores with alterations in the GerD and SpoVA proteins, which are important in spore germination. J Bacteriol 193:2301–2311. doi: 10.1128/JB.00122-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sorg JA, Sonenshein AL. 2010. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol 192:4983–4990. doi: 10.1128/JB.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sorg JA, Sonenshein AL. 2009. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J Bacteriol 191:1115–1117. doi: 10.1128/JB.01260-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sorg JA, Sonenshein AL. 2008. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol 190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howerton A, Ramirez N, Abel-Santos E. 2011. Mapping interactions between germinants and Clostridium difficile spores. J Bacteriol 193:274–282. doi: 10.1128/JB.00980-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeño-Tárraga AM, Wang HW, Holden MTG, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 22.Francis MB, Allen CA, Shrestha R, Sorg JA. 2013. Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLoS Pathog 9:e1003356. doi: 10.1371/journal.ppat.1003356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masayama A, Hamasaki K, Urakami K, Shimamoto S, Kato S, Makino S, Yoshimura T, Moriyama M, Moriyama R. 2006. Expression of germination-related enzymes, CspA, CspB, CspC, SleC, and SleM, of Clostridium perfringens S40 in the mother cell compartment of sporulating cells. Genes Genet Syst 81:227–234. doi: 10.1266/ggs.81.227. [DOI] [PubMed] [Google Scholar]
- 24.Shimamoto S, Moriyama R, Sugimoto K, Miyata S, Makino S. 2001. Partial characterization of an enzyme fraction with protease activity which converts the spore peptidoglycan hydrolase (SleC) precursor to an active enzyme during germination of Clostridium perfringens S40 spores and analysis of a gene cluster involved in the activity. J Bacteriol 183:3742–3751. doi: 10.1128/JB.183.12.3742-3751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banawas S, Korza G, Paredes-Sabja D, Li Y, Hao B, Setlow P, Sarker MR. 2015. Location and stoichiometry of the protease CspB and the cortex-lytic enzyme SleC in Clostridium perfringens spores. Food Microbiol 50:83–87. doi: 10.1016/j.fm.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 26.Miyata S, Moriyama R, Miyahara N, Makino S. 1995. A gene (sleC) encoding a spore-cortex-lytic enzyme from Clostridium perfringens S40 spores; cloning, sequence analysis and molecular characterization. Microbiology 141:2643–2650. doi: 10.1099/13500872-141-10-2643. [DOI] [PubMed] [Google Scholar]
- 27.Paredes-Sabja D, Sarker MR. 2010. Effect of the cortex-lytic enzyme SleC from non-food-borne Clostridium perfringens on the germination properties of SleC-lacking spores of a food poisoning isolate. Can J Microbiol 56:952–958. doi: 10.1139/w10-083. [DOI] [PubMed] [Google Scholar]
- 28.Paredes-Sabja D, Setlow P, Sarker MR. 2009. SleC is essential for cortex peptidoglycan hydrolysis during germination of spores of the pathogenic bacterium Clostridium perfringens. J Bacteriol 191:2711–2720. doi: 10.1128/JB.01832-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fimlaid KA, Jensen O, Donnelly ML, Francis MB, Sorg JA, Shen A. 2015. Identification of a novel lipoprotein regulator of Clostridium difficile spore germination. PLoS Pathog 11:e1005239. doi: 10.1371/journal.ppat.1005239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Francis MB, Allen CA, Sorg JA. 2015. Spore cortex hydrolysis precedes dipicolinic acid release during Clostridium difficile spore germination. J Bacteriol 197:2276–2283. doi: 10.1128/JB.02575-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donnelly ML, Fimlaid KA, Shen A. 2016. Characterization of Clostridium difficile spores lacking either SpoVAC or dipicolinic acid synthetase. J Bacteriol 198:1694–1707. doi: 10.1128/JB.00986-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tovar-Rojo F, Chander M, Setlow B, Setlow P. 2002. The products of the spoVA operon are involved in dipicolinic acid uptake into developing spores of Bacillus subtilis. J Bacteriol 184:584–587. doi: 10.1128/JB.184.2.584-587.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cartman ST, Kelly ML, Heeg D, Heap JT, Minton NP. 2012. Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcdC genotype and toxin production. Appl Environ Microbiol 78:4683–4690. doi: 10.1128/AEM.00249-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paredes-Sabja D, Setlow P, Sarker MR. 2009. The protease CspB is essential for initiation of cortex hydrolysis and dipicolinic acid (DPA) release during germination of spores of Clostridium perfringens type A food poisoning isolates. Microbiology 155:3464–3472. doi: 10.1099/mic.0.030965-0. [DOI] [PubMed] [Google Scholar]
- 35.Barela TD, Sherry AD. 1976. A simple, one-step fluorometric method for determination of nanomolar concentrations of terbium. Anal Biochem 71:351–357. doi: 10.1016/S0003-2697(76)80004-8. [DOI] [PubMed] [Google Scholar]
- 36.Heeg D, Burns DA, Cartman ST, Minton NP. 2012. Spores of Clostridium difficile clinical isolates display a diverse germination response to bile salts. PLoS One 7:e32381. doi: 10.1371/journal.pone.0032381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carlson PE Jr., Kaiser AM, McColm SA, Bauer JM, Young VB, Aronoff DM, Hanna PC. 2015. Variation in germination of Clostridium difficile clinical isolates correlates to disease severity. Anaerobe 33:64–70. doi: 10.1016/j.anaerobe.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhattacharjee D, Francis MB, Ding X, McAllister KN, Shrestha R, Sorg JA. 2015. Reexamining the germination phenotypes of several Clostridium difficile strains suggests another role for the CspC germinant receptor. J Bacteriol 198:777–786. doi: 10.1128/JB.00908-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang S, Shen A, Setlow P, Li YQ. 2015. Characterization of the dynamic germination of individual Clostridium difficile spores using Raman spectroscopy and differential interference contrast microscopy. J Bacteriol 197:2361–2373. doi: 10.1128/JB.00200-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Francis MB, Sorg JA. 2016. Detecting cortex fragments during bacterial spore germination. J Vis Exp. doi: 10.3791/54146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paredes-Sabja D, Setlow P, Sarker MR. 2011. Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol 19:85–94. doi: 10.1016/j.tim.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 42.Wahome PG, Setlow P. 2006. The synthesis and role of the mechanosensitive channel of large conductance in growth and differentiation of Bacillus subtilis. Arch Microbiol 186:377–383. doi: 10.1007/s00203-006-0152-2. [DOI] [PubMed] [Google Scholar]
- 43.Wahome PG, Setlow P. 2008. Growth, osmotic downshock resistance and differentiation of Bacillus subtilis strains lacking mechanosensitive channels. Arch Microbiol 189:49–58. doi: 10.1007/s00203-007-0292-z. [DOI] [PubMed] [Google Scholar]
- 44.Wahome PG, Cowan AE, Setlow B, Setlow P. 2009. Levels and localization of mechanosensitive channel proteins in Bacillus subtilis. Arch Microbiol 191:403–414. doi: 10.1007/s00203-009-0465-z. [DOI] [PubMed] [Google Scholar]
- 45.Booth IR. 2014. Bacterial mechanosensitive channels: progress towards an understanding of their roles in cell physiology. Curr Opin Microbiol 18:16–22. doi: 10.1016/j.mib.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang XC, Liu Z, Li J. 2016. From membrane tension to channel gating: a principal energy transfer mechanism for mechanosensitive channels. Protein Sci 25:1954–1964. doi: 10.1002/pro.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cowan AE, Koppel DE, Setlow B, Setlow P. 2003. A soluble protein is immobile in dormant spores of Bacillus subtilis but is mobile in germinated spores: implications for spore dormancy. Proc Natl Acad Sci U S A 100:4209–4214. doi: 10.1073/pnas.0636762100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kevorkian Y, Shirley DJ, Shen A. 2016. Regulation of Clostridium difficile spore germination by the CspA pseudoprotease domain. Biochimie 122:243–254. doi: 10.1016/j.biochi.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams CM, Eckenroth BE, Putnam EE, Doublié S, Shen A. 2013. Structural and functional analysis of the CspB protease required for Clostridium spore germination. PLoS Pathog 9:e1003165. doi: 10.1371/journal.ppat.1003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paredes-Sabja D, Udompijitkul P, Sarker MR. 2009. Inorganic phosphate and sodium ions are cogerminants for spores of Clostridium perfringens type A food poisoning-related isolates. Appl Environ Microbiol 75:6299–6305. doi: 10.1128/AEM.00822-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paredes-Sabja D, Setlow P, Sarker MR. 2009. Role of GerKB in germination and outgrowth of Clostridium perfringens spores. Appl Environ Microbiol 75:3813–3817. doi: 10.1128/AEM.00048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Osmolytes delay DPA release by C. difficile M68 spores. C. difficile M68 spores were suspended in buffer containing terbium chloride and glycine alone (filled circles) or supplemented with taurocholate (filled triangles) or taurocholate and 38% osmolyte (open triangles) (sorbitol [A], trehalose [B], or sucrose [C]). For clarity, every fifth data point is plotted, and the data represent the average results from three independent experiments. Error bars represent the standard deviations from the means. Download Figure S1, TIF file, 0.8 MB (819.7KB, tif) .
Copyright © 2016 Francis and Sorg.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.