It is increasingly evident that new environmentally sustainable agricultural practices will be necessary to support an estimated human population of 9 billion by 2050. Considering the predicted climate change-associated events such as flooding, drought, increased temperatures, and increased soil salinity, it is critical to broaden our study and use of cereals and small grains. Millets offer significant potential toward meeting these goals. The genus Setaria comprises more than one hundred species distributed in a wide range of habitats in Asia, Africa, South America, and North America. Millets, including foxtail millet (Setaria italica), pearl millet (Pennisetum glaucum) and proso millet (Panicum milaceum), are consumed by millions of humans across the world, as well as used as forage and bird feed. Recently, foxtail millet and its wild-progenitor, green foxtail millet (S. viridis), have emerged as model plant systems for studying C4 grass biology and other agronomic traits (Doust et al., 2009; Li and Brutnell, 2011; Lata et al., 2013; Mandadi et al., 2014; Xianmin et al., 2014; Brutnell, 2015; Brutnell et al., 2015). Here, we comment on the significance of S. italica and S. viridis as model systems for agronomic and translational research related to improvement of nutritional quality, biotic and abiotic stress responses, photosynthetic efficiency and biomass potential of C4 grasses.
Foxtail millet is in the Paniceae tribe (sub-family Panicoideae of the Poaceae), which also contains pearl millet and proso millet. Foxtail millet was domesticated from green foxtail in northern China ca. 8000 years ago (Barton et al., 2009), and has been widely cultivated in arid and semi-arid regions of Asia, Africa and the Americas (Lata et al., 2013). It is one of the most resilient cereal crops, with good yields in dry and marginal land with minimal agricultural inputs. Foxtail millet also adapts well to adverse weather conditions such as low and unpredictable precipitation (Lata et al., 2013; Muthamilarasan and Prasad, 2015). Furthermore, it appears to be resilient to multiple abiotic and biotic stresses such as drought (Zhang et al., 2005; Meng et al., 2009; Lata et al., 2011), salinity (Ardie et al., 2015) and fungal diseases (Xu et al., 2011). Because global climate change will have significant adverse effects on the production of other major cereals, foxtail-, pearl- and proso-millets are increasingly attractive alternatives for small grain production (Tadele, 2016).
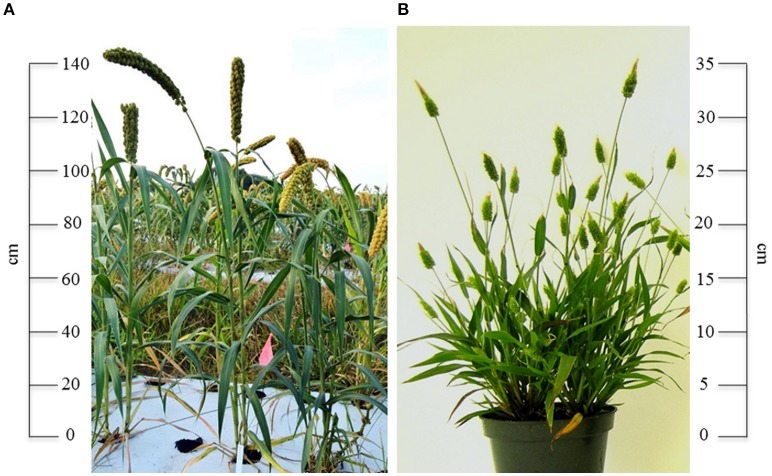
Green foxtail millet has also come into the spotlight in recent years as an attractive alternative to more established grass model systems such as rice and maize (Figure 1). S. viridis offers several research advantages, including short stature (10–30 cm), rapid life cycle (6–9 weeks), prolific seed production (~13,000 seeds per plant), self-compatibility, small genome size (~395 Mb), diploid genetics (2n = 18), ability to be grown in controlled environments under relatively low light levels, and amenability to transformation (Devos et al., 1998; Doust et al., 2009; Brutnell et al., 2010; Li and Brutnell, 2011; Wang et al., 2011; Xianmin et al., 2014; Saha and Blumwald, 2016). Like maize, green and foxtail millets are C4 plants, but they have smaller genomes and are true diploids. In combination with sorghum (Sorghum bicolor), S. italica and S. viridis are proving valuable in studying C4 photosynthesis, with the long-term goal of engineering C4 traits into important C3 crops such as rice and wheat.
Figure 1.
(A) Setaria italica and (B) S. viridis are emerging model species for millet and C4 grass biology. Photo credits: John Hodge and Jesse D. Pyle.
The S. italica and S. viridis system offer many genetic resources, including sequenced draft genomes (Bennetzen et al., 2012; Zhang et al., 2012) (Setaria viridis v1.1, DOE-JGI, http://phytozome.jgi.doe.gov) and a high-density haplotype map of genome variation (Jia et al., 2013). Comparisons of the arrangement of the nine chromosomes of S. italica and S. viridis with the corresponding chromosomes of rice and sorghum reveal relatively few rearrangements, and the genome sequence has been used to guide the assembly of the polyploid genome of its close relative, Panicum virgatum (switchgrass), a promising bioenergy feedstock (Daverdin et al., 2015). The high-quality genome sequence of foxtail millet has also allowed in-depth analyses of transposon family dynamics and locations that reveal hitherto unsuspected variation between transposon families in insertion site preference and turnover (Bennetzen et al., 2016). Mutant populations have been characterized in both foxtail millet and green foxtail millet, and the identity of candidate genes was revealed by novel high-throughput sequencing approaches (Li et al., 2016; Liu et al., 2016; Martins et al., 2016; Xue et al., 2016). S. italica and S. viridis have been used in the characterization of important agronomic traits, including yield-related architectural traits such as height, branching, biomass, flowering time and domestication-related traits such as shattering (Qian et al., 2012; Jia et al., 2013; Mauro-Herrera et al., 2013; Wang et al., 2013; Doust et al., 2014; Gupta et al., 2014; Layton and Kellogg, 2014; Qie et al., 2014; Fahlgren et al., 2015; Fang et al., 2016; Hodge and Kellogg, 2016; Liu et al., 2016; Mauro-Herrera and Doust, 2016).
In plants, salicylic acid (SA), jasmonic acid (JA) and ethylene (ET) are key defense hormones that mediate signaling during plant-microbe interaction. Prevailing evidence, primarily based on studies of dicot plants, suggest that SA and JA/ET pathways are antagonistic to each other during plant-microbe interactions. Recently, in conjunction with Brachypodium distachyon (a C3 grass), S. viridis was used to characterize the dynamics of C3 vs. C4 plant defense signaling responses against a diverse group of grass-infecting viruses (Scholthof, 1999; Mandadi and Scholthof, 2012; Mandadi et al., 2014). This study showed that SA and JA/ET crosstalk does exist in S. viridis during grass-virus interactions; however, there were also unique C4 host-dependent signaling responses. S. italica was also used to dissect grass defense signaling pathways modulated during an incompatible interaction with a fungal pathogen, Uromyces setariae-italicae (Li et al., 2015). Together, these studies demonstrate the utility of S. italica and S. viridis for fundamental research pertaining to plant-microbe interactions.
S. italica and S. viridis are also very useful for translational research related to bioenergy traits. In addition to plant-derived sugars it has been shown that cell-wall derived cellulose, hemicellulose and lignin are sustainable sources for bioenergy production. Owing to their superior carbon-assimilation and photosynthetic pathways, several C4 grasses such as switchgrass, Miscanthus, and energycane have emerged as feedstocks for plant ligno-cellulosic biomass (Brutnell et al., 2010). Efforts are underway to engineer C4 photosynthetic pathways into C3 crops (e.g., rice) to improve photosynthetic capacity and yield (von Caemmerer et al., 2012; Karki et al., 2013). However, the genomes and genetics of the leading bioenergy grasses are complex, and are a hindrance in deciphering the mechanisms of C4 photosynthesis, and ultimately to engineer them into C3 grasses. Both S. italica and S. viridis are C4 grasses and their genomes are highly syntenic to other Panicoid grasses (Kumari et al., 2013) and, in this context, these model plants offer powerful tools to understand the genetics of biomass production of panicoid bioenergy grasses, as well as to limn the evolution of C4 vs. C3 traits (Doust et al., 2009; Li and Brutnell, 2011; Lata et al., 2013; Mandadi et al., 2014; Brutnell et al., 2015; Muthamilarasan and Prasad, 2015; Martin et al., 2016).
In addition to enabling research into agronomic traits, S. italica and S. viridis are valuable models in the study of phytonutrient pathways pertaining to small grain millets. For example, S. italica is a good source of calories and essential micronutrients, and has higher nutritive value than major cereal grains like rice, wheat and sorghum. S. italica also contains high levels of proteins, dietary fibers, vitamins, anti-oxidants and non-starchy polysaccharides with low glycemic index, when compared to rice, wheat and sorghum (Taylor et al., 2006; Suma and Urooj, 2012; Amadou et al., 2013; Muthamilarasan and Prasad, 2015; Muthamilarasan et al., 2015). Moreover, over fifty phenolic compounds belonging to classes such as hydroxybenzoic acids, hydroxycinnamic acids and flavonoids were identified from S. italica (Chandrasekara and Shahidi, 2011b). In in vitro studies, phenolic compounds derived from S. italica, exhibited health promoting properties including antioxidant and free radical quenching, which helps in boosting immunity and inhibiting cancer cell proliferation (Dykes and Rooney, 2006; Chandrasekara and Shahidi, 2010, 2011a,b, 2012; Muthamilarasan and Prasad, 2015). Notwithstanding these reported nutritional qualities, certain anti-nutritional constraints were also reported for S. italica. These include the presence of toxic substances, some of which might be harmful to human health. For instance, some foxtail millet grains contain goitrogens that suppress thyroid activity which may have a role in goiter—an enlargement of the thyroid gland (Gaitan et al., 1989; Gluchoff-Fiasson et al., 1989; Elnour et al., 2000). The major goitrogenic and antithyroid compounds isolated were C-glycosylflavones (Gaitan et al., 1989; Gluchoff-Fiasson et al., 1989). Furthermore, in vivo experiments with vitexin (one of the three major C-glycosylflavones) in rats revealed evidence of antithyroid activity (Gaitan et al., 1995). In this context, further research is needed to carefully delineate the nutritional and anti-nutritional properties of phytochemicals present in S. italica and perhaps other small grain millets. The availability of S. italica and S. viridis genomic and genetic resources should enable research into identifying and inactivating such anti-nutritional compounds using novel genome-editing technologies (e.g., CRISPR-CAS9) to enhance S. italica nutritional traits.
Given the importance of S. italica as a food crop and in the study of grass biology, we suggest that S. italica, and its wild-progenitor, S. viridis, are promising translational research models to study nutritional pathways, abiotic and biotic stress resistance pathways, as well as to advance C4 photosynthesis and bioenergy research.
Author contributions
SP, SI, AD, KS, and KM contributed to the design, preparation, and editing of the manuscript.
Funding
This manuscript was supported by funds from USDA-NIFA-AFRI (2016-67013-24738) to KS and KM, Texas A&M AgriLife Research Bioenergy/Bioproducts Grant (124738-96210) to KM, and NSF Plant Genome (IOS-1339332) to AD.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Amadou I., Le G. W., Amza T., Sun J., Shi Y. H. (2013). Purification and characterization of foxtail millet-derived peptides with antioxidant and antimicrobial activities. Food Res. Int. 51, 422–428. 10.1016/j.foodres.2012.12.045 [DOI] [Google Scholar]
- Ardie S. W., Khumaida N., Nur A., Fauziah N. (2015). Early identification of salt tolerant foxtail millet (Setaria italica L. Beauv). Procedia Food Sci. 3, 303–312. 10.1016/j.profoo.2015.01.033 [DOI] [Google Scholar]
- Barton L., Newsome S. D., Chen F. H., Wang H., Guilderson T. P., Bettinger R. L. (2009). Agricultural origins and the isotopic identity of domestication in northern China. Proc. Natl. Acad. Sci. U.S.A. 106, 5523–5528. 10.1073/pnas.0809960106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Schmutz J., Wang H., Percifield R., Hawkins J., Pontaroli A. C., et al. (2012). Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 30, 555–561. 10.1038/nbt.2196 [DOI] [PubMed] [Google Scholar]
- Bennetzen J., Park M., Wang H., Zhou H. (2016). LTR retrotransposon dynamics and specificity in Setaria italica, in Genetics and Genomics of Setaria, eds Doust A., Diao X.(Cham: Springer International Publishing; ). Available online at: http://www.springer.com/us/book/9783319451039#aboutBook [Google Scholar]
- Brutnell T. P. (2015). Model grasses hold key to crop improvement. Nat. Plants 1, 15062 10.1038/NPLANTS.2015.62 [DOI] [Google Scholar]
- Brutnell T. P., Bennetzen J. L., Vogel J. P. (2015). Brachypodium distachyon and Setaria viridis: model genetic systems for the grasses. Annu. Rev. Plant Biol. 66, 465–485. 10.1146/annurev-arplant-042811-105528 [DOI] [PubMed] [Google Scholar]
- Brutnell T. P., Wang L., Swartwood K., Goldschmidt A., Jackson D., Zhu X.-G., et al. (2010). Setaria viridis: a model for C4 photosynthesis. Plant Cell 22, 2537–2544. 10.1105/tpc.110.075309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekara A., Shahidi F. (2010). Content of insoluble bound phenolics in millets and their contribution to antioxidant capacity. J. Agric. Food Chem. 58, 6706–6714. 10.1021/jf100868b [DOI] [PubMed] [Google Scholar]
- Chandrasekara A., Shahidi F. (2011a). Antiproliferative potential and DNA scission inhibitory activity of phenolics from whole millet grains. J. Funct. Foods 3, 159–170. 10.1016/j.jff.2011.03.008 [DOI] [Google Scholar]
- Chandrasekara A., Shahidi F. (2011b). Determination of antioxidant activity in free and hydrolyzed fractions of millet grains and characterization of their phenolic profiles by HPLC-DAD-ESI-MSn. J. Funct. Foods 3, 144–158. 10.1016/j.jff.2011.03.007 [DOI] [Google Scholar]
- Chandrasekara A., Shahidi F. (2012). Bioaccessibility and antioxidant potential of millet grain phenolics as affected by simulated in vitro digestion and microbial fermentation. J. Funct. Foods 4, 226–237. 10.1016/j.jff.2011.11.001 [DOI] [Google Scholar]
- Daverdin G., Bahri B. A., Wu X., Serba D. D., Tobias C., Saha M. C., et al. (2015). Comparative relationships and chromosome evolution in switchgrass (Panicum virgatum) and its genomic model, foxtail millet (Setaria italica). Bioenergy Res. 8, 137–151. 10.1007/s12155-014-9508-7 [DOI] [Google Scholar]
- Devos K. M., Wang Z. M., Beales J., Gale M. D., Sasaki T. (1998). Comparative genetic maps of foxtail millet (Setaria italica) and rice (Oryza sativa). Theor. Appl. Genet. 96, 63–68. 10.1007/s001220050709 [DOI] [Google Scholar]
- Doust A. N., Kellogg E. A., Devos K. M., Bennetzen J. L. (2009). Foxtail millet: a sequence-driven grass model system. Plant Physiol. 149, 2003–2005. 10.1104/pp.108.129627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doust A. N., Lukens L., Olsen K. M., Mauro-Herrera M., Meyer A., Rogers K. (2014). Beyond the single gene: how epistasis and gene-by-environment effects influence crop domestication. Proc. Natl. Acad. Sci. U.S.A. 111, 6178–6183. 10.1073/pnas.1308940110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykes L., Rooney L. W. (2006). Sorghum and millet phenols and antioxidants. J. Cereal Sci. 44, 236–251. 10.1016/j.jcs.2006.06.007 [DOI] [Google Scholar]
- Elnour A., Hambraeus L., Eltom M., Dramaix M., Bourdoux P. (2000). Endemic goiter with iodine sufficiency: a possible role for the consumption of pearl millet in the etiology of endemic goiter. Am. J. Clin. Nutr. 71, 59–66. [DOI] [PubMed] [Google Scholar]
- Fahlgren N., Feldman M., Gehan M. A., Wilson M. S., Shyu C., Bryant D. W., et al. (2015). A versatile phenotyping system and analytics platform reveals diverse temporal responses to water availability in Setaria. Mol. Plant 8, 1520–1535. 10.1016/j.molp.2015.06.005 [DOI] [PubMed] [Google Scholar]
- Fang X., Dong K., Wang X., Liu T., He J., Ren R., et al. (2016). A high density genetic map and QTL for agronomic and yield traits in foxtail millet [Setaria italica (L.) P. Beauv.]. BMC Genomics 17:336. 10.1186/s12864-016-2628-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitan E., Cooksey R. C., Legan J., Lindsay R. H. (1995). Antithyroid effects in vivo and in vitro of vitexin: a C-glucosylflavone in millet. J. Clin. Endocrinol. Metab. 80, 1144–1147. 10.1210/jcem.80.4.7714083 [DOI] [PubMed] [Google Scholar]
- Gaitan E., Lindsay R., Reichert R., Ingbar S., Cooksey R., Legan J., et al. (1989). Antithyroid and goitrogenic effects of millet: role of C-glycosylflavones. J. Clin. Endocrinol. Metab. 68, 707–714. 10.1210/jcem-68-4-707 [DOI] [PubMed] [Google Scholar]
- Gluchoff-Fiasson K., Jay M., Viricel M. R. (1989). Flavone O- and C-glycosides from Setaria italica. Phytochemistry 28, 2471–2475. 10.1016/S0031-9422(00)98008-7 [DOI] [Google Scholar]
- Gupta S., Kumari K., Muthamilarasan M., Parida S. K., Prasad M. (2014). Population structure and association mapping of yield contributing agronomic traits in foxtail millet. Plant Cell Rep. 33, 881–893. 10.1007/s00299-014-1564-0 [DOI] [PubMed] [Google Scholar]
- Hodge J. G., Kellogg E. A. (2016). Abscission zone development in Setaria viridis and its domesticated relative, Setaria italica. Am. J. Bot. 103, 998–1005. 10.3732/ajb.1500499 [DOI] [PubMed] [Google Scholar]
- Jia G., Huang X., Zhi H., Zhao Y., Zhao Q., Diao X., et al. (2013). A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat. Genet. 45, 957–961. 10.1038/ng.2673 [DOI] [PubMed] [Google Scholar]
- Karki S., Rizal G., Quick W. P. (2013). Improvement of photosynthesis in rice (Oryza sativa L.) by inserting the C4 pathway. Rice 6:28. 10.1186/1939-8433-6-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari K., Muthamilarasan M., Misra G., Gupta S., Subramanian A., Parida S. K., et al. (2013). Development of eSSR-markers in Setaria italica and their applicability in studying genetic diversity, cross-transferability and comparative mapping in millet and non-millet species. PLoS ONE 8:e67742. 10.1371/journal.pone.0067742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata C., Bhutty S., Bahadur R. P., Majee M., Prasad M. (2011). Association of an SNP in a novel DREB2-like gene SiDREB2 with stress tolerance in foxtail millet [Setaria italica (L.)]. J. Exp. Bot. 62, 3387–3401. 10.1093/jxb/err016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata C., Gupta S., Prasad M. (2013). Foxtail millet: a model crop for genetic and genomic studies in bioenergy grasses. Crit. Rev. Biotechnol. 33, 328–343. 10.3109/07388551.2012.716809 [DOI] [PubMed] [Google Scholar]
- Layton D. J., Kellogg E. A. (2014). Morphological, phylogenetic, and ecological diversity of the new model species Setaria viridis (Poaceae: Paniceae) and its close relatives. Am. J. Bot. 101, 539–557. 10.3732/ajb.1300428 [DOI] [PubMed] [Google Scholar]
- Li P., Brutnell T. P. (2011). Setaria viridis and Setaria italica, model genetic systems for the Panicoid grasses. J. Exp. Bot. 62, 3031–3037. 10.1093/jxb/err096 [DOI] [PubMed] [Google Scholar]
- Li W., Tang S., Zhang S., Shan J., Tang C., Chen Q., et al. (2016). Gene mapping and functional analysis of the novel leaf color gene SiYGL1 in foxtail millet Setaria italica (L.) P. Beauv. Physiol. Plant. 157, 24–27. 10.1111/ppl.12405 [DOI] [PubMed] [Google Scholar]
- Li Z. Y., Wang N., Dong L., Bai H., Quan J. Z., Liu L., et al. (2015). Differential gene expression in foxtail millet during incompatible interaction with Uromyces setariae-italicae. PLoS ONE 10:e0123825. 10.1371/journal.pone.0123825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Tang S., Jia G., Schnable J. C., Su H., Tang C., et al. (2016). The C-terminal motif of SiAGO1b is required for the regulation of growth, development and stress responses in foxtail millet (Setaria italica (L.) P. Beauv). J. Exp. Bot. 67:erw135. 10.1093/jxb/erw135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandadi K. K., Pyle J. D., Scholthof K.-B. G. (2014). Comparative analysis of antiviral responses in Brachypodium distachyon and Setaria viridis reveal conserved and unique outcomes among C3 and C4 plant defenses. Mol. Plant Microbe Interact. 27, 1277–1290. 10.1094/MPMI-05-14-0152-R [DOI] [PubMed] [Google Scholar]
- Mandadi K. K., Scholthof K.-B. G. (2012). Characterization of a viral synergism in the monocot Brachypodium distachyon reveals distinctly altered host molecular processes associated with disease. Plant Physiol. 160, 1432–1452. 10.1104/pp.112.204362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A. P., Palmer W. M., Brown C., Abel C., Lunn J. E., Furbank R. T., et al. (2016). A developing Setaria viridis internode: an experimental system for the study of biomass generation in a C4 model species. Biotechnol. Biofuels 9, 45. 10.1186/s13068-016-0457-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins P. K., Mafra V., de Souza W. R., Ribeiro A. P., Vinecky F., Basso M. F., et al. (2016). Selection of reliable reference genes for RT-qPCR analysis during developmental stages and abiotic stress in Setaria viridis. Sci. Rep. 6:28348. 10.1038/srep28348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro-Herrera M., Doust A. N. (2016). Development and genetic control of plant architecture and biomass in the Panicoid grass, Setaria. PLoS ONE 11:e0151346. 10.1371/journal.pone.0151346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro-Herrera M., Wang X., Barbier H., Brutnell T. P., Devos K. M., Doust A. N. (2013). Genetic control and comparative genomic analysis of flowering time in Setaria (Poaceae). G3 3, 283–295. 10.1534/g3.112.005207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q., Guan Z., Feng B., Chai Y., Hu Y. (2009). Principal component analysis and fuzzy clustering on drought-tolerance related traits of foxtail millet (Setaria italica). Sci. Agric. Sin. 42, 2667–2675. 10.3864/j.issn.0578-1752.2009.08.005 [DOI] [Google Scholar]
- Muthamilarasan M., Dhaka A., Yadav R., Prasad M. (2015). Exploration of millet models for developing nutrient rich graminaceous crops. Plant Sci. 242, 89–97. 10.1016/j.plantsci.2015.08.023 [DOI] [PubMed] [Google Scholar]
- Muthamilarasan M., Prasad M. (2015). Advances in Setaria genomics for genetic improvement of cereals and bioenergy grasses. Theor. Appl. Genet. 128, 1–14. 10.1007/s00122-014-2399-3 [DOI] [PubMed] [Google Scholar]
- Qian J., Jia G., Zhi H., Li W., Wang Y., Li H., et al. (2012). Sensitivity to gibberellin of dwarf foxtail millet varieties. Crop Sci. 52, 1068–1075. 10.2135/cropsci2011.04.0192 [DOI] [Google Scholar]
- Qie L., Jia G., Zhang W., Schnable J., Shang Z., Li W., et al. (2014). Mapping of quantitative trait locus (QTLs) that contribute to germination and early seedling drought tolerance in the interspecific cross Setaria italica x Setaria viridis. PLoS ONE 9:e101868. 10.1371/journal.pone.0101868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha P., Blumwald B. (2016). Spike-dip transformation of Setaria viridis. Plant J. 86, 89–101. 10.1111/tpj.13148 [DOI] [PubMed] [Google Scholar]
- Scholthof K.-B. G. (1999). A synergism induced by satellite panicum mosaic virus. Mol. Plant. Microbe Interact. 12, 163–166. 10.1094/MPMI.1999.12.2.163 [DOI] [Google Scholar]
- Suma P. F., Urooj A. (2012). Antioxidant activity of extracts from foxtail millet (Setaria italica). J. Food Sci. Technol. 49, 500–504. 10.1007/s13197-011-0300-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadele Z. (2016). Drought adaptation in millets, in Abiotic and Biotic Stress in Plants - Recent Advances and Future Perspectives, eds Shanker A. K., Shanker C.(Rijeka: InTech; ), 639–662. Available online at: http://www.intechopen.com/books/abiotic-and-biotic-stress-in-plants-recent-advances-and-future-perspectives [Google Scholar]
- Taylor J. R. N., Schober T. J., Bean S. R. (2006). Novel food and non-food uses for sorghum and millets. J. Cereal Sci. 44, 252–271. 10.1016/j.jcs.2006.06.009 [DOI] [Google Scholar]
- von Caemmerer S., Quick W. P., Furbank R. T. (2012). The development of C4 rice: current progress and future challenges. Science 336, 1671–1672. 10.1126/science.1220177 [DOI] [PubMed] [Google Scholar]
- Wang J., Wang Z., Yang H., Yuan F., Guo E., Tian G., et al. (2013). Genetic analysis and preliminary mapping of a highly male-sterile gene in foxtail millet (Setaria italica L. Beauv.) using SSR markers. J. Integr. Agric. 12, 2143–2148. 10.1016/S2095-3119(13)60392-5 [DOI] [Google Scholar]
- Wang M.-Z., Pan Y.-L., Li C., Liu C., Zhao Q., Ao G.-M., et al. (2011). Culturing of immature inflorescences and Agrobacterium-mediated transformation of foxtail millet (Setaria italica). African J. Biotechnol. 10, 16466–16479. 10.5897/AJB10.2330 [DOI] [Google Scholar]
- Xianmin D., James S., Bennetzen J. L., Jiayang L. (2014). Initiation of Setaria as a model plant. Front. Agric. Sci. Eng. 1:16 10.15302/J-FASE-2014011 [DOI] [Google Scholar]
- Xu W., Wei L., Qu W., Liang Z., Wang J., Peng X., et al. (2011). A novel antifungal peptide from foxtail millet seeds. J. Sci. Food Agric. 91, 1630–1637. 10.1002/jsfa.4359 [DOI] [PubMed] [Google Scholar]
- Xue C. X., Zhi H., Fang X., Liu X., Tang S., Chai Y., et al. (2016). Characterization and fine mapping of SiDWARF2 (D2) in foxtail millet. Crop Sci. 56, 95–103. 10.2135/cropsci2015.05.0331 [DOI] [Google Scholar]
- Zhang G., Liu X., Quan Z., Cheng S., Xu X., Pan S., et al. (2012). Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 30, 549–554. 10.1038/nbt.2195 [DOI] [PubMed] [Google Scholar]
- Zhang J. P., Yan W. M., Feng B. Y., Jia J. P., Ying W. G. (2005). Rapid evaluation on drought tolerance of foxtail millet at seedling stage. J. Plant Genet. Resour. 6, 59–62. [Google Scholar]