Abstract
Grasses produce seeds on spikelets, a unique type of inflorescence. Despite the importance of grass crops for food, the genetic mechanisms that control spikelet development remain poorly understood. In this study, we used m34-z, a new mutant allele of the rice (Oryza sativa) E-class gene OsMADS34, to examine OsMADS34 function in determining the identities of glumes (rudimentary glume and sterile lemma) and grain size. In the m34-z mutant, both the rudimentary glume and sterile lemma were homeotically converted to the lemma-like organ and acquired the lemma identity, suggesting that OsMADS34 plays important roles in the development of glumes. In the m34-z mutant, most of the grains from the secondary panicle branches (spb) were decreased in size, compared with grains from wild-type, but no differences were observed in the grains from the primary panicle branches. The amylose content and gel consistency, and a seed-setting rate from the spb were reduced in the m34-z mutant. Interesting, transcriptional activity analysis revealed that OsMADS34 protein was a transcription repressor and it may influence grain yield by suppressing the expressions of BG1, GW8, GW2, and GL7 in the m34-z mutant. These findings revealed that OsMADS34 largely affects grain yield by affecting the size of grains from the secondary branches.
Keywords: spikelet, rudimentary glume, sterile lemma, grain size, OsMADS34, transcriptional repressor, rice (Oryza sativa L.)
Introduction
In most flowering plants, flowers consist of sepals, petals, stamens, and pistils, which are arranged in concentric whorls (Ohmori et al., 2009). Genetic and morphological studies of floral homeotic mutants in eudicots, including Arabidopsis thaliana, Antirrhinum majus, and Petunia hybrida, have established the classical ABC model of floral development, and subsequently the ABCDE model, which can also partly explain floral development in grass species (Coen and Meyerowitz, 1991; Jeon et al., 2000; Nagasawa et al., 2003; Ditta et al., 2004; Yamaguchi et al., 2006; Dreni et al., 2007; Li H. et al., 2010). Most of the genes in the ABCDE model encode MIKCC-type MADS-domain transcription factors. The temporal and spatial expression patterns of the A, B, C, D, and E-class genes, and their complicated protein interactions, determine the identity and patterning of floral organs (Coen and Meyerowitz, 1991; Pelaz et al., 2000; Theissen and Saedler, 2001; Li H. et al., 2011; Kobayashi et al., 2012; Yun et al., 2013; Hu Y. et al., 2015). In the extended ABCDE model, E-class (SEPALLATA, SEP) genes affect the development of floral organs by interacting with and modulating the expression of floral organ-related genes (Jeon et al., 2000; Pelaz et al., 2000; Ferrario et al., 2003; Malcomber and Kellogg, 2004; Liu et al., 2009).
Arabidopsis thaliana has four functionally redundant E-class genes (SEP1, SEP2, SEP3, and SEP4) that determine floral meristem fate and floral organ identity (Pelaz et al., 2000; Ditta et al., 2004; Li H. et al., 2010). Rice has at least five E-class genes: LEAFY HULL STERILE1/OsMADS1 (LHS1/OsMADS1), OsMADS5, OsMADS7/OsMADS45, OsMADS8/OsMADS24, and OsMADS34/PAP2 (Jeon et al., 2000; Malcomber and Kellogg, 2005; Kater et al., 2006; Yamaguchi and Hirano, 2006; Gao et al., 2010; Kobayashi et al., 2010). LHS1/OsMADS1 determines the identities of four whorls of floral organs and affects the determinacy of the floral meristem (Jeon et al., 2000; Hu Y. et al., 2015). OsMADS5 loss-of-function mutants show no obvious phenotypic abnormities (Gao et al., 2010). OsMADS7 and OsMADS8 are involved in the regulation of flowering time, floral organ identity, and floral meristem determinacy (Pelucchi et al., 2002; Cui et al., 2010). Silencing of OsMADS1, OsMADS5, OsMADS7, and OsMADS8 causes homeotic transformation of the palea, lodicule, stamen, and pistil into leaf-like structures (Cui et al., 2010). The osmads34 mutant displays an abnormal inflorescence with an elongated sterile lemma, suggesting that OsMADS34 controls spikelet and inflorescence morphology mainly by mainly regulating the identity of the sterile lemma and number of branches of the panicle (Gao et al., 2010; Kobayashi et al., 2010; Lin et al., 2014; Zhang and Yuan, 2014).
As a model monocot plant, rice has unique floral architecture that differs from those of eudicots (Ren et al., 2013). The spikelet is the floral unit in rice and comprises one floret and two pairs of glume-like organs, rudimentary glumes and sterile lemmas (glumes). Generally, the rudimentary glume and sterile lemma are regarded as severely reduced bracts, but their origin has been widely debated (Schmidt and Ambrose, 1998; Ambrose et al., 2000; Hong et al., 2010; Ren et al., 2013). Map-based cloning approaches have helped to elucidate how these organs are specified in the rice spikelet. LONG STERILE LEMMA/ELONGATED EMPTY GLUME (G1/ELE) belongs to a plant-specific gene family that encodes an unknown domain protein and is strongly expressed in the sterile lemma primordia. In the g1/ele mutant, the sterile lemma was homeotically transformed into a lemma-like organ (Yoshida et al., 2009; Hong et al., 2010). OsMADS34/PAP2 is important for retaining normal sterile lemma identity (Gao et al., 2010; Kobayashi et al., 2010). In the osmads34/pap2 mutant, normal sterile lemmas were not observed at sites where longer glume-like organs were present. Furthermore, expression of the lemma marker gene DROOPING LEAF (DL) was detectable in the glume-like organs, indicating that the sterile lemma had acquired the lemma identity (Lin et al., 2014). The EXTRA GLUME 1 (EG1) and ABERRANT SPIKELET AND PANICLE 1 (ASP1) genes determine the identity of the sterile lemma. In the eg1 and asp1 mutants, the sterile lemma was elongated and had lemma identity, and the asp1 mutant also showed enlarged rudimentary glumes and the epidermal structure of rudimentary glumes was similar to that of sterile lemmas, suggesting that the identity was altered (Li et al., 2009; Yoshida et al., 2012). The other class of genes comprises FRIZZY PANICLE (FZP), SUPERNUMERARY BRACT (SNB), OsINDETERMINATE SPIKELET1 (OsIDS1), and MULTI-FLORET SPIKELET1 (MFS1), which belong to the APETALA2/ethylene responsive (AP2/ERF) gene family and determine the identities of the rudimentary glumes and/or sterile lemma. Loss of function of FZP and SNB resulted in extra rudimentary glumes in the mutants, but no sterile lemmas were found in the corresponding position (Komatsu et al., 2003; Lee et al., 2007). A mutation of OsIDS1 and MFS1 caused the sterile lemmas to be converted into bract-like organs, which were similar to rudimentary glumes (Lee and An, 2012; Ren et al., 2012, 2013). Although several genes have been successfully isolated and characterized, the identities and origins of the highly derived grass-specific glumes, are still controversial, thus it is necessary to identify more corresponding mutants and isolate these genes involved in regulation of these characteristic organs.
In this study, we discovered a new mutant allele of OsMADS34 (m34-z). The m34-z mutant had a unique mutation different from the reported osmads34 mutants, and m34-z caused different phenotypic defects. The m34-z mutant had more primary panicle branches (ppb), fewer secondary panicle branches (spb), shorter panicles, and enlarged sterile lemmas, consistent with the phenotypes of the reported osmads34 mutants. However, the m34-z mutant also exhibited elongated rudimentary glumes, small grains, low amylose content (AC) and gel consistency (GC), and a low seed-setting rate from the spb. In addition, DL was expressed in the rudimentary glume and sterile lemma in the m34-z mutant. These results revealed that both the rudimentary glume and sterile lemma were converted to the lemma-like organs and acquired the lemma identity, indicating that the rudimentary glume, sterile lemma, and lemma may be homologous organs. Our findings also showed that OsMADS34 is a transcriptional repressor that negatively regulates the expression of genes involved in grain yield and glumes fate. These results indicate that OsMADS34 plays important roles in the determination of organ identity and affects grain yield and quality.
Materials and Methods
Plant Materials
The m34-z mutant was a spontaneous mutant whose genetic background was a japonica cultivar, Zhonghua (ZH11). The m34-z mutant was crossed with the typical indica cultivar Nan Jing 6 (NJ6) to construct the mapping population. The obtained F1 seeds were sown and transplanted as individual plants to generate the F2 plants for gene mapping. ZH11 was used as the wild-type plants for phenotypic analysis. All plants were grown in paddies at the China National Rice Research Institute, Hangzhou and in Lingshui, Hainan Province, China.
Map-Based Cloning of OsMADS34
To create the mapping population, the m34-z mutant was crossed with NJ6 and 962 F2 plants showing the mutational phenotype were obtained. For initial gene mapping, simple sequence repeat (SSR) markers were developed from public rice databases in the Rice Genomic Research Program and the Gramene websites1,2. Fine mapping was performed using single nucleotide polymorphism (SNP) makers from comparisons of genomic sequences between Nipponbare (a japonica cultivar) and 9311 (an indica cultivar; Guo et al., 2014). The sequences of the primers used are shown in Supplementary Table 1.
Microscopy Observations
The paraffin section and scanning electron microscopy (SEM) analyses were carried out as previously described in Ren et al. (2016).
Characterization of Pollen Sterility
Ten anthers were randomly selected, placed on the slide, and mashed. Pollens were stained with 1% I2-KI solution and photographed with a NIKON ECLIPSE 90i microscope.
Grain Quality Determination
The GC, gelatinization temperature (GT), and AC were determined as previously described (Su et al., 2011).
RNA Isolation and Expression Analysis
Total RNA was extracted from roots, culms, leaves, inflorescences with different lengths, developing seeds, and all floral organs of the wild-type and m34-z mutant using the RNeasy Plant Mini Kit (Axygen). cDNA was obtained by reverse transcription using the SuperScript III Reverse Transcriptase Kit (Invitrogen) with genomic DNA digestion (Takara) using 2 μg total RNA in a 25 μL reaction volume. qPCR was carried out with the StepOne-Plus System (Applied Biosystems) using the SYBR Green PCR Master Mix kit (Promega). At least three biological replicates were performed for each tissue.
Complementation Tests
The full coding sequence (CDS) of OsMADS34 driven by the Cauliflower mosaic virus 35S promoter was inserted into the binary vector pCAMBIA1301 to generate the recombinant pCA1301-C plasmid. The pCA1301-C plasmid was transformed into Agrobacterium tumefaciens strain LBA4404 and the positive bacterial strains were introduced into the m34-z mutants using the A. tumefaciens-mediated transformation method (Rao et al., 2015). The primers used are listed in Supplementary Table 1.
GUS Staining
Plant samples (proOsMADS34-GUS) were stained with a solution containing 50 mM NaPO4 buffer, 1 mM 5-bromo-4-chloro-3-indolyl-β-D-GlcA, 0.4 mM K3Fe(CN)6, 0.4 mM K4Fe(CN)6, and 0.1% (v/v) Triton X-100 (Rao et al., 2015) and incubated at 37°C in the dark for 6–12 h. Chlorophyll was removed from the tissues using an ethanol series. The primer sequences used are listed in Supplementary Table 1.
Subcellular Localization
The coding region of OsMADS34 without the stop codon was amplified from ZH11 using the primers OsMADS34OE-1F and OsMADS34OE-1R, which contain SalI sites. Next, the fragment was fused into the 35S-GFP (S65T)-NOS (pCA1301) vector to generate the OsMADS34-GFP recombinant vector using the In-Fusion HD Cloning Kit (Takara). Then, the plasmids of GFP (negative control), AFD1-GFP (positive control), and OsMADS34-GFP were transformed into rice protoplasts and transiently expressed (Ren et al., 2015). After 816 h of incubation at 25–28°C, green fluorescent signals were observed using an OLYMPUS IX71 confocal microscope. The primer sequences used are listed in Supplementary Table 1.
Transcriptional Activity
Transcription activation tests were conducted using the Matchmaker GAL4 Two-Hybrid System 3 (Clontech). The full-length sequence of OsMADS34 from ZH11 was amplified. For the positive control, the full-length sequence of OsMADS15 from ZH11 was amplified (Wang et al., 2010). The two target fragments were inserted into the vector pGBKT7 to fuse the GAL4 DNA-binding domain (BD) using the In-Fusion HD Cloning Kit (Takara). All vectors were transformed into yeast strain AH109 and the clones were diluted to an OD600 of 0.5. Then, 1 μl of liquid culture was added to tryptophan-, histidine-, and adenine-negative synthetic dropout medium (Hu J. et al., 2015). The transcriptional activity of OsMADS34 was investigated using the dual luciferase reporter assay system in Arabidopsis protoplasts and the relative luciferase activity was detected as previously described (Wu et al., 2013). The primers used are listed in Supplementary Table 1.
Results
Phenotypic Defects of the m34-z Mutant Spikelet
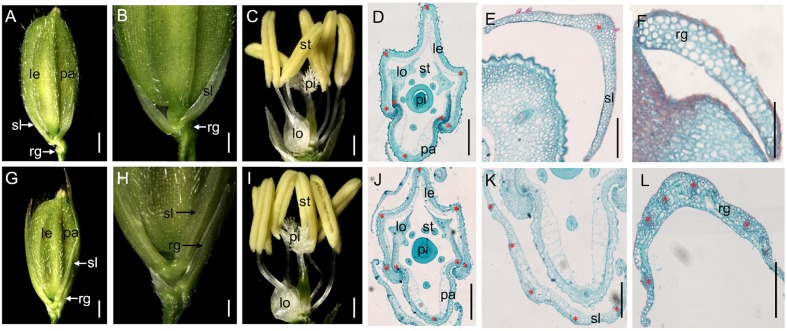
A normal wild-type rice spikelet consists of two pairs of vestigial glumes (rudimentary glumes and sterile lemmas), which are generated from the spikelet meristem, and one terminal floret comprising a lemma, a palea, two lodicules, six stamens, and a pistil (Figures 1A–D).
FIGURE 1.
Phenotypes of spikelets in the wild-type and the m34-z mutant. (A) Wild-type spikelet. (B) Partial magnification in (A). (C) Wild-type floret. (D) Histological analysis of the wild-type floret. (E) Histological analysis of the sterile lemma in the wild-type spikelet. (F) Histological analysis of the rudimentary glume in the wild-type spikelet. (G) m34-z mutant spikelet. (H) Partial magnification in (G). (I) m34-z mutant floret. (J) Histological analysis of the m34-z mutant floret. (K) Histological analysis of the sterile lemma in the m34-z mutant spikelet. (L) Histological analysis of the rudimentary glume in the m34-z mutant spikelet. rg, rudimentary glume; sl, sterile lemma; le, lemma; pa, palea; lo, lodicule; st, stamen; pi, pistil. Red stars represent vascular bundles. Bars = 1000 μm in (A,B,G), and H; 500 μm in (C,I); and 100 μm in (D–F,J–L).
In the vegetative phase, we observed no obvious defects in the m34-z mutant. However, in the reproductive phase, we observed significant abnormalities in the m34-z mutant after heading. First, 56% of the m34-z mutant spikelets showed longer rudimentary glumes which were different from the reported phenotypes of the other osmads34 mutants. The wild-type rudimentary glumes averaged about 0.5 mm in length, but the sizes ofm34-z mutant rudimentary glumes were various from about 1.8– 7.9 mm in length (Figures 1A,B,G,H,L; Table 1). In the m34-z mutant spikelets with serious defects, the rudimentary glumes resembled sterile lemmas or lemmas and were similar in size to the wild-type lemmas (Table 1). Second, 92% of the m34-z mutant spikelets developed larger sterile lemmas that were indistinguishable from the lemmas of the wild-type or the m34-z mutant (Figures 1G,E,K; Table 1).
Table 1.
Comparisons of organ size in the wild type and m34–z mutant.
| Rg (mm) | SI (mm) | Le (mm) | Pa (mm) | |
|---|---|---|---|---|
| WT-p | 0.3–0.5 | 2.0 ± 0.4 | 7.3 ± 0.3 | 6.7 ± 0.4 |
| m34-z-p | 1.8–7.9 | 7.9 ± 0.8 | 7.4 ± 0.2 | 6.9 ± 0.3 |
| WT-s | 0.3–0.5 | 1.9 ± 0.3 | 7.2 ± 0.4 | 6.8 ± 0.2 |
| m34-z-s | 1.9–7.6 | 6.2 ± 0.5 | 6.0 ± 0.3 | 5.8 ± 0.3 |
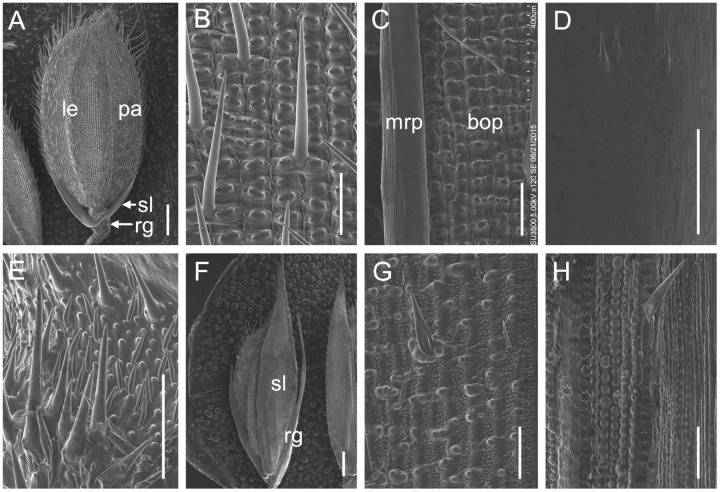
We also investigated the floral organs of four whorls and these organs appeared normal in the m34-z mutant (Figures 1A,C,D,G,I,J). We performed paraffin section and SEM analysis on the structures of the spikelets in the m34-z mutant and wild-type plants. The wild-type lemma and palea [which comprises the marginal regions of the palea (mrp) and the body of the palea (bop)] developed five and three vascular bundles, respectively (Figures 1D and 2C). The wild-type lemma and bop had four cell layers, including non-silicified cells, spongy parenchymatous cells, fibrous sclerenchyma, and silicified cells (Supplementary Figure 1), and the mrp exhibited a smooth epidermis but lacked silicified epicuticular cells (Figure 2C). In the wild-type spikelets, the sterile lemma developed one vascular bundle, showed a smooth epidermis and regularly arranged cells (Figures 1E and 2D), and rare trichomes were observed on the epidermis (Figure 2D). The epidermis of the rudimentary glumes of the wild-type showed irregularly arranged cells with numerous, small protrusions, and trichomes (Figure 2E). No obvious vascular bundles were observed in the rudimentary glumes of the wild-type (Figure 1F). In contrast, the sterile lemmas of the m34-z mutant were elongated and developed five vascular bundles (Figures 1G,K and 2F), and they exhibited a similar histological structure to that of the wild-type lemma or bop including protrusions and trichomes (Figures 2B,C,G). In the m34-z mutant spikelets, the rudimentary glumes were elongated, had five vascular bundles, and contained large protrusions and trichomes (Figures 1L and 2F,H), similar to that of the wild-type lemma or bop.
FIGURE 2.
scanning electron microscopy (SEM) analysis of glumes in the wild-type and the m34-z mutant at heading stage. (A–E) Wild-type spikelet. (B) Epidermal surface of the wild-type lemma. (C) Epidermal surface of the wild-type palea. (D) Epidermal surface of the wild-type sterile lemma. (E) Epidermal surface of the wild-type rudimentary glume. (F–H) m34-z mutant spikelet. (G) Epidermal surface of the m34-z mutant sterile lemma. (H) Epidermal surface of the m34-z mutant rudimentary glume. rg, rudimentary glume; sl, sterile lemma; le, lemma; pa, palea; bop, body of palea; mrp, marginal region of palea. Bars = 1000 μm in (A,F) and 100 μm in (B–E,G–H).
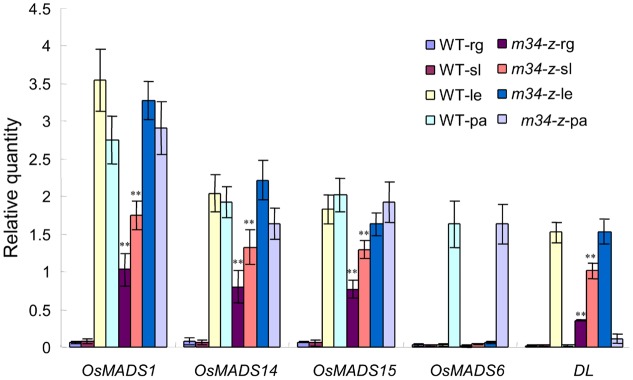
Next, we investigated the expressions of the lemma marker gene DL, the hull (lemma and palea) marker genes OsMADS1, OsMADS14, and OsMADS15, and the palea marker gene OsMADS6 in the m34-z mutant sterile lemmas and rudimentary glumes. In the wild-type, the transcripts of OsMADS1, OsMADS14, and OsMADS15 were found in the lemma and palea. DL and OsMADS6 was mainly expressed in the lemma and palea, respectively. However, no signals of all these genes were observed in the sterile lemma and rudimentary glume. Whereas, the expression of OsMADS1, OsMADS14, OsMADS15, and DL increased in the m34-z mutant compared with wild-type, but transcripts of OsMADS6 were not detected in the sterile lemma and rudimentary glume of m34-z mutant (Figure 3). These results indicated that the rudimentary glumes and sterile lemmas of the m34-z mutant were converted to lemma-like organs and partly acquired the identity of the lemma but not the identity of the palea.
FIGURE 3.
Relative expression levels of genes involved in floral organ development in the wild-type and the m34-z mutant spikelet. rg, rudimentary glume; sl, sterile lemma; le, lemma; pa, palea; WT, wild-type. Error bars indicate SD. ∗∗Significant difference at P < 0.01 compared with the wild-type by Student’s test.
Abnormal Early Spikelet Development in the m34-z Mutant
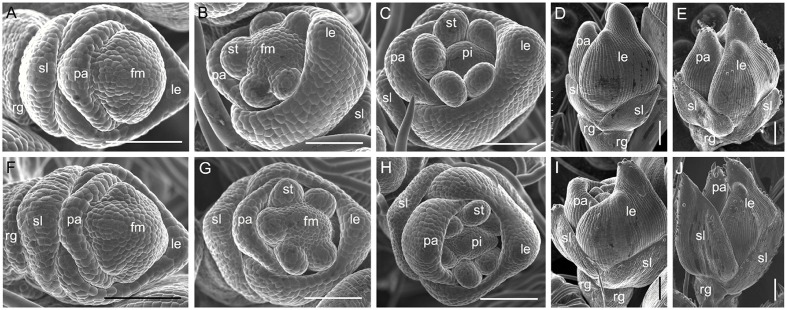
We analyzed young spikelets at different developmental stages to further investigate spikelet development in the m34-z mutant. During the spikelet 4 stage (Sp4) of development in the wild-type, the sterile lemma and rudimentary glume primordia were formed and the palea and lemma primordia were developing (Figure 4A). At the Sp5 and Sp6 stages, six stamen primordia were found in the wild-type spikelet and the stamen primordium on the lemma side was delayed (Figure 4B). At these stages, the growth of the rudimentary glume ceased and the sterile lemma continued growing. During the Sp7 and Sp8 stages in the wild-type spikelet, the pistil primordium was formed and the sterile lemma further differentiated, becoming much longer than the rudimentary glume (Figures 4C,D). In contrast, the sterile lemma of the m34-z mutant spikelets showed obvious morphological differences from that of the wild-type. At Sp4 through Sp8, the m34-z mutant spikelets displayed larger sterile lemmas than those of the wild-type (Figures 4F–I). After the Sp8 stage, we found a few small protrusions on the epidermis of the wild-type and m34-z spikelets. And the sterile lemma of the m34-z mutant was dramatically larger than that of the wild-type and the size of the sterile lemma of the m34-z mutant was comparable to the lemma of the wild-type or m34-z mutant (Figures 4E,J). However, we found no obvious differences between the rudimentary glumes of the wild-type and the m34-z mutant at from Sp4 through Sp8 (Figures 4A–J). Also, we did not observe significant defects in floral organs in the m34-z mutant, including the lemma, palea, lodicule, stamen, and pistil (Figures 4A–C,F–H). These results revealed that OsMADS34 affects the initiation and enlargement of the sterile lemma but does not influence the development of floral organs. Because longer rudimentary glumes were observed in the wild-type at the heading stage, we speculated that OsMADS34 may be involved in regulation of the rudimentary glume at later stages.
FIGURE 4.
Spikelets at early developmental stages in the wild-type and the m34-z mutant. (A–E) wild-type spikelet. (A) Sp4, (B) Sp5-6, (C) Sp7, (D) Sp8, (E) After Sp8, (F–I) m34-z mutant spikelet, (F) Sp4, (G) Sp5-6, (H) Sp7, (I) Sp8, (J) After Sp8. fm, floral meristem; rg, rudimentary glume; sl, sterile lemma; le, lemma; pa, palea; st, stamen; pi, pistil. Bars = 100 μm.
The m34-z Mutant Has Reduced Grain Yield and Quality
We compared agronomic traits between the wild-type and the m34-z mutant at grain maturation. The m34-z mutant displayed shorter panicles, more ppb, and fewer spb compared to the wild-type (Supplementary Figures 2B,C). A lower seed-setting rate was also found in the m34-z mutant. The total seed-setting rate was only 73% in the m34-z mutant, whereas it reached up to 86% in the wild-type (Supplementary Figure 2E). The seed-setting rate of the ppb in the m34-z mutant was comparable with that in the wild-type. However, the seed-setting rate of the spb in the m34-z mutant was 65%, whereas the seed-setting rate of the spb in the wild-type was 84% (Supplementary Figure 2E). This finding indicated that the low seed-setting rate in the m34-z mutant could be attributed to the lower seed-setting rate of the spb.
Next, an I2-KI test showed that the viability of the pollen from the stamens of the ppb of the m34-z mutant spikelets was not altered, but the viability of the pollen from the spb was decreased (Figures 5E,J,O,T; Supplementary Figure 2D), which was consistent with the low seed-setting rate of the spb in the m34-z mutant. The grains from the spb were smaller in the m34-z mutant compared to the wild-type, but the grains from the ppb in the m34-z mutant were normal in size (Figures 5A–C,F–H,K–M,P–R; Supplementary Figure 2A). The average lengths of the wild-type grains and brown rice (grains from which the hull was removed) were 7.0 and 5.6 mm, respectively, while the average lengths of the m34-z mutant grains and brown rice were 6.0 and 4.5 mm, respectively (Figures 5K,L,P,Q; Supplementary Figures 2A and 3). The average widths of the m34-z mutant grains and brown rice were similar to those of the wild-type (Supplementary Figure 3). Moreover, the 1,000-grain weight and weight of 1,000 brown rice from the spb of the m34-z mutant were markedly decreased compared to the wild-type, but the 1,000-grain weight and weight of 1,000 brown rice from the ppb were not changed (Supplementary Figure 3).
FIGURE 5.
Phenotypic observations of grain and pollen viability in the wild-type and the m34-z mutant. (A–E) Phenotypes of the grain and pollen viability from the primary panicle branches (ppb) in the wild-type. (F–J) Phenotypes of the grain and pollen viability from the ppb in the m34-z mutant. (K–O) Phenotypes of the grain and pollen viability from the secondary panicle branches (spb) in the wild-type. (P–T) Phenotypes of the grain and pollen viability from the spb in the m34-z mutant. (A,F,K,P) grain size. (B,G,L,Q) size of brown rice. (C–D,H–I,M–N,R–S) epidermal surface of brown rice. E, J, O, and T, pollen viability. Bars = 1000 μm in (A–C,F–H,K–M), and P-R; 50 μm in (D,I,N,S); 100 μm in (E,J,O,T).
To investigate the cellular basis for the smaller grains from the spb of the m34-z mutant, we measured the cell size from the middle part of the lemmas by SEM. The average cell lengths and widths of the lemmas from the ppb of the m34-z mutant were similar to those of the wild-type (Figures 6A–F). However, the average cell lengths were reduced in the lemmas from the spb of the m34-z mutant compared to the wild-type, but the average cell widths were unchanged (Figures 6A–F). We also investigated cell number in the outer epidermis of the lemmas and found no differences in total cell number along the longitudinal axis of the lemmas from the ppb and spb of the wild-type and m34-z mutant (Figures 6G–K). However, the number of cells per millimeter along the longitudinal axis of the lemmas from the spb of the m34-z mutant was significantly higher than that of the wild-type (Figures 6G–J,L).
FIGURE 6.
OsMADS34 influences cell expansion. (A) Inner epidermal cells of hulls from the ppb in the wild-type. (B) Inner epidermal cells of hulls from the ppb in the m34-z mutant. (C) Inner epidermal cells of hulls from the spb in the wild-type. (D) Inner epidermal cells of hulls from the spb in the m34-z mutant. (E) Cell length. (F) Cell width. (G) Grain from the ppb in the wild-type. (H) Grain from the ppb in the m34-z mutant. (I) Grain from the spb in the wild-type. (J) Grain from the spb in the m34-z mutant. (K) Total cell number along the longitudinal axis of the lemma. (L) Total cell number per millimeter along the longitudinal axis of the lemma. (M) Expression analysis of cell expansion-related genes in young panicles. WT-p, grains from the ppb in the wild-type; WT-s, grains from the spb in the wild-type; m34-z-p, grains from the ppb in the m34-z mutant; m34-z-s, grains from the spb in the m34-z mutant. The red boxes indicate the middle part of the lemma in (G,H,I,J). Bars = 50 μm in (A–D). Error bars indicate SD. ∗∗Significant difference at P < 0.01 compared with the wild-type by Student’s test.
Next, we examined the transcript levels of 35 genes involved in the regulation of cell cycle and cell expansion in rice. Among them, the expression of six cell expansion-related genes was decreased in the spb of the m34-z mutant and no differences were detected in the ppb of the m34-z mutant (Figure 6M). These results suggest that OsMADS34 mainly controls grain size on the spb by regulating cell expansion but not cell number. GN1, DEP1, DEP2, DEP3, APO1, and NAL1 are closely associated with panicle architecture in rice (Ashikari et al., 2005; Ikeda et al., 2007; Huang et al., 2009; Li F. et al., 2010; Qiao et al., 2011; Fujita et al., 2013). We investigated the expression levels of these genes in young panicles of the m34-z mutant and the wild-type. DEP1, DEP2, and DEP3 were up-regulated compared to the control, and the expression levels of GN1a, APO1, and NAL1 were not changed in the m34-z mutant (Supplementary Figure 4). We also detected the expression levels of genes related to grain size in the wild-type and the m34-z mutant (Song et al., 2007; Wang et al., 2008, 2012; Mao et al., 2010; Li Y. et al., 2011; Chen et al., 2013; Hu J. et al., 2015; Liu L. et al., 2015; Liu S. et al., 2015; Wang S. et al., 2015; Wang Y. et al., 2015). Compared with the wild-type, BG1, GW2, GW8, and GL7/GW7 were up-regulated in the m34-z mutant (Supplementary Figure 4), indicating that OsMADS34 negatively regulates the expression of these genes involved in grain size. These findings supported the phenotypic observations and the hypothesis that OsMADS34 may influence panicle architecture and grain size by negatively regulating the expression.
Next, we studied the epidermis of brown rice. Compared with the wild-type, the epidermis of brown rice from the spb of the m34-z mutant was easily wrinkled (Figures 5C,D,H,I,M,N,R,S), but the epidermis of brown rice from the spb in the m34-z mutant showed rare protrusion-like structures (Figures 5M,N,R,S). No differences in the epidermis of brown rice from the ppb were detected between the wild-type and the m34-z mutant (Figures 5C,D,H,I). Next, we examined the GC, GT, and amylase content (AC) of the grains from the wild-type and the m34-z mutant. In the grains from the spb of the m34-z mutant, AC was reduced by 17% compared to that in the wild-type, the GC was slightly reduced compared to that in the wild-type, but GT was not altered (Supplementary Table 2). No significant differences were found in the GC, GT, and AC of grains from the ppb (Supplementary Table 2). Thus, the mutation in OsMADS34 in the m34-z mutant results in reduced grain quality from the secondary branches.
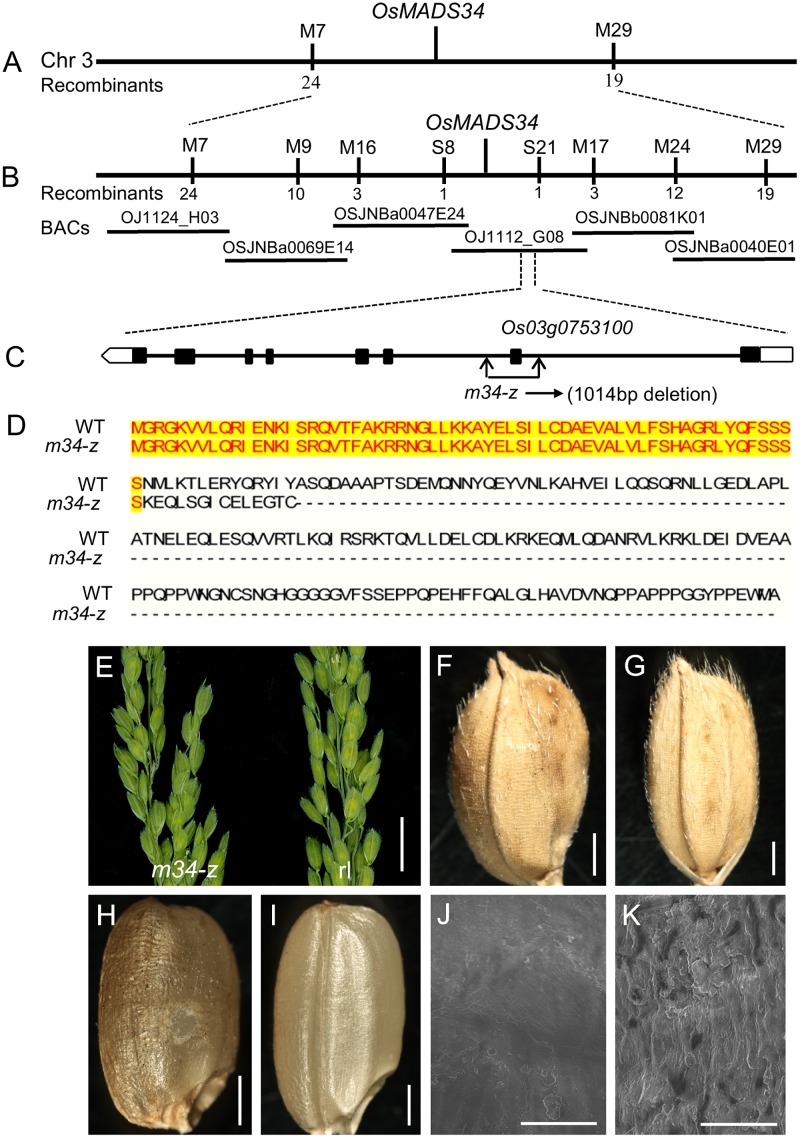
Identification of OsMADS34 As Responsible for the m34-z Phenotype
To clone the locus responsible for the m34-z mutant phenotype, we performed a cross between the m34-z mutant and indica cultivar NJ6. Among the F2 plants, 2,736 displayed the normal phenotype and 962 plants displayed the m34-z mutant phenotype, a segregation ratio of approximately 3:1, indicating that the mutant phenotype was controlled by a single recessive nuclear gene. We used the 962 recessive mutant plants for the mapping population. Among the 215 SSR markers used in this study, which were evenly distributed throughout the 12 chromosomes, 130 were polymorphic between the two parental lines, and the mutated locus was preliminarily mapped on chromosome 3 between the markers M7 and M29 (Figure 7A). For fine mapping of OsMADS34, 36 SNP markers were used and six markers displayed polymorphisms (Figure 7B). The location of OsMADS34 was narrowed to a 78 kb distance between the two markers S8 and S21 (Figure 7B). We identified a 1,014 bp deletion in Os03g0753100 (OsMADS34) by sequencing analysis, which triggered a premature translation stop (Figures 7C,D). To determine whether Os03g0753100 was causally linked to the m34-z mutant phenotypes, the coding region of OsMADS34 driven by the Cauliflower mosaic virus 35S promoter was transformed into calli derived from m34-z mutant seeds. Eighteen transgenic lines were obtained and all phenotypic defects were rescued in each line, including the elongated rudimentary glumes and sterile lemmas, and the reduced seed set from the spb (Figures 7E–K; Supplementary Figures 5A,B).
FIGURE 7.
Identification of OsMADS34 gene as the causal gene for the the m34-z mutant. (A–C) Map position of the OsMADS34 locus. The relative position of the BAC clone is shown. Genomic structure of OsMADS34. The mutated site of the m34-z mutant is shown. (D) OsMADS34 encodes a 239 amino acid protein, while the mutation of OsMADS34 in the m34-z mutant encodes a 76 amino acid protein due to a coding frame-shift. (E–K) Complementation test. All mutant phenotypes (deformed spikelets, reduced grains, and abnormal epidermal surface) were rescued in the transgenic plants. (E) Deformed spikelets were recovered in the rescued lines. (F,H) Grains from the spb in the m34-z mutant. (G,I) Reduced grains from the spb in the m34-z mutant were rescued in the transgenic lines. (J) Epidermal surface of brown rice from the spb in the m34-z mutant. (K) Epidermal surface of brown rice from the spb in the m34-z mutant was similar with that of the wild-type in the transgenic lines. Bars = 1 cm in (E); 1000 μm in (F–I); 50 μm in (J–K).
Expression Patterns, Subcellular Localization, and Transcriptional Activity Analysis
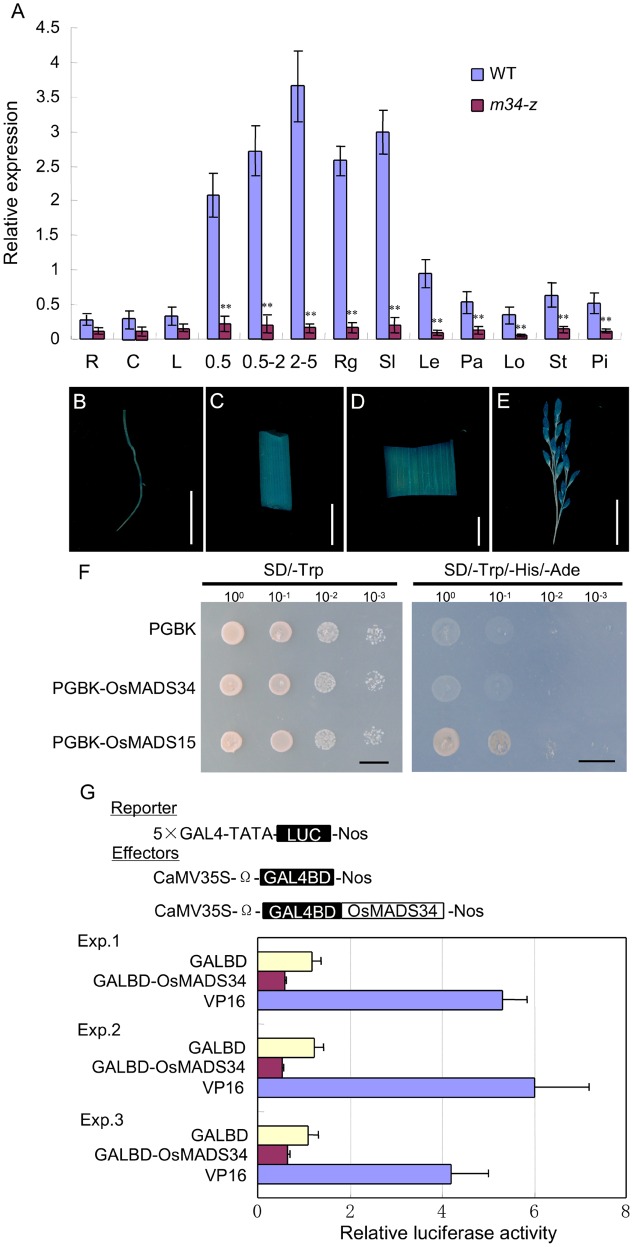
We investigated the expression of OsMADS34 in the wild-type and m34-z mutant to determine its expression patterns. OsMADS34 transcripts appeared in all examined tissues and organs including roots, culms, leaves, panicles of different lengths, developing seeds, rudimentary glumes, sterile lemmas, lemmas, paleae, lodicules, stamens, and pistils in the wild-type (Figure 9A). And the expression level was higher in young panicles, developing seeds, rudimentary glumes, sterile lemmas, and lemmas. However, much lower expressions were found in tested vegetative and reproductive tissues in the m34-z mutant.
FIGURE 9.
Expression patterns and transcriptional activity analysis. (A) OsMADS34 expression in different tissues detected by qPCR. (B) GUS staining of root. (C) GUS staining of culm. (D) GUS staining of leaf. (E) GUS staining of spikelet. (F,G) Transcriptional activity test. R, root; C, culm; L, leaf; Rg, rudimentary glume; Sl, sterile lemma; Le, lemma; Pa, palea; Lo, lodicule; st, stamen; pi, pistil; 0.5 cm, young panicles (≤0.5 cm); 0.5–2 cm, young panicles (0.5–2 cm); 2–5 cm, young panicles (2–5 cm); 1D, 1 day after pollination; 4D, 4 days after pollination; 7D, 7 days after pollination; 10D, 10 days after pollination. Bars = 1 cm in (B–G). Error bars indicate SD. ∗∗Significant difference at P < 0.01 compared with the wild-type by Student’s test.
To investigate the tissue specificity of OsMADS34 expression, we fused the OsMADS34 promoter region with the β-glucuronidase (GUS) reporter gene and transformed the construct into ZH11. GUS signals from proOsMADS34-GUS were detected in the root, culm, leaf, and panicle (Figures 9B–E). However, the signals were weak in the root, culm, and leaf, and strong in the panicle (Figures 9B–E). In previous studies, in situ hybridization revealed that the OsMADS34 mRNA mainly accumulated in the various stages of inflorescence development (Kobayashi et al., 2010). Subsequently, strong OsMADS34 signals were detected in the spikelet and floral primordia, such as rudimentary glumes, sterile lemmas, and four whorls of floral organs (Kobayashi et al., 2010). These results were consistent with our qPCR analysis and GUS staining, and supported the phenotypic observations.
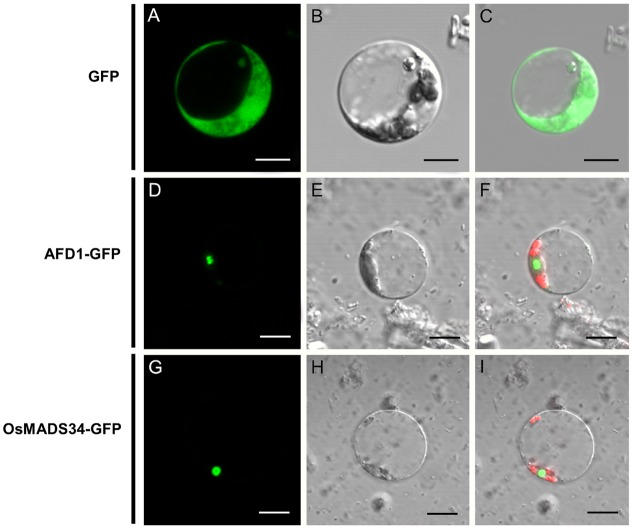
We also examined the subcellular localization of the OsMADS34 protein. The fusion proteins, including OsMADS34-GFP, AFD1-GFP (positive nuclear protein control, Ren et al., 2015), and single GFP, were transiently expressed in rice protoplasts. In the cells that were transformed with single GFP, the green fluorescence was visible in whole cells and the signals surrounded the cell nucleus, cytoplasm, and cell membrane (Figures 8A–C). In the cells that were transformed with OsMADS34-GFP and AFD1-GFP, the green fluorescence was exclusively found in the nuclei (Figures 8D–I), suggesting that OsMADS34 is predominantly a nuclear protein.
FIGURE 8.
Subcellular localization of the OsMADS34 protein. (A–C) GFP fusion protein. (A) differential interference contrast (DIC) image; (B) bright-field image; (C) merged image of GFP fusion protein. (D–F) AFD1-GFP. (D) DIC image; (E) bright-field image; (F) merged image of AFD1-GFP fusion protein. (G–I) OsMADS34-GFP. (G) DIC image; (H) bright-field image; (I) merged image of OsMADS34-GFP fusion protein. Bars = 5 μm in (B–G).
Next, we assayed the transcriptional activation activity of OsMADS34 protein. The coding regions for OsMADS34 and the known transcriptional activator OsMADS15 were fused to the DNA-BD of yeast GAL4. The empty pGBKT7 vector and BD-OsMADS15 were regarded as the negative control and positive control, respectively (Wang et al., 2010). BD-OsMADS15 enabled transformed yeast cells to grow on histidine-deficient medium, but yeast cells harboring BD-OsMADS34 or the empty pGBKT7 vector could not survive on the histidine-deficient medium (Figure 9F), suggesting that OsMADS34 does not have transcription activation activity.
We further analyzed the transcriptional activity using a dual luciferase reporter assay in transient assays in Arabidopsis protoplasts. We used a luciferase (LUC) reporter gene that contains five copies of binding sites for GAL4. As in the yeast assays, the coding region of OsMADS34 was fused in-frame to the GAL4 DNA-BD, but the construct was driven by the constitutive 35S promoter of Cauliflower mosaic virus (BD-OsMADS34; Figure 9G). The GAL4-BD was used as the negative control and VP16 was used as a transcriptional activator (Wu et al., 2013). Three effectors, GAL4-BD, BD-OsMADS34, and VP16, were expressed transiently in Arabidopsis protoplasts. OsMADS34 showed less luciferase activity compared with GAL4-BD and VP16 (Figure 9G). These results demonstrated that OsMADS34 functions as a transcriptional repressor.
OsMADS34 Affects the Expression of Genes Related to Glume Development
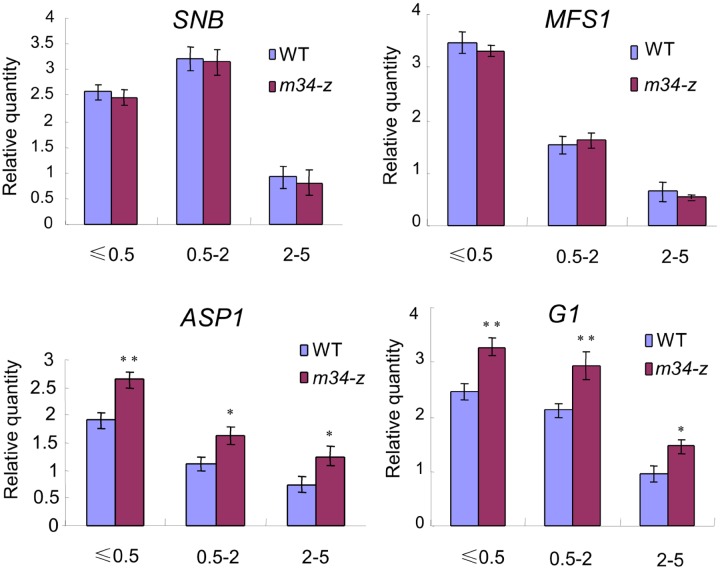
Because the m34-z mutant exhibited defects in glume development, we investigated if OsMADS34 regulates the expression of genes known to be associated with glume (rudimentary glume and sterile lemma) specification and spikelet development in rice. We examined the transcript levels of G1, ASP1, MFS1, and SNB in young panicles less than 0.5 cm, between 0.5 and 2 cm, and between 2 and 5 cm in length from the wild-type and m34-z mutant (Lee et al., 2007; Yoshida et al., 2009, 2012; Hong et al., 2010; Lee and An, 2012; Ren et al., 2013). In the wild-type, abundant levels of G1 and ASP1 transcripts were found in the panicles less than 2 cm long, and their transcript levels decreased markedly in the panicles with a length of 2–5 cm (Figure 10). Higher levels of expression of G1 and ASP1 were detected in the m34-z mutant in all tested panicles compared to the wild-type (Figure 10). However, the expression levels of SNB and MFS1 were not altered in the m34-z mutant (Figure 10). These results revealed that the abnormal glumes in the m34-z mutant may be related to altered levels of transcription of glume and spikelet development-related genes, and OsMADS34 may determine glumes fate by negatively modulating the expressions of G1 and ASP1.
FIGURE 10.
Expression of genes related to glume development at different stages. 0.5 cm, young panicles (≤0.5 cm); 0.5–2 cm, young panicles (0.5–2 cm); 2–5 cm, young panicles (2–5 cm). Error bars indicate SD. **Significant difference at P < 0.01 compared with the wild-type by Student’s test; *Significant difference at P < 0.05 compared with the wild-type by Student’s test.
Discussion
OsMADS34 Affects Grain Size and Quality in Rice
Grain size and weight are important agronomic traits for yield and quality in cereal crops (Zuo and Li, 2013). Although several genes that control rice grain shape (GW2, GW8, GS5, GL7/GW7, BG1, and DSG1) have been cloned (Song et al., 2007; Li Y. et al., 2011; Wang et al., 2012; Liu L. et al., 2015; Wang S. et al., 2015; Wang Y. et al., 2015), the complicated genetic mechanisms and regulatory networks by which grain shape is determined remain unknown.
The m34-z mutant produced small, short grains from the spb, but normal grains from the ppb. Meantime, qPCR analysis showed that the expressions of BG1, GW8, GW2, and GL7 were dramatically increased in the m34-z spb and no obvious differences were observed in m34-z ppb compared with the wild-type, which supported abnormal phenotypes in the m34-z mutant. These results suggest that OsMADS34 may be a regulatory factor that modulates grain size in the spb in rice. Cell number and cell size determine organ size via cell proliferation and cell expansion, respectively (Jiang et al., 2012). In the m34-z mutant, the hulls from the spb had smaller cells. No difference in cell number was found between the wild-type and the m34-z mutant. qPCR analyses showed that OsMADS34 regulated the expression of cell expansion-related genes, in agreement with the phenotypic observations. These findings indicated that OsMADS34 affects grain size by regulating cell expansion. Also, the m34-z mutant produced shorter panicles, more ppb, and fewer spb which mimicked the phenotype of osmads34-1 mutant (Gao et al., 2010), implying that OsMADS34 influences rice panicle architecture. Interesting, transcriptional activity analysis revealed that OsMADS34 protein was a transcription repressor and the expressions of several genes involved in the regulation of grain size and panicle shape were obviously up-regulated in the m34-z mutant. These findings suggested that OsMADS34 may influence grain yield by suppressing the expressions of related genes, and high expressions of BG1, GW8, GW2, and GL7 in the m34-z mutant may be important factors for smaller grain from the m34-z spb (Figure 10; Supplementary Figure 4). In addition, eating and cooking quality (ECQ) is mainly affected by the AC, GC, and GT (Tian et al., 2009), and these qualities were altered in the grains from the spb in the m34-z mutant, indicating that the mutation of OsMADS34 affected the ECQ. These phenotypic differences between the m34-z and earlier reported mutants may be due to the different genetic backgrounds and the different mutated sites in the target gene. The nucleotides in osmads34-1 were changed from TCA to AAG, causing a frame-shift mutation (Gao et al., 2010). No obvious expression change of OsMADS34 was observed in osmads34-1 compared with the wild-type. pap2-1 and los were from an insertion of Tos17 and T-DNA in the OsMADS34, respectively (Kobayashi et al., 2010; Lin et al., 2014). While m34-z was a natural mutant allele of OsMADS34, had a 1014-bp deletion, resulting in a premature translation stop, and much lower expressions were detected in m34-z compared with the wild-type. Meantime, the m34-z mutant was derived from the japonica variety ZH11, but the osmads34-1 and pap2-1 mutants were isolated from japonica variety 9552 and Nipponbare, respectively. These results may be responsible for different phenotypic defects that were not found in the reported osmads34 mutants. Taken together, these findings showed that OsMADS34 not only influences grain size, grain yield, and panicle architecture, but also affects ECQ in rice. This work provides new insight into the functions of OsMADS34 and MADS-box family genes, which are involved in the regulation of floral development in rice.
OsMADS34 Determines Glume Fate in Rice
In grass, glumes (rudimentary glume and sterile lemma) are unique structures and account for some of the dramatic morphological variations in spikelets (Zanis, 2007; Li et al., 2009). In the m34-z mutant, most of the spikelets developed larger rudimentary glumes and sterile lemmas, compared with wild-type spikelets. The rudimentary glume and sterile lemma in the m34-z mutant exhibited similar protrusions and trichomes, and had a similar number of vascular bundles as wild-type. However, no abnormities were observed in the rudimentary glumes of reported osmads34 mutants. The expression of OsMADS1, OsMADS14, OsMADS15, and DL was completely absent in both the rudimentary glumes and sterile lemmas of the wild-type, but these genes were expressed in the rudimentary glumes and sterile lemmas of the m34-z mutant. Transcripts of OsMADS6 were found only in the paleae in the wild-type and the m34-z mutant. These results indicated that both rudimentary glumes and sterile lemmas acquired the lemma identity. Meantime, G1 and ASP1 were maily expressed in rudimentary glumes and/or sterile lemmas, and restrained the elongation of glumes (Yoshida et al., 2009, 2012). G1 and ASP1 transcripts were increased in the m34-z mutant, which were consistent of enlarged glumes. Together, with the abundant expression of OsMADS34 in the glumes, our results revealed that OsMADS34 not only determined the sterile lemma fate, but also controlled the identity of rudimentary glume.
A putative ancestor of Oryza species may have had a spikelet that contained a terminal floret and two lateral florets (Arber, 1934; Takeoka et al., 1993; Kellogg, 2009; Ren et al., 2013). The two lateral florets may have degenerated into the lemmas and the sterile lemma may be morphological modification of the lemmas during evolution (Arber, 1934; Takeoka et al., 1993; Kellogg, 2009; Ren et al., 2013; Lin et al., 2014). In the m34-z, g1/ele, eg1, and asp1 mutants, the sterile lemmas are elongated and transformed into lemma-like organs, acquiring the lemma identity, which supports this hypothesis. Interestingly, the rudimentary glumes were enlarged and their identity was changed in the m34-z and asp1 mutants. The epidermal structure of the rudimentary glumes in the asp1 mutant was similar to that of the wild-type sterile lemma, whereas the epidermal structure of the rudimentary glumes in the m34-z mutant resembled that of the wild-type lemma. In addition, the sterile lemma in the mfs1 mutant was transformed into a rudimentary glume-like organ and acquired rudimentary glume identity, which indicates that the rudimentary glume and sterile lemma may be homologous organs and that the rudimentary glume and sterile lemma may be severely reduced bract structures (Schmidt and Ambrose, 1998; Terrell et al., 2001; Hong et al., 2010; Ren et al., 2013). In maize and wheat, the bract-like glume organs are similar to the lemma in size and structure (Kellogg, 2001; Yoshida et al., 2009; Ren et al., 2013), whereas it is severely reduced and equivalent to the rudimentary glume in Oryza (Takeoka et al., 1993; Bommert et al., 2005; Li et al., 2009; Yoshida et al., 2009; Hong et al., 2010; Ren et al., 2013). Therefore, these results further suggest that the rudimentary glume, sterile lemma, and the lemma may be homologous organs.
OsMADS34 Is a Grass-Specific E-Function Gene
In grass, functional analysis and morphological evolutionary studies have shown that OsMADS34 plays an important role in the spikelet development and has distinct functions from the E-class genes in Arabidopsis (Gao et al., 2010; Zhang and Yuan, 2014). The transcripts of E-class genes are mainly detected in floral organs in Arabidopsis, but OsMADS34 also belongs to the E-class MADS-box family and is expressed in all vegetative and reproductive organs (Rounsley et al., 1995; Pelaz et al., 2000; Gao et al., 2010) (Figure 9A). In addition, loss of function of OsMADS34 leads to various defects, including dwarf phenotypes and abnormal inflorescence architecture and spikelet morphology (Gao et al., 2010; Kobayashi et al., 2010) (Figures 1 and 2; Supplementary Figure 2B–C), which are different from the defects that occur due to loss of function of other E-class genes, including OsMADS1, OsMADS5, OsMADS7, and OsMADS8 (Pelucchi et al., 2002; Cui et al., 2010). The osmads34 osmads1 double mutant shows more defects of the inner floral organs than those of the single mutants, indicating that OsMADS34 and OsMADS1 redundantly regulate the identity of the inner three whorls of floral organs, but independently control spikelet and inflorescence morphology in rice (Gao et al., 2010). In this study, the m34-z mutant exhibited uniquely larger rudimentary glumes and the rudimentary glume had acquired the lemma identity (Figures 1 and 2), which was different from the reported phenotype of the osmads34 mutants. Interestingly, our results also showed that OsMADS34 affected grain size by promoting cell expansion in the hulls. OsMADS34 negatively regulated the expression of genes that are closely associated with grain yield (Supplementary Figure 4), suggesting that OsMADS34 may affect grain yield by suppressing the expression of related genes. These findings reveal that OsMADS34 exhibits unique functions that are distinct from the reported functions of OsMADS34 and other SEP homologs in rice and Arabidopsis. In grasses, further research into the functions of OsMADS34 and its orthologs will facilitate a better understanding of the genetic and evolutionary mechanisms and regulatory networks of spikelet development, which may provide a new opportunity to improve grain yield and quality in rice breeding.
Author Contributions
Experimental design: QQ and LG; Experiments: DR, YR, YL, ZL, QX, LW, ZQ, JH, and GC; Data analysis: DX, DZ, GZ, LZ, ZG, and GD; Manuscript preparation: DR and QQ.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer JG and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31401464, 31461143014, and 31521064). This work was supported by the National Science and Technology Major Project (2016ZX08009003-003-008), Zhejiang Provincial Natural Science Foundation of China (LY16C130001). We gratefully thank Prof. Zhukuan Chen and Prof. Jinsong Zhang from the Institute of Genetics and Developmental Biology (Beijing, China) for providing the vectors of the transcriptional activity test.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01853/full#supplementary-material
References
- Ambrose B., Lerner D., Ciceri P., Padilla C., Yanofsky M., Schmidt R. (2000). Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5 569–579. 10.1016/S1097-2765(00)80450-5 [DOI] [PubMed] [Google Scholar]
- Arber A. (1934). The Gramineae: A Study of Cereal, Bamboo, and Grasses. Cambridge: Cambridge University Press. [Google Scholar]
- Ashikari M., Sakakibara H., Lin S., Yamamoto T., Takashi T., Nishimura A., et al. (2005). Cytokinin oxidase regulates rice grain production. Science 309 741–745. 10.1126/science.1113373 [DOI] [PubMed] [Google Scholar]
- Bommert P., Satoh-Nagasawa N., Jackson D., Hirano H. Y. (2005). Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol. 46 69–78. 10.1093/pcp/pci504 [DOI] [PubMed] [Google Scholar]
- Chen Y., Xu Y., Luo W., Li W., Chen N., Zhang D., et al. (2013). The F-box protein osFBK12 targets OsSAMS1 for degradation and affects pleiotropic phenotypes, including leaf senescence, in rice. Plant Physiol. 163 1673–1685. 10.1104/pp.113.224527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E., Meyerowitz E. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353 31–37. 10.1038/353031a0 [DOI] [PubMed] [Google Scholar]
- Cui R., Han J., Zhao S., Su K., Wu F., Du X., et al. (2010). Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J. 6 767–781. 10.1111/j.1365-313X.2009.04101.x [DOI] [PubMed] [Google Scholar]
- Ditta G., Pinyopich A., Robles P., Pelaz S., Yanofsky M. (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14 1935–1940. 10.1016/j.cub.2004.10.028 [DOI] [PubMed] [Google Scholar]
- Dreni L., Jacchia S., Fornara F., Fornari M., Ouwerkerk P., An G., et al. (2007). The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J. 52 690–699. 10.1111/j.1365-313X.2007.03272.x [DOI] [PubMed] [Google Scholar]
- Ferrario S., Immink R., Shchennikova A., Busscher-Lange J., Angenent G. (2003). The MADS box gene FBP2 is required for SEPALLATA function in petunia. Plant Cell 15 914–925. 10.1105/tpc.010280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita D., Trijatmiko K., Taglea A., Sapasap M., Koide Y., Sasaki K., et al. (2013). NAL1 allele from a rice landrace greatly increases yield in modern indica cultivars. Proc. Natl. Aacd. Sci. U.S.A. 10 20431–20436. 10.1073/pnas.1310790110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Liang W., Yin C., Ji S., Wang H., Su X., et al. (2010). The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol. 153 728–740. 10.1104/pp.110.156711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Gao Z., Qian Q. (2014). Application of resequencing to rice genomics, functional genomics and evolutionary analysis. Rice 7:4 10.1186/s12284-014-0004-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L., Qian Q., Zhu K., Tang D., Huang Z., Gao L., et al. (2010). ELE restrains empty glumes from developing into lemmas. J. Genet. Genomics 37 101–115. 10.1016/S1673-8527(09)60029-1 [DOI] [PubMed] [Google Scholar]
- Hu J., Wang Y., Fang Y., Zeng L., Xu J., Yu H., et al. (2015). A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant 8 1455–1465. 10.1016/j.molp.2015.07.002 [DOI] [PubMed] [Google Scholar]
- Hu Y., Liang W., Yin C., Yang X., Ping B., Li A., et al. (2015). Interactions of OsMADS1 with floral homeotic genes in rice flower development. Mol. Plant 8 1366–1384. 10.1016/j.molp.2015.04.009 [DOI] [PubMed] [Google Scholar]
- Huang X., Qian Q., Liu Z., Sun H., He S., Luo D., et al. (2009). Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41 494–497. 10.1038/ng.352 [DOI] [PubMed] [Google Scholar]
- Ikeda K., Ito M., Nagasawa N., Kyozuka J., Nagato Y. (2007). Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate. Plant J. 51 1030–1040. 10.1111/j.1365-313X.2007.03200.x [DOI] [PubMed] [Google Scholar]
- Jeon J., Jang S., Lee S., Nam J., Kim C., Lee S., et al. (2000). leafy hull sterile1 Is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12 871–884. 10.1105/tpc.12.6.871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Bao L., Jeong S., Kim S., Xu C., Li X., et al. (2012). XIAO is involved in the control of organ size by contributing to the regulation of signaling and homeostasis of brassinosteroids and cell cycling in rice. Plant J. 70 398–408. 10.1111/j.1365-313X.2011.04877.x [DOI] [PubMed] [Google Scholar]
- Kater M., Dreni L., Colombo L. (2006). Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J. Exp. Bot. 57 3433–3444. 10.1093/jxb/erl097 [DOI] [PubMed] [Google Scholar]
- Kellogg E. (2001). Evolutionary history of the grasses. Plant Physiol. 125 1198–1205. 10.1104/pp.125.3.1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg E. (2009). The evolutionary history of Ehrhartoideae, Oryzeae, and Oryza. Rice 2 1–14. 10.1007/s12284-009-9022-2 [DOI] [Google Scholar]
- Kobayashi K., Maekawa M., Miyao A., Hirochika H., Kyozuka J. (2010). PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol. 51 47–57. 10.1093/pcp/pcp166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Yasuno N., Sato Y., Yoda M., Yamazaki R., Kimizu M., et al. (2012). Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 24 1848–1859. 10.1105/tpc.112.097105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Chujo A., Nagato Y., Shimamoto K., Kyozuka J. (2003). FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 130 3841–3850. 10.1242/dev.00564 [DOI] [PubMed] [Google Scholar]
- Lee D., An G. (2012). Two AP2 family genes, Supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J. 69 445–461. 10.1111/j.1365-313X.2011.04804.x [DOI] [PubMed] [Google Scholar]
- Lee D., Lee J., Moon S., Park S., An G. (2007). The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. Plant J. 49 64–78. 10.1111/j.1365-313X.2006.02941.x [DOI] [PubMed] [Google Scholar]
- Li F., Liu W., Tang J., Chen J., Tong H., Hu B., et al. (2010). Rice DENSE AND ERECT PANICLE 2 is essential for determining panicle outgrowth and elongation. Cell Res. 20 838–849. 10.1038/cr.2010.69 [DOI] [PubMed] [Google Scholar]
- Li H., Liang W., Hua Y., Zhu L., Yin C., Xu J., et al. (2011). Rice MADS6 interacts with the floral homeotic genes SUPERWOMAN1, MADS3, MADS58, MADS3, and DROOPING LEAF in specifying floral organ identities and meristem fate. Plant Cell 23 2536–2552. 10.1105/tpc.111.087262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liang W., Jia R., Yin C., Zong J., Kong H., et al. (2010). The AGL6-like gene OsMADS6 regulates floral organ and meristem identities in rice. Cell Res. 20 299–313. 10.1038/cr.2009.143 [DOI] [PubMed] [Google Scholar]
- Li H., Xue D., Gao Z., Yan M., Xu W., Xin Z., et al. (2009). A putative lipase gene EXTRA GLUME1 regulates both empty-glume fate and spikelet development in rice. Plant J. 57 593–605. 10.1111/j.1365-313X.2008.03710.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Fan C., Xing Y., Jiang Y., Luo L., Sun L., et al. (2011). Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 243 1266–1270. 10.1038/ng.977 [DOI] [PubMed] [Google Scholar]
- Lin X., Wu F., Du X., Shi X., Liu Y., Liu S., et al. (2014). The pleiotropic SEPALLATA-like gene OsMADS34 reveals that the ‘empty glumes’ of rice (Oryza sativa) spikelets are in fact rudimentary lemmas. New Phytol. 202 689–702. 10.1111/nph.12657 [DOI] [PubMed] [Google Scholar]
- Liu C., Xi W., Shen L., Tan C., Yu H. (2009). Regulation of floral patterning by flowering time genes. Dev. Cell 16 711–722. 10.1016/j.devcel.2009.03.011 [DOI] [PubMed] [Google Scholar]
- Liu L., Tong H., Xiao Y., Che R., Xu F., Hu B., et al. (2015). Activation of Big Grain 1 significantly improves grain size by regulating auxin transport in rice. Proc. Natl. Aacd. Sci. U.S.A. 112 11102–11107. 10.1073/pnas.1512748112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Hua L., Dong S., Chen H., Zhu X., Jiang J., et al. (2015). OsMAPK6, a mitogen-activated protein kinase, influences rice grain size and biomass production. Plant J. 84 672–681. 10.1111/tpj.13025 [DOI] [PubMed] [Google Scholar]
- Malcomber S., Kellogg E. (2004). Heterogeneous expression patterns and separate roles of the SEPALLATA gene LEAFY HULL STERILE1 in grasses. Plant Cell 16 1692–1706. 10.1105/tpc.021576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcomber S., Kellogg E. (2005). SEPALLATA gene diversification: brave new whorls. Trends Plant Sci. 10 427–435. 10.1016/j.tplants.2005.07.008 [DOI] [PubMed] [Google Scholar]
- Mao H., Sun S., Yao J., Wang C., Yu S., Xu C., et al. (2010). Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Aacd. Sci. U.S.A. 107 19579–19584. 10.1073/pnas.1014419107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa N., Miyoshi M., Sano Y., Satoh H., Hirano H., Sakai H., et al. (2003). SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130 705–718. 10.1242/dev.00294 [DOI] [PubMed] [Google Scholar]
- Ohmori S., Kimizu M., Sugita M., Miyao A., Hirochika H., Uchida E., et al. (2009). MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 21 3008–3025. 10.1105/tpc.109.068742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S., Ditta G., Baumann E., Wisman E., Yanofsky M. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405 200–203. 10.1038/35012103 [DOI] [PubMed] [Google Scholar]
- Pelucchi N., Fornara F., Favalli C., Masiero S., Lago C., Pè M., et al. (2002). Comparative analysis of rice MADS-box genes expressed during flower development. Sex. Plant Reprod. 15 113–122. 10.1007/s00497-002-0151-7 [DOI] [Google Scholar]
- Qiao Y., Piao R., Shi J., Lee S., Jiang W., Kim B., et al. (2011). Fine mapping and candidate gene analysis of dense and erect panicle 3 , DEP3, which confers high grain yield in rice (Oryza sativa L.). Theor. Appl. Genet. 122 1439–1449. 10.1007/s00122-011-1543-6 [DOI] [PubMed] [Google Scholar]
- Rao Y., Yang Y., Xu J., Li X., Leng Y., Dai L., et al. (2015). EARLY SENESCENCE1 encodes a SCAR-LIKE PROTEIN2 that affects water loss in rice. Plant Physiol. 169 1225–1239. 10.1104/pp.15.00991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Li Y., Wang Z., Xu F., Guo S., Zhao F., et al. (2012). Identification and gene mapping of a multi-floret spikelet 1 (mfs1) mutant associated with spikelet development in rice. J. Integr. Agric. 11 1574–1579. 10.1016/S2095-3119(12)60160-9 [DOI] [Google Scholar]
- Ren D., Li Y., Zhao F., Sang X., Shi J., Wang N., et al. (2013). MULTI-FLORET SPIKELET1, which encodes an AP2/ERF protein, determines spikelet meristem fate and sterile lemma identity in rice. Plant Physiol. 162 872–884. 10.1104/pp.113.216044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Rao Y., Huang L., Leng Y., Hu J., Lu M., et al. (2016). Fine mapping identifies a new QTL for brown rice rate in rice (Oryza Sativa L.). Rice 9 4 10.1186/s12284-016-0076-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Rao Y., Wu L., Xun Q., Li Z., Yu H., et al. (2015). The pleiotropic ABNORMAL FLOWER AND DWARF1 affects plant height, floral development and grain yield in rice. J. Integr. Plant Biol. 58 529–539. 10.1111/jipb.12441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsley S., Ditta G., Yanofsky M. (1995). Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 7 1259–1269. 10.1105/tpc.7.8.1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R., Ambrose B. (1998). The blooming of grass flower development. Curr. Opin. Plant Biol. 1 60–67. 10.1016/S1369-5266(98)80129-5 [DOI] [PubMed] [Google Scholar]
- Song X., Huang W., Shi M., Zhu M., Lin H. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39 623–630. 10.1038/ng2014 [DOI] [PubMed] [Google Scholar]
- Su Y., Rao Y., Hu S., Yang Y., Gao Z., Zhang G., et al. (2011). Map-based cloning proves qGC-6, a major QTL for gel consistency of japonica/indica cross, responds by Waxyin rice (Oryza sativa L.). Theor. Appl. Genet. 123 859–867. 10.1007/s00122-011-1632-6 [DOI] [PubMed] [Google Scholar]
- Takeoka Y., Shimizu M., Wada T. (1993). “Panicles,” in Science of the Rice Plant, Vol. I, eds Matsuo T., Hoshikawa K. (Tokyo: Nobunkyo; ), 295–326. [Google Scholar]
- Terrell E., Peterson P., Wergin W. (2001). Epidermal features and spikelet micromorphology in Oryza and related genera (Poaceae: Oryzeae). Smithsonian Contr. Bot. 91 1–50. 10.5479/si.0081024X.91 [DOI] [Google Scholar]
- Theissen G., Saedler H. (2001). Plant biology. Floral quartets. Nature 409 469–471. 10.1038/35054172 [DOI] [PubMed] [Google Scholar]
- Tian Z., Qian Q., Liu Q., Yan M., Liu X., Yan C., et al. (2009). Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc. Natl. Aacd. Sci. U.S.A. 106 21760–21765. 10.1073/pnas.0912396106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E., Wang J., Zhu X., Hao W., Wang L., Li Q., et al. (2008). Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 40 1370–1374. 10.1038/ng.220 [DOI] [PubMed] [Google Scholar]
- Wang K., Tang D., Hong L., Xu W., Huang J., Li M., et al. (2010). DEP and AFO regulate reproductive habit in rice. PLoS Genet. 6:e1000818 10.1371/journal.pgen.1000818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Li S., Liu Q., Wu K., Zhang J., Wang S., et al. (2015). The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 47 949–954. 10.1038/ng.3352 [DOI] [PubMed] [Google Scholar]
- Wang S., Wu K., Yuan Q., Liu X., Liu Z., Lin X., et al. (2012). Control of grain size,shape and quality by OsSPL16 in rice. Nat. Genet. 44 950–955. 10.1038/ng.2327 [DOI] [PubMed] [Google Scholar]
- Wang Y., Xiong G., Hu J., Jiang L., Yu H., Xu J., et al. (2015). Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 47 944–948. 10.1038/ng.3346 [DOI] [PubMed] [Google Scholar]
- Wu X., Tang D., Li M., Wang K., Cheng Z. (2013). Loose Plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot Gravitropism, regulates plant architecture in rice. Plant Physiol. 161 317–329. 10.1104/pp.112.208496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Hirano H. (2006). Function and diversification of MADS-box genes in rice. Scientific World J. 6 1923–1932. 10.1100/tsw.2006.320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Lee D., Miyao A., Hirochika H., An G., Hirano H. (2006). Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell 18 15–28. 10.1105/tpc.105.037200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A., Ohmori Y., Kitano H., Taguchi-Shiobara F., Hirano H. (2012). ABERRANT SPIKELET AND PANICLE1, encoding a TOPLESS-related transcriptional co-repressor, is involved in the regulation of meristem fate in rice. Plant J. 70 327–339. 10.1111/j.1365-313X.2011.04872.x [DOI] [PubMed] [Google Scholar]
- Yoshida A., Suzaki T., Tanaka W., Hirano H. (2009). The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proc. Natl. Aacd. Sci. U.S.A. 106 20103–20108. 10.1073/pnas.0907896106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun D., Liang W., Dreni L., Yin C., Zhou Z., Kater M., et al. (2013). OsMADS16 genetically interacts with OsMADS3 and OsMADS58 in specifying floral patterning in rice. Mol. Plant 6 743–756. 10.1093/mp/sst003 [DOI] [PubMed] [Google Scholar]
- Zanis M. (2007). Grass spikelet genetics and duplicate gene comparisons. Int. J. Plant Sci. 168 93–110. 10.1086/509787 [DOI] [Google Scholar]
- Zhang D., Yuan Z. (2014). Molecular control of grass inflorescence development. Annu. Rev. Plant Biol. 65 14.1–14.26 10.1146/annurev-arplant-050213-040104 [DOI] [PubMed] [Google Scholar]
- Zuo J., Li J. (2013). Molecular dissection of complex agronomic traits of rice: a team effort by Chinese scientists in recent years. Natl. Sci. Rev. 1 253–276. 10.1093/nsr/nwt004 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.