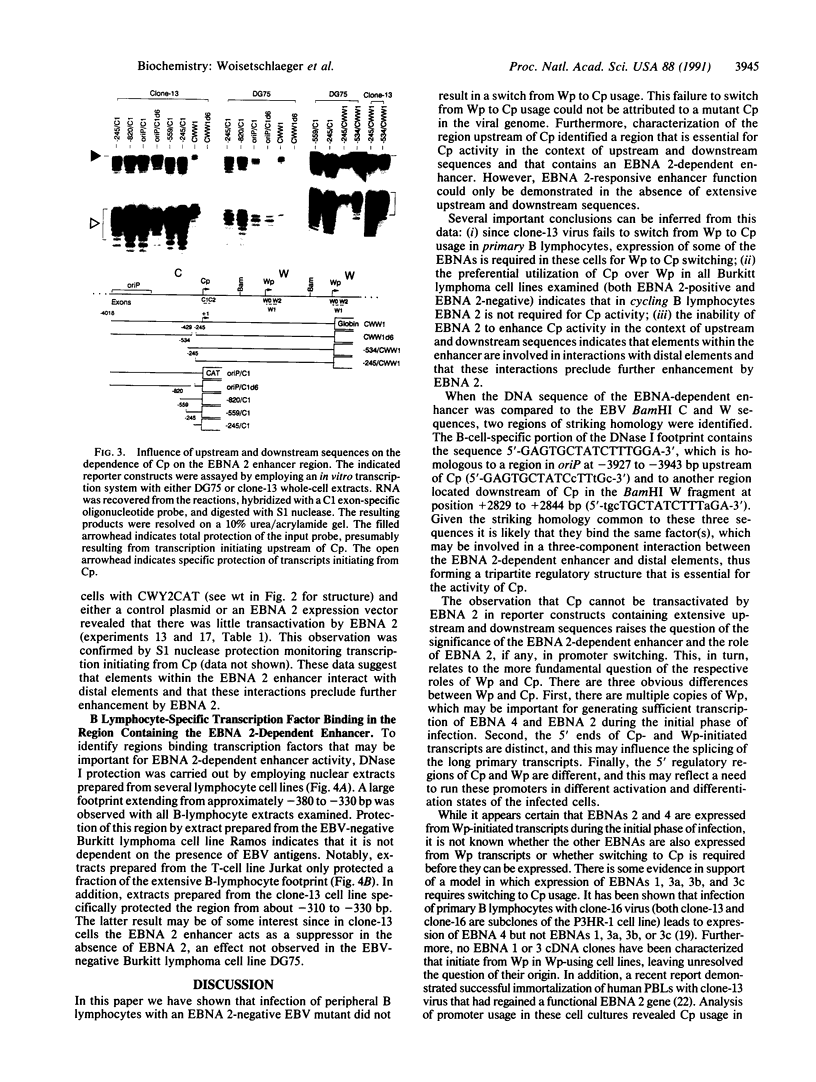
Abstract
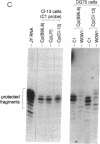
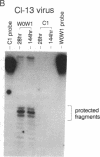
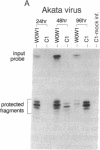
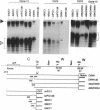
During latent Epstein-Barr virus (EBV) infection of human B lymphocytes, six viral nuclear antigen (EBNAs) are expressed from long primary transcripts by means of alternative splicing and alternative polyadenylylation sites. These transcripts initiate from one of two promoters, Cp or Wp, that function in a mutually exclusive fashion. Wp is exclusively utilized during the initial stages of infection of primary B lymphocytes, followed by a switch to Cp usage. These studies have been extended to show that (i) a mutant EBV strain lacking the gene encoding EBNA 2 fails to switch from Wp to Cp usage in primary B lymphocytes, although the virus contains a functional Cp; (ii) a region from -429 to -245 base pairs upstream of Cp is essential for Cp activity in B lymphocytes, but only in the context of upstream and downstream sequences; (iii) this region contains an EBNA 2-dependent enhancer; and (iv) DNase I protection employing nuclear extracts from B and T lymphocytes revealed a B-cell-specific footprint in the region of the EBNA 2-dependent enhancer. These results support a model for viral promoter switching during the initial stages of infection in which Wp activity leads to the expression of EBNA 2, followed by activation of Cp through the EBNA 2-dependent enhancer.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Ben-Bassat H., Goldblum N., Mitrani S., Goldblum T., Yoffey J. M., Cohen M. M., Bentwich Z., Ramot B., Klein E., Klein G. Establishment in continuous culture of a new type of lymphocyte from a "Burkitt like" malignant lymphoma (line D.G.-75). Int J Cancer. 1977 Jan;19(1):27–33. doi: 10.1002/ijc.2910190105. [DOI] [PubMed] [Google Scholar]
- Brandsma J., Miller G. Nucleic acid spot hybridization: rapid quantitative screening of lymphoid cell lines for Epstein-Barr viral DNA. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6851–6855. doi: 10.1073/pnas.77.11.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemington E., Speck S. H. Identification of phorbol ester response elements in the promoter of Epstein-Barr virus putative lytic switch gene BZLF1. J Virol. 1990 Mar;64(3):1217–1226. doi: 10.1128/jvi.64.3.1217-1226.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J., Walker L., Guy G., Brown G., Rowe M., Rickinson A. Control of human B-lymphocyte replication. II. Transforming Epstein-Barr virus exploits three distinct viral signals to undermine three separate control points in B-cell growth. Immunology. 1986 Aug;58(4):591–595. [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai T. W., Horikoshi M., Roeder R. G., Green M. R. Analysis of the role of the transcription factor ATF in the assembly of a functional preinitiation complex. Cell. 1988 Sep 23;54(7):1043–1051. doi: 10.1016/0092-8674(88)90119-5. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt W., Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature. 1989 Aug 3;340(6232):393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- Jeang K. T., Hayward S. D. Organization of the Epstein-Barr virus DNA molecule. III. Location of the P3HR-1 deletion junction and characterization of the NotI repeat units that form part of the template for an abundant 12-O-tetradecanoylphorbol-13-acetate-induced mRNA transcript. J Virol. 1983 Oct;48(1):135–148. doi: 10.1128/jvi.48.1.135-148.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., Mitchell P., Tjian R. Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell. 1987 Jun 19;49(6):741–752. doi: 10.1016/0092-8674(87)90612-x. [DOI] [PubMed] [Google Scholar]
- Rabson M., Gradoville L., Heston L., Miller G. Non-immortalizing P3J-HR-1 Epstein-Barr virus: a deletion mutant of its transforming parent, Jijoye. J Virol. 1982 Dec;44(3):834–844. doi: 10.1128/jvi.44.3.834-844.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricksten A., Olsson A., Andersson T., Rymo L. The 5' flanking region of the gene for the Epstein-Barr virus-encoded nuclear antigen 2 contains a cell type specific cis-acting regulatory element that activates transcription in transfected B-cells. Nucleic Acids Res. 1988 Sep 12;16(17):8391–8410. doi: 10.1093/nar/16.17.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricksten A., Svensson C., Welinder C., Rymo L. Identification of sequences in Epstein-Barr virus DNA required for the expression of the second Epstein-Barr virus-determined nuclear antigen in COS-1 cells. J Gen Virol. 1987 Sep;68(Pt 9):2407–2418. doi: 10.1099/0022-1317-68-9-2407. [DOI] [PubMed] [Google Scholar]
- Rooney C., Howe J. G., Speck S. H., Miller G. Influence of Burkitt's lymphoma and primary B cells on latent gene expression by the nonimmortalizing P3J-HR-1 strain of Epstein-Barr virus. J Virol. 1989 Apr;63(4):1531–1539. doi: 10.1128/jvi.63.4.1531-1539.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skare J., Strominger J. L. Cloning and mapping of BamHi endonuclease fragments of DNA from the transforming B95-8 strain of Epstein-Barr virus. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3860–3864. doi: 10.1073/pnas.77.7.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung N. S., Kenney S., Gutsch D., Pagano J. S. EBNA-2 transactivates a lymphoid-specific enhancer in the BamHI C promoter of Epstein-Barr virus. J Virol. 1991 May;65(5):2164–2169. doi: 10.1128/jvi.65.5.2164-2169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada K., Ono Y. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J Virol. 1989 Jan;63(1):445–449. doi: 10.1128/jvi.63.1.445-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhorst C., Parham P., Mann D. L., Strominger J. L. Structure of HLA antigens: amino-acid and carbohydrate compositions and NH2-terminal sequences of four antigen preparations. Proc Natl Acad Sci U S A. 1976 Mar;73(3):910–914. doi: 10.1073/pnas.73.3.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woisetschlaeger M., Strominger J. L., Speck S. H. Mutually exclusive use of viral promoters in Epstein-Barr virus latently infected lymphocytes. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6498–6502. doi: 10.1073/pnas.86.17.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woisetschlaeger M., Yandava C. N., Furmanski L. A., Strominger J. L., Speck S. H. Promoter switching in Epstein-Barr virus during the initial stages of infection of B lymphocytes. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1725–1729. doi: 10.1073/pnas.87.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]