Abstract
Nonalcoholic fatty liver disease (NAFLD) is characterized by the accumulation of triglycerides (TG) as lipid droplets in the liver. Although lipid-metabolizing enzymes are considered important in NAFLD, the involvement of phospholipase D1 (PLD1) has not yet been studied. Here, we show that the genetic ablation of PLD1 in mice induces NAFLD due to an autophagy defect. PLD1 expression was decreased in high-fat diet-induced NAFLD. Subsequently, PLD1 deficiency led to an increase in hepatic TGs and liver weight. Autophagic flux was blocked in Pld1−/− hepatocytes, with decreased β-oxidation rate, reduced oxidation-related gene expression, and swollen mitochondria. The dynamics of autophagy was restored by treatment with the PLD product, phosphatidic acid (PA) or adenoviral PLD1 expression in Pld1−/− hepatocytes, confirming that lysosomal PA produced by PLD1 regulates autophagy. Notably, PLD1 expression in Pld1−/− liver significantly reduced hepatic lipid accumulation, compared with Pld1−/− liver. Thus, PLD1 plays an important role in hepatic steatosis via the regulation of autophagy.
Nonalcoholic fatty liver disease (NAFLD) is a chronic liver disease and a major hepatic health problem worldwide1. NAFLD is characterized by hepatic macrovesicular steatosis without any obvious cause of secondary fat accumulation, such as significant alcohol consumption2. It is strongly connected to metabolic syndromes such as obesity, hypertension, dyslipidemia, and insulin resistance3. Despite its prevalence and importance, the underlying mechanisms of NAFLD induction are poorly characterized.
One of the primary factors for hepatic steatosis is an imbalance of the lipid flux in the liver4. Various factors result in lipid accumulation in the liver, e.g., increased lipolysis in adipose tissues and/or high dietary fat intake, increased de novo hepatic lipid synthesis, a decrease in fat oxidation, and decreased hepatic very low-density lipoprotein (VLDL) secretion4,5. Furthermore, an autophagy defect in the liver induces hepatic steatosis, which is accompanied by a reduced β-oxidation rate6. Several genetic studies have shown that deficiencies in fatty acid-metabolizing enzymes result in NAFLD, and elevated levels of diacylglycerol (DAG) are an indicator of NAFLD-induced insulin resistance. These findings underscore the importance of lipid-metabolizing enzymes in NAFLD and subsequent insulin resistance7,8. Despite the importance of DAG in NAFLD and insulin resistance, the roles of other DAG-convertible lipids in NAFLD are not clear.
DAG is converted to phosphatidic acid (PA) by DAG kinases and the reverse reaction is catalyzed by PA phosphatases9. PA is a lipid second messenger involved in membrane transport and several important signaling cascades including mammalian target of rapamycin (mTOR). In addition, several PA species may be involved in insulin signaling10,11. Di-16:0 PA dampens protein kinase B (Akt) phosphorylation in insulin-stimulated hepatocytes by disrupting the interaction between mTOR and rapamycin-insensitive companion of mTOR (rictor)10. Additionally, 16:0/18:1 PA and 18:1/20:4 PA enhance hepatic glucose production in AGPAT2−/− mice by elevating the expression of glucose-6-phosphatase and phosphoenolpyruvate carboxykinase, resulting in hyperglycemia11. However, the functions of PA species from different enzymatic reactions in hepatic steatosis have not been demonstrated.
PLD1 hydrolyzes phosphatidylcholine (PC) to produce PA, which contains fatty acid chains with one or two unsaturated bonds12. The PA species produced by PLD1 (primarily 16:0/18:1 PA, 18:0/18:1 PA, and di-18:1 PA) activate mTOR complex 1 (mTORC1) in mitogenic stimulation, suggesting the unique role of PLD1-produced PA species in mTORC1 regulation13. In addition, the involvement of PLD1 in liver disease and insulin signaling was previously reported; nuclear ARF dependent PLD activity increases during S-Phase of rat liver regeneration14, and PLD1 plays a role in the development and progression of rat liver fibrosis15,16. Additionally, PLD1 is activated by insulin in rat hepatocytes17 and regulates insulin-stimulated fusion of localized glucose transporter type 4 (GLUT4)-containing vesicles to the plasma membrane, resulting in an increase in glucose uptake by the adipocyte18. Nevertheless, the precise roles of PLD1 and its product, PA in NAFLD and insulin resistance have not been examined.
Herein, we used Pld1−/− mice to investigate the role of PLD1 in NAFLD and its consequent insulin resistance. Our results strongly suggested that PLD1 deficiency impairs autophagy, resulting in the accumulation of lipids in the liver, without affecting insulin resistance.
Results
PLD1 is downregulated in NAFLD
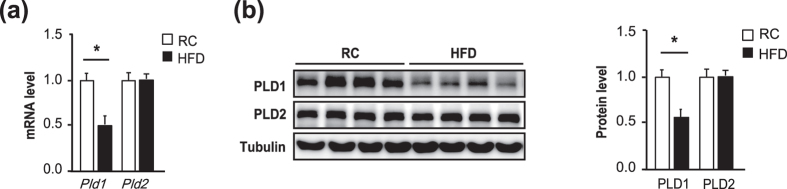
To examine the involvement of PLD in NAFLD, we compared the expression levels of PLD between the liver and other metabolism-related organs, such as skeletal muscle, epididymal fat, and brown adipose tissue (BAT). PLD1 and PLD2 were highly expressed in the liver, but not in other organs (Supplementary Fig. 1). Next, we compared Pld expression levels between the liver of high-fat diet (HFD)-fed mice with hepatic steatosis and mice fed regular chow (RC) without hepatic steatosis to validate the relevance of PLD in NAFLD. The Pld1 transcript (Fig. 1a) and protein (Fig. 1b) levels were significantly lower in mice with HFD-induced hepatic steatosis than in mice fed RC, whereas Pld2 expression did not differ between the groups. Thus, PLD1, but not PLD2, could be related to hepatic steatosis.
Figure 1. Pld1 expression is decreased in HFD-induced hepatic steatosis.
(a) C57BL/6 mice were fed either RC or a HFD for 4 weeks (n = 10 per group) starting at the age of 13 weeks and fasted overnight prior to collection of the liver. mRNA was isolated from the liver for qRT-PCR. (b) Mice were maintained as in (a) and the lysates were analyzed by western blot. Western blot results were analyzed by densitometry to obtain the relative ratio of either PLD1 or PLD2 to tubulin. The data are presented as means ± SE or representative blots from 3 to 5 independent experiments. *P < 0.05 versus the RC group. Abbreviations: HFD, high-fat diet; PLD2, phospholipase D2; RC, regular chow; SE, standard error; qRT-PCR quantitative real-time polymerase chain reaction.
PLD1 deficiency induces NAFLD
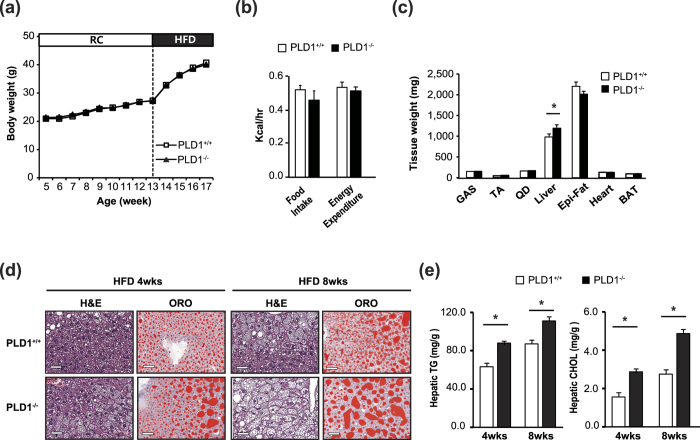
We used Pld1−/− mice to examine the role of PLD1 in hepatic steatosis. We used Pld1+/+ littermates as control throughout the study. The elimination of PLD1 protein expression in liver tissues was confirmed (Supplementary Fig. 2A), as shown previously19. Pld1−/− mice were fed HFD for 4 weeks starting at 13 weeks of age. The body weight of Pld1−/− mice did not differ from that of their Pld1+/+ littermates fed both, RC and a HFD (Fig. 2a). There were no differences in food intake and energy expenditure, as measured by CLAMS (Fig. 2b). Additionally, there was no difference in body composition (body weight, fat, and muscle) between Pld1+/+ mice and Pld1−/− mice (Supplementary Fig. 2B). To further analyze body composition, we measured the weight of individual tissues. As shown in Fig. 2c, the liver was significantly heavier in Pld1−/− mice than Pld1++ mice, whereas the weights of other tissues did not differ between mice. A histological analysis using hematoxylin and eosin (H&E) and Oil Red O staining showed enhanced intracellular vacuolation and lipid accumulation in both 4- and 8-week HFD-fed Pld1−/− mice (Fig. 2d). Consistent with these observations, hepatic TG and cholesterol increased by 26.5% and 60.4%, respectively, in 4-week HFD-fed Pld1−/− mice as well as by 22.4% and 75%, respectively, in 8-week HFD-fed Pld1−/− mice (Fig. 2e). In addition, there was no difference in the size of adipocytes in the epididymal fat (Supplementary Fig. 2C), suggesting that lipid accumulation was specific to the liver. Moreover, alanine transaminase (ALT) and aspartate transaminase (AST) activity, which are two of the most reliable markers of hepatocellular injury20, were 45.6% and 23.4% higher, respectively, in the serum of Pld1−/− mice than in Pld1+/+ mice, indicating that liver dysfunction occurred in the Pld1−/− mice (Table 1). These data indicate that PLD1 deficiency specifically induces lipid accumulation in the liver.
Figure 2. Pld1−/− mice have hepatic steatosis.
(a) Pld1+/+ and Pld1−/− littermates were fed HFD for 4 weeks starting at the age of 13 weeks (n = 9–11 per group). Body weights were measured. (b) The mice were fed HFD for 2 weeks starting at the age of 13 weeks. The food intake and energy expenditure of the mice were measured using indirect calorimetry over a 24-h period. (c–e) Pld1+/+ and Pld1−/− littermates were fed HFD for either 4 or 8 weeks starting at the age of 13 weeks (n = 9–11 per group). (c) The weights of each tissue were measured. (d) The livers were isolated and frozen sections were stained with H&E (left) and ORO (right). Representative images were obtained at 200× magnification. Scale bar, 50 μm. (e) Hepatic triglycerides and cholesterol were quantitatively analyzed. The data are presented as means ± SE or representative images from 3 to 5 independent experiments. *P < 0.05 versus the Pld1+/+ mice. Abbreviations: BAT, brown adipose tissue; GAS, gastrocnemius; HFD, high-fat diet; H&E, hematoxylin and eosin; ORO, Oil Red O; SE, standard error; TA, tibialis anterior; QD, quadriceps.
Table 1. Plasma lipid analyses.
| Group | WT | KO |
|---|---|---|
| (n = 9) | (n = 11) | |
| FFA (mEq/L) | 0.69 ± 0.04 | 0.74 ± 0.03 |
| TG (mg/dL) | 74.1 ± 4.9 | 64.9 ± 3.0 |
| Total Cholesterol (mg/dL) | 159.9 ± 6.4 | 159.3 ± 4.7 |
| HDL-Chol (mg/dL) | 108.8 ± 4.3 | 112.3 ± 2.4 |
| LDL-Chol (mg/dL) | 25.4 ± 1.9 | 27.6 ± 2.1 |
| ALT (U/L) | 47.4 ± 4.5 | 69.2 ± 7.9* |
| AST (U/L) | 75.3 ± 3.6 | 92.9 ± 6.7* |
*P < 0.05, when compared to the Pld1+/+.
Fat oxidation is decreased in the liver of Pld1 −/− mice
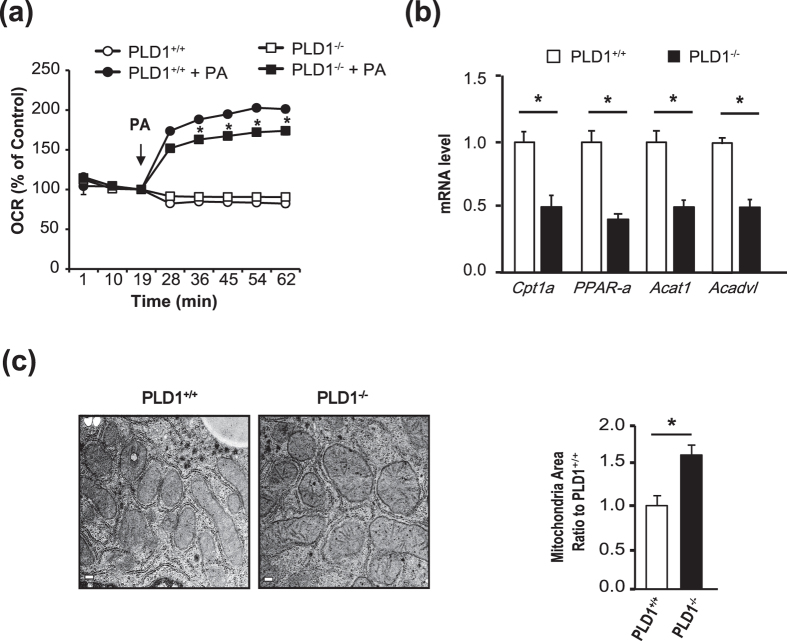
Hepatic lipid content is controlled by a balance between hepatic lipid uptake, lipid synthesis, fat oxidation (or lipolysis), and lipid export21, suggesting that NAFLD is caused by an imbalance in lipid supply and demand. To investigate the mechanism by which PLD1 deficiency induces NAFLD, we tested whether the Pld1−/− mice had a lipid flux defect. We did not observe differences in the expression of the relevant genes between Pld1−/− and Pld1+/+ mice (Supplementary Fig. 3A). The TG levels in Pld1−/− mice after the injection of the lipoprotein lipase inhibitor poloxamer during fasting were comparable to those in their Pld1+/+ littermates (Supplementary Fig. 3B), indicating that VLDL secretion was normal in Pld1−/− mice. Furthermore, the levels of genes involved in lipid synthesis and PA metabolism were not significantly altered in the Pld1−/− mice (Supplementary Fig. 3C). Next, we examined the oxygen consumption rate (OCR) in primary hepatocytes to assess mitochondrial function. As shown in Fig. 3a, the OCR did not differ between Pld1+/+ and Pld1−/− hepatocytes in the basal state; however, it was significantly lower in Pld1−/− hepatocytes than in Pld1+/+ hepatocytes after palmitic acid loading. Furthermore, the expression levels of fatty acid oxidation-related genes were significantly decreased in Pld1-null primary hepatocytes (Fig. 3b). Pld1−/− livers had swollen mitochondria under fasting conditions and a significant increase was observed in the mitochondrial area in Pld1−/− compared with Pld1+/+ mice (Fig. 3c). Together, these results strongly suggest that lipid accumulation in the Pld1-null liver results from decreased fat oxidation and impaired mitochondrial function.
Figure 3. The fat oxidation rate is reduced in the livers of Pld1−/− mice.
(a) Primary hepatocytes were isolated from Pld1+/+ and Pld1−/− littermates, serum starved, and treated with bovine serum albumin/palmitate complexes for 2 h. The oxygen consumption rate was measured at the indicated time points (n = 5 per each group). (b) The Pld1+/+ and Pld1−/− littermates were fed a HFD for 4 weeks starting at the age of 13 weeks (n = 9–11 per group) and qRT-PCR was performed. (c) The mice were maintained as described in (b) and the livers were collected. The mitochondria in the livers from Pld1+/+ and Pld1−/− littermates were visualized by transmission electron microscopy. The mitochondrial area was quantitatively measured using ImageJ (n = 3 mice per group). At least 15 pictures per group were evaluated. Scale bar, 0.2 μm. The data are presented as means ± SE or representative images from 3 to 5 independent experiments. *P < 0.05 versus the Pld1+/+ group. Abbreviations: HFD, high-fat diet; PA, palmitic acid; qRT-PCR, quantitative real-time polymerase chain reaction; SE, standard error.
Pld1 −/− mice have an autophagy defect
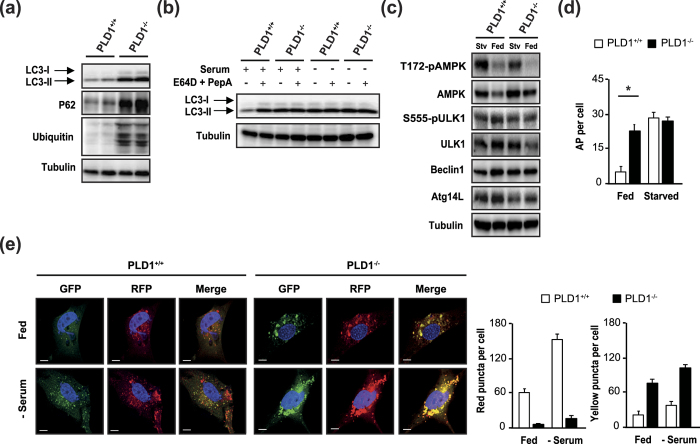
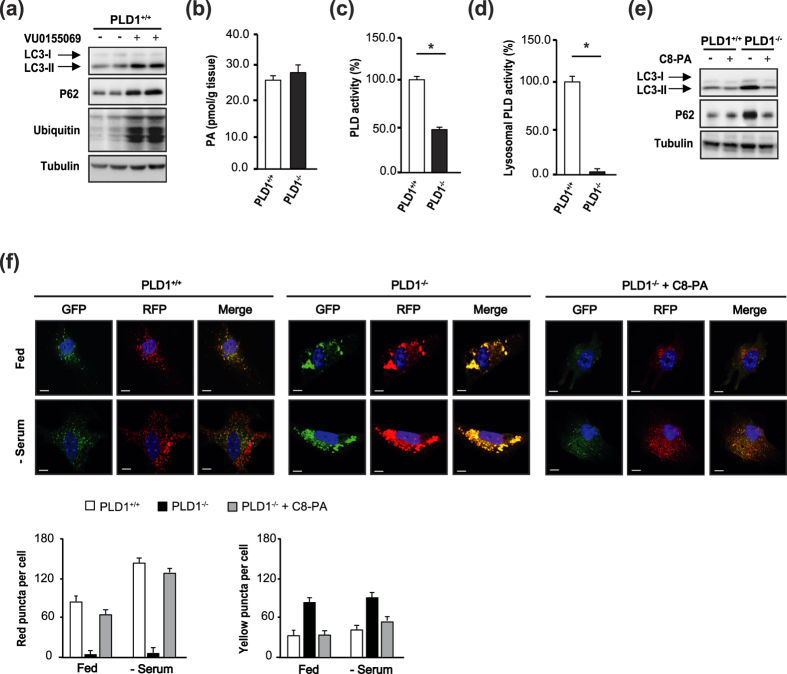
Autophagy regulates hepatic lipid stores in vivo, and defects in lipolysis are accompanied by defects in autophagy6. Autophagy-related gene (ATG)-deleted mouse models exhibit mitochondrial swelling and reduced respiration in the liver22, similar to our observations in Pld1−/− liver. In addition, PLD1 is involved in macroautophagy19. Therefore, we examined autophagy flux in Pld1−/− mice to determine the mechanism underlying NAFLD. Surprisingly, microtubule-associated protein 1 light chain 3 alpha (LC3)-I/II, P62, and polyubiquitinated protein expression increased in hepatocytes isolated from Pld1−/− mice (Fig. 4a), indicating a decrease in autophagic protein degradation. To validate the defect in autophagic degradation in Pld1−/− hepatocytes, we treated cells with the lysosomal protease inhibitors E64D and pepstatin A (PepA). The LC3-I/II levels in Pld1+/+ hepatocytes were increased by E64D and PepA treatment in the serum-fed state to levels similar to those observed for serum starvation (Fig. 4b). In contrast, the elevated LC3-I/II levels in Pld1−/− hepatocytes in the serum-fed state were not increased upon serum starvation or by E64D and PepA treatment, indicating that autophagic flux was impaired in Pld1−/− mice (Fig. 4b). However, PLD1 deficiency did not alter autophagy signaling, as shown by the expression level of autophagic regulators such as ATG14L and Beclin1, and the phosphorylation of ULK1 at serine 555 and AMPK at threonine 172 (Fig. 4c). These results suggest that the PLD1 deficiency did not regulate autophagy by regulating the autophagic signaling. In addition, an electron microscopy analysis of the liver showed that autolysosomes (dense structures with membranes) were reduced in Pld1−/− mice and autophagosomes (AP) with lipids (light structures with membranes) were primarily observed in Pld1−/− mice (Supplementary Fig. 4). In agreement with these results, the number of APs was higher in liver of fed Pld1−/− mice (Fig. 4d), indicating that PLD1 deficiency decreased the fusion of APs to lysosomes, resulting in AP accumulation. To confirm the defect in autolysosome formation in Pld1−/− mouse embryonic fibroblasts (MEFs), we attempted to differentiate between APs and autolysosomes using an RFP-GFP-LC3 construct. Because RFP is more stable than GFP under acidic conditions, only RFP maintains its fluorescence upon AP and lysosome fusion, resulting in an increase in red puncta. Red puncta were observed in Pld1+/+ MEFs in the serum-fed state, establishing a basal autolysosome level (Fig. 4e). Serum starvation increased the numbers of both red and yellow (merge of green and red) puncta, indicating increases in both APs and autolysosomes. However, the Pld1−/− MEFs contained only a few red puncta and many large-sized yellow aggregates in both serum-fed and -starved states (Fig. 4e). These results demonstrate that Pld1−/− MEFs had a defect in the fusion of autolysosomes, rather than in the formation of APs.
Figure 4. Autophagy is impaired in the livers of Pld1−/− mice.
(a) Primary hepatocytes were isolated from Pld1+/+ and Pld1−/− littermates, cultured in a serum-fed state, and the cell lysates were analyzed by western blotting. (b) Primary hepatocytes isolated as described in (a) were treated with or without E64D (10 μg/ml) and PepA (10 μg/ml) under either serum-fed or serum-starved conditions for 24 h. The cell lysates were analyzed by western blot. (c) Livers were collected from Pld1+/+ and Pld1−/− littermates were fed HFD for 4 weeks (n = 9–11 per group) under either fed or starved conditions for 24 h. Liver lysates were analyzed by western blot. (d) Livers were collected as described in (c). APs were quantitatively analyzed in transmission electron microscopic images of the liver. (e) Pld1+/+ and Pld1−/− MEF were infected with RFP-GFP-LC3B (60 particles per cell) for 24 h and then assigned to either serum-fed or serum-starved conditions. Representative confocal microscopy images are shown. GFP-negative/RFP-positive and GFP-positive/RFP-positive puncta were counted using ZEN software; more than 80–90 cells were included for each of 3 independent experiments. Scale bar, 20 μm. The data are presented as means ± SE or representative blots from 3 to 5 independent experiments. *P < 0.05 versus Pld1+/+ mice. Abbreviations: GFP, green fluorescence protein; HFD; MEF, mouse embryonic fibroblasts; PepA, pepstatin A; RFP, red fluorescence protein; SE, standard error.
The PLD1 product PA regulates autophagy
To determine whether PLD1 activity is required to regulate autophagy in the liver, we treated primary hepatocytes with the PLD1-specific inhibitor VU0155069. As shown in Fig. 5a, LC3-I/II, P62, and polyubiquitinated proteins were elevated in VU0155069-treated Pld1+/+ hepatocytes, consistent with the observations in Pld1−/− hepatocytes. Next, we tested whether PLD1 deficiency indeed decreased PA levels in cells. Unexpectedly, neither PA (Fig. 5b, Supplementary Fig. 5A) nor other anionic phospholipid levels (Supplementary Fig. 5B) were altered in Pld1−/− liver as compared to controls. We measured PLD activity in the liver and lysosomal fractions of the liver (the primary site of PLD1 localization23 and autophagolysosome formation). PLD activity in Pld1−/− liver was not completely eliminated owing to PLD2 activity (Fig. 5c). Notably, PLD activity was almost completely dampened in the lysosomal fraction of the Pld1−/− liver (Fig. 5d), suggesting that PA in lysosomes is predominantly produced by PLD1, and not by PLD2, and PLD1-produced PA may play a critical role in the regulation of autophagy. To confirm that PA is required for autophagic flux, we treated Pld1−/− hepatocytes in the serum-fed state with C8-PA. C8-PA treatment restored the autophagic flux in Pld1−/− hepatocytes, as evidenced by decreases in LC3-I/II and P62 to levels similar to those of Pld1+/+ hepatocytes (Fig. 5e). Consistent with this observation, C8-PA treatment increased the number of red puncta in the Pld1−/− MEFs and reduced the number of large yellow aggregates (Fig. 5f), confirming that PA is critical for autophagy.
Figure 5. PA restores autophagy.
(a) Primary hepatocytes were isolated from Pld1+/+ and Pld1−/− littermates, treated with 5 μM VU0155069 in serum-fed conditions for 24 h, and the lysates were analyzed by western blot. (b) The Pld1+/+ and Pld1−/− littermates were fed HFD for 4 weeks starting at the age of 13 weeks (n = 3 per group), livers were collected, and total phosphatidic acid levels were measured by LC-MS/MS. (c,d) The mice were fed as described in (b). (c) The lysates and (d) the lysosomal fractions of livers were isolated and a PLD assay was performed using a PLD assay kit. (e) Primary hepatocytes were isolated from Pld1+/+ and Pld1−/− littermates and treated with or without 300 μM C8-PA in serum-fed conditions for 24 h; the lysates were analyzed by western blot. (f) Pld1+/+ and Pld1−/− MEFs were infected with RFP-GFP-LC3B (60 particles per cell) for 24 h. Pld1+/+ MEFs, Pld1−/− MEFs, and Pld1−/− MEFs treated with 300 μM C8-PA were maintained in either serum-fed or serum-starved conditions for 24 h. Representative confocal microscopy images from 3–5 independent experiments are shown. The average numbers of GFP-negative/RFP-positive or GFP- and RFP-positive cells were counted using ZEN software from three independent experiments (60 cells per experiment). The data are presented as means ± SE or representative blots from 3 to 5 independent experiments. *P < 0.05 versus Pld1+/+ mice. Abbreviations: C8-PA, C8-phosphatidic acid; GFP, green fluorescence protein; HFD, high-fat diet; LC-MS/MS, liquid chromatography–mass spectrometry/mass spectrometry; MEF, mouse embryonic fibroblasts; RFP, red fluorescence protein; SE, standard error.
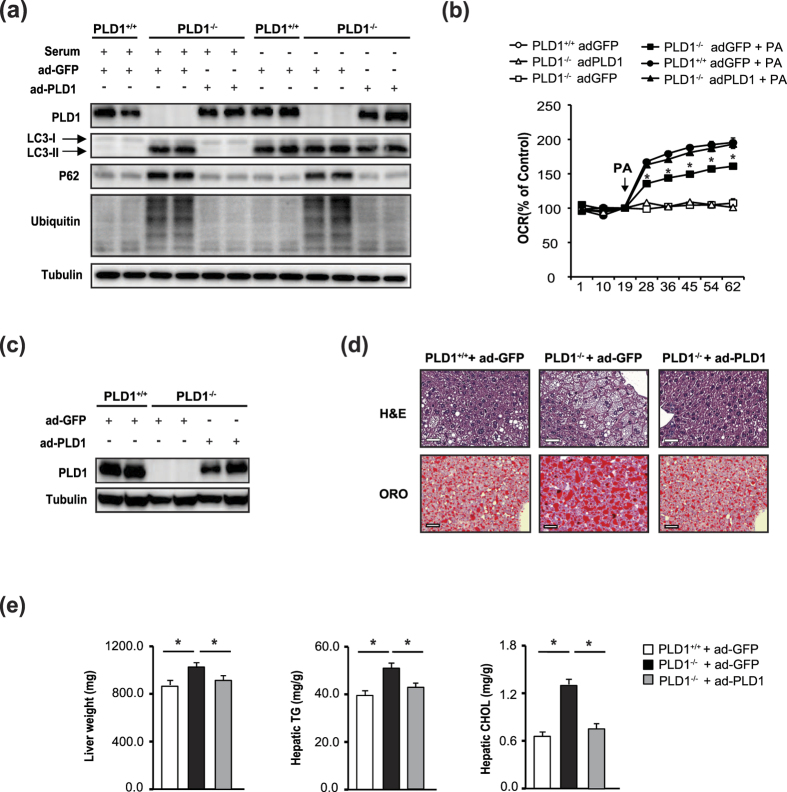
Hepatic PLD1 expression in Pld1 −/− mice attenuates hepatic steatosis
To clarify the role of PLD1 in autophagy and hepatic steatosis, we expressed PLD1 using adenoviral gene transfer. We used adenoviral GFP as a control to remove non-specific effects of adenoviral infection. Consistent with previous reports in which autophagic flux decreased24,25, LC3-I/II, P62, and ubiquitinated proteins in Pld1−/− hepatocytes were elevated in both serum-fed and serum-starved conditions, indicating the blockage of autophagic flux (Fig. 6a). In serum-starved conditions, autophagic flux in Pld+/+ hepatocytes increased; LC3-I/II increased, and there were no changes in the levels of P62 and ubiquitinated proteins compared to serum-fed conditions (Fig. 6a). However, PLD1 expression restored autophagic flux to a level similar to that in Pld1+/+ hepatocytes in both serum-fed and -starved conditions (Fig. 6a). PLD1 expression in Pld1−/− hepatocytes dampened the elevations in LC3-I/II, P62, and ubiquitinated protein expression in serum-fed conditions, and P62 and ubiquitinated proteins in serum-starved conditions, suggesting that PLD1 expression in Pld1−/− hepatocytes restored autophagic flux (Fig. 6a). Consistent with the augmentation of autophagy in PLD1-expressing Pld1−/− hepatocytes, the OCR was recovered in these cells (Fig. 6b), indicating that the defect in fat oxidation might be related to the autophagic defect in Pld1−/− hepatocytes. When PLD1 expression was restored in Pld1−/− livers using adenoviral gene transfer via tail vein injection (Fig. 6c), hepatic lipid accumulation was dramatically reduced (Fig. 6d), accompanied by significant decreases in liver weight, hepatic TGs, and hepatic cholesterol (Fig. 6e). Despite the successful PLD1 expression by adPLD1 in Pld1−/− livers, adPLD1 did not further induce PLD1 expression in Pld1+/+ livers, and was not accompanied by any changes in liver weight, hepatic TG, or cholesterol (Supplementary Fig. 6A,B). These results indeed confirm that PLD1 is a critical regulator of hepatic lipid accumulation by controlling autophagy.
Figure 6. Hepatic PLD1 expression in Pld1−/− mice attenuates hepatic steatosis.
(a) Primary hepatocytes were isolated from Pld1+/+ and Pld1−/− littermates, infected with adGFP or adPLD1 for 24 h, and treated with or without serum for 24 h. Western blot was performed with cell lysates. (b) Primary hepatocytes were infected with adGFP or adPld1 for 24 h, serum starved, and then treated with bovine serum albumin/palmitate complexes for 2 h. The OCR was measured at the indicated time points. (c) After feeding the mice with a HFD for one week beginning at 13 weeks, the mice were injected with adGFP or adPld1 (1 × 109 PFU) by tail vein injection. At 12 days post-injection, mice were sacrificed for analyses. Mouse liver lysates were analyzed by western blotting. (d) Mice were maintained as described in (c) and the livers were isolated, and frozen sections were stained with H&E (left) and ORO (right). Representative images were obtained at 200× magnification from 3–5 independent experiments. Scale bar, 50 μm. (e) Mice were maintained as described in (c); the weights of livers from mice were measured and hepatic triglycerides and cholesterol were quantitatively analyzed. The data are presented as means ± SE or representative blots from 3 to 5 independent experiments. *P < 0.05 versus Pld1+/+ mice. Abbreviations: adGFP, adenovirus-expressing green fluorescence protein; adPld1, adenovirus-expressing phospholipase D1; H&E, hematoxylin and eosin; HFD,; OCR, oxygen consumption rate; PA, palmitic acid; PFU, plaque-forming units; SE, standard error.
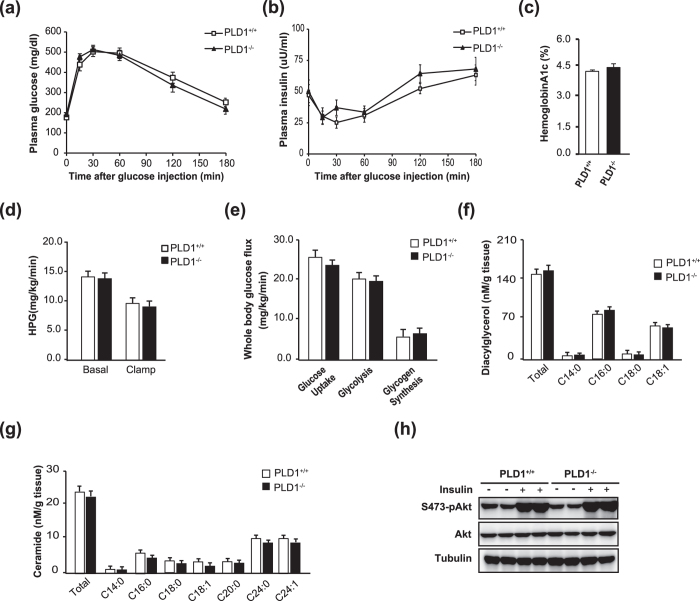
Pld1 −/− mice exhibit normal insulin sensitivity
Hepatic steatosis is associated with insulin resistance, which is a major factor in the pathogenesis of type 2 diabetes26. To explore the role of hepatic steatosis in insulin resistance in Pld1−/− mice, we analyzed the glucose response using an intraperitoneal glucose tolerance test (IPGTT). Glucose clearance was similar in Pld1+/+ and Pld1−/− mice (Fig. 7a). In addition, there were no significant differences in insulin levels during IPGTT (Fig. 7b) or hemoglobin A1c (Fig. 7c). To examine the differences in whole body and tissue-specific insulin sensitivities between Pld1+/+ and Pld1−/− mice, a hyperinsulinemic-euglycemic clamp study was conducted. The steady-state blood glucose level (Supplementary Fig. 7A), glucose infusion rate (Supplementary Fig. 7B), hepatic glucose production rate (Fig. 7d), and the rates of whole body glucose uptake, glycolysis, and glycogen synthesis (Fig. 7e) in Pld1−/− mice were not significantly different from those in Pld1+/+ mice. To better understand normal insulin sensitivity despite hepatic steatosis in Pld1−/− mice, we analyzed lipid metabolite levels in the livers of Pld1+/+ and Pld1−/− mice because elevated ceramide and DAG levels are associated with insulin resistance27,28,29. As shown in Fig. 7f,g, neither total DAG nor ceramide showed differences between the Pld1−/− and Pld1+/+ mice. Finally, insulin administration similarly increased Akt phosphorylation in the liver (Fig. 7h), skeletal muscle, and white adipose tissue (Supplementary Fig. 7C), confirming the comparable insulin sensitivity in Pld1−/− and Pld1+/+ mice. Taken together, our results demonstrate that hepatic steatosis in Pld1−/− mice is not coupled with insulin resistance.
Figure 7. Hepatic steatosis in Pld1−/− mice is dissociated from insulin resistance.
The mice were fed HFD for 4 weeks at 13 weeks. (a,b) The mice were fasted overnight and injected with glucose (IP, 1 g/kg body weight). (a) The plasma glucose level was analyzed at the indicated time points. (b) Plasma insulin was measured during IPGTT. (c) Hemoglobin A1c was measured in whole blood from mice. (d) Hepatic glucose production and (e) the whole body glucose flux were measured before and during clamp. (f) The ceramide and (g) diacylglycerol levels in the livers of Pld1+/+ and Pld1−/− mice were measured by LC/MS. (h) The mice were fasted overnight, injected with either saline or insulin (IP, 1.5 units/kg; n = 5 per group), and sacrificed 10 min after injection. Liver lysates were analyzed by western blot. The results are shown as means ± SE or representative blots from 3 to 5 independent experiments. *P < 0.05 versus Pld1+/+ mice. Abbreviations: IPGTT, intraperitoneal glucose tolerance test; LC/MS, liquid chromatography–mass spectrometry; SE, standard error.
Discussion
Although PLD has known roles in cancer and inflammation30, the relevance of PLD1 in NAFLD remains unclear. We provide the first evidence that PLD1 is a critical determinant of NAFLD. Specifically, the loss of PLD1 impaired autophagic flux via defects in the fusion between APs and lysosomes, resulting in NAFLD. Furthermore, we showed that PLD1 deficiency-induced NAFLD was not associated with obesity or insulin sensitivity.
Autophagy involves the delivery of intracellular cargo to lysosomes for degradation31. Basal autophagy performs a housekeeping function by removing damaged components. During nutrient starvation, autophagy supplies nutrients for survival by degrading proteins, lipids, and organelles. Hepatic autophagy breaks down lipid droplets to provide free fatty acids for oxidation to produce ATP in response to an increase in lipid availability or nutrient deprivation6. Defective autophagy in mice results in larger lipid droplets and increases hepatic triglyceride levels and gross liver size31, in agreement with our observations in Pld1−/− mice (Fig. 2). Fat accumulation in Pld1−/− liver was comparable to that in Vps34−/− 25 and Atg5−/− livers22. Indeed, autophagic flux and the fat oxidation (β-oxidation) rate were significantly reduced, whereas de novo fat synthesis, fat uptake, and VLDL secretion were unchanged in Pld1−/− mice. Decreased β-oxidation rate and swollen mitochondria, which were observed in Pld1−/− livers, are correlated with defective autophagy is other mouse models25, suggesting that defective autophagy in the Pld1−/− mice caused the mitochondrial abnormalities and subsequent lipid accumulation.
Previous studies have shown that PLD1 is a positive regulator of autophagy19,32, even though an independent study suggested that PLD1 negatively regulates autophagy via the coordination of important molecules in the autophagic pathway, AMPK-mTOR-ULK1 and Vps34/Beclin 133. Dall’Armi et al. proposed that PLD1 functions as a regulator of AP dynamics during starvation-induced autophagy19. They observed that a significant pool of PLD1 localizes on LAMP1-positive late endosomes/lysosomes in normal medium, consistent with previous studies23,34. They found that under nutrient starved conditions, PLD1 is transferred from late endosomes/lysosomes to APs, and functions during maturation as an effector of Vps34, a class III PI3 kinase. Unlike their study, in which PLD1 localization on APs is important for AP maturation during autophagy, we found that PLD1 activity on lysosomes is critical for the regulation of autophagy in the liver. We observed that the decrease in lysosomal PLD activity in Pld1−/− livers dampened the fusion between APs and lysosomes, suggesting that PLD1-generated PA on lysosomes is required for AP-lysosome fusion (Figs 4 and 5). Indeed, PA treatment in Pld1-null hepatocytes increased fusion between the AP and the lysosome, as shown by the increase in the number of autophagolysosomes and decreases in LC3-II, P62, and ubiquitinated protein levels (Fig. 5). Nevertheless, it is possible that PA on APs is required for AP-lysosome fusion together with PA on lysosome, which is worthy of further investigation.
Recently, autophagy activation in POMC neurons was reported to elicit lipid utilization in peripheral tissues such as BAT and liver tissue, by activating both lipophagy and cytosolic lipases, suggesting that autophagy in the central nervous system regulates lipid accumulation in peripheral tissues35. They showed that autophagy signaling from the brain increases the expression of ATGs in BAT and liver tissues, and AMPK-induced ULK1 phosphorylation in BAT. PLD1 inhibition in the brain decreases autophagic flux32; therefore, it is possible that PLD1 depletion in the brain prompts lipid accumulation in Pld1−/− liver. However, we observed a defect in the fusion process between APs and lysosomes, without changes in the expression of Atg14L or Beclin1, or in the phosphorylation of AMPK and ULK1 (Fig. 4c), implying that lipid accumulation in Pld1−/− livers is not caused by autophagic defects from the central nervous system.
Based on the decrease in lysosomal PA in Pld1−/− mice, transient cellular PA kinetics36, and the absence of other lipid-converting enzymes in lysosomes37,38, the conversion of PC to PA by PLD1 may be the main determinant of lysosomal PA levels, rather than the conversion of other lipids such as DAG. Indeed, other PA-generating enzymes, such as GPAT/AGPAT and DGK, are localized on the endoplasmic reticulum and mitochondrial membranes37 and are unlikely to regulate the lysosomal PA level. In contrast to the lack of differences in PA levels observed in our deficiency model, the overexpression of Pld1 in a whole cell system increases the total PA level39. Elevated PA levels in Pld1-overexpressing hepatocytes inhibit Akt by disrupting mTORC239, which localizes to the plasma membrane40. Based on the different localization patterns of mTORC2 and PLD1, the lysosomal deficiency of PA probably does not regulate insulin-stimulated Akt via mTORC2.
NAFLD is associated with hepatic insulin resistance, which is a major risk factor for type 2 diabetes36. One potential mechanism underlying fat-induced hepatic insulin resistance is the accumulation of various lipid metabolites (especially DAG and ceramide) in the liver. This accumulation may activate several protein kinase cascades such as protein kinase C (PKC)-ε, resulting in the disruption of insulin signaling41,42,43,44. In our study, total content of DAG and ceramide, which are the most probable candidate lipid metabolites associated with insulin resistance, did not differ between Pld1−/− and Pld1+/+ liver (Fig. 7f,g). As expected, Akt phosphorylation in Pld1−/− liver did not differ from that in Pld1+/+ liver (Fig. 7h). These results provide another example of the dissociation between hepatic steatosis and insulin resistance and explain the underlying mechanisms45,46,47,48,49. Recently, Trujillo Viera et al. reported that Pld1−/− mice consume more food due to defects in the hypothalamus, which results in obesity50. They observed obvious obesity in 20-week old Pld1−/− mice, followed by increase in free fatty acid (FFA) levels and glucose levels in the blood of Pld1−/− mice. Hence, they proposed that elevation of FFA decreased insulin sensitivity in Pld1−/− mice. However, in our current study, we did not observe any changes in FFA (Table 1) and glucose levels (Supplementary Fig. 7A) in the blood of Pld1−/− mice or in insulin sensitivity in Pld1−/− liver (Fig. 7d,e,h) of Pld1−/− mice fed HFD for 4 weeks starting at 13 weeks of age. Since Trujillo Viera et al. proposed that FFA elevation due to obesity induced insulin resistance, we believe that lack of obesity in Pld1−/− mice in our study was responsible for the difference in our findings. The precise reasons for the discrepancies between our data and those of Trujillo Viera et al. are difficult to be certain of; all we can say is that they studied the mice at different age from ours and on a different diet formulation (Regular Chow versus High Fat Diet) and laboratory environment.
In summary, the results of this study demonstrate that a decrease in PA production by lysosomal PLD1 inhibits autolysosome formation in the Pld1−/− liver, leading to hepatic steatosis, without affecting insulin sensitivity. Thus, deciphering the specific function of lysosomal PLD1 is essential for the identification of new therapeutic strategies for hepatic steatosis.
Materials And Methods
Animals
Pld1−/− mice were generated and characterized as previously described19. Littermates that were homozygous for the PLD1 allele were used as controls. Mice were housed under controlled temperature (22 °C ± 2 °C) and 12-h light-dark cycle, and fed HFD (60 kcal% fat diet for 4 and 8 weeks; D12492 obtained from Research Diet Incorporation). All experimental protocols for animals, maintenance and care, were conducted according to Gachon University Animal Care guidelines. All animal procedures were approved by the Center of Animal Care and Use, Lee Gil Ya Cancer and Diabetes Institute, Gachon University and the Institutional Animal Care and Use Committee (IACUC) (Permission number: LCDI-2016-0008).
Basal study
Fat and lean body mass were assessed using an 1H minispec system (Bruker BioSpin) before and after 4 weeks on HFD. Activity, food consumption, and energy expenditure were evaluated using a metabolic monitoring system (CLAMS; Columbus Instruments, Columbus) for 4 days (2 days of acclimation followed by 2 days of measurement) before and after 2 weeks on HFD, as previously described by Choi et al.51. For the glucose tolerance test and lipoprotein inhibitor assay, mice were fasted overnight before either intraperitoneal injection of glucose (1.5 g/kg body weight) or Poloxamer 407 (1 g/kg body weight). Blood glucose and insulin were analyzed from tail-vein blood collected at the indicated time points. To examine the insulin signaling pathway, insulin (1.0 U/kg body weight) was injected into the peritoneal cavity. Liver, muscle, and epididymal fat were isolated 10 min after the injection. For the analyses of gene expression and lipid metabolites, mice were fasted for 16 h and the blood and tissue samples were immediately collected.
Hyperinsulinemic-euglycemic clamp
To assess whole body and hepatic insulin sensitivity, we performed a hyperinsulinemic-euglycemic clamp study as previously described by Choi et al.52. Indwelling catheters were placed in the internal jugular vein 7 days before the hyperinsulinemic-euglycemic clamp studies. After overnight fasting, [3-3H] glucose (Perkin Elmer) was infused at a rate of 0.05 μCi/min for 2 h to measure the basal glucose turnover. After 2 h, a hyperinsulinemic-euglycemic clamp was used for 150 min with an infusion of insulin (3 mU/kg/min). Blood samples (10 μl) were collected at the indicated time points to analyze plasma glucose, and 20% dextrose was infused at various rates to keep plasma glucose at basal levels (120 mg/dl). To assess whole body glucose fluxes, [3-3H] glucose was infused at a rate of 0.1 μCi/min throughout the clamps. To estimate insulin-stimulated glucose uptake and metabolism in peripheral tissues, 2-deoxy-d-[1-14C] glucose (Perkin Elmer) was administered as a bolus at 125 min throughout the clamps. To determine 3H and 14C activity levels in the plasma, blood samples were taken at the basal period and during the last 60 min of the clamp.
Isolation of lysosomal fractions and PLD activity assay
Lysosomal fractions were isolated from the liver using a Lysosome Isolation Kit (LYSISO1, Sigma Aldrich) according to the manufacturer instructions. PLD activity was measured using Phospholipase D Assay Kit (MAK137, Sigma Aldrich) according to the manufacturer instructions.
Preparation and culture of primary hepatocytes and mouse embryonic fibroblasts (MEFs)
To prepare MEFs, 13–15-d post-coital mouse embryos were minced and digested with trypsin. The cells were collected and cultured in Dulbecco’s modified Eagle’s medium with 4.5 mg/l glucose, 10% fetal bovine serum [FBS], 100 U/ml penicillin, and 100 μg/ml streptomycin. To prepare primary hepatocytes, 8 to 10-week-old male mice were anesthetized and their livers were isolated. Primary hepatocytes were prepared by 2-step perfusion using liver perfusion medium and liver digestion buffer (Life Technologies, 17701038, 17703034) as previously described53. Primary hepatocytes were cultured on tissue culture dishes in M199 medium (Sigma Aldrich, M4530) containing 10% FBS, 100 U/ml penicillin and streptomycin, and 10 nM dexamethasone.
Measurement of oxygen consumption rate
The rate of oxygen consumption (OCR) was measured using an extracellular Flux Analyzer XF-24 (Seahorse Biosciences). Primary mouse hepatocytes were seeded into 24-well plates (2 × 104/plate) and cultured overnight in M199 media with 10% FBS. Cells washed with Krebs-Henseleit buffer (111 mM NaCl, 4.7 mM KCl, 2 mM MgSO4, 1.2 mM Na2HPO4, 2.5 mM glucose, and 0.5 mM carnitine) and incubated in the same buffer for 1 h at 37 °C. Cells were then loaded onto the XF24 analyzer to measure the OCR in short and repeated intervals. After injection with BSA (17 μM) or BSA-complexed palmitic acid (final concentration 150 μM), mixing, waiting and OCR were measured at indicated time points.
Western blot analyses
Cells and tissue proteins were lysed in cell lysis buffer (Cell Signaling Technology, 9803) containing phosphatase inhibitors and protease inhibitors (Sigma Aldrich, P5726, P2714). Twenty micrograms of protein were used for western blot. The proteins were resolved on sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes. Antibodies were applied and incubated as per the manufacturer instructions. The horseradish peroxidase-conjugated secondary antibodies were detected using an enhanced chemiluminescent substrate (Millipore, P90720). The images were captured using an LAS4000 luminescent image analyzer (Fujifilm). Band intensities were quantified using Image J software (NIH).
Primary antibodies against the following were used: LC3 (Novus Biologicals, NB100-2220), ubiquitin (Covance, MMS-258R), and β-tubulin, (Abcam, ab108342), PLD2 (Santa Cruz Biotechnology, sc-25513), PLD1 (Cell Signaling Technology, #3832), p62/SQSTM1(Cell Signaling Technology, #5114), Akt (Cell Signaling Technology, #9272S), phospho-Akt (Ser473) (Cell Signaling Technology, #9271S), ULK1(Cell Signaling Technology, #8054), phospho-ULK1 (Ser555) (Cell Signaling Technology, #5869), AMPK (Cell Signaling Technology, #2532), phospho-AMPK (Thr172) (Cell Signaling Technology, #2535), Beclin1 (Bethyl laboratories, A302-567A), and Atg14L (Sigma Aldrich, A6358).
RNA isolation and quantitative real time polymerase chain reaction (PCR)
Total RNA from cells and tissues was extracted using TRIzol reagent RNA extraction kit (Life Technologies, 15596026) according to the manufacturer instructions and used to synthesize cDNA, using the TOPscriptTM RT DryMIX kit (dT18 plus) (Enzynomics, RT200). Real-time PCR analysis was performed with ABI7300 (Applied Biosystems Incorporation) using TOPrealTM qPCR 2X PreMIX (SYBR Green with high ROX) (Enzynomics, RT501S). To normalize the expression of the genes, cyclophilin A was used. The list of primer sequences is shown in Supplementary Table 1.
Lipid analyses
To extract ceramide, DAG and PA, 20 mg of liver was homogenized in cold methanol/phosphate-buffered saline (2:1 v/v) for ceramide and cold chloroform/methanol (2:1 v/v) for DAG and PA. Organic phase were separated from aqueous phase by adding chloroform and water. After centrifugation, the organic layer was collected, dried under nitrogen flow, and reconstituted with 0.1% formic acid in methanol for ceramide, hexane/methylene chloride/ether (95/4.5/0.5 v/v/v) for DAG, and methanol for PA. The internal standards for ceramide, DAG, and PA are C17:0 ceramide (500 ng/ml), 1, 3-dipenta decanoin and phosphatidic acid (C10:0/10:0), respectively. Diacylglycerol was separated from triacylglycerol using preconditioned columns (Waters SepPak Cartridge, WAT020845) and eluted with hexane/ethyl acetate (85/15 v/v) under a low negative pressure. Ceramide and DAG were measured by LC-MS/MS 4000 Q TRAP (Applied Biosystems). Phosphatidic acid was analyzed by Agilent 6490 Triple Quadrupole LC/MS System (Agilent Technologies). Anionic phospholipids (PI, cardiolipin, PS and phosphoinositides) were measured using HPLC combined with suppressed conductivity (Dionex), as previously described54.
Metabolic measurements
Blood glucose during hyperinsulinemic-clamp and glucose tolerance test was measured using the GM9 Glucose Analyzer (Analox Instruments). Lipids in tissue were extracted using the method of Bligh and Dyer55 and hepatic triglyceride, cholesterol and plasma NEFA were measured by colorimetric assay kits from Wako (Wako Pure Chemical Industries, 279-75401). Plasma triglyceride, total cholesterol, low-density lipoprotein (LDL), high-density lipoprotein (HDL), ALT, and AST were measured using a Cobas c111 analyzer (Roche). Plasma insulin was measured using Mouse Ultrasensitive Insulin ELISA kits (ALPCO, 80-INSMSU-E01).
Histology
Liver was collected and frozen in optimal cutting temperature embedding medium (Cell Path, KMA-0100-00A). Paraffin-embedded 5 μm thick sections were stained with H&E (Sigma Aldrich, H9627). Frozen liver sections were stained with Oil red O. The images were visualized by phase-contrast microscopy.
Fluorescent images
Cells were cultured on Lab-Tek II Chamber Slides (Nalgene Nunc International, Rochester, 154534). The PremoTM Autophagy Tandem Sensor RFP-GFP-LC3B Kit (Life Technologies, P36239) was used to monitor autophagy flux. The cells were treated, followed by fixation in a 4% formaldehyde solution. Fluorescent images were obtained using the Laser Scanning Microscope 700 (Carl Zeiss).
Transmission electron microscopy images
Liver tissues were quickly isolated from anaesthetized mice; they were then immediately fixed with 2.5% glutaraldehyde in 0.1 M phosphate-buffered saline for at least 2 h at 4 °C. The tissues were post-fixed at room temperature with 1% osmium tetroxide for 1.5 h. Tissue sections with a thickness of approximately 70–80 nm were prepared with an ultramicrotome (Leica) and collected on copper grids. The samples were analyzed with a Cryo-Tecnai F20 (Field Electron and Ion).
Statistical analyses
All data are presented as means ± standard error (SE). The significance of the differences in mean values between the Pld1+/+ and Pld1−/− mice was evaluated using unpaired Student’s t-test.
Additional Information
How to cite this article: Hur, J. H. et al. Phospholipase D1 deficiency in mice causes nonalcoholic fatty liver disease via an autophagy defect. Sci. Rep. 6, 39170; doi: 10.1038/srep39170 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We are very grateful to Kyeong-Hoon Jeong for maintaining the PLD1-null mice. We thank Yun Kyeong Nam and Kyu Hwan Shim for their technical assistance. This work was supported by the Bio and Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF-2014M3A9D5A01073886 to CC) and Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry for Health and Welfare, Korea (HI15C0987 and HI14C1135 to CC). This work was also supported by NIH grants R21 AG045020 (to GDP) and P50 AG008702 (pilot grant to CDA).
Footnotes
Author Contributions J.H.H., M.S.Y., D.S.M. and C.S.C. designed the study, were involved in all aspects of the experiments, and drafted the manuscript. S.Y.P., C.D.A., J.S.L., G.D.P., and H.Y.L. contributed to experiments and interpretation of data.
References
- Sanyal A. J. NASH: A global health problem. Hepatol Res 41, 670–674, doi: 10.1111/j.1872-034X.2011.00824.x (2011). [DOI] [PubMed] [Google Scholar]
- Chalasani N. et al. The diagnosis and management of non-alcoholic fatty liver disease: practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology 55, 2005–2023, doi: 10.1002/hep.25762 (2012). [DOI] [PubMed] [Google Scholar]
- Marchesini G. et al. Nonalcoholic fatty liver, steatohepatitis, and the metabolic syndrome. Hepatology 37, 917–923, doi: 10.1053/jhep.2003.50161 (2003). [DOI] [PubMed] [Google Scholar]
- Cohen J. C., Horton J. D. & Hobbs H. H. Human fatty liver disease: old questions and new insights. Science 332, 1519–1523, doi: 10.1126/science.1204265 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagle C. A., Klett E. L. & Coleman R. A. Hepatic triacylglycerol accumulation and insulin resistance. J Lipid Res 50 Suppl, S74–79, doi: 10.1194/jlr.R800053-JLR200 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. et al. Autophagy regulates lipid metabolism. Nature 458, 1131–1135, doi: 10.1038/nature07976 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbo T. & Shulman G. I. Lipid-induced hepatic insulin resistance. Aging (Albany NY) 5, 582–583 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumashiro N. et al. Cellular mechanism of insulin resistance in nonalcoholic fatty liver disease. Proc Natl Acad Sci USA 108, 16381–16385, doi: 10.1073/pnas.1113359108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster D. A. Phosphatidic acid and lipid-sensing by mTOR. Trends Endocrinol Metab 24, 272–278, doi: 10.1016/j.tem.2013.02.003 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. et al. Glycerolipid signals alter mTOR complex 2 (mTORC2) to diminish insulin signaling. Proc Natl Acad Sci USA 109, 1667–1672, doi: 10.1073/pnas.1110730109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankella S., Garg A., Horton J. D. & Agarwal A. K. Hepatic gluconeogenesis is enhanced by phosphatidic acid which remains uninhibited by insulin in lipodystrophic Agpat2 −/− mice. J Biol Chem 289, 4762–4777, doi: 10.1074/jbc.M113.530998 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt T. R., McDermott M., Saqib K. M., Shimwell N. & Wakelam M. J. Phospholipase D1b and D2a generate structurally identical phosphatidic acid species in mammalian cells. The Biochemical journal 360, 707–715 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon M. S. et al. Rapid mitogenic regulation of the mTORC1 inhibitor, DEPTOR, by phosphatidic acid. Mol Cell 58, 549–556, doi: 10.1016/j.molcel.2015.03.028 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banno Y. et al. Nuclear ADP-ribosylation factor (ARF)- and oleate-dependent phospholipase D (PLD) in rat liver cells. Increases of ARF-dependent PLD activity in regenerating liver cells. J Biol Chem 272, 5208–5213 (1997). [DOI] [PubMed] [Google Scholar]
- Zhu X. et al. The Role of Phospholipase D1 in Liver Fibrosis Induced by Dimethylnitrosamine In Vivo. Digestive Diseases and Sciences 59, 1779–1788, doi: 10.1007/s10620-014-3130-6 (2014). [DOI] [PubMed] [Google Scholar]
- Seo H. Y. et al. Phospholipase D1 decreases type I collagen levels in hepatic stellate cells via induction of autophagy. Biochem Biophys Res Commun 449, 38–43, doi: 10.1016/j.bbrc.2014.04.149 (2014). [DOI] [PubMed] [Google Scholar]
- Donchenko V., Zannetti A. & Baldini P. M. Insulin-stimulated hydrolysis of phosphatidylcholine by phospholipase C and phospholipase D in cultured rat hepatocytes. Biochim Biophys Acta 1222, 492–500 (1994). [DOI] [PubMed] [Google Scholar]
- Huang P., Altshuller Y. M., Hou J. C., Pessin J. E. & Frohman M. A. Insulin-stimulated plasma membrane fusion of Glut4 glucose transporter-containing vesicles is regulated by phospholipase D1. Mol Biol Cell 16, 2614–2623, doi: 10.1091/mbc.E04-12-1124 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Armi C. et al. The phospholipase D1 pathway modulates macroautophagy. Nat Commun 1, 142, doi: 10.1038/ncomms1144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko S. H., Baeg M. K., Han K. D., Ko S. H. & Ahn Y. B. Increased liver markers are associated with higher risk of type 2 diabetes. World J Gastroenterol 21, 7478–7487, doi: 10.3748/wjg.v21.i24.7478 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. J., Samuel V. T., Petersen K. F. & Shulman G. I. The role of hepatic lipids in hepatic insulin resistance and type 2 diabetes. Nature 510, 84–91, doi: 10.1038/nature13478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura A. et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev 25, 795–800, doi: 10.1101/gad.2016211 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon M. S., Du G., Backer J. M., Frohman M. A. & Chen J. Class III PI-3-kinase activates phospholipase D in an amino acid-sensing mTORC1 pathway. The Journal of cell biology 195, 435–447, doi: 10.1083/jcb.201107033 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M. et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. The Journal of cell biology 169, 425–434, doi: 10.1083/jcb.200412022 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaber N. et al. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci USA 109, 2003–2008, doi: 10.1073/pnas.1112848109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinella M. E. Nonalcoholic fatty liver disease: a systematic review. JAMA 313, 2263–2273, doi: 10.1001/jama.2015.5370 (2015). [DOI] [PubMed] [Google Scholar]
- Turinsky J., O’Sullivan D. M. & Bayly B. P. 1,2-Diacylglycerol and ceramide levels in insulin-resistant tissues of the rat in vivo. J Biol Chem 265, 16880–16885 (1990). [PubMed] [Google Scholar]
- Samuel V. T., Petersen K. F. & Shulman G. I. Lipid-induced insulin resistance: unravelling the mechanism. Lancet 375, 2267–2277, doi: 10.1016/S0140-6736(10)60408-4 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magkos F. et al. Intrahepatic diacylglycerol content is associated with hepatic insulin resistance in obese subjects. Gastroenterology 142, 1444–1446 e1442, doi: 10.1053/j.gastro.2012.03.003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D. W., Choi K. Y. & Min do S. Functional regulation of phospholipase D expression in cancer and inflammation. J Biol Chem 289, 22575–22582, doi: 10.1074/jbc.R114.569822 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz J. D. & White E. Autophagy and metabolism. Science 330, 1344–1348, doi: 10.1126/science.1193497 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae E. J. et al. Phospholipase D1 regulates autophagic flux and clearance of alpha-synuclein aggregates. Cell Death Differ 21, 1132–1141, doi: 10.1038/cdd.2014.30 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang Y. H., Choi K. Y. & Min D. S. Phospholipase D-mediated autophagic regulation is a potential target for cancer therapy. Cell Death Differ 21, 533–546, doi: 10.1038/cdd.2013.174 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown F. D. et al. Phospholipase D1 localises to secretory granules and lysosomes and is plasma-membrane translocated on cellular stimulation. Current Biology 8, 835–838, doi: http://dx.doi.org/10.1016/S0960-9822(98)70326-4 (1998). [DOI] [PubMed] [Google Scholar]
- Martinez-Lopez N. et al. Autophagy in the CNS and Periphery Coordinate Lipophagy and Lipolysis in the Brown Adipose Tissue and Liver. Cell Metab 23, 113–127, doi: 10.1016/j.cmet.2015.10.008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Vilella-Bach M., Bachmann R., Flanigan A. & Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science 294, 1942–1945, doi: 10.1126/science.1066015 (2001). [DOI] [PubMed] [Google Scholar]
- Takeuchi K. & Reue K. Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis. Am J Physiol Endocrinol Metab 296, E1195–1209, doi: 10.1152/ajpendo.90958.2008 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topham M. K. & Epand R. M. Mammalian diacylglycerol kinases: molecular interactions and biological functions of selected isoforms. Biochim Biophys Acta 1790, 416–424, doi: 10.1016/j.bbagen.2009.01.010 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. et al. Inhibited insulin signaling in mouse hepatocytes is associated with increased phosphatidic acid but not diacylglycerol. J Biol Chem 290, 3519–3528, doi: 10.1074/jbc.M114.602789 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partovian C., Ju R., Zhuang Z. W., Martin K. A. & Simons M. Syndecan-4 regulates subcellular localization of mTOR Complex2 and Akt activation in a PKCalpha-dependent manner in endothelial cells. Mol Cell 32, 140–149, doi: 10.1016/j.molcel.2008.09.010 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel V. T. & Shulman G. I. Mechanisms for insulin resistance: common threads and missing links. Cell 148, 852–871, doi: 10.1016/j.cell.2012.02.017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z. & Lazar M. A. Dissociating fatty liver and diabetes. Trends Endocrinol Metab 24, 4–12, doi: 10.1016/j.tem.2012.09.005 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaurasia B. & Summers S. A. Ceramides - Lipotoxic Inducers of Metabolic Disorders. Trends Endocrinol Metab 26, 538–550, doi: 10.1016/j.tem.2015.07.006 (2015). [DOI] [PubMed] [Google Scholar]
- Erion D. M. & Shulman G. I. Diacylglycerol-mediated insulin resistance. Nat Med 16, 400–402, doi: 10.1038/nm0410-400 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. X. et al. Antisense oligonucleotide reduction of DGAT2 expression improves hepatic steatosis and hyperlipidemia in obese mice. Hepatology 42, 362–371, doi: 10.1002/hep.20783 (2005). [DOI] [PubMed] [Google Scholar]
- Monetti M. et al. Dissociation of hepatic steatosis and insulin resistance in mice overexpressing DGAT in the liver. Cell Metab 6, 69–78, doi: 10.1016/j.cmet.2007.05.005 (2007). [DOI] [PubMed] [Google Scholar]
- Minehira K. et al. Blocking VLDL secretion causes hepatic steatosis but does not affect peripheral lipid stores or insulin sensitivity in mice. J Lipid Res 49, 2038–2044, doi: 10.1194/jlr.M800248-JLR200 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijboer A. C. et al. Sixteen hours of fasting differentially affects hepatic and muscle insulin sensitivity in mice. J Lipid Res 46, 582–588, doi: 10.1194/jlr.M400440-JLR200 (2005). [DOI] [PubMed] [Google Scholar]
- Matsuzaka T. et al. Crucial role of a long-chain fatty acid elongase, Elovl6, in obesity-induced insulin resistance. Nat Med 13, 1193–1202, doi: 10.1038/nm1662 (2007). [DOI] [PubMed] [Google Scholar]
- Trujillo Viera J., El-Merahbi R., Nieswandt B., Stegner D. & Sumara G. Phospholipases D1 and D2 Suppress Appetite and Protect against Overweight. PloS one 11, e0157607, doi: 10.1371/journal.pone.0157607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C. S. et al. Continuous fat oxidation in acetyl-CoA carboxylase 2 knockout mice increases total energy expenditure, reduces fat mass, and improves insulin sensitivity. Proc Natl Acad Sci USA 104, 16480–16485, doi: 10.1073/pnas.0706794104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C. S. et al. Overexpression of uncoupling protein 3 in skeletal muscle protects against fat-induced insulin resistance. J Clin Invest 117, 1995–2003, doi: 10.1172/JCI13579 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. A. et al. Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature 494, 256–260, doi: 10.1038/nature11808 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman D. E. et al. Oligomeric amyloid-beta peptide disrupts phosphatidylinositol-4,5-bisphosphate metabolism. Nat Neurosci 11, 547–554, doi: 10.1038/nn.2100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh E. G. & Dyer W. J. A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37, 911–917, doi: 10.1139/o59-099 (1959). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.