Abstract
Acute promyelocytic leukemia (APL) is a hematopoietic malignancy that is known with its special cytogenetic feature. Several studies have surveyed expression signature of microRNAs (miRNAs) in APL patients, especially patients who are treated with conventional therapy of this disease. Using miRNAs as diagnostic or prognostic biomarkers in various cancers has been widely studied. Currently, most studies are focusing on exploiting miRNAs as therapeutic tools, and promising progress has been achieved in this field. Recently, studies in the field of miRNA-based therapy in APL have been started.
Keywords: Acute promyelocytic leukemia, cancer therapy, microRNAbased therapy
INTRODUCTION
According to the French–American–British classification, acute promyelocytic leukemia (APL) is an M3 subtype of acute myeloblastic leukemia (AML) and accounts for 10–15% all types of AML. Now, however, in the World Health Organization classification system, APL is classified as APL with t(15;17)(q22;q12); promyelocytic leukemia-retinoic acid receptor α (PML-RARα). In this malignancy, arrest of leukocyte differentiation takes place on promyelocyte stage.[1,2] Because of the high mortality rate from this disease, APL has ever been considered the most deadly type of AML.[3] Since the identification of this cancer, there have been many attempts to treat it which consists mainly of chemotherapy, all-trans retinoic acid (ATRA) and recently arsenic trioxide (As2O3 or ATO). These are used alone or in combination with each other. Despite significant progress in this context, in some cases, these strategies for treatment are not effective. With the advent of microRNAs (miRNAs), hopes for treatment of many diseases, especially cancer, increased.[4] They is a class of evolutionary conserved single-stranded noncoding RNAs with approximately 19–25 nucleotides which have a key function in posttranscriptional gene expression regulation in processes such as proliferation, development, differentiation, and apoptosis.[5] miRNAs modulate a major part of the human genome and increasing evidence suggests that expression of them are deregulated in diseases, especially in cancers.[6] The aim of this review article is to compare the role of miRNAs as therapeutic tools for cancers such as APL with conventional therapy of this disease.
CHARACTERISTIC OF ACUTE PROMYELOCYTIC LEUKEMIA
This rare but deadly leukemia involves some specific chromosomal rearrangements that in almost most cases (>98%) are a balanced reciprocal translocation between chromosomes 15 and 17, which leads to the creation of fusion protein between the PML gene and the RARα gene.[7,8] Generated fusion protein has properties of an incorporation that facilitate a block in myeloid differentiation by suppressing the transcription of retinoid acid (RA) responsive genes.[9] This constitutive suppression is conducted by recruiting abnormal transcription factors and epigenetic enzymes such as histone deacetylases and DNA methyltransferase on retinoic acid target gene promoters.[10,11]
CONVENTIONAL ACUTE PROMYELOCYTIC LEUKEMIA THERAPIES
Initially, treatment of APL was performed by chemotherapy with an anthracycline (daunorubicin, idarubicin, or others) and cytarabine arabinoside as frontline treatment similar to all other subtypes of AML.[12] This approach achieved complete remission (CR) in 75–80% of patients[13] but associated with a high early death rate for some reasons such as deterioration of preexisting disseminated intravascular coagulation because of the release of azure granules in the malignant cells. Despite a high sensitivity to anthracycline, merely about 45% are cured only through standard chemotherapy.[14]
In recent decades, the introduction of ATRA has revolutionized APL therapy. The current standard treatment for APL is based on a combination of ATRA with anthracycline-based chemotherapy.[15] After starting induction therapy with ATRA, one problem that may occur in some patients is symptoms of differentiation syndrome (DS). The combination of ATRA and chemotherapy, therefore, has a major challenge because of death induction due to infection and DS.[16]
In 1971, researchers at the Harbin Medical University in China found that As2O3 or ATO, a traditional Chinese medicine, has significant impact on the treatment of APL.[17]
Unlike ATRA that targets RARα moiety of fusion protein, ATO targets the PML moiety, and both of them degrade PML/RARα.[18] In animal models, synergistic effect was revealed for clearing APL cells.[19] The ATO alone or combination of ATO and ATRA is more effective in inducing CR in newly diagnosed and relapsed patients with low and intermediate risk. In high-risk cases of APL, ATRA/chemotherapy remains the preferred approach.[20] Two major obstacles for using of ATO treatment is that it is much more expensive than chemotherapy and partly associated with toxicity. In a study, it was revealed that using ATRA in combination with the ATO has less hematologic toxicity and less infections than ATRA plus chemotherapy, although coupled with more hepatic toxicity.[16]
BIOGENESIS AND MECHANISM OF MICRORNA
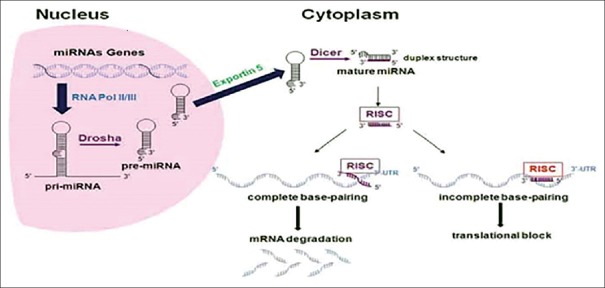
miRNA biogenesis starts through transcription of either particular genes or introns by RNA polymerase II.[21,22,23,24] The generated transcript is a long primary miRNA (pri-miR) which folds into a hairpin structure and similar to other mRNAs is capped with 7-methyl-guanosine on the 5′ end and with a polyadenylation tail on the 3′ end. Then an enzyme called Drosha with its cofactor, DiGeorge syndrome critical region 8, processes pri-miR to produce ~70 nucleotide precursor miR (pre-miR).[25]
Complex of exportin 5 and Ran-GTP recognizes this pre-miR and transport it into the cytoplasm. In the cytoplasm, a double-stranded RNA-specific RNaseIII endonuclease (Dicer) cleaves pre-miR's terminal loop by its dsRNA binding partner, resulting in a mature miRNA [Figure 1].
Figure 1.
Schematic representation of biogenesis and mechanism of microRNA[27]
Mature miRNA is then unwound and incorporated into the RNA-induced silencing complex (RISC). miRNAs via the RISC bind to 3′-untranslated region of the target mRNA(s) and acts as endogenous suppressors of gene expression.[26] If the miRNA binds incompletely to the target mRNA, represses translation of mRNA, whereas if the binding is almost complete, the miRNA induces degradation of the mRNA.[27] Seed region, nucleotides 3-8 from the 5′ end of the mature miRNA, is the crucial binding location for translational repression.[28] Some miRNAs might indirectly regulate gene expression through the targeting of transcription factors and other miRNAs directly target 5′ region, ribonucleoproteins, and promoters.[29] Some miRNAs activate, rather than inhibit, gene expression of their targets by binding directly to the 5′ UTR.[30]
THE FUNCTIONS OF MICRORNAS IN CANCER
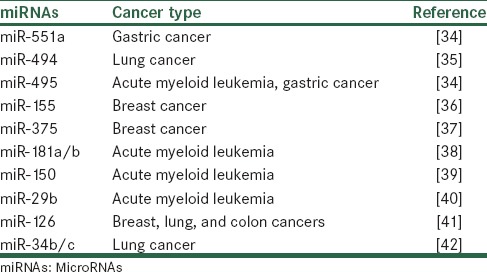
In the last decades, researchers have found that the expression patterns of miRNAs are dysregulated in various cancers.[31,32] Increasing evidence show that miRNAs can act as a tumor suppressor or an oncogene. miRNAs that act like tumor suppressor genes include a group of these noncoding RNA whose loss of function promotes cancer development.[33] This category of miRNAs act similar to protein-coding tumor suppressor genes. They are involved in many cancers [Table 1].
Table 1.
Examples of tumor suppressor microRNAs reported to be downregulated
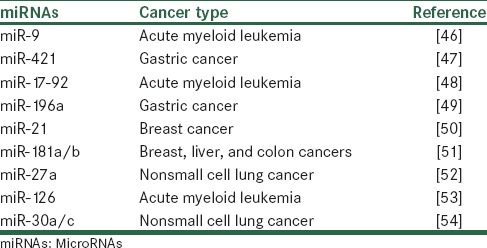
Oncogenic miRNAs (oncomiRs) are a category of miRNAs that upregulation of them lead to an acceleration of tumor formation and development.[43] The two hallmarks of cancer that are mostly affected by oncomiRs are metastasis and proliferation of tumor cells.[44] OncomiRs through targeting of mRNAs which are encoded by tumor suppressor genes can affect tumorigenesis.[45] Recently, many oncomiRs have been known to be dysregulated in various tumors. Some examples of them are summarized in Table 2.
Table 2.
Examples of oncogenic microRNAs reported to be upregulated
THE ROLE OF MICRORNAS IN NORMAL HEMATOPOIESIS
The important role of miRNAs in hematopoiesis was found by identifying three murine hematopoietic tissues-specific expressed miRNAs, miR-181a, miR-142s, and miR-223. miR-142s expression was found to be limited to B-lineage and myeloid lineages, and miR-223 was found to be expressed in myeloid lineages. Considerably, deregulation of expression of miR-223 or miR-142 leads to 30–50% increase of T-lineage cells, but do not affect the numbers of B-lineage or myeloid cells. It was revealed that miR-181a preferentially expresses in the B-lineage and ectopic expression of this miRNA resulted in a doubling of B-lineage cells.[55] Altogether, these results show that miRNAs might have an important role in the differentiation of hematopoietic cells.
A very strong relation between the miRNAs expression profile and differentiation stage of murine hematopoietic cell populations was revealed by comparison of cell populations at different stages of differentiation. However, the correlation between precursor cells and their corresponding mature cell populations were much less.[56] This verifies the importance role of miRNAs in the differentiation and keeping of hematopoietic maturation stages.
Some studies have revealed the expression of pri-miR-155 in lymphoid tissues and T and B cells.[57,58] In lymphoid tissues, confirmation of expression of the B-cell integration cluster (BIC) locus-derived miR-155 in tonsil and lymph node was confirmed by Northern blotting.[59] By applying anti-CD3 in combination with anti-CD28, stimulation of CD4+ T-cells result in a sharp rise in expression of BIC.[60] The results propose a correlation between induction of high BIC/miR-155 and activation of T and B-cells.
miR-221 and miR-222 were found to be downregulated in erythropoietic cells at different stages of erythrocyte differentiation and maturation. Both miRNAs are identical at the first eight nucleotides of the 5′ sites (seed region) that indicate overlapping in their target genes. Through prediction studies, it was revealed that KIT receptor is a target for both miR-221 and miR-222. C-KIT play an important role in the proliferation control of primitive hematopoietic and erythropoietic cells, and it was revealed that following erythrocyte differentiation, the expression of miR-221/miR-222 and KIT are inversely correlated.[61]
In two studies, researchers exploited generation of conditional DICER-1 knockout alleles to clarify the general role of miRNAs in murine T-cell differentiation.[62,63] Both studies revealed decreased viability of T-cells.
MICRORNAS IN ACUTE PROMYELOCYTIC LEUKEMIA
By studying an APL model system, linking of miR-223 expression to the retinoic acid signaling pathway and CCAAT/enhancer binding protein-α in normal human granulopoiesis was obtained.[64] miRNAs expression profiling of AML patients has shown that by analyzing miRNA signatures, we can distinguish between cytogenetic subtypes of AML.[53,65,66] Dixon-McIver et al. identified a special expression profile of miRNAs in APL patients. It included upregulation of miR-127, miR-154 *, miR-299, miR-323, miR-368, and miR-370 which their genes located in the 14q32 and nine other miRNAs were downregulated.[65] The two other studies also determined two different signatures of a gene cluster.[53,66]
Some roles of miRNAs in APL were identified by studies that surveyed the effect of conventional treatment on expression of miRNAs. By using RA treatment, it was revealed that RA without affecting the expression of miR-142 or miR-181a, exclusively increase the expression of miR-223.[67] While ectopic expression of miR-223 increases differentiation of NB4 cells, knock down the miR-223 inhibits the effect of RA on the differentiation of cells.[64] De Marchis et al. showed that miR-342 mediates ATRA effect on APL differentiation in combination with PU.1 and interferon regulatory factor proteins.[68] In another study, it was revealed that during ATRA therapy in APL patients and APL cell lines, some miRNAs are differentially expressed.[69] Zhong et al. revealed that during ATRA induction, miR-146a modulate proliferation of APL cells through transforming growth factor-beta 1/Smad signal transduction pathway which inactivation of it is related to human leukemia.[70] One study showed that ATO can modulate 88 cancer-related miRNAs.[71] Construction of a dysregulated miRNA network was performed in a study that examined the effect of ATO on apoptosis of APL cells.[72]
MICRORNA-BASED CANCER THERAPY
miRNA-based therapy mainly depends on nucleic acid-based strategies. The aim of these strategies is to restore the normal function of miRNAs. Targeting overexpressed oncomiRs is conducted mainly by two methods: Antisense oligonucleotides (antagomiRs) and miRNA sponges.
Antisense molecules that are chemically modified and have a bridge between the 2′-O and 4′-C at each nucleotide are called locked nucleic acid (LNA) oligonucleotides and are used for synthesizing anti-miRNA nucleic acids.[73,74]
miRNA antagonists (antagomiRs) contain 3′-conjugated cholesterol residues, 2′-O-methylation of ribose residues, and substitutionary of phosphodiester bonds through phosphorothioate linkages.[75] AntagomiRs affect cancer-related pathway through binding and inhibiting oncomiRs. In some studies, the effectiveness of this method has been surveyed.[76,77]
miRNA sponges are produced from transgenes within cells and have complementary binding sites to seed sequences of target miRNAs. This advantage gives them the ability to inhibit a family of miRNAs.[78] It has been revealed that using miR-9 sponges, the activity of this miRNA is reduced to 50%.[79]
miRNA replacement therapy or miRNA mimics method mostly has conducted in animal models. In this method, malfunctioning tsmiRs are replaced.[80] In the field of cancer treatment, exploiting MRX34 is the first example of miRNA-based cancer therapy. MRX34 is a synthetic miR-34a mimic that is transferred by liposomal nanoparticles.[81] miR-34a is a tsmiR downstream of p53 and using replacement strategy, it counteracts chemoresistance and self-renewal of cancer cells.[82] In a study, introduction of lentiviral Let-7 miRNA into self-renewing breast cancer cells lead to an increase in Let-7 level.[83] In other studies, it was shown that we can restore downregulated tsmiRs miR-205, miR-126, miR-335, and miR-451 by using miRNA replacement therapy.[84,85]
Few studies have been conducted on the use of miRNA therapy in APL. Inhibition of miR-92a by LNA in HL-60 (human APL cell line) induced apoptosis and inhibited cell proliferation through the expression of p63 and following the recovery of cellular pathways which are modulated by p63 protein.[86,87,88]
CONCLUSION
In the past decade with the advent of the term “personalized medicine,” targeted-therapy of diseases, in particular, cancer has been taken into consideration. As well as in hematological malignancies such as APL attempts have been made. The emersion of ATRA made changes in this area of therapy and, in fact, ATRA is the first example of targeted therapy in cancers. However, despite the efforts that have been made to improve the quality of APL treatment such as ATO-based drugs, still we need methods that are more efficient. miRNA-based therapy is a new and attractive method in this field. As previously mentioned, several in vitro studies have been conducted. This method has its limitation which should be considered, such as the paucity of preclinical and clinical studies. Our knowledge about miRNA is still in its infancy, methods that are used for delivery and entry into cells have problems and “off-target effect” of this technique because of any miRNA has many targets and its expression is regulated by several genes. Therefore, prior to use in clinical, many of the problems related to it must be resolved. Undoubtedly, with establishing miRNA-based therapy in the clinic as an effective therapeutic tool, we can prevent many of the side effects of chemical drugs, especially those which are used in chemotherapy.
Financial support and sponsorship
Isfahan University of Medical Sciences.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
This study was conducted with the financial support of Isfahan University of Medical Sciences, IRAN.
REFERENCES
- 1.Li J, Zhu H, Hu J, Mi J, Chen S, Chen Z, et al. Progress in the treatment of acute promyelocytic leukemia: Optimization and obstruction. Int J Hematol. 2014;100:38–50. doi: 10.1007/s12185-014-1603-1. [DOI] [PubMed] [Google Scholar]
- 2.Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: Rationale and important changes. Blood. 2009;114:937–51. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 3.Chang H, Kuo MC, Shih LY, Dunn P, Wang PN, Wu JH, et al. Clinical bleeding events and laboratory coagulation profiles in acute promyelocytic leukemia. Eur J Haematol. 2012;88:321–8. doi: 10.1111/j.1600-0609.2011.01747.x. [DOI] [PubMed] [Google Scholar]
- 4.Garofalo M, Leva GD, Croce CM. MicroRNAs as anti-cancer therapy. Curr Pharm Des. 2014;20:5328–35. doi: 10.2174/1381612820666140128211346. [DOI] [PubMed] [Google Scholar]
- 5.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 6.Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mistry AR, Pedersen EW, Solomon E, Grimwade D. The molecular pathogenesis of acute promyelocytic leukaemia: Implications for the clinical management of the disease. Blood Rev. 2003;17:71–97. doi: 10.1016/s0268-960x(02)00075-9. [DOI] [PubMed] [Google Scholar]
- 8.Wang ZY, Chen Z. Acute promyelocytic leukemia: From highly fatal to highly curable. Blood. 2008;111:2505–15. doi: 10.1182/blood-2007-07-102798. [DOI] [PubMed] [Google Scholar]
- 9.de Thé H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–84. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 10.Di Croce L, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M, et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295:1079–82. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- 11.Licht JD. Acute promyelocytic leukemia – Weapons of mass differentiation. N Engl J Med. 2009;360:928–30. doi: 10.1056/NEJMcibr0810371. [DOI] [PubMed] [Google Scholar]
- 12.Sanz MA, Jarque I, Martín G, Lorenzo I, Martínez J, Rafecas J, et al. Acute promyelocytic leukemia. Therapy results and prognostic factors. Cancer. 1988;61:7–13. doi: 10.1002/1097-0142(19880101)61:1<7::aid-cncr2820610103>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 13.Cunningham I, Gee TS, Reich LM, Kempin SJ, Naval AN, Clarkson BD. Acute promyelocytic leukemia: Treatment results during a decade at Memorial Hospital. Blood. 1989;73:1116–22. [PubMed] [Google Scholar]
- 14.Fenaux P, Wang ZZ, Degos L. Treatment of acute promyelocytic leukemia by retinoids. Curr Top Microbiol Immunol. 2007;313:101–28. doi: 10.1007/978-3-540-34594-7_7. [DOI] [PubMed] [Google Scholar]
- 15.Sanz MA, Grimwade D, Tallman MS, Lowenberg B, Fenaux P, Estey EH, et al. Management of acute promyelocytic leukemia: Recommendations from an expert panel on behalf of the European LeukemiaNet. Blood. 2009;113:1875–91. doi: 10.1182/blood-2008-04-150250. [DOI] [PubMed] [Google Scholar]
- 16.Sanz MA, Iacoboni G, Montesinos P. Acute promyelocytic leukemia: Do we have a new front-line standard of treatment? Curr Oncol Rep. 2013;15:445–9. doi: 10.1007/s11912-013-0339-z. [DOI] [PubMed] [Google Scholar]
- 17.Cyranoski D. Arsenic patent keeps drug for rare cancer out of reach of many. Nat Med. 2007;13:1005. doi: 10.1038/nm0907-1005. [DOI] [PubMed] [Google Scholar]
- 18.de Thé H, Le Bras M, Lallemand-Breitenbach V. The cell biology of disease: Acute promyelocytic leukemia, arsenic, and PML bodies. J Cell Biol. 2012;198:11–21. doi: 10.1083/jcb.201112044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jing Y, Wang L, Xia L, Chen GQ, Chen Z, Miller WH, et al. Combined effect of all-trans retinoic acid and arsenic trioxide in acute promyelocytic leukemia cells in vitro and in vivo. Blood. 2001;97:264–9. doi: 10.1182/blood.v97.1.264. [DOI] [PubMed] [Google Scholar]
- 20.Cull EH, Altman JK. Contemporary treatment of APL. Curr Hematol Malig Rep. 2014;9:193–201. doi: 10.1007/s11899-014-0205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee J, Li Z, Brower-Sinning R, John B. Regulatory circuit of human microRNA biogenesis. PLoS Comput Biol. 2007;3:e67. doi: 10.1371/journal.pcbi.0030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pacifico F, Crescenzi E, Mellone S, Iannetti A, Porrino N, Liguoro D, et al. Nuclear factor-{kappa} B contributes to anaplastic thyroid carcinomas through up-regulation of miR-146a. J Clin Endocrinol Metab. 2010;95:1421–30. doi: 10.1210/jc.2009-1128. [DOI] [PubMed] [Google Scholar]
- 23.Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y, et al. Structure and activity of putative intronic miRNA promoters. RNA. 2010;16:495–505. doi: 10.1261/rna.1731910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bortolin-Cavaillé ML, Dance M, Weber M, Cavaillé J. C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucleic Acids Res. 2009;37:3464–73. doi: 10.1093/nar/gkp205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price C, Chen J. MicroRNAs in cancer biology and therapy: Current status and perspectives. Genes Dis. 2014;1:53–63. doi: 10.1016/j.gendis.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeyaseelan K, Herath WB, Armugam A. MicroRNAs as therapeutic targets in human diseases. Expert Opin Ther Targets. 2007;11:1119–29. doi: 10.1517/14728222.11.8.1119. [DOI] [PubMed] [Google Scholar]
- 27.Rota R, Ciarapica R, Giordano A, Miele L, Locatelli F. MicroRNAs in rhabdomyosarcoma: Pathogenetic implications and translational potentiality. Mol Cancer. 2011;10:120. doi: 10.1186/1476-4598-10-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fazi F, Nervi C. MicroRNA: Basic mechanisms and transcriptional regulatory networks for cell fate determination. Cardiovasc Res. 2008;79:553–61. doi: 10.1093/cvr/cvn151. [DOI] [PubMed] [Google Scholar]
- 29.Yang Z, Wang L. Regulation of microRNA expression and function by nuclear receptor signaling. Cell Biosci. 2011;1:31. doi: 10.1186/2045-3701-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–71. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 31.Michael MZ, O’ Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–91. [PubMed] [Google Scholar]
- 32.McManus MT. MicroRNAs and cancer. Semin Cancer Biol. 2003;13:253–8. doi: 10.1016/s1044-579x(03)00038-5. [DOI] [PubMed] [Google Scholar]
- 33.Tie J, Fan D. Big roles of microRNAs in tumorigenesis and tumor development. Histol Histopathol. 2011;26:1353–61. doi: 10.14670/HH-26.1353. [DOI] [PubMed] [Google Scholar]
- 34.Li Z, Cao Y, Jie Z, Liu Y, Li Y, Li J, et al. miR-495 and miR-551a inhibit the migration and invasion of human gastric cancer cells by directly interacting with PRL-3. Cancer Lett. 2012;323:41–7. doi: 10.1016/j.canlet.2012.03.029. [DOI] [PubMed] [Google Scholar]
- 35.Romano G, Acunzo M, Garofalo M, Di Leva G, Cascione L, Zanca C, et al. MiR-494 is regulated by ERK1/2 and modulates TRAIL-induced apoptosis in non-small-cell lung cancer through BIM down-regulation. Proc Natl Acad Sci U S A. 2012;109:16570–5. doi: 10.1073/pnas.1207917109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gasparini P, Lovat F, Fassan M, Casadei L, Cascione L, Jacob NK, et al. Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc Natl Acad Sci U S A. 2014;111:4536–41. doi: 10.1073/pnas.1402604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward A, Balwierz A, Zhang JD, Küblbeck M, Pawitan Y, Hielscher T, et al. Re-expression of microRNA-375 reverses both tamoxifen resistance and accompanying EMT-like properties in breast cancer. Oncogene. 2013;32:1173–82. doi: 10.1038/onc.2012.128. [DOI] [PubMed] [Google Scholar]
- 38.Li Z, Huang H, Li Y, Jiang X, Chen P, Arnovitz S, et al. Up-regulation of a HOXA-PBX3 homeobox-gene signature following down-regulation of miR-181 is associated with adverse prognosis in patients with cytogenetically abnormal AML. Blood. 2012;119:2314–24. doi: 10.1182/blood-2011-10-386235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang X, Huang H, Li Z, Li Y, Wang X, Gurbuxani S, et al. Blockade of miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell. 2012;22:524–35. doi: 10.1016/j.ccr.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–8. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Du YY, Lin YF, Chen YT, Yang L, Wang HJ, et al. The cell growth suppressor, mir-126, targets IRS-1. Biochem Biophys Res Commun. 2008;377:136–40. doi: 10.1016/j.bbrc.2008.09.089. [DOI] [PubMed] [Google Scholar]
- 42.Okada N, Lin CP, Ribeiro MC, Biton A, Lai G, He X, et al. A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes Dev. 2014;28:438–50. doi: 10.1101/gad.233585.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krutovskikh VA, Herceg Z. Oncogenic microRNAs (OncomiRs) as a new class of cancer biomarkers. Bioessays. 2010;32:894–904. doi: 10.1002/bies.201000040. [DOI] [PubMed] [Google Scholar]
- 44.Kaboli PJ, Rahmat A, Ismail P, Ling KH. MicroRNA-based therapy and breast cancer: A comprehensive review of novel therapeutic strategies from diagnosis to treatment. Pharmacol Res. 2015;97:104–21. doi: 10.1016/j.phrs.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 45.Reshmi G, Pillai MR. Beyond HPV: Oncomirs as new players in cervical cancer. FEBS Lett. 2008;582:4113–6. doi: 10.1016/j.febslet.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 46.Chen P, Price C, Li Z, Li Y, Cao D, Wiley A, et al. miR-9 is an essential oncogenic microRNA specifically overexpressed in mixed lineage leukemia-rearranged leukemia. Proc Natl Acad Sci U S A. 2013;110:11511–6. doi: 10.1073/pnas.1310144110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou H, Xiao B, Zhou F, Deng H, Zhang X, Lou Y, et al. MiR-421 is a functional marker of circulating tumor cells in gastric cancer patients. Biomarkers. 2012;17:104–10. doi: 10.3109/1354750X.2011.614961. [DOI] [PubMed] [Google Scholar]
- 48.Wong P, Iwasaki M, Somervaille TC, Ficara F, Carico C, Arnold C, et al. The miR-17-92 microRNA polycistron regulates MLL leukemia stem cell potential by modulating p21 expression. Cancer Res. 2010;70:3833–42. doi: 10.1158/0008-5472.CAN-09-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsai KW, Liao YL, Wu CW, Hu LY, Li SC, Chan WC, et al. Aberrant expression of miR-196a in gastric cancers and correlation with recurrence. Genes Chromosomes Cancer. 2012;51:394–401. doi: 10.1002/gcc.21924. [DOI] [PubMed] [Google Scholar]
- 50.Mandal CC, Ghosh-Choudhury T, Dey N, Choudhury GG, Ghosh-Choudhury N. miR-21 is targeted by omega-3 polyunsaturated fatty acid to regulate breast tumor CSF-1 expression. Carcinogenesis. 2012;33:1897–908. doi: 10.1093/carcin/bgs198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y, Yu Y, Tsuyada A, Ren X, Wu X, Stubblefield K, et al. Transforming growth factor-ß regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM. Oncogene. 2011;30:1470–80. doi: 10.1038/onc.2010.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Acunzo M, Romano G, Palmieri D, Laganá A, Garofalo M, Balatti V, et al. Cross-talk between MET and EGFR in non-small cell lung cancer involves miR-27a and Sprouty2. Proc Natl Acad Sci U S A. 2013;110:8573–8. doi: 10.1073/pnas.1302107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT, et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci U S A. 2008;105:15535–40. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moch H, Lukamowicz-Rajska M. miR-30c-2-3p and miR-30a-3p: New pieces of the jigsaw puzzle in HIF2a regulation. Cancer Discov. 2014;4:22–4. doi: 10.1158/2159-8290.CD-13-0897. [DOI] [PubMed] [Google Scholar]
- 55.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–6. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 56.Monticelli S, Ansel KM, Xiao C, Socci ND, Krichevsky AM, Thai TH, et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005;6:R71. doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van den Berg A, Kroesen BJ, Kooistra K, de Jong D, Briggs J, Blokzijl T, et al. High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosomes Cancer. 2003;37:20–8. doi: 10.1002/gcc.10186. [DOI] [PubMed] [Google Scholar]
- 58.Tam W. Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA. Gene. 2001;274:157–67. doi: 10.1016/s0378-1119(01)00612-6. [DOI] [PubMed] [Google Scholar]
- 59.Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–9. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 60.Haasch D, Chen YW, Reilly RM, Chiou XG, Koterski S, Smith ML, et al. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell Immunol. 2002;217:78–86. doi: 10.1016/s0008-8749(02)00506-3. [DOI] [PubMed] [Google Scholar]
- 61.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci U S A. 2005;102:18081–6. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cobb BS, Nesterova TB, Thompson E, Hertweck A, O’Connor E, Godwin J, et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–73. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–9. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–31. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 65.Dixon-McIver A, East P, Mein CA, Cazier JB, Molloy G, Chaplin T, et al. Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PLoS One. 2008;3:e2141. doi: 10.1371/journal.pone.0002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Löwenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 2008;111:5078–85. doi: 10.1182/blood-2008-01-133355. [DOI] [PubMed] [Google Scholar]
- 67.Nervi C, Fazi F, Rosa A, Fatica A, Bozzoni I. Emerging role for microRNAs in acute promyelocytic leukemia. Curr Top Microbiol Immunol. 2007;313:73–84. doi: 10.1007/978-3-540-34594-7_5. [DOI] [PubMed] [Google Scholar]
- 68.De Marchis ML, Ballarino M, Salvatori B, Puzzolo MC, Bozzoni I, Fatica A. A new molecular network comprising PU.1, interferon regulatory factor proteins and miR-342 stimulates ATRA-mediated granulocytic differentiation of acute promyelocytic leukemia cells. Leukemia. 2009;23:856–62. doi: 10.1038/leu.2008.372. [DOI] [PubMed] [Google Scholar]
- 69.Garzon R, Pichiorri F, Palumbo T, Visentini M, Aqeilan R, Cimmino A, et al. MicroRNA gene expression during retinoic acid-induced differentiation of human acute promyelocytic leukemia. Oncogene. 2007;26:4148–57. doi: 10.1038/sj.onc.1210186. [DOI] [PubMed] [Google Scholar]
- 70.Zhong H, Wang HR, Yang S, Zhong JH, Wang T, Wang C, et al. Targeting Smad4 links microRNA-146a to the TGF-beta pathway during retinoid acid induction in acute promyelocytic leukemia cell line. Int J Hematol. 2010;92:129–35. doi: 10.1007/s12185-010-0626-5. [DOI] [PubMed] [Google Scholar]
- 71.Ghaffari SH, Bashash D, Dizaji MZ, Ghavamzadeh A, Alimoghaddam K. Alteration in miRNA gene expression pattern in acute promyelocytic leukemia cell induced by arsenic trioxide: A possible mechanism to explain arsenic multi-target action. Tumour Biol. 2012;33:157–72. doi: 10.1007/s13277-011-0259-1. [DOI] [PubMed] [Google Scholar]
- 72.Liang H, Li X, Wang L, Yu S, Xu Z, Gu Y, et al. MicroRNAs contribute to promyelocyte apoptosis in As2O3-treated APL cells. Cell Physiol Biochem. 2013;32:1818–29. doi: 10.1159/000356615. [DOI] [PubMed] [Google Scholar]
- 73.Chabot S, Teissié J, Golzio M. Targeted electro-delivery of oligonucleotides for RNA interference: siRNA and antimiR. Adv Drug Deliv Rev. 2015;81:161–8. doi: 10.1016/j.addr.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 74.Davalos A, Suarez Y. MiRNA-based therapy: From bench to bedside. Pharmacol Res. 2013;75:1–2. doi: 10.1016/j.phrs.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 75.Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, Marcusson EG, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28:341–7. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma F, Zhang J, Zhong L, Wang L, Liu Y, Wang Y, et al. Upregulated microRNA-301a in breast cancer promotes tumor metastasis by targeting PTEN and activating Wnt/ß-catenin signaling. Gene. 2014;535:191–7. doi: 10.1016/j.gene.2013.11.035. [DOI] [PubMed] [Google Scholar]
- 77.McCubrey JA, Davis NM, Abrams SL, Montalto G, Cervello M, Libra M, et al. Targeting breast cancer initiating cells: Advances in breast cancer research and therapy. Adv Biol Regul. 2014;56:81–107. doi: 10.1016/j.jbior.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 78.Ebert MS, Sharp PA. MicroRNA sponges: Progress and possibilities. RNA. 2010;16:2043–50. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–56. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70:7027–30. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bouchie A. First microRNA mimic enters clinic. Nat Biotechnol. 2013;31:577. doi: 10.1038/nbt0713-577. [DOI] [PubMed] [Google Scholar]
- 82.Bader AG. miR-34 – A microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 84.Nickel A, Stadler SC. Role of epigenetic mechanisms in epithelial-to-mesenchymal transition of breast cancer cells. Transl Res. 2015;165:126–42. doi: 10.1016/j.trsl.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 85.Kota SK, Balasubramanian S. Cancer therapy via modulation of micro RNA levels: A promising future. Drug Discov Today. 2010;15:733–40. doi: 10.1016/j.drudis.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 86.Sharifi M, Salehi R, Gheisari Y, Kazemi M. Inhibition of microRNA miR-92a induces apoptosis and necrosis in human acute promyelocytic leukemia. Adv Biomed Res. 2014;3:61. doi: 10.4103/2277-9175.125826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sharifi M, Salehi R, Gheisari Y, Kazemi M. Inhibition of microRNA miR-92a induces apoptosis and inhibits cell proliferation in human acute promyelocytic leukemia through modulation of p63 expression. Mol Biol Rep. 2014;41:2799–808. doi: 10.1007/s11033-014-3134-5. [DOI] [PubMed] [Google Scholar]
- 88.Sharifi M, Salehi R, Gheisari Y, Kazemi M. Inhibition of MicroRNA miR-92a inhibits cell proliferation in human acute promyelocytic leukemia. Turk J Haematol. 2013;30:157–62. doi: 10.4274/Tjh.2012.0171. [DOI] [PMC free article] [PubMed] [Google Scholar]