Abstract
Molecular details of the recognition of disordered antigens by their cognate antibodies have not been studied as extensively as folded protein antigens and much is still unknown. To follow the conformational changes in the antibody and cross-talk between its subunits and with antigens, we performed molecular dynamics (MD) simulations of the complex of Fab and prion-associated peptide in the apo and bound forms. We observed that the inter-chain disulfide bond in constant domains restrains the conformational changes of Fab, especially the loops in the CH1 domain, resulting in inhibition of the cross-talk between Fab subdomains that thereby may prevent prion peptide binding. We further identified several negative and positive correlations of motions between the peptide and Fab constant domains, which suggested structural cross-talks between the constant domains and the antigen. The cross-talk was influenced by the inter-chain disulfide bond, which reduced the number of paths between them. Importantly, network analysis of the complex and its bound water molecules observed that those water molecules form an integral part of the Fab/peptide complex network and potential allosteric pathways. On-going work focuses on developing strategies aimed to incorporate these new network communications—including the associated water molecules—toward the grand challenge of antibody design.
Keywords: antibody–antigen interaction, bound water molecules, community analysis, disulfide bond, dynamic network, motion correlation
Introduction
Protein conformational fluctuations and dynamics in water are intrinsic thermodynamic phenomena, and the distributions of the states on the energy landscape are determined by statistical thermodynamics; however, they are optimized to perform biological functions (Wei et al., 2016). The antibody variable regions are necessarily flexible to enable recognition of a range of diversified targets. Antibody–antigen recognition is associated with structural transitions of the antibody through its inherent flexibility (Keskin, 2007; Thielges et al., 2008; Li et al., 2014). The variable domains, especially complementarity determining regions (CDRs), mainly control the specificity and affinity (Mian et al., 1991), while the constant domains modulate the isotype/effector (Torres and Casadevall, 2008) and independently the variable and constant domains’ functions.
Recent studies indicated that besides the variable domains, the constant domain also plays an essential role in the antigen binding (Pritsch et al., 1996; Adachi et al., 2003; Dam et al., 2008; Tudor et al., 2012; Li et al., 2015). There is communication between the variable domains of the light and heavy chains (Pellequer et al., 1999) and distant communication between the variable and constant domains (Janda and Casadevall, 2010). Redistribution of flexibility in stabilizing antibody fragment mutants was found in anti-lymphotoxin-beta receptor antibody (Li et al., 2014). Evidence also suggests allosteric effects during antibody–antigen recognition (Sela-Culang et al., 2013). For instance, Pritsch et al. (1996) suggested that antibodies with identical variable domains, but different isotypes, have significantly different affinities when binding to tubulin. Oda et al. (2003) showed that the binding of antigen causes conformational changes in the protein G and protein A binding sites on the heavy chain constant domains. A recent study surveyed over 100 crystal structures of antibodies in either the apo or the bound form, and found that distant loops from CH1-1 undergo significant fluctuation upon antigen binding and this fluctuation is common among these structures (Sela-Culang et al., 2012).
Water molecules are critical in protein structure, dynamics and function (Mattos, 2002; Bellissent-Funel et al., 2016) and thus in antibody–antigen recognition (Rudolph and Wilson, 2002). Recent studies suggested that certain water molecules are conserved near some essential hydrophilic residues, and these water molecules are implicated in the communication between distant sites (Buhrman et al., 2010; Kearney et al., 2014). In our recent survey of antibody recognition of amyloidogenic peptides, we also found that water molecules can play an important structural role in bridging interactions between antibodies and peptide antigens (Ma and Nussinov, 2016). However, although recent comparative studies of antibody–antigen structures provided insights into the antibody–antigen recognition, the structural and dynamical mechanisms behind the potential allosteric effects in inter-domain cross-talk and the role of bound water molecules—at atomic level detail—still elusive.
Neurodegenerative diseases (ND) are caused by progressive loss of structure and function of neurons, including neuronal death. Commonly studied NDs include Alzheimer, Parkinson, Huntington, prion (‘mad cow’) diseases and Down's syndrome. The dominant common feature is protein aggregation that leads to neuronal loss. Immunotherapies using antibodies and antibody fragments to target protein aggregation in neurodegenerative diseases have been widely tested as the disease-modifying treatment (De Genst et al., 2014). Among these neurodegenerative diseases, the prion disease has long been recognized to have a protein-only self-propagation infectious mechanism. Anti-prion antibodies and new vaccines have been tested over more than a decade, aiming to break the immune tolerance to the prion protein (Rovis and Legname, 2014), including an antibody that can specifically neutralize PrPSc (Taschuk et al., 2015).
In this study, we used molecular dynamics (MD) simulations, evolutionary analysis, motion correlation analysis and dynamic network to theoretically study the communication between antibody subdomains, their cross-talk with the antigen and themselves and the participation of water molecules in this process, focusing on a Fab/prion peptide complex in the bound and apo forms (Kanyo et al., 1999). Experimentally, it was found that in the apo form, the antibody has a disulfide bond connecting the C-terminal Cys214 of the light chain and Cys125 on the CH1-1 loop of the heavy chain, while this inter-chain disulfide bond (inter-SS bond) was broken in the bound form. Our results indicated that in this specific case of Fab/peptide complex, inter-subdomain correlations are important for peptide binding and the inter-chain disulfide bond between the light and heavy chains reduces the communications among Fab subdomains as well as those between the peptide and the Fab subdomains. Our network optimal path analysis suggests that water molecules become members of the Fab/peptide complex network and increase the frequency of communication between the peptide and potential allosteric sites.
Materials and methods
Molecular modeling and simulations
The structures of the bound and apo forms of the Fab/peptide complex are based on crystal structures Protein Data Bank archive (PDB) IDs 1cu4 and 1cr9, respectively (Kanyo et al., 1999). There is an inter-SS bond in the apo form but not in the bound form. The two unresolved N-terminal residues of the heavy chain in the bound form were modeled using MODELLER (Šali and Blundell, 1993; Fiser et al., 2000; Martí-Renom et al., 2000; Webb and Sali, 2014). The unbound form was obtained by manually removing the peptide in the bound (1cu4) structure. The inter-SS bond was either kept or removed from the bound and apo structures to consider the effects of the inter-SS bond, resulting in six systems. The isolated prion peptide was also simulated independently starting from the conformation in 1cu4 (Table I). As the non-sequential Kabat numbering scheme is used in the crystal structures, we renumber the residues for convenience in the simulation. For the light chain, heavy chain and the peptide, the N-termini and C-termini were charged as NH3 + and COO− groups, respectively.
Table I.
Summarization of the systems for the MD simulation in this study
| System no. | System | SS bond | Peptide | Total atoms | Cations | Anion | Water | Time (ns) |
|---|---|---|---|---|---|---|---|---|
| 1 | Apo | No | No | 62 121 | 55 | 52 | 18 482 | 100 |
| 2 | Apo | Yes | No | 62 119 | 55 | 52 | 18 482 | 100 |
| 3 | Bound | No | Yes | 60 083 | 51 | 50 | 17 751 | 100 |
| 4 | Bound | Yes | Yes | 60 081 | 51 | 50 | 17 751 | 100 |
| 5 | Unbound | Yes | No | 60 019 | 53 | 50 | 17 795 | 100 |
| 6 | Unbound | No | No | 60 017 | 53 | 50 | 17 795 | 100 |
| 7 | Peptide | N/A | N/A | 7410 | 6 | 8 | 2412 | 100 |
| 8 | Apo | No | No | 97 108 | 87 | 84 | 30 123 | 100 |
| 9 | Apo | Yes | No | 97 031 | 87 | 84 | 30 098 | 100 |
| 10 | Bound | No | Yes | 102 518 | 92 | 91 | 31 869 | 100 |
| 11 | Bound | Yes | Yes | 102 516 | 92 | 91 | 31 869 | 100 |
There are only a limited number of crystallized water molecules in the bound structure less than in the apo structure due to the poor resolution (Kanyo et al., 1999). To better compare the systems in the simulations, all crystal water molecules were deleted. The systems were then solvated by TIP3P water molecules, and sodium and chlorides were added to neutralize the system to a total concentration of ~150 mM. The resulting solvated systems were energy minimized for 5000 conjugate gradient steps, with the protein fixed and water molecules and counterions allowed to move, followed by additional 5000 conjugate gradient steps, where all atoms were allowed to move. In the equilibration stage, each system was gradually relaxed by performing a series of dynamic cycles, in which the harmonic restraints on proteins were gradually removed to optimize the protein–water interactions. In the production stage, all simulations were performed using the constant pressure and constant temperature ensemble at 310 K. All MD simulations were performed using the NAMD software (Kale et al., 1999) with CHARMM27 force field (MacKerell et al., 1998). MD trajectories were saved by every 2 ps for analysis. To reduce the statistical noise, the systems of 1cu4, 1cu4 with the inter-SS bond, 1cr9 and 1cr9 without the inter-SS bond (Table I) were repeated independently. The initial structure of each individual repeat system was minimized by using different energy minimization protocols. The systems were then re-solvated randomly by water molecules and ions. The initial velocity distribution of each repeat system was also set differently. Thus, the repeat simulations were started from alternate conformations. A summary of all simulation systems is listed in Table I.
Binding energy evaluation
To evaluate the binding energy between Fab and the prion peptide, the trajectory for each bound and apo system was extracted from the last 20 ns of explicit solvent MD without water molecules and ions. The solvation energies of all systems were calculated using the generalized Born method with molecular volume (GBMV) (Lee et al., 2003) after 500 steps of energy minimization to relax the local geometries caused by the thermal fluctuations that occurred in the MD simulations. In the GBMV calculation, the dielectric constant of water is set to 80 and no distance cutoff is used. The binding energy between two Fab and the prion peptide was calculated by .
Evolutionary analysis
The sequence of Fab (PDB ID: 1cu4) was used to generate relative conservation scores for each amino-acid position using the Bayesian method implemented in the ConSurf server (Landau et al., 2005). The sequence homologs of Fab were identified after three iterations of CS-BLAST algorithm against UNIREF90 database. The selected sequence homologs with an E-value of <0.0001 were then aligned using Multiple Alignment using Fast Fourier Transform (Katoh et al., 2002). The resulting Multiple Sequence Alignment (500 sequences with ≥35% similarity) was used to calculate the conservation scores, which ranged from 1 (variable) to 9 (highly conserved). The Fab residues were color coded based on conservation scores obtained.
Correlation analysis
Correlations between all the residues in the six systems were analyzed for the entire 100-ns MD trajectory (25 000 frames) using the normalized covariance to characterize the correlation in motion of protein residues (Ichiye and Karplus, 1991; Hünenberger et al., 1995; Tai et al., 2001; Young et al., 2001), ranging from −1 to 1. If two residues move in the same (opposite) direction in the majority of the frames, the motion is considered as (anti-)correlated, and the correlation value is close to −1 or 1. If the correlation value between two residues is close to zero, then they are generally uncorrelated. The correlations evaluation was performed by using CARMA (Glykos, 2006).
Weighted network, community analysis, optimal/suboptimal paths in Fab/peptide systems
A network is defined as a set of nodes with connecting edges. The nodes in this work represent the amino-acid residues and the essential bound water molecules. An edge between two nodes was defined if any heavy atoms from the two residues/water molecules are within 4.5 Å of each other in over 75% of the analyzed frames. Neighboring residues in sequence are not considered to be in contact because they will form numerous trivial suboptimal paths in the weighted network. The dynamical networks were constructed based on the 100-ns trajectories. To study the water effect on the network, water molecules were deleted from the system and analyzed separately. The communities within a network were defined as the substructures of the network in which the nodes are more heavily interconnected to each other than to other nodes. The community was identified by the Girvan–Newman algorithm (Girvan and Newman, 2002). For simplicity and clarity, communities with member number <10 and edges with betweenness <1000 were not considered. The length of a path between two distant nodes is the sum of the edges weights between consecutive nodes along the path. The shortest path is obtained by optimization of this length. Suboptimal paths are determined in addition to the shortest path to measure the path degeneracy of the network. The network, community and optimal/suboptimal paths analysis of Fab/peptide systems was performed by using NetworkView (Eargle and Luthey-Schulten, 2012) module in VMD.
Results and discussion
Structural features of the apo and bound forms of the Fab–prion peptide complex
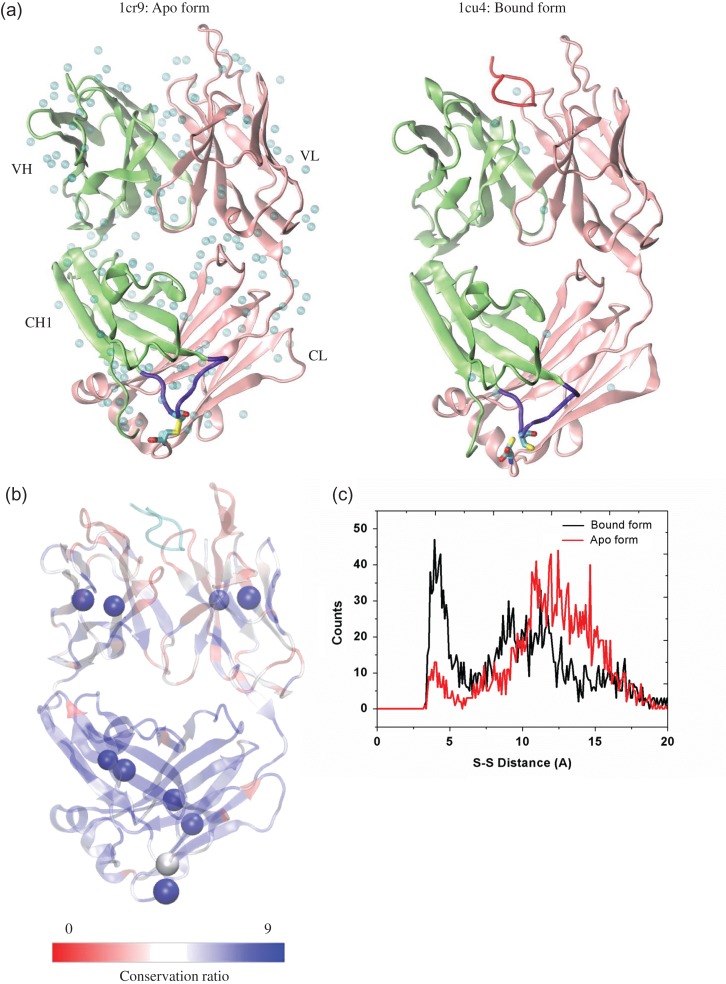
We first study the structural change between the apo and bound forms of the Fab/peptide complex. Based on the two crystal structures, there is 1.08 Å overall root mean square deviation (RMSD) change in Fab upon binding a peptide (Fig. 1a). There is an inter-chain disulfide bond (inter-SS bond) between the C-terminal Cys214 of the light chain and Cys125 on the CH1-1 loop of the heavy chain in the apo form, which was broken in the bound form. To evaluate the conservation of this inter-SS bond in immunoglobulins, we surveyed 500 Fab sequences with >35% sequence similarity and performed a multiple alignment. We found that the C-terminal cysteine of the light chain is highly conserved (score of 9), while the cysteine on the heavy chain CH1-1 loop is much less conserved (score of 5) (Fig. 1b). The CH1-1 loop, where the inter-chain disulfide bond is located, has a significant conformational change between the Apo and bound antibody structures (Kanyo et al., 1999). Forming or breaking the inter-chain disulfide bond may constrain or relax the conformation of the flexible CH1-1 loop.
Fig. 1.
Structural evaluation of the simulation systems. (a) Crystal structure of Fab 3F4 in apo form (PDB:1cr9) and the complex (PDB: 1cu4) with its cognate peptide (SHaPrP104-113). Light chain, heavy chain, the cognate peptide, CH1-1 loop, the inter-chain disulfide bond and the water molecules in the crystal are colored in pink, lime, red, purple, yellow and cyan, respectively. (b) Conservation ratio of the Fab residues mapping onto the Fab/peptide complex in this study. Residues with low up to high conservation ratio are colored from red to blue. The cysteines formed intra- and inter-disulfide bonds were represented by beads, and the peptide (conservation not evaluated) was colored by cyan. (c) Distribution of the distance between the sulfur atoms from light chain Cys214 and heavy chain Cys125 in the bound form and apo form of Fab without inter-chain disulfide bond.
To consider this effect, we tested both keeping and breaking the inter-SS bond for the apo and bound systems, resulting in four systems. To reduce the statistical noise, we repeated the simulations of the four systems with alternative starting configurations and initial velocity distributions. Two unbound systems were also simulated by removing the prion peptide. Details of these 10 systems were summarized in Table I. The backbone RMSD of the 10 Fab-related systems (Figs S1 and S2) showed that there were larger fluctuations of the Fab in the apo or the unbound form than in the bound form, and most systems reach equilibrium after ~50 ns simulation. For the apo form without disulfide bond, the 2D RMSD plot indicated that a new conformation cluster appeared around 35–40 ns (Fig. S2d), and the disulfide bond suppressed this structural variation. We further evaluated the distance distribution of two Cys sulfur atoms in the bound and apo systems, without forming covalent disulfide bond. The distances between the sulfur atoms during the 100-ns simulation in these two systems range from 3.2 to ~17.0 Å, which is larger than the S–S bond length of 2.05 Å. It is interesting to note that non-bonded cysteines still largely remained at close contact distance, indicating that a disulfide bond is not needed to keep the interaction between these two regions harboring Cys residues (Fig. 1c). However, without peptide binding in the apo form, these two Cys residues are separated by as much as 10–15 Å. Therefore, our simulation explained why the apo form crystal structure has to be in reduced form with a disulfide to keep the heavy chain and the light chain interaction in the region.
Inter-chain disulfide bond reduces the communication between the subdomains of the light and heavy chains
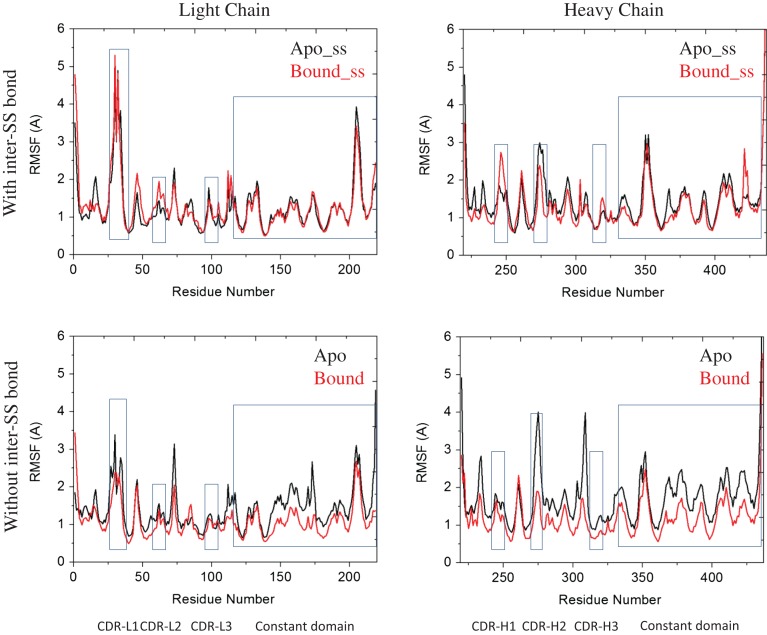
We first evaluate the mobility change of loops of Fab upon prion peptide binding with/without the inter-SS bond by using root-mean-square fluctuation (RMSF) values (Fig. 2). We found that most of the loop regions showed RMSF >2 Å, including CDR loops, VL-2/VH-2 loops and CH1-1 loop. When the inter-SS bond is broken, the RMSFs of the CDR loops are reduced upon peptide binding and the constant region loops also respond. Interestingly, when the inter-SS bond forms, the RMSFs of Fab in the apo and bound forms remained unchanged, indicating that Fab did not respond to the peptide binding and that there were less cross-talks between Fab variable and constant domains.
Fig. 2.
Residue-based RMSF of the systems studied in this work during the 100-ns MD simulation the rectangle and square showed the RMSF of CDR loops, and constant domain respectively.
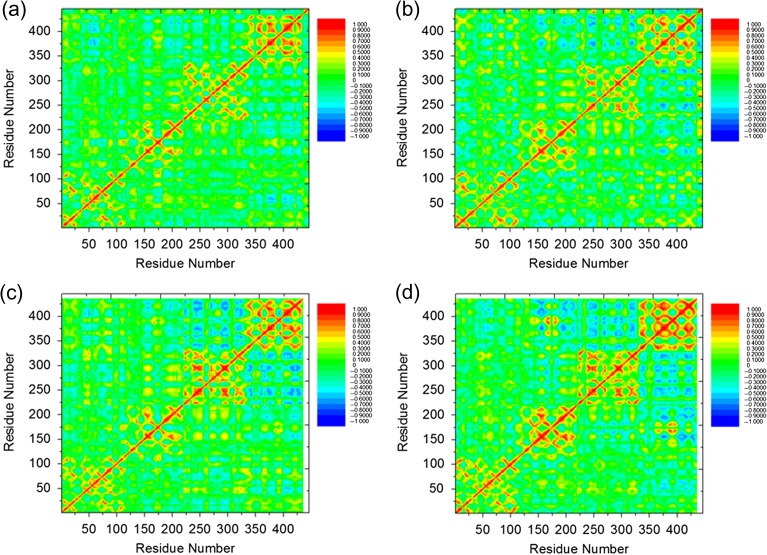
Cross-talk between antibody subdomains is essential for antibody–antigen interactions. Li et al. (2015) probed the flexibility/rigidity response of three sets of antigen–Fab complexes upon affinity maturation by MD simulation sampling coupled with distance constraints. The plots of six antigen–Fab complexes showed that the motions of Fab subdomains are often correlated. To study the inter-subdomain cross-talk with or without this inter-chain disulfide bond, we measured the degree of coupled motion in the Fab/peptide complex by normalizing the cross-correlation matrix of atomic fluctuations over time frames of 0–100, 20–60 and 60–100 ns. For all systems, there were four major local correlations along the diagonal, representing the four subdomains, i.e. VL, CL, VH and CH1 domains (Fig. 3 and Fig. S3). Besides these, there are couplings between distant parts of the complex in some of these systems; most are anti-correlation. In the two bound form systems, there is stronger long-distance anti-correlation between subdomains without than with the inter-SS bond. The most negatively correlated subdomains are VH and CH-1 domains. In the two apo form systems, there is much stronger correlation among all Fab subdomains, and the motion correlations between subdomains are not sensitive to the inter-SS bond. By comparing the structures in the bound and apo forms, the binding of the peptide generally decreases the inter-domain correlation (Fig. 3).
Fig. 3.
Correlation analysis (Cij) of the motion during a 100-ns MD simulation of the Fab/peptide complex with the inter-chain disulfide bond in (a) bound and (c) Apo forms and without the inter-chain disulfide bond in (b) bound and (d) Apo forms. Residues with highly (anti)correlated motion are red (blue).
Many studies have shown the conformational diversity of a certain antibody. This diversity is essential for antigen binding (James et al., 2003; Keskin, 2007). There are equilibrium intermediate conformations that are more favorable for antigen binding than the apo form. To search the intermediate states between the apo and bound forms, the unbound systems, in which the peptide was removed manually, were simulated. The structural properties, e.g. RMSD, and the motion correlation of the intermediate structures are very similar to those of the apo state. Moreover, the intermediate structures also showed stronger correlation among all Fab subdomains than those bound structures and the intermediate structures are not sensitive to the inter-SS bond, which is similar to the apo form structures. This indicates that the intermediate structures share similar structural and dynamical properties with the apo structures.
To evaluate the effects of the inter-chain SS bond on peptide binding, we evaluated the decomposed energy, total energy of each individual system and the binding energy between the prion peptide and Fab (Table II). The binding energy (−158.3 kcal/mol in the first run and −176.8 kcal/mol in the repeat run) without the inter-chain SS bond is essentially lower (more favorable) than that (2.3 kcal/mol in the first run and −40.9 kcal/mol in the repeat run) with the inter-chain SS bond (Table III). Moreover, Fab showed lower energy with the inter-chain SS bond in the apo/unbound form and lower energy without the inter-chain SS bond in the bound form. From the energy landscape standpoint, this explains that in crystal structures, the inter-chain SS bond is formed in the apo structure (1cr9) but is broken in bound structure (1cu4).
Table II.
Summarization of energy decomposition of simulation systems using GBMV
| No. | System | SS | Peptide | aEVDW | bEelec | cEint | dEsol | eEtotal |
|---|---|---|---|---|---|---|---|---|
| 1 | Apo | No | No | −1994.1 ± 16 | −5997.9 ± 116.3 | 3434.4 ± 19 | −5272.9 ± 107.7 | −9830.4 ± 35 |
| 2 | Apo | Yes | No | −2000.4 ± 19.6 | −6112.9 ± 112.4 | 3418.2 ± 16.6 | −5206.7 ± 106.1 | −9901.8 ± 36.5 |
| 3 | Bound | No | Yes | −2114.3 ± 21.3 | −6802.1 ± 123.6 | 3544.4 ± 17.4 | −4873 ± 101.3 | −10 245 ± 32.9 |
| 4 | Bound | Yes | Yes | −2082.8 ± 21.1 | −6438.2 ± 119.9 | 3503.3 ± 17.6 | −5138.1 ± 110.4 | −10 155.8 ± 31.8 |
| 5 | Unbound | No | No | −2049 ± 23.8 | −6354.6 ± 143 | 3352.1 ± 17 | −4904.6 ± 128.3 | −9956.1 ± 33.6 |
| 6 | Unbound | Yes | No | −2093.3 ± 15.9 | −6384.1 ± 96.5 | 3370 ± 17.6 | −4876.7 ± 84.8 | −9984 ± 25.5 |
| 7 | Peptide | N/A | N/A | −21.4 ± 3 | 71.6 ± 42 | 77.3 ± 5.2 | −383.7 ± 40.9 | −256.3 ± 4.6 |
| 8 | Apo | No | No | −2059 ± 18 | −6254.2 ± 98 | 3343.4 ± 18.4 | −4949 ± 84.5 | −9918.8 ± 32.2 |
| 9 | Apo | Yes | No | −2096.9 ± 15.5 | −6195.5 ± 123.7 | 3370.2 ± 18.4 | −5049.5 ± 104 | −9971.7 ± 31.4 |
| 10 | Bound | No | Yes | −2126.4 ± 20.4 | −6983.4 ± 107.6 | 3430.7 ± 17.6 | −4672.8 ± 98.4 | −10 351.9 ± 32.2 |
| 11 | Bound | Yes | Yes | −2125.9 ± 16.4 | −6679.6 ± 116.1 | 3432 ± 20.9 | −4895.3 ± 104 | −10 268.9 ± 30.8 |
The unit of the energy is kcal/mol.
aVan der Waals potential energy.
bElectric potential energy.
cInternal energy.
dSolvation energy.
eTotal energy.
Table III.
Binding energy between prion peptide and Fab with and without the inter-chain disulfide bond
| No SS | With SS | |
|---|---|---|
| Run 1 | −158.3 | 2.3 |
| Run 2 | −176.8 | −40.9 |
The binding energy is calculated by . The unit of the energy is kcal/mol.
Correlation of motion between the peptide and Fab suggests potential allosteric sites
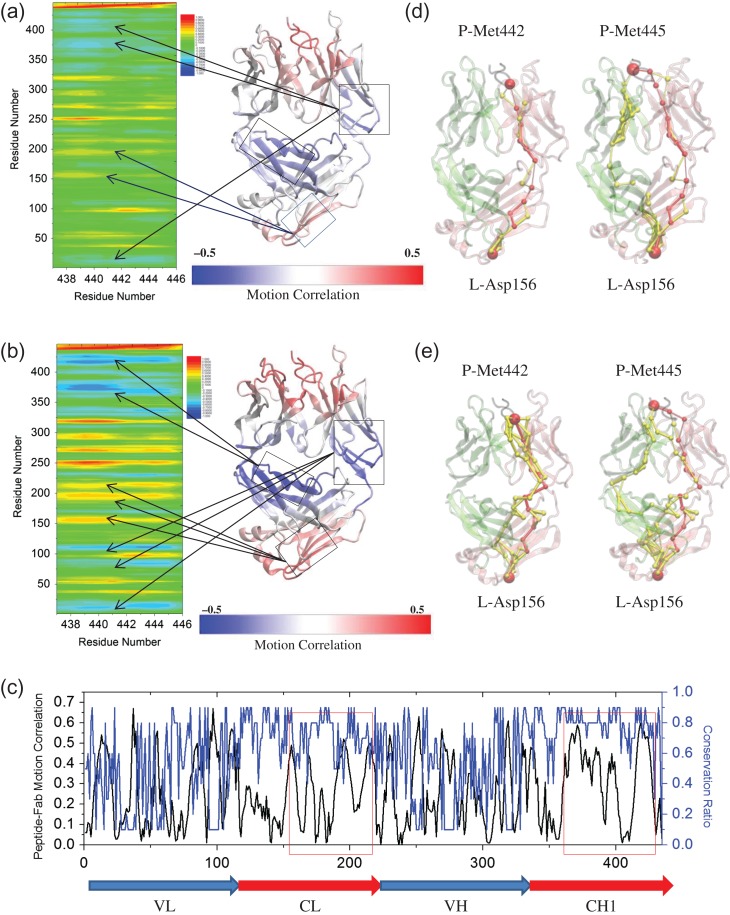
That antibody–antigen recognition is an allosteric process, which means that there is distant regulation and correlations of the motions are expected and well established (Sela-Culang et al., 2012). Along these lines, we evaluated the coupled motions between the peptide and Fab with/without the inter-SS bond in the bound systems. Besides the correlations of the CDR loops, the C-terminal residues of the light chain, which is >60 Å away from the peptide, showed considerable positive correlations with the peptide (Fig. 4). Generally, there were stronger correlations of motion without the inter-SS bond than with it (Fig. 4a and b), indicating that the disulfide bond restricts the motion and allosteric communications. The common anti-correlations in the two bound systems are from the CH1 domain and the hinge region between the variable and constant domains. The CH1-1 loop did not show significant correlation with the peptide (0.1–0.2). The motion of both light chain and heavy chain constant domains, i.e. CL and CH1 domains, which have high conservation ratio, is highly correlated with the peptide. Interestingly, the CL domain showed positive motion correlation while CH1 domain showed negative motion correlation (Fig. 4c).
Fig. 4.
Distant motion correlation and allosteric signals between prion peptide and Fab controlled by the inter-chain disulfide bond. Correlation analysis (Cij) of the motion between two essential methionines of prion peptide and Fab in the complex system (a) with and (b) without the inter-chain disulfide bond during a 100-ns MD simulation. The averaged Cij is obtained by averaging the Cij values from Met442 and Met445. Residues with highly correlated motion to loosely correlated motion, and to highly anti-correlated motion are colored from red, green/yellow, to blue. The correlation of the motion between peptide and each individual Fab residue is mapped onto the Fab structure from highly correlated (red) to loosely correlated (white), and to highly anti-correlated (blue). (c) The absolute values of peptide–Fab motion correlation (black line) and conservation ratio (blue line, 0–0.9, higher value indicates higher conservation) of each individual residue of the bound system without inter-SS bond. Optimal and suboptimal paths connecting Met442/Met445 of prion peptide to Asp156 (d) with and (e) without the inter-chain disulfide bond. Optimal and suboptimal paths are colored by red and yellow, respectively. The thickness of each edge is proportional to the number of suboptimal paths that cross it during the calculation. Light chain, heavy chain and the peptide are colored by pink, lime and black, respectively. Met442/Met445 and Asp156 are represented by red beads.
We used optimal path analysis to further elucidate how the information is transferred between the peptide and those distant—albeit strongly coupled—residues. Kanyo et al. (1999) reported that Met109 and Met112 (i.e. Met442 and Met445 in this work) of the prion peptide determine the specificity between peptide and Fab, and are crucial for the PrPC to PrPSc conformational transition. These two hydrophobic residues penetrated deep into distinct hydrophobic cavities formed by the heavy and light chain CDR loops. Given the crucial roles of these two residues in antigen binding, we evaluated the allosteric pathways between them and Fab. For the other end of the path, we chose Asp156 at the bottom of the CL domain which has considerable correlation (0.49) with the Mets of the peptide. Consistent with the inter-subdomain motion correlation and binding energy, systems without the inter-SS bond have more sub-optional paths than those with the inter-chain disulfide bond (Fig. 4d and e and Table IV). The communication pathways between Met442 and Asp156 mainly cross the light chain while the pathways between Met445 and Asp156 across both light and heavy chains. The suboptimal pathways cross either the CH1 domain or the hinge region between the light chain variable/constant domains. Both domains showed strong negative motion correlation with the peptide. In all four cases, the optimal pathway mainly crosses the light chain.
Table IV.
Number of suboptimal pathways from distant site Asp156 to Met442/445 of peptide in the Fab/peptide complex with and without the inter-chain disulfide bond
| Res no. | With water | SS bond | No. of paths |
|---|---|---|---|
| 442 | No | Yes | 28 |
| 442 | No | No | 39 |
| 445 | No | Yes | 96 |
| 445 | No | No | 125 |
| 442 | Yes | Yes | 28 |
| 442 | Yes | No | 61 |
| 445 | Yes | Yes | 28 |
| 445 | Yes | No | 31 |
Key bound water molecules participate in the Fab/peptide complex
Water molecules play essential roles in protein stability, dynamics and function (Bellissent-Funel et al., 2016). Recently, Srivastava et al. (2016) used mechanical perturbation to elucidate how mutations change the allosteric response through changes in the hydrogen-bond network. Due to the different hydrophobicity of mutated residues, they hypothesized that the binding/disassociation of local water molecules around these mutated residues affects the conformational dynamics of the antibody fragment systems.
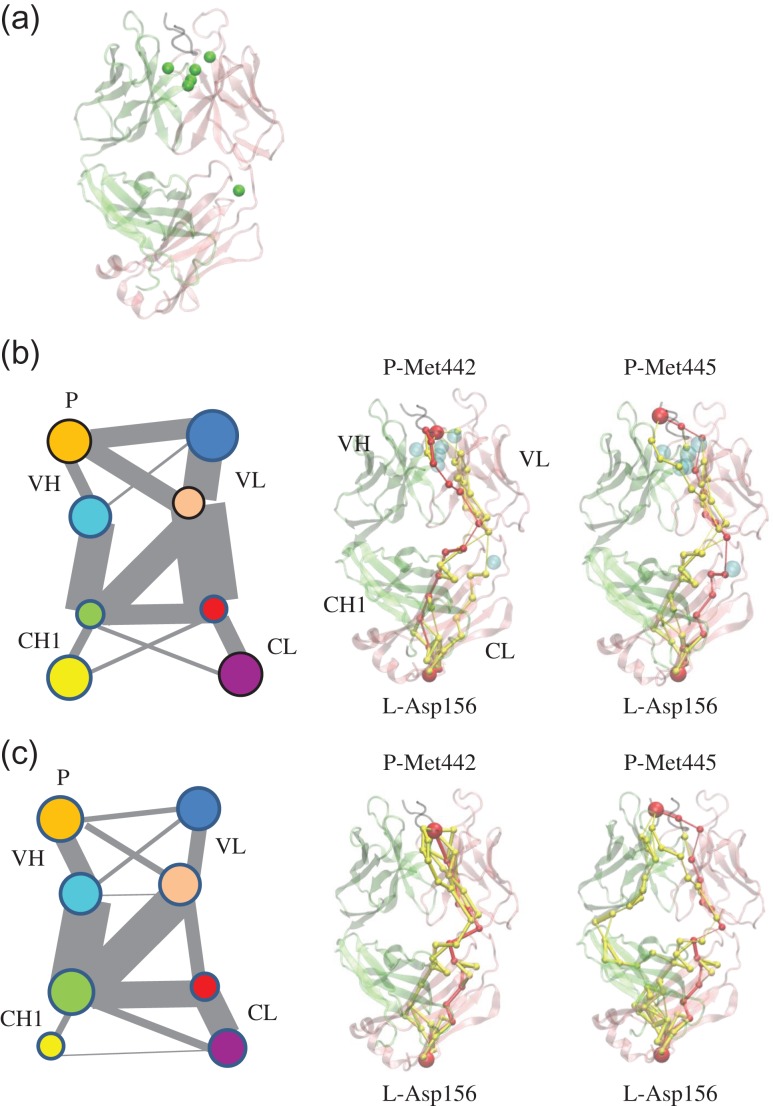
To consider the water effects, we studied the trajectories focusing on the water molecules, which stay within 4.5 Å of the protein atoms for 75% of the simulation time. In the two bound systems, most water molecules were on the interface between the peptide and Fab while in the two apo systems, there were almost no water molecules around the CDR. Besides the interface water, there are water molecules near the hinge region and between the light and heavy constant domains. However, there are no conserved water molecules in the four systems. We identified six water molecules (Fig. 5a) in the bound system without the inter-SS bond (1cu4), which meet the above criteria. To study their role in the dynamics of the Fab/peptide complex, we carried out community analysis of the network formed by the Fab/peptide complex and the water molecules. Members within the same community communicate with each other more frequently than with members from another community. The complex was divided into eight communities or four community pairs, which roughly represented the four subdomains of Fab, VH, VL, CH1 and CL. The associated water molecules participated in communities to which they were close (Fig. 5b). To study the role of water in the communication, we removed the water molecules and re-evaluated the communities: both community size and communications between the communities were modified (Fig. 5c).
Fig. 5.
Associated water molecules participate the antibody–antigen complex motion and allosteric signal pathway in the system of the bound structure without inter-SS bond (1cu4). (a) Associated water molecules (green beads) in the antibody–antigen complex. Community analysis (left panel) and allosteric signal analysis (middle and right panel) (b) with and (c) without the consideration of associated water molecules. The communities are represented in circles in different colors and the size of the communities is proportional to the diameter. The gray lines represented the communications between different communities and the communication strength (betweeness) is proportional to width of the lines. P, the prion peptide; VL, light chain variable domain; CL, light chain constant domain; VH, heavy chain variable domain; CH1, heavy chain constant domain-1. Light chain, heavy chain and the peptide are colored by pink, lime and black, respectively. Optimal and suboptimal pathways are colored by red and yellow, respectively. Met442/Met445, Asp156 and the associated water molecules are represented by beads.
We further evaluated the pathways between Met442/445 and Asp156 with the consideration of associated water molecules. We found that water molecules that participated in the protein network and certain additional water molecules can be on the suboptimal or optimal pathways. Though these water molecules were not on the shortest path (Fig. 5b and c), they dramatically influence (increase or decrease) the number of possible pathways (Table IV). These analyses showed that the associated local water molecules influenced the conformational dynamics of the antibody fragment systems and the allosteric communication between the prion peptide and the Fab constant domain.
Conclusion
Conformational change and subsequent cross-talks between subdomains are essential for antibody–antigen recognition. In this work, MD simulations of four systems derived from the bound and apo forms of the Fab/prion peptide complex were performed with/without the inter-chain disulfide bond between the C-terminal of light chain and the CH1-1 loop of the heavy chain. To evaluate the variance of the motion correlation and the network properties, we performed analysis in the time frames of 0–100, 20–60 and 60–100 ns. We did find variance of the motion correlation (Fig. S4) and the allosteric flow (Fig. S5) in all three frames; however, the variance is not significant within the 100-ns scale nor among the different time frames. We observed that the inter-SS bond constrains the conformational changes, which appear crucial for Fab–peptide binding and decreased the binding affinity between the prion peptide and Fab. Although the disulfide bond covalently links the C-terminal of the light chain and the CH1-1 loop of heavy chain, it results in the inhibition of inter-domain communications and thus significantly reduces the information transferred between the Fab subdomains. Through motion correlation analysis, we found that distant Fab domains may have significant motion correlation or anti-correlation with the peptide. These sites are potentially allosteric sites for Fab–peptide binding. The optimal path analysis indicated that the inter-SS bond dramatically decreased the possible paths between these sites and the peptide. Analysis of the role of water molecules in antibody–antigen recognition by network and community analysis suggests that bound water molecules become part of the Fab/peptide complex and their participation in the network enhanced the light–heavy chain and the variable-constant domain communications.
Based on the definition of IgG subtypes (Liu and May, 2012), the disulfide bond between light and heavy chains should form in IgG2a, IgG3 and IgG4 but not in the IgG1. In the structural protein data bank, we found several antibodies, IgG2a, IgG3 and IgG4, in the apo form and/or bound form with different antigens. Among these structures, some do not show electron density of this disulfide bond due to the high flexibility (Table V). In structures with electron density at this region, some formed the inter-chain disulfide bond and others did not. Although no strong correlation between the inter-chain disulfide bond and antigen binding, it is interesting that this disulfide bond does not always form. The Fab region in this study is a fragment of mouse IgG2a antibody 3F4 (Kanyo et al., 1999). IgG2a is the classical disulfide bond structure and IgG2b and IgG2a/b are two major non-classical structures with the inter-chain disulfide bonds being different from IgG2a. It was reported that IgG2a has either similar or higher affinity and biological activity than IgG2b (Dillon et al., 2008; Martinez et al., 2008). This suggests that the distribution of the inter-chain disulfide bonds influences the biological functions of IgG2. Our results indicate that in this specific case, the inter-chain disulfide bonds between the heavy and light chains influence the antibody function, e.g. binding affinity and allosteric communication. Future studies of the heavy–light chain disulfide bond functions in other IgG subtypes will further illuminate its behavior and function.
Table V.
Available structures of Fab or full antibodies in their bound or apo form with the potential formation of the inter-chain disulfide bond between CH-1 loop and C-terminal of light chain
| Cluster no. | PDB code | Apo/bound | Inter-chain SS | Template | Fragments |
|---|---|---|---|---|---|
| 1 | 1cr9 | Apo | Yes | IgG2a | Fab |
| 1 | 1cu4 | Bound | No | IgG2a | Fab |
| 2 | 1cfq | Apo | No | IgG2a | Fab |
| 2 | 1cfn | Bound | No | IgG2a | Fab |
| 2 | 1cfs | Bound | No | IgG2a | Fab |
| 2 | 1cft | Bound | No | IgG2a | Fab |
| 3 | 1bbd | Apo | No | IgG2a | Fab |
| 3 | 1a3r | Bound | Yes | IgG2a | Fab |
| 4 | 1mnu | Apo | Yes | IgG2a | Fab |
| 4 | 1mpa | Bound | Yes | IgG2a | Fab |
| 4 | 2mpa | Bound | Yes | IgG2a | Fab |
| 5 | 2hkh | Apo | Yes | IgG2a | Fab |
| 5 | 2hkf | Bound | Yes | IgG2a | Fab |
| 6 | 3bkm | Apo | No density | IgG2a | Fab |
| 6 | 3bkc | Apo | No density | IgG2a | Fab |
| 6 | 3bkj | Bound | No density | IgG2a | Fab |
| 6 | 3bae | Bound | No density | IgG2a | Fab |
| 7 | 1igt | Apo | Yes | IgG2a | Full |
| 8 | 4hdi | Apo | Yes | IgG3 | Fab |
| 9 | 3i50 | Bound | No density | IgG3 | Fab |
| 10 | 1uz6 | Apo | No density | IgG3 | Fab |
| 11 | 5DK3 | Apo | Yes | IgG4 | Full |
A major aim in immunotherapies is antibody design. This challenge has sparked innumerable approaches. We believe that detailed analysis of available crystal structures and elucidation of molecular fluctuations involved in allosteric communications could provide useful guidelines. Within this framework, conserved water molecules should not be overlooked as they can play important roles in information transfer. Here, we carried out a detailed analysis along these lines. Our simulations and residue contact network analysis revealed that the dispensable inter-chain disulfide bond restrains Fab's conformational changes, resulting in inhibition of the cross-talk between the Fab subdomains and thereby may prevent the prion peptide binding. The next step is incorporating nature's mode of recognition and allosteric control—affirmed here—in design strategies. We are now taking steps toward addressing this challenge focusing on amyloid-recognizing antibodies.
Supplementary data
Supplementary data are available at PEDS online.
Abbreviations
Fab, antigen-binding fragment; MD, molecular dynamics; VL, light chain variable domain; CL, light chain constant domain; VH, heavy chain variable domain; CH1, heavy chain constant domain-1; inter-SS bond, inter-chain disulfide bond.
Funding
This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health [under contract number HHSN261200800001E]. This research was supported (in part) by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. J.Z. was supported in part by the Intramural Research Program of the National Institutes of Health, National Institute on Deafness and Other Communication Disorders. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov).
References
- Adachi M., Kurihara Y., Nojima H., Takeda‐Shitaka M., Kamiya K. and Umeyama H. (2003) Protein Sci., 12, 2125–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellissent-Funel M.-C., Hassanali A., Havenith M., Henchman R., Pohl P., Sterpone F., van der Spoel D., Xu Y. and Garcia A.E. (2016) Chem. Rev., 116, 7673–7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhrman G., Holzapfel G., Fetics S. and Mattos C. (2010) Proc. Natl. Acad. Sci., 107, 4931–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dam T.K., Torres M., Brewer C.F. and Casadevall A. (2008) J. Biol. Chem., 283, 31366–31370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Genst E., Messer A. and Dobson C.M. (2014) Biochim. Biophys. Acta, 1844, 1907–1919. [DOI] [PubMed] [Google Scholar]
- Dillon T.M., Ricci M.S., Vezina C. et al. , (2008) J. Biol. Chem. 283, 16206–16215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eargle J. and Luthey-Schulten Z. (2012) Bioinformatics, 28, 3000–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiser A., Do R.K.G. and Šali A. (2000) Protein Sci., 9, 1753–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girvan M. and Newman M.E. (2002) Proc. Natl. Acad. Sci., 99, 7821–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glykos N.M. (2006) J. Comput. Chem., 27, 1765–1768. [DOI] [PubMed] [Google Scholar]
- Hünenberger P., Mark A. and Van Gunsteren W. (1995) J. Mol. Biol., 252, 492–503. [DOI] [PubMed] [Google Scholar]
- Ichiye T. and Karplus M. (1991) Proteins, 11, 205–217. [DOI] [PubMed] [Google Scholar]
- James L.C., Roversi P. and Tawfik D.S. (2003) Science, 299, 1362–1367. [DOI] [PubMed] [Google Scholar]
- Janda A. and Casadevall A. (2010) Mol. Immunol., 47, 1421–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale L., Skeel R., Bhandarkar M., et al. (1999) J. Comput. Phys. 151, 283–312. [Google Scholar]
- Kanyo Z.F., Pan K.M., Williamson R.A., Burton D.R., Prusiner S.B., Fletterick R.J. and Cohen F.E. (1999) J. Mol. Biol., 293, 855–863. [DOI] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K.i. and Miyata T. (2002) Nucleic Acids. Res., 30, 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney B.M., Johnson C.W., Roberts D.M., Swartz P. and Mattos C. (2014) J. Mol. Biol., 426, 611–629. [DOI] [PubMed] [Google Scholar]
- Keskin O. (2007) BMC. Struct. Biol., 7, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T. and Ben-Tal N. (2005) Nucleic Acids. Res., 33, W299–W302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.S., Feig M., Salsbury F.R. Jr and Brooks C.L. III (2003) J. Comput. Chem., 24, 1348–1356. [DOI] [PubMed] [Google Scholar]
- Li T., Tracka M.B., Uddin S., Casas-Finet J., Jacobs D.J. and Livesay D.R. (2014) PLoS ONE, 9, e92870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Tracka M.B., Uddin S., Casas-Finet J., Jacobs D.J. and Livesay D.R. (2015) PLoS Comput. Biol., 11, e1004327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. and May K. (2012) mAbs, 4, 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B., Zhao J. and Nussinov R. (2016) Biochim. Biophys. Acta., 1860, 2672–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell A.D., Bashford D., Bellott M. et al. , (1998) J. Phys. Chem. B. 102, 3586–3616. [DOI] [PubMed] [Google Scholar]
- Martí-Renom M.A., Stuart A.C., Fiser A., Sánchez R., Melo F. and Šali A. (2000) Annu. Rev. Biophys. Biomol. Struct., 29, 291–325. [DOI] [PubMed] [Google Scholar]
- Martinez T., Guo A., Allen M.J. et al. , (2008) Biochemistry 47, 7496–7508. [DOI] [PubMed] [Google Scholar]
- Mattos C. (2002) Trends Biochem. Sci., 27, 203–208. [DOI] [PubMed] [Google Scholar]
- Mian I.S., Bradwell A.R. and Olson A.J. (1991) J. Mol. Biol., 217, 133–151. [DOI] [PubMed] [Google Scholar]
- Oda M., Kozono H., Morii H. and Azuma T. (2003) Int. Immunol., 15, 417–426. [DOI] [PubMed] [Google Scholar]
- Pellequer J.L., Chen S.W., Roberts V.A., Tainer J.A. and Getzoff E.D. (1999) J. Mol. Recognit., 12, 267–275. [DOI] [PubMed] [Google Scholar]
- Pritsch O., Hudry-Clergeon G., Buckle M., Pétillot Y., Bouvet J.P., Gagnon J. and Dighiero G. (1996) J. Clin. Invest., 98, 2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovis T.L. and Legname G. (2014) Viruses, 6, 3719–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph M.G. and Wilson I.A. (2002) Curr. Opin. Immunol., 14, 52–65. [DOI] [PubMed] [Google Scholar]
- Šali A. and Blundell T.L. (1993) J. Mol. Biol., 234, 779–815. [DOI] [PubMed] [Google Scholar]
- Sela-Culang I., Alon S. and Ofran Y. (2012) J Immunol, 189, 4890–4899. [DOI] [PubMed] [Google Scholar]
- Sela-Culang I., Kunik V. and Ofran Y. (2013) Front Immunol., 4, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A., Tracka M.B., Uddin S., Casas-Finet J., Livesay D.R. and Jacobs D.J. (2016) Biophys. J., 110, 1933–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai K., Shen T., Börjesson U., Philippopoulos M. and McCammon J.A. (2001) Biophys. J., 81, 715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschuk R., Van der Merwe J., Marciniuk K., Potter A., Cashman N., Griebel P. and Napper S. (2015) Prion, 9, 292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thielges M.C., Zimmermann J.r., Yu W., Oda M. and Romesberg F.E. (2008) Biochemistry, 47, 7237–7247. [DOI] [PubMed] [Google Scholar]
- Torres M. and Casadevall A. (2008) Trends Immunol., 29, 91–97. [DOI] [PubMed] [Google Scholar]
- Tudor D., Yu H., Maupetit J., Drillet A.-S., Bouceba T., Schwartz-Cornil I., Lopalco L., Tuffery P. and Bomsel M. (2012) Proc. Natl. Acad. Sci., 109, 12680–12685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B. and Sali A. (2014) Curr. Protoc. Bioinformatics, 5.6. 1–5.6.32. [DOI] [PubMed] [Google Scholar]
- Wei G., Xi W., Nussinov R. and Ma B. (2016) Chem. Rev. 116, 6516–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.A., Gonfloni S., Superti-Furga G., Roux B. and Kuriyan J. (2001) Cell, 105, 115–126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.