Abstract
Dual stimuli-sensitive mixed polymeric micelles (MM) are developed for co-delivery of the endogenous tumor suppressor miRNA-34a and the chemotherapeutic agent doxorubicin (Dox) into cancer cells. The novelty of the system resides in two stimuli-sensitive prodrugs, a matrix metalloproteinase 2 (MMP2)-sensitive Dox conjugate and a reducing agent (glutathione, GSH)-sensitive miRNA-34a conjugate, self-assembled in a single particle decorated with a polyethylene glycol corona for longevity and a cell-penetrating peptide (TATp) for enhanced intracellular delivery. The MMP2-sensitivity of the system results in 3-fold higher cytotoxicity in MMP2-overexpressing HT1080 cells compared to low MMP2-expressing MCF7 cells. Cellular internalization of Dox increases by more than 70% after inclusion of TATp to the formulation. MMP2-sensitive MM also inhibits proliferation and migration of HT1080 cells. Moreover, GSH-sensitive MM allows for an efficient downregulation of Bcl2, survivin, and notch1 (65%, 55% and 46%, respectively) in HT1080 cells. Combination of both conjugates in dual sensitive MM reduces HT1080 cell viability to 40% and expression of Bcl2 and survivin. Finally, 50% cell death is observed in 3D models of tumor mass. The results confirm the potential of the MM to co-deliver miRNA-34a and doxorubicin triggered by dual stimuli inherent of tumor tissues.
Keywords: stimuli-sensitive, mixed micelles, miRNA-34a, doxorubicin, spheroid
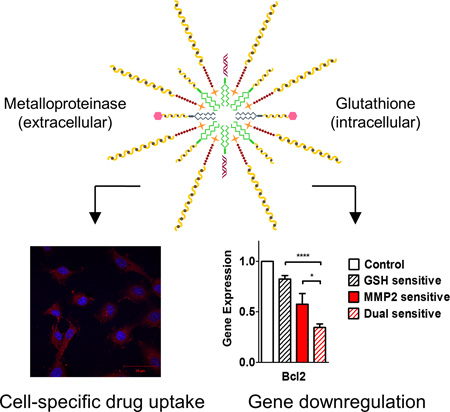
Graphical abstract
The designed system is composed of two stimuli-sensitive conjugates assembled in a particle decorated with a PEG corona and a cell-penetrating peptide. It selectively releases doxorubicin and miRNA-34a into the target tissue after cleavage by extracellular proteinases and an intracellular reductive environment, which promotes decrease in cell viability and downregulation of genes involved in tumor progression.
1. Introduction
Development of novel “smart” pharmaceutical preparations with increased activity and sensitivity to local biological stimuli is an important part of medical biotechnology. The preparations based on natural biological materials are of special interest, including the use of nucleic acids as promoters of RNA interference (RNAi).[1–4]
Double-stranded, small, non-coding microRNAs (miRNA) efficiently regulate gene expression.[1,2] Specifically, miRNA-34a is a p53-induced tumor suppressor commonly downregulated in cancer cells.[3, 5] It induces apoptosis and inhibits cell proliferation and migration through targeting a variety of oncogenes, such as Bcl2, notch1 and survivin.[6, 7]
Combination of miRNA-34a and chemotherapeutic drugs is a promising approach to improve survival and clinical outcome of the therapies currently used to treat cancer. In fact, synergistic effects of RNAi by miRNA-34a and DNA intercalation by doxorubicin (Dox) have been reported in sensitive[8] and resistant[6] tumor cell lines. Such strategy could not only promote same efficacy by using much lower doses of the chemotherapeutic drug but also lead to consequent decrease in off-site toxicity compared to the single treatment.
However, systemic delivery of small nucleotides to tumor tissues remains an important challenge to RNAi. Increasing their longevity in the bloodstream for tumor accumulation while protecting them from nucleases is a crucial step for successful miRNA transfection. Oligonucleotide internalization by the cells is also inefficient due to their negative charge. Finally, their action intracellularly depends on efficient endosomal escape and subsequent lysosomal degradation.[9, 10]
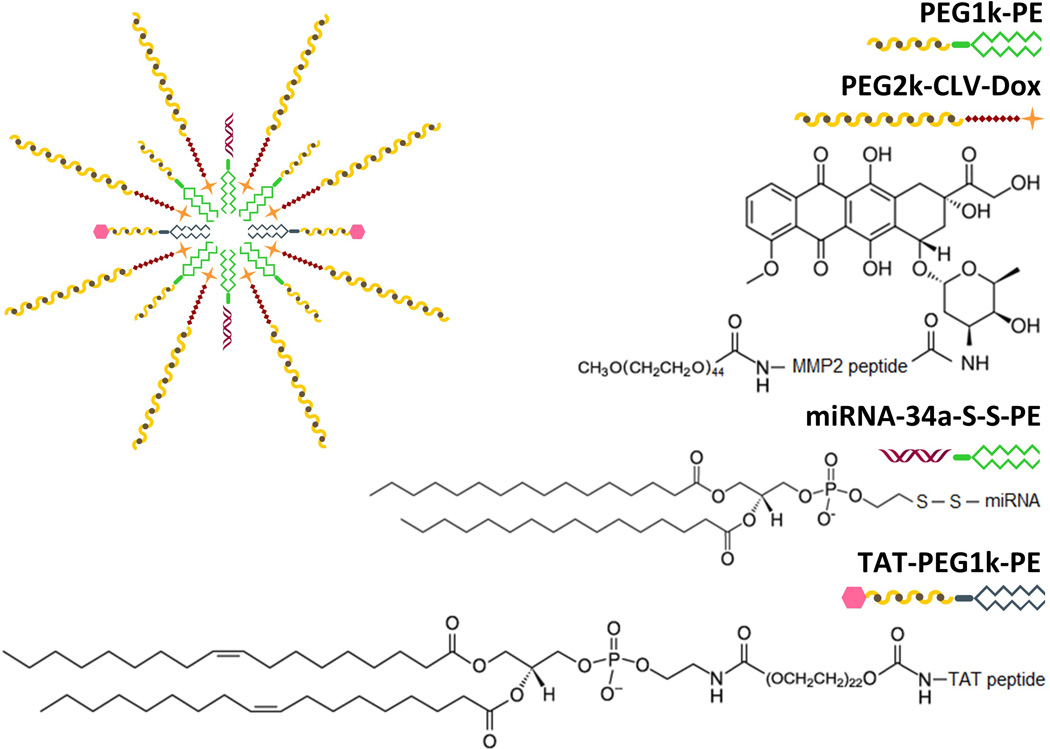
To overcome these challenges and co-deliver miRNA-34a and Dox to cancer cells, we designed a novel dual stimuli-sensitive nanopharmaceutical preparation. The system includes three functional conjugates that self-assemble to form polymeric mixed micelles (MM).
The first conjugate is a MMP2-sensitive Dox prodrug (PEG2k-CLV-Dox), where Dox is covalently linked to a peptide sensitive to matrix metalloproteinase 2 (MMP2) and modified with a long PEG chain (2,000 Da), which promotes the release of the drug after cleavage by MMP2 enzyme and allows long circulation of the particle in the bloodstream due to presence of PEG. MMP2 production is upregulated in many types of tumors, where they facilitate invasion, metastasis, and tumor progression by cleavage of most components of the extracellular matrix.[11] Sensitivity to this endopeptidase has driven the delivery of chemotherapeutic drugs, such as Dox and paclitaxel, by polymeric and liposomal nanosystems.[12–14]
The second conjugate (miRNA-34a-S-S-PE) sensitive to the reducing agent glutathione (GSH) was synthesized by linking miRNA-34a to a phospholipid (PE) moiety through a disulfide bond. The high intracellular concentration of GSH in cancer cells allows the reduction of the S-S bond and the release of free miRNA-34a inside the cells.[15, 16] Earlier, siRNA-S-S-PE conjugate was efficiently incorporated in PEG-PE micelles via its hydrophobic lipid moiety. The stability of micelle-incorporated siRNA against nucleolytic degradation dramatically increases.[17] At the same time, the system releases the intact siRNA upon the reduction by GSH and promotes downregulation of target genes in vivo.[18]
Finally, the third conjugate (TAT-PEG1k-PE) is formed by a cysteine-modified cell-penetrating TAT peptide (Cys-TATp) linked to a short PEG chain (1,000 Da) modified with a PE moiety. TATp promotes cellular internalization mainly through macropinocytosis, although direct penetration seems to play an important role when the peptide is attached to a hydrophobic cargo.[19, 20] The use of a longer PEG chain (2,000 Da) in the Dox conjugate was designed to protect the cationic proprieties of TATp, a known cause of non-specific cell binding.[21]
In the presence of PEG1k-PE, commonly used as a micellar backbone, these conjugates self-assemble in unique dual sensitive MM (Figure 1). Here we describe the synthesis and characterization of the conjugates as well as the production of the MM. Studies in MMP2-overexpressing HT1080 cells (human soft tissue sarcoma)[22] and low MMP2-expressing MCF7 cells (human breast carcinoma)[23] were carried out to prove the concept of dual stimuli-sensitivity as a trigger for simultaneous delivery of drug and miRNA in 2D and 3D cell culture models. Acquired multidrug resistance due to exposure to chemotherapeutics, including Dox, is one of the leading causes of the high resistance rate and low clinical response of soft tissue sarcomas to therapy.[24] The regulation of intrisic apoptosis pathway is one of the main causes for such resistance.[25, 26] Thus, co-delivering Dox and miRNA-34a efficiently to sarcoma tissues becomes a relevant strategy to prevent acquired drug resistance and increase response to therapy.
Figure 1.
Schematic representation of the drug delivery strategy and proposed chemical structures of the synthesized conjugates.
2. Results and Discussion
2.1 Synthesis and characterization of the conjugates
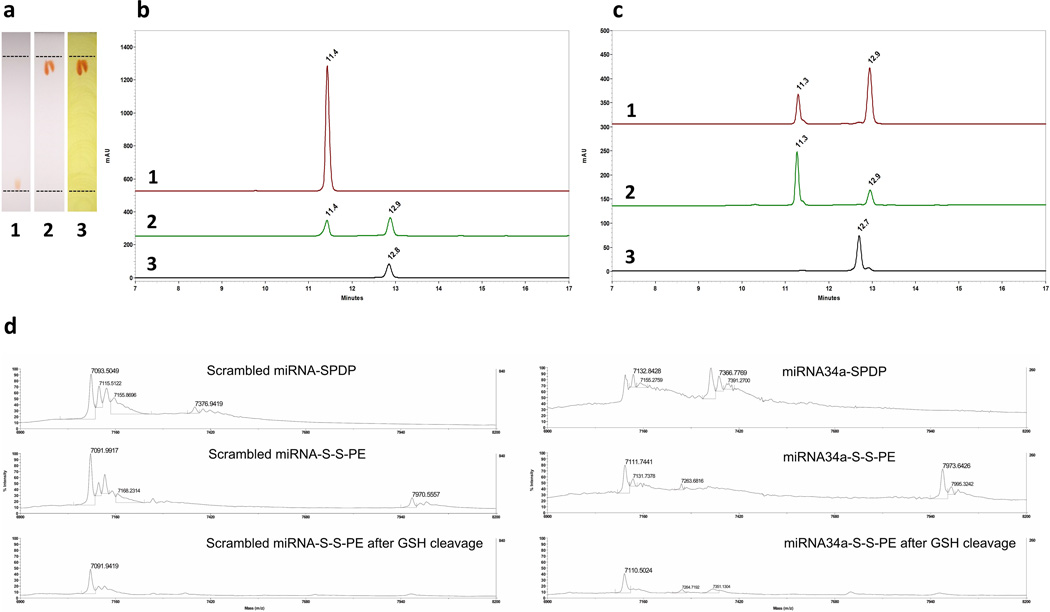
To synthesize the MMP2-sensitive Dox conjugate, a PEG2k-CLV conjugate, previously described,[27] was activated by N-hydroxysuccinimide (NHS) esterification and reacted with Dox. Characterization by TLC and RP-HPLC showed efficient formation of the conjugate and elimination of unreacted components by purification under dialysis (Figure 2a–b). Conjugation of Dox to PEG2k-CLV significantly increased the retention factor of the product compared to the free drug. Additionally, the formation of PEG2k-CLV-Dox was confirmed with Dragendorff’s reagent staining of the newly formed red spot on the plate. In the HPLC chromatograms, a well-defined peak related to PEG2k-CLV-Dox conjugate at tR ≈ 12.9 minutes was visualized, in contrast to disappearance of the peak of free Dox at tR ≈ 11.4 minutes. Reaction yield was approximately 50% in mass, and drug loading around 15 µg of Dox per mg of conjugate. PEG2k-CLV-Dox showed critical micelle concentration (CMC) of 5.6 × 10−6 M (Figure S1a). The MMP2-sensitivity of the conjugate was evaluated by HPLC after incubating micelles formed at 5 mg mL−1 in aqueous solution with human MMP2. After 3 hours of incubation with the enzyme, PEG2k-CLV-Dox conjugate was significantly cleaved. In contrast, at the same conditions, the MMP2-insensitive/uncleavable Dox conjugate (PEG2k-UNCLV-Dox) remained intact (Figure 2c).
Figure 2.
Characterization of the conjugates. a, Tracking of Dox conjugate synthesis by TLC analysis. (1) Free Dox (red spot), (2) PEG2k-CLV-Dox (red spot) and (3) PEG2k-CLV-Dox stained with Dragendorff’s reagent (orange spots are related to PEG moiety). b, RP-HPLC chromatograms of (1) Free Dox, (2) PEG2k-CLV-Dox immediately after synthesis and (3) PEG2k-CLV-Dox purified by dialysis. c, RP-HPLC chromatograms of (1) PEG2k-CLV-Dox after 1h and (2) 3h of exposition to MMP2 enzyme; (3) PEG2k-UNCLV-Dox after incubation overnight with MMP2 enzyme. d, MALDI-TOF mass spectra of free miRNAs and miRNA conjugates before and after cleavage by GSH.
miRNA-34a-S-S-PE was synthesized based on a S-S coupling reaction previously described to stabilize and deliver siRNA.[17, 28] After purification by desalting column, drug loading was approximately 350 µg of miRNA-34a per mg of conjugate. The GSH-sensitivity of the conjugate was analyzed by MALDI-TOF mass-spectrometry (MS). The disulfide bond was stable under the analysis conditions. Lipid conjugation on the SDPD-modified sense strand of the miRNA was confirmed by the shift of its peak from ≈ 7370 m z−1 to ≈ 7970 m z−1. After incubation with GSH, almost complete disappearance of the lipid-conjugated sense strand peak was observed (Figure 2d). The antisense strand peak (non-modified) remained unchanged (≈ 7100 m z−1) throughout the process, which indicates that the activity was preserved during the conjugation and GSH-induced cleavage.
TAT-PEG1k-PE was synthesized by maleimide (MAL) crosslinking reaction between TATp and MAL-PEG1k-PE. The purification by dialysis efficiently eliminated free components of the reaction (Figure S2). Significant decrease of the retention factor of the product compared to free MAL-PEG1k-PE stained with molybdenum blue suggested its conjugation with TATp. The conjugate showed CMC of 3.7 × 10−7 M (Figure S1b).
The hydration of a film composed of all conjugates and PEG1k-PE allowed the formation of MM. We suggest that hydrophobic interactions between the PE moieties of the conjugates and anthracycline rings of Dox drive their assembly into MM in aqueous surroundings.
First, MMP2-sensitive MM were produced to evaluate the effects of the formulation against HT1080 and MCF7 cells. Then MMP2/GSH-sensitive MM, or dual sensitive MM, were prepared for the studies on the action of miRNA-34a. In this case, the molar ratio of Dox conjugate in the formulation was reduced by 7-fold to avoid that the effect of the drug masked the action of miRNA-34a. The study of the molar and weight ratios between conjugates revealed, in all cases, MM with a mean size of about 15 nm (Table S1, Figure S3a) and narrow size distribution (P.I. ≤ 0.2). Additionally, no peak related to free miRNA conjugate (≈ 7370 m z−1) was seen after analysis of miRNA-34a MM by MALDI-TOF (Figure S3b), suggesting 100% of encapsulation. The miRNA protection was confirmed by no changes in the system and absence of nucleic acid-related peaks after exposure to RNases.
2.2 MMP2-triggered cell-specific effect of MMP2-sensitive MM
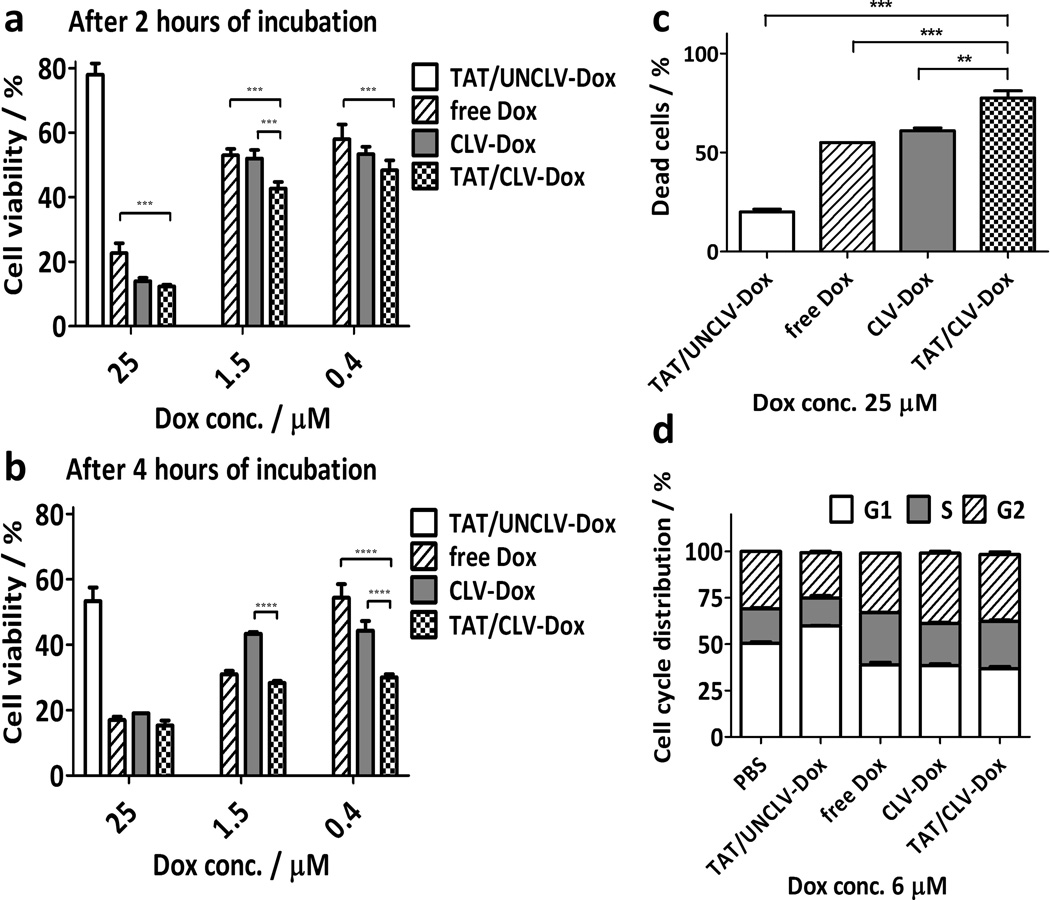
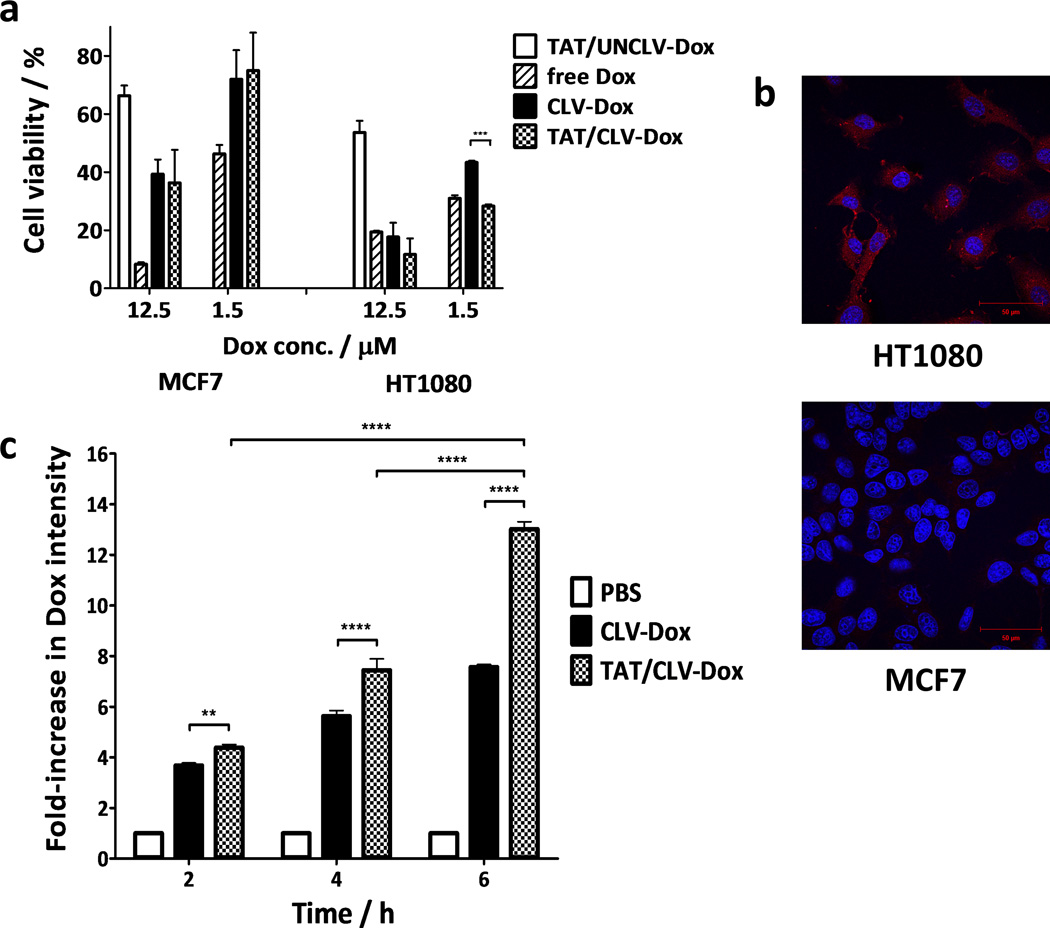
The cytotoxicity of the MM containing PEG2k-CLV-Dox and its uncleavable counterpart was evaluated in HT1080 cells known to overexpress MMP2.[22, 29] The treatment with (TAT/CLV-Dox MM) and without (CLV-Dox MM) inclusion of TAT-PEG1k-PE to the formulations promoted similar cytotoxic effect at high concentrations of Dox (25 µM). However, at lower Dox doses (1.5 and 0.4 µM), TATp significantly potentiated the effect of the drug (Figure 3a–b). Same potentiation was shown in the analysis by laser scanning cytometry (Figure 3c), when even at high Dox concentration the treatment with TAT/CLV-Dox led to the highest percentage of dead cells (≈ 75%). MM containing the uncleavable conjugate at the tested doses and incubation times showed the lowest cytotoxic effect as well as percentage of dead cells, due to the presence of PEG on the surface of the particles, which hampers their uptake by the cells.[30] Several studies have shown that the presence of TATp can not only facilitate the transport of the system into the cells but also mediate endosomal escape with a consequent increase in the intracellular drug concentration.[15, 31, 32]
Figure 3.
In vitro effect of MMP2-sensitive MM in HT1080 cells. a–b, Cell viability after treatment with MMP2-sensitive MM (n = 9 treatments). c–d, Apoptosis and cell cycle distribution, respectively, after treatment with MMP2-sensitive MM (n = 3 treatments). p-values were obtained by comparison of groups indicated. ****p ≤ 0.0001, ***p ≤ 0.001 and **p ≤ 0.01. Error bars represent mean ± SD.
To evaluate if the release of Dox by the cleavage of the conjugate could affect the mechanism of its action, the analysis of the cell cycle distribution was assessed by laser scanning cytometry (Figure 3d). After 24 hours of incubation with both TAT/CLV-Dox and CLV-Dox MM, the number of cells in the G2/S phases increased similarly to free Dox. On the other hand, the majority of cells were found in G1 phase after the treatment with TAT/UNCLV-Dox. These results indicate that, although Dox was chemically modified through the conjugation with PEG2k-CLV, its action after the cleavage of the conjugate remained similar to free Dox, i.e. the inhibition of proliferation and consequent cell death were due to the arrest of the cells in G2/S phases.
As the next step, the cytotoxic effect of MM was evaluated in HT1080 and MCF7 cells to confirm its correlation with the MMP2 expression. HT1080 cells showed MMP2 levels in the extracellular medium 6-fold higher than MCF7 cells (Figure S4a). In agreement with this finding, the treatment of HT1080 cells with TAT/CLV-Dox MM at higher dose was 3-fold more cytotoxic than the non-sensitive MM. Such difference decreased for 1.7-fold when low MMP2-expressing MCF7 cells were treated (Figure 4a). In addition, the penetration of the micelles into the cells was qualitatively analyzed by confocal scanning laser microscopy. After 2 hours of exposure to TAT/CLV-Dox MM, Dox fluorescence (red) was observed in the cytoplasm of HT1080 cells (Figure 4b). Much lower intensity was detected in same cells treated with TAT/UNCLV-Dox MM as well as in MCF7 cells treated with any of the formulations (Figure S4b). The higher the intensity of Dox fluorescence inside the nuclei of the cells, the lower was the level of Hoechst staining. This is the result of quenching of the Hoechst fluorescence by Fluorescence Resonance Energy Transfer (FRET). Such effect was clear in the cells treated with free Dox (Figure S5). The Forster equations for FRET are well known. We applied one component, the loss of fluorescence of the donor molecule, as a metric to determine the efficacy of the formulations. Among all groups treated with MM, FRET was observed only in HT1080 cells treated with TAT/CLV-Dox MM, which suggests higher nuclear localization of Dox and confirms that the MM selectively release Dox from the MMP2-sensitive conjugate because of higher levels of MMP2 in HT1080 cells.
Figure 4.
Cell-specific effect of MMP2-sensitive MM. a, Cell viability of HT1080 and MCF7 cells treated with MMP2-sensitive MM (n = 9 treatments). b, Confocal images of HT1080 and MCF7 cells treated with TAT/CLV-Dox MM. In the image, red is associated to Dox fluorescence and blue to the cell nuclei stained with Hoechst. Magnification = 63×. Scale bar = 50 µm. c, Quantitative analysis of Dox uptake in HT1080 cells treated with MMP2-sensitive MM (n = 3 treatments). p-values were obtained by comparison of groups indicated. ****p ≤ 0.0001, ***p ≤ 0.001, **p ≤ 0.01 and *p ≤ 0.05. Error bars represent mean ± SD.
To check if the increased cytotoxicity of TAT/CLV-Dox MM towards HT1080 cells was correlated with an enhanced uptake of the formulation due to presence of TATp, cellular uptake analysis was performed by FACS (Figure 4c). The effect of TATp was evident and time-dependent. Dox intensity following exposure to TAT/CLV-Dox MM for 2 and 6 hours, was 1.2 and 1.7-fold higher compared to CLV-Dox MM, respectively.
2.3 Cell behavior analysis in a 2D model
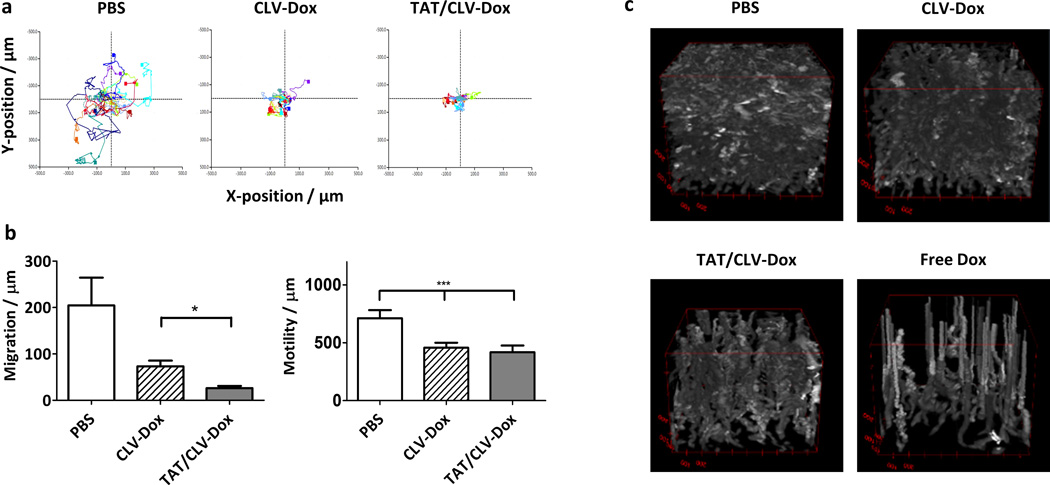
The effect of TAT/CLV-Dox MM on the dynamics of HT1080 cells was investigated by the time-lapse holographic imaging cytometry. Cell migration is a critical step in tumor invasion and metastasis. The trajectories of HT1080 cells non-treated and treated with CLV-Dox or TAT/CLV-Dox MM were analyzed (Figure 5a). A total of 10 representative cells were selected and their positions were plotted in Cartesian plots for 12 hours. The non-treated cells were quite motile. However, the exposure to 10 µM of Dox delivered by both, TAT/CLV-Dox and CLV-Dox MM induced a marked inhibition of cell motility and migration (Figure 5b). TAT/CLV-Dox MM induced the highest decrease in cell migration and also inhibited the mesenchymal-amoeboid transition observed in PBS and CLV-Dox groups (videos not shown). The transition between cellular motility shapes is a key factor to help cancer cells to migrate and invade healthy tissues.[33–36]
Figure 5.
Time-lapse cytometry analysis of HT1080 cells after treatment with MMP2-sensitive MM. a, Cell trajectory plot. b, Quantitative assessment of cell motility and migration. c, 4D plots of cell proliferation over the time of analysis. p-values were obtained by comparison of groups indicated. ***p ≤ 0.001 and *p ≤ 0.05. Error bars represent mean ± SD (n = 10 individual cells per treatment).
4D plots of the imaging fields, with the 48-hour time tracking being the z-axis parameter and cell thickness represented as brightness, were shown (Figure 5c). In the non-treated group (PBS), cell proliferation was evident, with cells reaching confluence by the end of the evaluation. The bright tracks are cells undergoing mitosis. In contrast, in the free Dox treatment, there was total abrogation of proliferation and, in the later stages, cell motility. The upward rising spires are the tracks of post-mortem cellular degradation. The treatment with CLV-Dox and TAT/CLV-Dox MM more closely resembled the control and free Dox groups, respectively.
2.4 GSH-triggered effect of dual sensitive MM
RNAi-based treatment of HT1080 cells promoted up to 40% decrease in levels of survivin protein, known to negatively regulate apoptosis by inhibition of caspase activation.[37] Knockdown of Bcl2 mRNA expression have also induced apoptosis in soft tissue sarcoma model as well as overexpression of notch1 conferred cell chemoresistance in a p53-dependent manner.[38, 39]
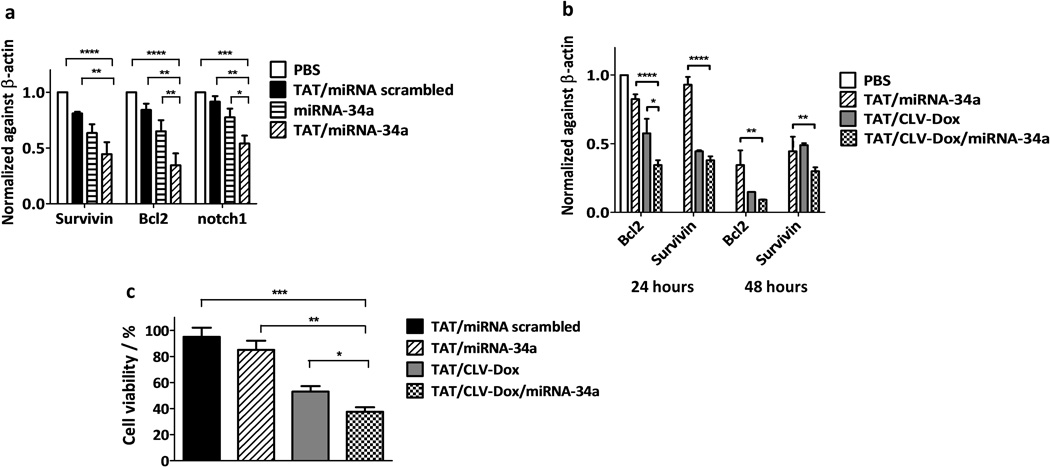
With this in mind, we investigated the ability of the GSH-sensitive MM to efficiently deliver intact miRNA-34a into HT1080 cells by qRT-PCR quantification of oncogene-related mRNA. The relative mRNA levels of Bcl2, survivin and notch1 in cells treated with TAT/miRNA-34a MM showed the highest decrease, compared to the non-treated group, by about 65%, 55% and 45%, respectively (Figure 6a). Also, significantly higher decrease of the mRNA levels of both Bcl2 and notch1 was found after the treatment with TAT/miRNA-34a MM compared to the formulations without TATp or with scrambled miRNA.
Figure 6.
Effect of MM in gene expression and cell viability of HT1080 cells. a, mRNA levels of Bcl2, survivin and notch1 after treatment with GSH-sensitive MM at miRNA-34a concentration of 100 nM (n = 3 treatments). b, mRNA levels of Bcl2 and survivin after treatment with dual sensitive MM at miRNA-34a/Dox concentration of 100 nM and 5 µM, respectively (n = 3 treatments). c, Cell viability in 2D monolayer model after treatment with dual sensitive MM at miRNA-34a/Dox concentration of 100 nM and 5 µM, respectively (n = 3 treatments). p-values were obtained by comparison of groups indicated. ****p ≤ 0.0001, ***p ≤ 0.001, **p ≤ 0.01 and *p ≤ 0.05. Error bars represent mean ± SD.
The most effectively downregulated mRNAs, Bcl2 and survivin, were evaluated after treatment with the combination of both stimuli-sensitive conjugates incorporated into the same MM. After 48 hours of treatment with TAT/CLV-Dox/miRNA-34a MM, mRNA levels of Bcl2 and survivin showed 91% and 70% of reduction, respectively (Figure 6b). Strong Bcl2 downregulation has a great impact in overcoming cellular growth highly stimulated in sarcomas through PI3K/Akt pathway triggered by hyperactivation of insulin-like growth factor-1 receptors.[40] Treatment with TAT/CLV-Dox MM also promoted gene downregulation, despite the absence of miRNA-34a (Figure 6b). It has been shown that Dox itself is able to downregulate Bcl2 and activate cytochrome c-dependent apoptosis pathway due to the DNA damage.[24] Moreover, dual sensitive MM reduced HT1080 cell viability to around 40%, which means 15% more efficiency compared to group treated with only TAT/CLV-Dox MM (Figure 6c). Overall, the data confirms the dual effect of the delivery system on both, mRNA levels and cellular viability on soft tissue sarcoma cells.
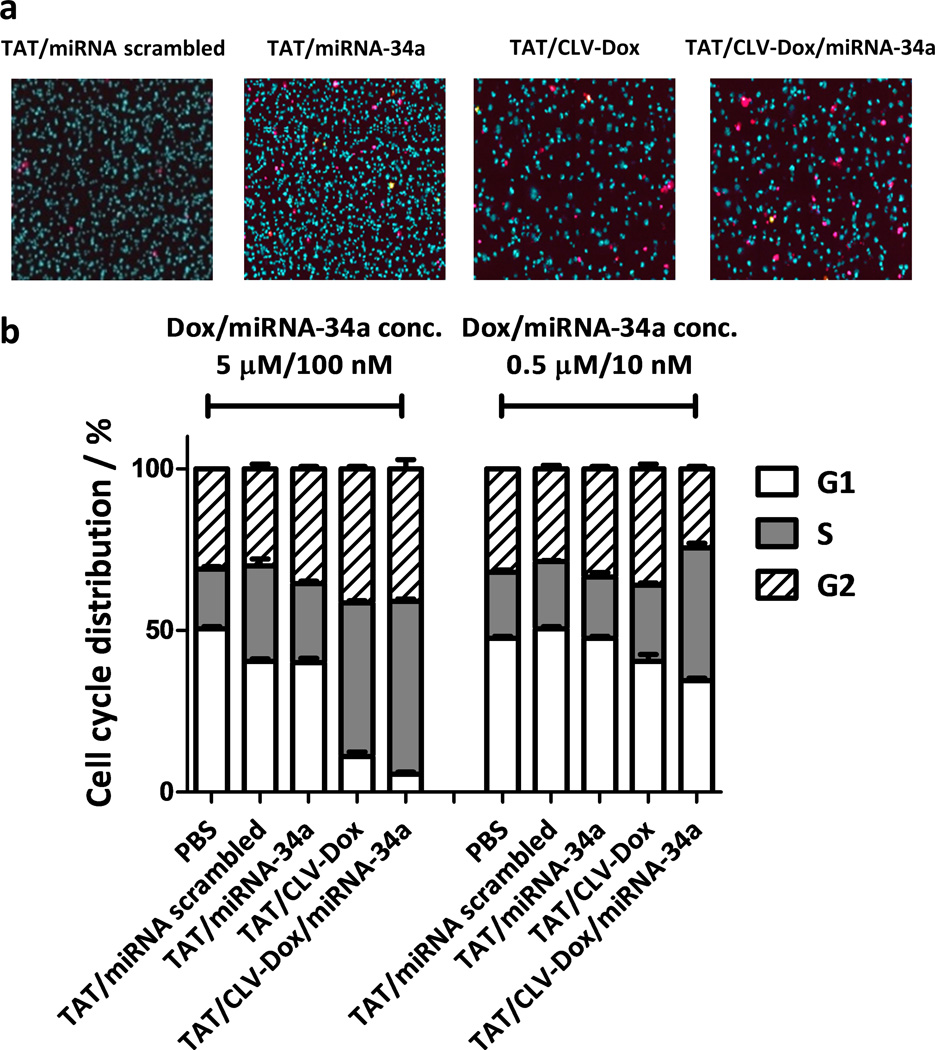
The apoptotic effect and the cell-cycle distribution after the treatment with the dual sensitive MM were evaluated by laser scanning cytometry. Compared to a single treatment with TAT/miRNA-34a and TAT/CLV-Dox MM, the dual sensitive MM resulted in 3.5- and 2.5-fold higher number of dead cells, respectively (Figure 7a, Figure S6). Additionally, after 48 hours of treatment at 5 µM Dox and 100 nM miRNA-34a concentrations, the amount of cells in S phase significantly increased for both TAT/CLV-Dox and TAT/CLV-Dox/miRNA-34a groups compared to controls. At 10-fold lower doses of Dox and miRNA-34a, the increase of cell number in S phase was detected only after the combined treatment (Figure 7b, Figure S7). These results suggest an enhancement of the anti-proliferative effect when the dual sensitive formulation is used. In all the cases, the treatment with scrambled miRNA did not show any significant effect.
Figure 7.
Effect of MM in apoptosis and cell cycle distribution of HT1080 cells. a, Laser scanning cytometry images after treatment with dual sensitive MM at miRNA-34a/Dox concentration of 50 nM and 2.5 mM, respectively. Magnification = 40×. Dead cells are stained in magenta and nuclei of live cells are stained in blue. b, Cell cycle distribution after treatment with dual sensitive MM at high and low concentrations of miRNA-34a and Dox. Error bars represent mean ± SD (n = 3 treatments).
It is important to note that TAT/CLV-Dox/miRNA-34a MM demonstrated both, silencing activity and anti-proliferative effects even in presence of serum. It is well known that neither free miRNA nor miRNA delivered by commercial transfection agents, such as lipofectamine and its derivatives, have activity in presence of serum.[41]
2.5 Effect of dual sensitive MM in 3D spheroid model
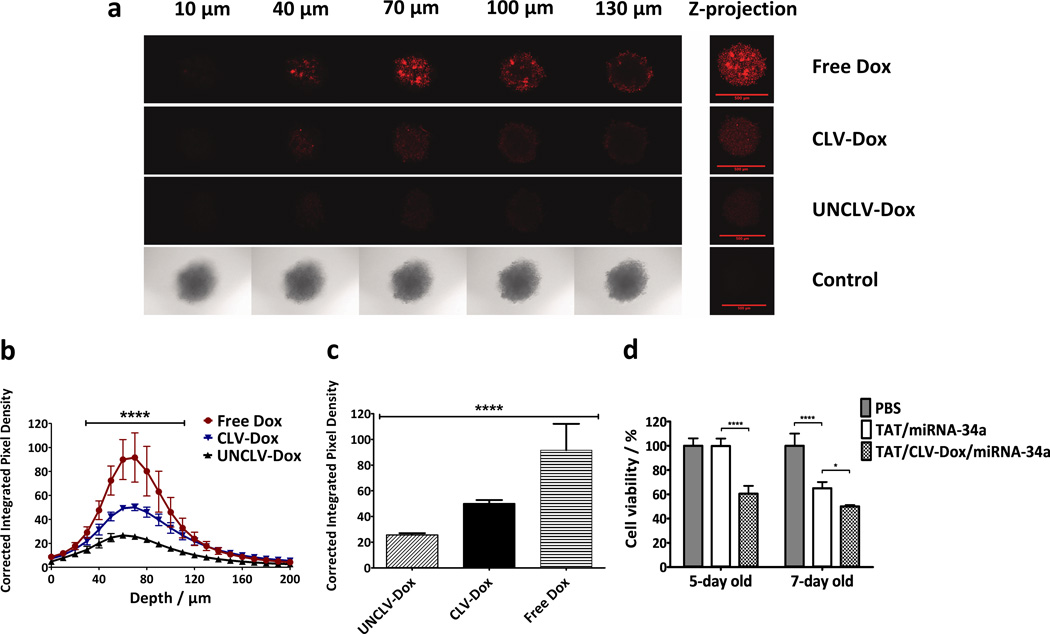
The 3D spheroid model of tumor mass has been proposed as a mimic of in vivo tumors and used extensively to evaluate nanopreparations as an alternative to preclinical trials.[42] To study the MMP2-sensitivity of PEG2k-CLV-Dox conjugate in the tumor mass, we developed a MMP2-overexpressing 3D spheroid model of HT1080 cells. After the treatment, the fluorescent signal of free Dox in all layers of the spheroids was higher compared to the conjugates, due to its immediate availability in the system and consequent higher and deeper penetration (Figure 8a–c). Still, the sensitivity of the conjugate to MMP2 cleavage was confirmed by higher and deeper penetration signals after the treatment with PEG2k-CLV-Dox compared to the uncleavable control. The PEG chains not detached from the PEG2k-UNCLV-Dox conjugate increase hydrophilicity and size of the molecule, which hinders its internalization in the tumor mass.
Figure 8.
Effect of MMP2-sensitive Dox conjugate and MM in HT1080 spheroid model. a, Dox distribution throughout the spheroids at different layers of depth. Maximum pixel intensity of Z-projection images represent the total Dox fluorescence in each layer of the spheroids. Scale bar = 500 µm. b, Corrected integrated pixel densities as an indicator of Dox intensity in the core area of each optical slice of the spheroids. The difference between all groups were significant from 40 to 110 µm (n = 5, ****p ≤ 0.0001, Two-way ANOVA with Tukey’s multiple comparisons test). c, Dox fluorescence signal at 70 µm depth of the spheroids in B (n = 5 spheroids). d, Cell viability in 3D spheroid model after treatment with dual sensitive MM at miRNA-34a/Dox concentration of 100 nM and 10 µM, respectively (n = 5 spheroids per treatment). p-values were obtained by comparison of groups indicated. ****p ≤ 0.0001, ***p ≤ 0.001, **p ≤ 0.01 and *p ≤ 0.05. Error bars represent mean ± SD.
After checking the ability of Dox conjugate to penetrate the tumor mass on its own, the cytotoxicity of the MM was tested in the 3D spheroid model. TAT/miRNA-34a MM had no effect in 5-day old, but promoted 35% of cytotoxicity in 7-day old spheroids (Figure 8d). It has been shown that the characteristics of 3D multicellular tumor spheroids are subject to their maturity, which could affect response to the treatment.[43] On the other hand, the addition of MMP2 sensitive Dox conjugate to the system worked in concert with miRNA-34a and significantly increased the cytotoxicity in both cases. This result is specially interesting due to the fact that chemotherapy resistance related to oncogene protein levels are more expressed in matured spheroids.[44–46] It is also in agreement with previously report of reversal of the sphere formation and chemoresistance of sarcoma models by downregulation of survivin.[47]
Thus, our findings suggest that the dual sensitive MM can penetrate into the solid tumor mass due to MMP2-sensitive PEG detachment and release functional miRNA-34a due to GSH-sensitive cleavage of the conjugate inside the cells.
3. Conclusion
Development of multi stimuli-sensitive drug delivery systems is a promising approach to improve co-delivery of chemotherapeutic drugs and miRNA into tumor cells. In this work, we have clearly proved the possibility of preparing combination pharmaceutical nanopreparations based on nucleic acids and simultaneously responding to multiple local stimuli. Significantly higher cytotoxicity of the developed system in MMP2-overexpressing HT1080 cells compared to low MMP2-expressing MCF7 cells showed that sensitivity to the enzymatic activity can be used as a trigger to release the drug in a specific target tissue. The benefits of cell-penetrating function (TATp) for the cellular uptake of nanopreparations was clearly confirmed by the increase in Dox association with HT1080 cells. We did also confirmed the possibility of modifying miRNAs with hydrophobic moieties to overcome pharmacokinetic drawbacks and efficiently delivered functional miRNA-34a into tumor cells to promote downregulation of oncogenic genes, such as Bcl2, survivin and notch1. Finally, we showed a significant decrease in cell viability in a 3D tumor mass, reinforcing the functionality of the dual sensitive nanopreparation even in models which better mimics an in vivo environment.
4. Experimental Section
Materials
Methoxy(polyethylene glycol)-2000-succinimidyl valerate (PEG2k-NHS) and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide (polyethylene glycol)-1000] (MAL-PEG1k-PE) were purchased from Laysan Bio, Inc. (AL, U.S.). 1,2-Distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-1000] (ammonium salt) (PEG1k-PE) and 1,2-dipalmitoyl-sn-glycero-3-phosphothioethanol (sodium salt) (PE-SH) were purchased from Avanti Polar Lipids, Inc. (AL, U.S.). Doxorubicin hydrochloride salt was purchased from LC Laboratories (MA, U.S.). miRNA34a, miRIDIAN microRNA hsa-miR-34a-5p mimic (C-300551-07) and a miRNA mimic negative control (CN-002000-01) modified at the 3’-end of the passenger strand with N-succinimidyl 3-(2-pyridyldithio) propionate (SPDP) group were purchased from GE Healthcare Dharmacon, Inc. (CO, U.S.). Cys-TAT peptide (CYGRKKRRQRRR), MMP2-cleavable (GPLGIAGQ) and uncleavable (GGGPALIQ) peptides were synthesized by Tufts University Core Facility (MA, U.S.). N,N’-dicyclohexylcarbodiimide (DCC) and N-hydroxysuccinimide (NHS) were purchased from Sigma-Aldrich (MO, U.S.). CellTiter-Glo® Luminescent Cell Viability Assay was purchased from Promega (Fitchburg, WI). All other chemicals were of analytical, HPLC or PCR grades. HT1080 and MCF7 cell lines were obtained from the American Type Culture Collection, ATCC (VA, U.S.). Cells were cultured in MEM and DMEM medium, respectively, containing 10% fetal bovine serum (FBS), 10,000 unit mL−1 penicillin G, 10 mg mL−1 streptomycin sulfate and 25 µg mL−1 amphotericin B (complete medium) in a humidified atmosphere of 95% air and 5% CO2 at 37°C.
Synthesis of MMP2-sensitive Dox conjugate
A PEG2k-CLV-Dox conjugate was synthesized as following description. To introduce the MMP2-cleavable peptide into the long chain of PEG2k, the primary amine of the peptide was reacted with the NHS group of modified PEG at 1.2:1 molar ratio, respectively, by mixing MMP2 peptide solution 4.3 mg mL−1 in PBS pH 7.4 and PEG2k-NHS solution 10 mg mL−1 in same buffer for 24 hours at 4°C. Reaction was followed by purification under dialysis against water (Spectra/Por® 7 Pre-treated Dialysis Tubing, MWCO 2,000) for 24 hours at 4°C. Purified conjugate (PEG2k-CLV) was freeze-dried overnight and powder was stored at −20°C.
To produce PEG2k-CLV-Dox, the purified PEG2k-CLV was dispersed in DMF at 2.5 mg mL−1 in presence of a 10-fold molar excess of each of two coupling reagents, DCC and NHS. Reaction mixture was stirred for 1 hour at room temperature to allow complete activation of carboxyl group of PEG2k-CLV. A 2.5 mg mL−1 Dox solution was prepared in DMF containing 2.0 µL mL−1 of trimethylamine. 1.3 molar excess of Dox was added to the reaction mixture. The reaction proceeded overnight, under light protection, nitrogen atmosphere and continuous stirring at room temperature. The conjugate was purified by dialysis: 1h against water, 24h against water/methanol (1:1, v/v) and finally 2h against water (Spectra/Por® 7 Pre-treated Dialysis Tubing, MWCO 2,000) at room temperature. Conjugate was filtered on a 1.2 mm nylon membrane to eliminate by-products of the reaction and consecutively freeze-dried overnight. The powder was stored at −20°C. A Dox conjugate non-sensitive to the MMP2 cleavage was synthesized as negative control following the same protocol. The reaction was monitored by TLC (chloroform/methanol, 7:3, v/v), using Dragendorff’s reagent as staining for PEG moiety (Figure 2a). RP-HPLC was also used to verify complete absence of free Dox after purification (Figure 2b). System was equipped with a C18 column (XBridge, 250 × 4.6 mm, 5µm), fluorescent detector (λex/em = 450/550 nm) and UV at λ = 485 nm. The chromatographic conditions were as follows: 0.1% TFA in acetonitrile (solvent A) and 0.1% TFA in water (solvent B). 0–15 min: 5%–75% A, 15–16 min: 75%–5% A, 16–20 min: 5% A. Flow rate: 1.0 mL min−1 at room temperature. Drug loading was determined by fluorescence measured at λex/em = 485/590 nm in a BioTek Multi-Mode microplate reader.
Synthesis of GSH-sensitive miRNA-34a conjugate
The miRNA-34a was conjugated to a PE moiety via a disulfide linkage as previously described,[17, 28] with slight modifications. Briefly, an aqueous solution of the SPDP-activated miRNA-34a (20 nmol in 120 µL of RNase-free water) was added dropwise to a solution of PE-SH (2 µmol in 146 µL of chloroform). DMSO was added in a ratio 5:1 DMSO:CHCl3 v/v and the reaction was carried out for 48 hours at room temperature under continuous stirring. Unreacted reagents were removed using a dextran desalting column. 10 aliquots of 1 mL each were collected and freeze-dried overnight. Aliquots containing miRNA-34a-S-S-PE conjugate in powder were hydrated in 1 mL of RNase-free water and filtered through 0.2 µm membrane filters to remove traces of PE-SH. The conjugate was stored at −80°C. Drug loading was determined by Nanodrop 2000c Spectrophotometer (Thermo Scientific, DE, U.S.A.) at λ = 280 nm. A miRNA negative control was modified following the same protocol.
Synthesis of TAT-PEG1k-PE conjugate
For the synthesis of TAT-PEG1k-PE conjugate, 7 mL of PE-PEG1k-MAL solution 2.5 mg mL−1 in methanol were mixed with 1 mL of Cys-TATp solution 20 mg mL−1 in methanol (1:1, molar ratio). The reaction mixture was stirred overnight, under nitrogen atmosphere at room temperature. The conjugate was purified by dialysis against water (Spectra/Por® 7 Pre-treated Dialysis Tubing, MWCO 2,000) for 24h to remove unreacted TATp. Purified conjugate was freeze-dried overnight and powder was stored at −20°C. The reaction and purification processes were monitored by TLC (chloroform/methanol, 7:3, v/v) using Dragendorff’s reagent as staining for PEG moiety, Ninhydrin reagent as staining for TATp and Molybdenum Blue reagent as staining for PE moiety (Figure S2).
Preparation of dual sensitive MM
The MMP2-sensitive MM (TAT/CLV-Dox) were prepared by hydration of a thin polymeric film.[13, 27] Briefly, PEG1k-PE, PEG2k-CLV-Dox and TAT-PEG1k-PE were dispersed in chloroform at 60:35:5 molar ratio, respectively. The organic solvent was removed under reduced pressure and the resulting film was hydrated with PBS pH 7.4. The system was gently vortexed for 5 minutes to allow self-assembly of the components into MM. The final concentrations of PEG1k-PE, PEG2k-CLV-Dox and TAT-PEG1k-PE in the formulation were 10, 10 and 1.4 mg mL−1, respectively. The non-sensitive MM (TAT/UNCLV-Dox) and the MM without TATp (CLV-Dox) were prepared similarly.
To produce the dual sensitive MM (TAT/CLV-Dox/miRNA-34a), PEG1k-PE, PEG2k-CLV-Dox and TAT-PEG1k-PE were dispersed in chloroform and mixed at 90:5:5 molar ratio, respectively. The organic solvent was removed under reduced pressure and the resulting film was hydrated with a solution of miRNA-34a-S-S-PE in phosphate buffer pH 7.4 at 66 µg mL−1 of miRNA. The system was gently vortexed for 5 minutes to allow self-assembly of the components into MM. The final concentrations of PEG1k-PE, PEG2k-CLV-Dox and TAT-PEG1k-PE in the formulation were 50, 5 and 5 mg mL−1, respectively. The weight ratio of PEG1k-PE/miRNA-34a-S-S-PE was 750:1, respectively. MM without TATp and Dox (miRNA-34a), with TATp but no Dox (TAT/miRNA-34a) or containing a scrambled miRNA (TAT/miRNA scrambled) were prepared similarly.
Characterization of dual sensitive MM
Size of the MM was measured at 20°C using Zetasizer Nano Z (Malvern, United Kingdom). Briefly, triplicate of each sample was diluted in ultrapure water (1:1 v/v) and analyzed by detector at 90° angle. Results were expressed in Z-average and Polydispersity Index (P.I.) was used as measure of the particle size distribution (Table S1). Unimodal distribution was confirmed by observation of single particle size peak (Figure S3a).
Cytotoxicity of the MM
HT1080 and MCF7 cells were seeded in 96-well plates at 5 × 103 cells per well and incubated for 24 hours before treatments. To study the cytotoxicity of MMP2-sensitive MM, HT1080 and MCF7 cells were treated for 2 and 4 hours at a Dox initial concentration of 25 µM in serum complete medium. After treatment, complete medium was replaced and cells were incubated for 24 hours. Cell viability (Figure 3a–b, Figure 4a) was determined by Cell Titer-Blue assay. Briefly, 5 µL of Cell Titer-Blue reagent was diluted in 50 µL of complete medium per well and incubated with treated cells at 37°C for 2 hours. Thereafter, the fluorescence was recorded at λex/em = 560/590 nm by a BioTek Multi-Mode microplate reader. Cells treated with free Dox and TAT/UNCLV-Dox MM were used as controls. To study the effect of dual sensitive MM on cell viability, HT1080 cells were seeded in 96-well plates at 3 × 103 cells per well and incubated for 24 hours before treatments. After 24 hours, cells were treated for 4 hours at an initial concentration of miRNA-34a and Dox of 200 nM and 10 µM in serum complete medium, respectively. After treatment, complete medium was replaced and cells were incubated for 48 hours. Cell viability was determined by Cell Titer-Blue assay (Figure 6c). Cells treated with TAT/miRNA scrambled MM were used as controls. The experiments were done in triplicate for three samples of each formulation.
Analysis of apoptosis and cell cycle distribution
HT1080 cells were seeded in black polystyrene 96-well plates (Corning, NY, U.S.A.) at 3 × 103 cells per well 24 hours prior to experiment. Cells were treated with MMP2-sensitive MM at 25 µM Dox starting concentration for 2 hours or dual sensitive MM at 200 nM/10 µM miRNA-34a and Dox starting concentrations, respectively, for 4 hours in serum complete medium. After treatment, complete medium was replaced and cells incubated for 24 hours (MMP2-sensitive) or 48 hours (dual sensitive). Consecutively, three markers, Hoechst 33342 (5 µg mL−1, for nucleic acid), Yo-Pro (0.12 µg mL−1, for early apoptosis) and propidium iodide (1 µg mL−1, for late apoptosis/necrosis), diluted in complete medium were added to cells. After 30 minutes of incubation at 37°C, stained cells were analyzed by laser scanning cytometer (CompuCyte Corp., MA, U.S.A.) (Figure 7a). A 40× objective lens was used with 0.2554 µm spatial resolution in two pass scanning. In the first pass, a 405 nm laser was used to excite Hoechst and fluorescence was collected through a 440/30 bandpass filter. In the second pass, a 488 nm argon laser was used with a 515/30 bandpass filter for green Yo-Pro fluorescence and a 650 nm longpass filter for red propidium iodide fluorescence. Cells were segmented using Hoechst fluorescence and total cellular DNA fluorescence was quantified. Live single cells were gated based on their combined DNA content and nuclear area (Figure 3c, Figure S6). For the analysis of cell cycle distribution cells were gated into G1, S and G2 phases depending on their DNA content (Figure 3d, Figure 7b, Figure S7). Phantom contours, a random segmentation technique, was used for analysis. Sampling elements with a 3 µm diameter and a minimum spacing of 6 µm were applied to each of the image fields. The total fluorescence intensity of the sampling elements was determined for each of the colors used in the analysis and the data was plotted as scattergrams and histograms, with gating and analysis capabilities analogous to flow cytometry. We have found that this method is immune to segmentation errors and provides the percentage of the scan area covered on an area basis, for example live cells vs. dead cells. The experiments were done in triplicate.
Cellular uptake of the MMP2-sensitive MM
HT1080 and MCF7 cells were seeded on coverslips at 5 × 103 cells per coverslip. After 24 hours, cells were treated with MMP2-sensitive MM for 2 hours at 10 µM of Dox concentration in serum complete medium. Consecutively, medium was removed and cells were incubated in methanol (80% v/v) for 5 min at 4°C. Following wash trice with cold PBS pH 7.4, cells were stained with Hoechst 33342 (5 µg mL−1) for 15 min at room temperature. The coverslips were mounted on glass slides with Fluoromount-G (Fisher Scientific, MA, U.S.A.) medium and sealed using a nail lacquer. Images from the slides were captured with a Zeiss LSM 700 inverted confocal microscope (Carl Zeiss Co. Ltd., Jena, Germany) (Figure 4b, Figure S4b). A quantitative comparison of cellular uptake of the formulations after treatment with MMP2-sensitive MM was done by fluorescence-activated cell sorting (FACS). HT1080 cells were seeded in 12-well plates at 1 × 105 cells/well, 24 hours prior to experiment. Cells were treated for 2, 4 and 6 hours at 10 µM Dox concentration in complete medium. After treatment, cells were collected, washed twice with cold PBS and resuspended prior to analysis by FACS. 488 nm blue laser was used for excitation and FL2 channel was used for recording of 10,000 gated events (Figure 4c). The experiments were done in triplicate.
Time-lapse cytometry analysis
For the analysis in a 2D migratory model, HT1080 cells were seeded in glass bottom 35 mm culture Petri dishes (MatTek Corp., MA, U.S.A.) at a density of 5 × 104 cells per dish. After 24 hours, cells were treated with MMP2-sensitive MM for 4 hours at a Dox concentration of 10 µM in serum complete medium. Consecutively, treatment-containing medium was replaced with fresh complete medium and the unlabeled cells were imaged with a Phase Holographic Imaging HM4™ system (Phase Holographic Imaging, Lund, Sweden).
Computer software was used to produce 2D images of the sample thickness within the field of integration (0.5 mm2 with a 20× objective lens). An automated stage allowed multiple collection of fields of view over time. For this study, we used 5 minute time-intervals for periods of 48 hours. Images were segmented and a list of features, including cell volume, thickness and area were obtained, along with cell positional information. Data was presented as Cartesian plots of cell tracks (Figure 5a). Ten representative cells were selected at the onset of the analysis and followed for a 12-hour period. Values for the migration and motility of the cells were calculated with the HM4™ software (Figure 5b).
Methods for producing 4D plots (X position, Y position, cell thickness and time) were developed. Images from the time course were exported to ImageJ version 1.42 software (NIH, MD, U.S.A.) and treated as an image stack. The 3D imaging plug-in was used to create a volume rendering of the cellular motion over time of analysis (Figure 5c). The brightness threshold was adjusted to view all cells within the imaging area or only the thicker cells, a feature that allows identification of mitosis, apoptosis and transition between mesenchymal and amoeboid cellular morphologies.[33–36]
Evaluation of Bcl2, survivin and notch1 mRNA expression by qRT-PCR analysis
HT1080 cells were seeded in 12-well plates (1.5 × 105 cells per well) 24 hours prior to the experiments. Cells were treated with dual sensitive MM in serum complete medium for 4 hours at a miRNA-34a/Dox concentration of 200 nM and 10 µM, respectively. Consecutively, complete medium was replaced and cells were incubated until 24 and 48-hour time points were reached. Bcl2, survivin and notch1 mRNA expression were assessed from cells by a quantitative real-time PCR method (qRT-PCR) (Figure 6a–b), as described previously.[48] Briefly, after each time point, cells were collected in a 1.5 mL tube containing 1 mL of cold PBS pH 7.4. Total RNA was isolated from cells using a Quick-RNA MiniPrep kit (Zymo Research, CA, U.S.) according to the manufacturer’s instructions. During RNA isolation, samples were treated with DNase, followed by RNA quantification using NanoDrop® ND-1000 Spectrophotometer (Thermo Scientific, DE, U.S.). cDNA synthesis and amplification was done by using 1 µg of the isolated RNA template, random hexamers and reverse transcriptase enzyme with a Verso cDNA synthesis kit (Thermo Scientific, DE, U.S.). qRT-PCR assay of triplicate samples treated with SYBR Green I Master® from Roche™ (Mannheim, Germany) was performed on a LightCycler® 480 qRT-PCR (Roche™, Mannheim, Germany) as described previously.[48] The following primer sequences were used:
survivin forward: 5’-AGAACTGGCCCTTCTTGGAGG-3’
survivin reverse: 5’-CTTTTTATGTTCCTCTATGGGGTC-3’
Bcl2 forward: 5’-CCTTGTGGATGACTGAGTACC-3’
Blc2 reverse: 5’AGCCAGGAGAAATCAAACAGAG-3’
notch1 forward: 5’GACAGCCTCAACGGGTACAA-3’
notch1 reverse: 5’-ATACACGTGCCCTGGTTCAG-3’
β-actin forward: 5’- CTCTTCCAGCCTTCCTTCCT-3’
β-actin reverse: 5’ AGCACTGTGTTGGCGTACAG-3’
All custom primers were designed using OligoPerfect™ Designer (Life Technologies, NY, U.S.) to have between 50–60% GC content, annealing temperature of ~ 60°C and 20-base length. No template controls (NTC) were run on each plate to verify that there was neither unspecific amplification nor the formation of primer dimers. Data were analyzed using the Roche RELATIVE quantification method Δ(ΔCt). The relative levels of analyzed mRNAs were normalized against mRNA level of an internal control gene (β-actin) performed in the same run. The relative expression levels were expressed as a percentage of the indicated control.
Cytotoxicity of MM in spheroids
The HT1080 tumor spheroids were prepared by modified liquid-overlay method as described previously.[42] Briefly, 50 µL of a sterile dispersion of agar in serum free medium (1.5% w/v) was added into each well of a 96-well flat bottom plate. After plates cool down under UV light, 1.5 × 104 HT1080 cells dispersed in 100 µL of serum complete medium were seeded in each well. The plates were centrifuged at 1500 rcf for 15 minutes at room temperature and spheroids were cultured until further tests. 50 µL of fresh complete medium were added every third day into the wells.
The cellular viability of the spheroids were determined after 48-hour treatment of 5- and 7-day old HT1080 spheroids at miRNA-34a/Dox concentration of 100 nM and 10 µM, respectively (Figure 8d). Five individually treated spheroids were collected as one replicate. Spheroids were washed two times with PBS and AccuMax® cell detachment solution was added to disperse them into single cells. Following addition of FBS, the cells were centrifuged and washed once with PBS pH 7.4. Complete EMEM:CellTiter-Glo® (1:1 v/v) mixture was added to the cells. After 30 minutes of incubation, 100 µL of the cell lysates were transferred into 96-well black-walled optical flat-bottom plates, and luminescence values were measured in a BioTek Multi-Mode microplate reader.
Statistical analysis
One-way ANOVA with Bonferroni correction was performed for comparison of groups using GraphPad Prism version 5.0 software (GraphPad Software, Inc, CA, U.S.A.). Other statistical methods were specified in the figure legends when they were used. Numerical data were expressed as mean ± SD. Any p value ≤ 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by the NIH Grant (1R21CA179286) to V. P. Torchilin and CAPES Foundation Scholarship (BEX 13275/13-5) to D. F. Costa. The authors thank Dr. W. Hartner for helpful comments on the preparation of the manuscript and Drs. R. W. Giese and P. Wang for assistance on the mass spectrometry analysis. G. Salzano and D. F. Costa contributed equally to this work.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Dr. Giuseppina Salzano, Center for Pharmaceutical Biotechnology and Nanomedicine, Northeastern University, Boston, MA, 02115, USA
Daniel F. Costa, Center for Pharmaceutical Biotechnology and Nanomedicine, Northeastern University, Boston, MA, 02115, USA CAPES Foundation, Ministry of Education of Brazil, Brasília, DF 70040-020, Brazil.
Dr. Can Sarisozen, Center for Pharmaceutical Biotechnology and Nanomedicine, Northeastern University, Boston, MA, 02115, USA
Ed Luther, Department of Pharmaceutical Sciences, Northeastern University, Boston, MA, 02115, USA.
George Mattheolabakis, Department of Pharmaceutical Sciences, Northeastern University, Boston, MA, 02115, USA.
Pooja P. Dhargalkar, Center for Pharmaceutical Biotechnology and Nanomedicine, Northeastern University, Boston, MA, 02115, USA
Prof. Dr. Vladimir P. Torchilin, Center for Pharmaceutical Biotechnology and Nanomedicine, Northeastern University, Boston, MA, 02115, USA Department of Biochemistry, Faculty of Science, King Abdulaziz University, Jeddah, 21589, Saudi Arabia.
References
- 1.Stefani G, Slack FJ. Nat. Rev. Mol. Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 2.Tong AW, Nemunaitis J. Cancer Gene Ther. 2008;15:341–355. doi: 10.1038/cgt.2008.8. [DOI] [PubMed] [Google Scholar]
- 3.Hammond SM. Cancer Chemother. Pharmacol. 2006;58:s63–s68. doi: 10.1007/s00280-006-0318-2. [DOI] [PubMed] [Google Scholar]
- 4.Bader AG, Brown D, Winkler M. Cancer Res. 2010;70:7027–7030. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siemens H, Jackstadt R, Kaller M, Hermeking H. Oncotarget. 2013;4:1399–1415. doi: 10.18632/oncotarget.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng X, Cao M, Zhang J, Hu K, Yin Z, Zhou Z, Xiao X, Yang Y, Sheng W, Wu Y, Zeng Y. Biomaterials. 2014;35:4333–4344. doi: 10.1016/j.biomaterials.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Geng D, Song X, Ning F, Song Q, Yin H. Int. J. Gynecol. Cancer. 2015;25:707–713. doi: 10.1097/IGC.0000000000000399. [DOI] [PubMed] [Google Scholar]
- 8.Marques SC, Ranjbar B, Laursen MB, Falgreen S, Bilgrau AE, Bødker JS, Jørgensen LK, Primo MN, Schmitz A, Ettrup MS, Johnsen HE, Bøgsted M, Mikkelsen JG, Dybkær K. Exp. Hematol. 2016;44:238–246. doi: 10.1016/j.exphem.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 9.Gandhi NS, Tekade RK, Chougule MB. J. Control Release. 2014;194:238–256. doi: 10.1016/j.jconrel.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navarro G, Pan J, Torchilin VP. Mol. Pharm. 2015;12:301–313. doi: 10.1021/mp5007213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egeblad M, Werb Z. Nat. Rev. Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 12.Zhu L, Kate P, Torchilin VP. ACS Nano. 2012;6:3491–3498. doi: 10.1021/nn300524f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu L, Wang T, Perche F, Taigind A, Torchilin VP. Proc. Natl. Acad. Sci. USA. 2013;110:17047–17052. doi: 10.1073/pnas.1304987110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tu Y, Zhu L. J. Control. Release. 2015;212:94–102. doi: 10.1016/j.jconrel.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 15.Balendiran GK, Dabur R, Fraser D. Cell Biochem. Funct. 2004;22:343–352. doi: 10.1002/cbf.1149. [DOI] [PubMed] [Google Scholar]
- 16.Estrela JM, Ortega A, Obrador E. Crit. Rev. Clin. Lab. Sci. 2006;43:143–181. doi: 10.1080/10408360500523878. [DOI] [PubMed] [Google Scholar]
- 17.Musacchio T, Vaze O, D'Souza G, Torchilin VP. Bioconjug. Chem. 2010;21:1530–1536. doi: 10.1021/bc100199c. [DOI] [PubMed] [Google Scholar]
- 18.Salzano G, Navarro G, Trivedi MS, De Rosa G, Torchilin VP. Mol. Cancer Ther. 2015;14:1075–1084. doi: 10.1158/1535-7163.MCT-14-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palm-Apergi C, Lönn P, Dowdy SF. Mol. Ther. 2012;20:695–697. doi: 10.1038/mt.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirose H, Takeuchi T, Osakada H, Pujals S, Katayama S, Nakase I, Kobayashi S, Haraguchi T, Futaki S. Mol. Ther. 2012;20:984–993. doi: 10.1038/mt.2011.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salzano G, Torchilin VP. Methods Mol. Biol. 2015;1324:357–368. doi: 10.1007/978-1-4939-2806-4_24. [DOI] [PubMed] [Google Scholar]
- 22.Maquoi E, Noël A, Frankenne F, Angliker H, Murphy G, Foidart JM. FEBS Lett. 1998;424:262–266. doi: 10.1016/s0014-5793(98)00187-2. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Lin T, Zhang W, Jiang Y, Jin H, He H, Yang VC, Chen Y, Huang Y. Theranostics. 2015;5:787–795. doi: 10.7150/thno.11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehnhardt M, Klein-Hitpass L, Kuhnen C, Homann HH, Daigeler A, Steinau HU, Roehrs S, Schnoor L, Steinstraesser L, Mueller O. BMC Cancer. 2005;5:74. doi: 10.1186/1471-2407-5-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slovak ML, Hoeltge GA, Dalton WS, Trent JM. Cancer Res. 1988;48:2793–2797. [PubMed] [Google Scholar]
- 26.Davidovich IA, Levenson AS, Levenson Chernokhvostov VV. Cancer Lett. 2004;211:189–197. doi: 10.1016/j.canlet.2004.03.026. [DOI] [PubMed] [Google Scholar]
- 27.Zhu L, Kate P, Torchilin VP. ACS Nano. 2012;6:3491–3498. doi: 10.1021/nn300524f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salzano G, Riehle R, Navarro G, Perche F, De Rosa G, Torchilin VP. Cancer Lett. 2014;343:224–231. doi: 10.1016/j.canlet.2013.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grant GM, Giambernardi TA, Grant AM, Klebe RJ. Matrix Biol. 1999;18:145–148. doi: 10.1016/s0945-053x(99)00003-7. [DOI] [PubMed] [Google Scholar]
- 30.Torchilin VP. Nat. Rev. Drug Discov. 2014;13:813–827. doi: 10.1038/nrd4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fawell S, Seery J, Daikh Y, Moore C, Chen LL, Pepinsky B, Barsoum J. Proc. Natl. Acad. Sci. USA. 1994;91:664–668. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torchilin VP, Levchenko TS. Curr. Protein Pept. Sci. 2003;4:133–140. doi: 10.2174/1389203033487298. [DOI] [PubMed] [Google Scholar]
- 33.Azorín E, Solano-Agama C, Mendoza-Garrido ME. Arch. Biochem. Biophys. 2012;528:148–155. doi: 10.1016/j.abb.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 34.Lees JG, Bach CT, Bradbury P, Paul A, Gunning PW, O'Neill GM. Oncogene. 2010;30:1241–1251. doi: 10.1038/onc.2010.516. [DOI] [PubMed] [Google Scholar]
- 35.Millerot-Serrurot E, Guilbert M, Fourré N, Witkowski W, Said G, Van Gulick L, Terryn C, Zahm JM, Garnotel R, Jeannesson P. Cancer Cell. Int. 2010;10:26. doi: 10.1186/1475-2867-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolf K, Mazo I, Leung H, Engelke K, von Andrian UH, Deryugina EI, Strongin AY, Bröcker EB, Friedl P. J. Cell Biol. 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cardoso AM, Trabulo S, Cardoso AL, Maia S, Gomes P, Jurado AS, Pedroso de Lima MC. Mol. Pharm. 2013;10:2653–2666. doi: 10.1021/mp400078h. [DOI] [PubMed] [Google Scholar]
- 38.Zeng H, Wu M, Botnen JH. J. Nutr. 2009;139:1613–1618. doi: 10.3945/jn.109.110320. [DOI] [PubMed] [Google Scholar]
- 39.Mungamuri SK, Yang X, Thor AD, Somasundaram K. Cancer Res. 2006;66:4715–4724. doi: 10.1158/0008-5472.CAN-05-3830. [DOI] [PubMed] [Google Scholar]
- 40.Zha J, Lackner MR. Clin. Cancer Res. 2010;16:2512–2517. doi: 10.1158/1078-0432.CCR-09-2232. [DOI] [PubMed] [Google Scholar]
- 41.Whitehead KA, Langer R, Anderson DG. Nat. Rev. Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarisozen C, Abouzeid AH, Torchilin VP. Eur. J. Pharm. Biopharm. 2014;88:539–550. doi: 10.1016/j.ejpb.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jimenez Valencia AM, Wu PH, Yogurtcu ON, Rao P, DiGiacomo J, Godet I, He L, Lee MH, Gilkes D, Sun SX, Wirtz D. Oncotarget. 2015;6:43438–43451. doi: 10.18632/oncotarget.5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Espinosa M, Ceballos-Cancino G, Callaghan R, Maldonado V, Patiño N, Ruíz V, Meléndez-Zajgla J. Cancer Lett. 2012;318:61–67. doi: 10.1016/j.canlet.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 45.Khaitan D, Chandna S, Arya MB, Dwarakanath BS. J. Transl. Med. 2006;4:12. doi: 10.1186/1479-5876-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perche F, Patel NR, Torchilin VP. J. Control. Release. 2012;164:95–102. doi: 10.1016/j.jconrel.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dioufa N, Tsokos M. FASEB J. 2015;29:S417.6. [Google Scholar]
- 48.Trivedi M, Shah J, Hodgson N, Byun HM, Deth R. Mol. Pharmacol. 2014;85:747–757. doi: 10.1124/mol.114.091728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.