Abstract
Copy number variants (CNVs) are risk factors in neurodevelopmental disorders, including autism, epilepsy, intellectual disability (ID) and schizophrenia. Childhood onset schizophrenia (COS), defined as onset before the age of 13 years, is a rare and severe form of the disorder, with more striking array of prepsychotic developmental disorders and abnormalities in brain development. Because of the well-known phenotypic variability associated with pathogenic CNVs, we conducted whole genome genotyping to detect CNVs and then focused on a group of 46 rare CNVs that had well-documented risk for adult onset schizophrenia (AOS), autism, epilepsy and/or ID. We evaluated 126 COS probands, 69 of which also had a healthy full sibling. When COS probands were compared with their matched related controls, significantly more affected individuals carried disease-related CNVs (P = 0.017). Moreover, COS probands showed a higher rate than that found in AOS probands (P<0.0001). A total of 15 (11.9%) subjects exhibited at least one such CNV and four of these subjects (26.7%) had two. Five of 15 (4.0% of the sample) had a 2.5–3 Mb deletion mapping to 22q11.2, a rate higher than that reported for adult onset (0.3–1%) (P<0.001) or autism spectrum disorder and, indeed, the highest rate reported for any clinical population to date. For one COS subject, a duplication found at 22q13.3 had previously only been associated with autism, and for four patients CNVs at 8q11.2, 10q22.3, 16p11.2 and 17q21.3 had only previously been associated with ID. Taken together, these findings support the well-known pleiotropic effects of these CNVs suggesting shared abnormalities early in brain development. Clinically, broad CNV-based population screening is needed to assess their overall clinical burden.
Keywords: CNV, genetics, neurodevelopment, schizophrenia
INTRODUCTION
Schizophrenia is a debilitating brain disease characterized by hallucinations, delusions, disordered thinking and emotional deficits, and with an approximately 1% prevalence worldwide.1 Although the onset of schizophrenia typically occurs during late adolescence and early adulthood, there are rare (estimated at 1/30 000 births), severe, possibly more homogenous cases with onset during childhood (defined as onset before the age of 13 years). Stratifying by age of onset has been useful in medicine, generally for identifying causal genetic variants.2,3
Our previous study identified a higher rate of large rare copy number variants (CNVs) interrupting genes in pathways of neuronal development and regulation in childhood-onset patients.4 This 2008 study, however, was carried out before there were many disorder-associated CNVs and with relatively small control populations.
Since that time numerous studies have reported various schizophrenia-associated large (>100 kb) rare (<1% in the general population) CNVs, including deletions or duplications on 1q21.1, 2p16.3 (NRXN1), 15q11.2, 15q11–q13, 15q13.3, 16p11.2, 16p13.1, 17p12 and 22q11.2.5–13 Consequently, large rare CNVs have been implicated in a variety of neurodevelopmental disorders such as autism, intellectual disability (ID) and epilepsy.14–20 A major theme in the genetics and genomics of neurodevelopmental disorders is that the same recurrent deletions/duplications at specific loci (2p16.3, 15q13.3, 16p11.2, for example) have been identified in patients with various phenotypes, whereas heterogeneous locus can lead to similar phenotypes.6,8,18–25
This study had three major aims:
to study our rare sample of childhood onset schizophrenia (COS) patients and their families to identify CNVs that have previously been associated with schizophrenia and/or other neurodevelopmental disorders. This approach allowed us to assign likely pathogenicity to previously uninterpretable risk regions, even in a relatively small patient cohort;
to evaluate the rate of 46 selected CNVs in our COS patients vs those found in their matched sibling controls, mitigating the confound of population structure in the case–control comparison; and
to compare the rates of these high-risk CNVs in COS probands to previously published large case–control cohorts of adult onset schizophrenia (AOS), autism and other neurodevelopmental phenotypes
MATERIALS AND METHODS
Patient population
Patients meeting the DSM-II IR/DSM-IV (Diagnostic and Statistical Manual of Mental Disorders, Third Edition Revised/Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition) criteria for schizophrenia with the onset of psychosis before the age of 13 years were recruited nationally. To address the concern of false positives resulting from inclusion of language disorders, we included only patients with clear positive symptoms (delusions or hallucinations) in this study. Medical or neurological disorders, or IQ under 70 were exclusionary criteria. Patients and their available first-degree relatives were interviewed for lifetime and current psychiatric disorders using structured psychiatric interviews and Autism Symptom Questionnaire.26,27 Diagnosis was confirmed with in-patient medication-free observation. A total of 361 patients were screened. This study was approved by the Institutional Review Board of The National Institute of Mental Health. All participants provided written assent/consent with written informed consent from a parent or legal guardian for minors.
Single-nucleotide polymorphism genotyping and quality control
Genomic DNA was purified from either peripheral blood leukocytes or Epstein–Barr virus-transformed immortalized lymphoblastoid cells using the QIAamp DNA Extraction Kit (Qiagen, Valencia, CA, USA) (Supplementary Table S1). Genotyping was performed using Illumina Human or HumanOmni 2.5S BeadChips using the Infinium Assay (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Single-nucleotide polymorphism calls were made using the Illumina GenomeStudio software (Illumina, San Diego, CA, USA).
We carried out three steps of quality of control (QC) using Plink v. 1.07 (http://pngu.mgh.harvard.edu/~purcell/plink)28 and removed samples from the analysis based on the following criteria: (1) samples with discordant sex information; (2) samples with genotyping call rate <97%; and (3) samples with excessive Mendelian errors or demonstrating cryptic relatedness by estimation of identity by descent.
Selection of disease-related CNVs
For the purpose of this screening, we set the following criteria for inclusion in our list of ‘well-documented neurodevelopmental risk CNVS’:
We first reviewed a large group of studies reporting disease-related CNVs for neurodevelopmental disorders (schizophrenia, autism spectrum disorder (ASD), ID and/or epilepsy). Two criteria had to be met: (1) they were identified by case–control in schizophrenia, ASD, ID and/or epilepsy and (2) the sample size of cases was over 300 and the sample size of controls was over 300.
After filtering, 25 studies (9 schizophrenia, 8 ASD, 5 ID and 3 epilepsy) met the criteria. The range of sample size for cases was 400–23 000. On the basis of these 25 studies, our final list of disease-related regions included 46 loci that had a significant P-value (Supplementary Table S2). Of 46 loci, 24, 4 and 8 were associated with only ID, autism and schizophrenia, respectively, and 1 CNV carried risk for all 4 selected neurodevelopmental disorders.
CNV detection and validation
All samples that passed single-nucleotide polymorphism QC procedures were included in the CNV analysis. CNVs were detected first using the CNVision software (http://www.cnvision.org/),24 which automates the detection of structural variation by identifying and then combining predictions from three algorithms: (1) PennCNV version 16 June 2011,29 (2) QuantiSNP v.2.330 and (3) GNOSIS (http://www.cnvision.org/).24 PennCNV and QuantiSNP are based on a hidden Markov model. GNOSIS uses a continuous distribution function employing a sliding window approach, identifying groups of intensity outliers determined by a comparison of patient data to that derived from the Illumina HapMap data set. On the basis of a previously published blinded assessment of the positive predictive value of using 1, 2 or 3 algorithm detection,24 only CNVs identified by two or more algorithms were carried forward through analysis. Additional details of CNV detection and confirmation are provided in Supplementary Information.
Stringent CNVs that passed all QC filters (⩾100 kb size, ⩾1 gene) were considered disease-related if they were within or had ⩾50% overlapping of their length with a disease-related CNV. For the comparison with AOS samples, the same procedures have been used.
All such CNVs were visually inspected with Nexus copy number (http://www.biodiscovery.com/). Given the high positive predictive value for transmitted CNVs, those observed in a family member with ⩾80% overlap by length of the CNV were considered validated. Because of the low prior probability for de novo CNVs, all de novo predictions were validated using one or more of the following approaches: Affymetrix 500K Mapping Array, Agilent 185 or 244K array comparitive genomic hybridization or real-time quantitative polymerase chain reaction with absolute quantification in triplicate. All genomic coordinates reported are genome build NCBI 36/hg18.
CNV analysis
Overall rates of disease-related CNVs were compared with the rates in their healthy control groups as reported. COS patients and healthy full siblings were computed and compared using Fisher’s exact test. Further, the overall rates of disease-related CNVs in COS probands were compared with the published results in large cohort studies using Fisher’s exact test with adjustment for multiple comparisons. Finally, CNVs were screened for AOS patients31,32 and the rate of disease-related CNVs was compared using Fisher’s exact test. All analyses were performed using the statistical package SAS V.9.2 (SAS Institute, Cary, NC, USA) and the two-sided P-values <0.05 were considered significant.
RESULTS
A total of 126 COS probands and 117 full siblings passed all the QC steps, and 19 full siblings were diagnosed with neurodevelopmental disorders including schizophrenia spectrum (n = 11), autism (n = 2) or ID (n = 1) or with bipolar disorder (n = 5). These were excluded from this study. For the remaining group, 69 COS patients had at least one healthy full sibling. We screened the selected 46 disease-related CNVs in COS samples, their healthy full siblings and publically available AOS samples. There were 86 families (68.3%) available for de novo prediction.
Analysis of CNVs in COS probands
Our findings on the rate of COS probands is summarized in Table 1. Among 126 COS patients, we discovered that 15 COS probands (11.9%) had at least one disease-related CNV (see Table 1).
Table 1.
Disease-related CNVs observed in patients with COS
| NSB ID | Chr. band | Start (hg18) | Stop (hg18) | Size (kb) | Type | Duplicated or deleted genes | Inheritance | Disease | Reported in 2008 |
|---|---|---|---|---|---|---|---|---|---|
| 1358a | 2p25.3 | 1 591 064 | 1 836 375 | 245 | Dup | 2 | Mother | SCZ | Yes |
| 534a | 2p25.3 | 1 720 133 | 1 827 317 | 107 | Dup | 2 | Unknown | SCZ | Yes |
| 581 | 2p16.3 | 50 025 162 | 50 136 989 | 112 | Del | 2 | Unknown | SCZ, ASD | Yes |
| 534a | 8q11.2 | 53 550 992 | 54 043 684 | 493 | Dup | 3 | Unknown | ID | No |
| 885 | 10q22.3 | 81 415 378 | 81 588 866 | 173 | Del | 5 | De novo | ID | No |
| 448 | 15q11.2 | 18 818 086 | 20 203 694 | 1386 | Del | 24 | Unknown | SCZ, ID, Epi | No |
| 1358a | 15q11.2 | 20 203 694 | 20 778 963 | 575 | Del | 13 | Mother | SCZ, ID, Epi | No |
| 1546a | 15q13.3 | 30 238 780 | 30 620 951 | 382 | Del | 26 | De novo | SCZ, ASD, ID, Epi | No |
| 498 | 15q13.3 | 30 238 780 | 30 713 368 | 475 | Del | 30 | Mother | SCZ, ASD, ID, Epi | No |
| 481 | 16p12.1 | 21 498 074 | 21 946 841 | 449 | Del | 7 | Father | ID | No |
| 676a | 16p11.2 | 29 502 984 | 30 107 306 | 604 | Dup | 15 | Father | SCZ, ASD | Yes |
| 2011 | 16p11.2 | 29 782 436 | 30 227 808 | 445 | Dup | 34 | Father | SCZ, ASD | Yes |
| 1546a | 17q21.3 | 41 321 621 | 41 706 070 | 384 | Dup | 4 | Father | ID | No |
| 1275 | 22q11.2 | 17 092 563 | 20 077 678 | 2985 | Del | 49 | De novo | SCZ, ID | No |
| 1220 | 22q11.2 | 17 224 632 | 19 842 333 | 2618 | Del | 48 | De novo | SCZ, ID | No |
| 537 | 22q11.2 | 17 257 787 | 19 855 248 | 2597 | Del | 46 | Unknown | SCZ, ID | No |
| 1804 | 22q11.2 | 17 257 787 | 19 963 350 | 2706 | Del | 47 | De novo | SCZ, ID | No |
| 3169 | 22q11.2 | 17 269 794 | 20 128 199 | 2858 | Del | 55 | De novo | SCZ, ID | No |
| 676a | 22q13.3 | 47 903 228 | 49 557 485 | 1654 | Dup | 4 | De novo | ASD | No |
Abbreviations: ASD, autism spectrum disorders; COS, childhood onset schizophrenia; CNV, copy number variants; Epi, epilepsy; ID, intellectual disability; SCZ, schizophrenia.
Individuals with two events.
As shown in Table 1, CNVs found in probands included deletions at 2q16.3, 10q22.3, 15q11.2, 15q13.3, 16p12.1 and 22q11.2, and duplications at 2q25.3, 8q11.2, 16p11.2, 17q21.3 and 22q13. Most strikingly, five patients (4.0%) carried a microdeletion in the chromosome 22q11.2 region, a recognized risk factor for schizophrenia, autism and other neurodevelopmental disorders.33–35 The frequency of this microdeletion in COS probands (4.0%) was significantly higher than the frequency in healthy controls (0.2% vs 4.0%, P<0.0001)36 and was also higher than the previously reported frequency in AOS patients (0.3–1%)37–39 (P<0.0001). Because 20% of our COS patients had prepsychotic autism spectrum disorder,40 we were particularly interested in the prepsychotic developmental pattern of our COS probands with the 22q11.2 deletion. While the 22q11.2 deletion has been associated with autism,34,35 it is of interest that none of our five 22q11.2 deletion subjects had previous autism symptomatology, indicating an independent pathway of development for this childhood schizophrenia subgroup. This supports the recent finding in 22q11.2 deletion AOS patients for whom early autism spectrum symptoms were also not observed.41
Further, COS patients had higher rates for three of our risk CNVs than those reported in previous AOS patients (2q25.3, 15q13.3 and16p11.2; Table 2).38,42,43
Table 2.
CNV loci are more frequent in COS patients than in AOS patients
| CNV | Rate in COS casesa | Rate in AOS cases | P-value | Reference |
|---|---|---|---|---|
| 2q25.3 | 2/126 | 11/5970 | 0.0007 | 43 |
| 15q13.3 del | 2/126 | 21/10 887 | 0.0007 | 38 |
| 16p11.2 dup | 2/126 | 26/8590 | 0.0114 | 38 |
| 22q11.2 del | 5/126 | 35/11 400 | <0.0001 | 38 |
Abbreviations: AOS, adult onset schizophrenia; COS, childhood onset schizophrenia; CNV, copy number variants.
The rates are presented as numbers of CNV carriers per number of all individuals.
As shown in Table 2, we identified five COS probands who carried CNVs previously reported only for ID (4) or autism (1); however, three of these represented ‘second hits’ for previously identified probands carrying schizophrenia-associated CNVs. Four of the five COS patients carried CNVs reported previously for ID (subject ID: 885, 481, 534 and 1546).44–47 These subjects, respectively, had full-scale IQs of 99, 94, 70 and 78; this is the first report that has identified these CNVs in schizophrenia. One (ID: 676) carried a CNV reported previously only for ASD.48 This subject had no prepsychotic pattern of autistic behavior (although 20% of COS patients do have this pattern)49 and, to our knowledge, this is the first report to associate these CNVs with schizophrenia.
We next sought to determine whether these disease-related CNVs were inherited or if they arose de novo in the COS probands using the parental data that were available for 11 probands (14 CNVs), who carried at least one disease-related CNV, 7 CNVs were confirmed as de novo (50.0%, 7/14), although these data were unavailable for five probands. Four of the 22q11.2 deletions were shown to be de novo, for one these data were unavailable.
Analysis of COS proband–healthy full sibling pairs
Since the ancestries of COS patients were highly heterogeneous, healthy siblings from the respective COS proband–full sibling pair were selected for comparison using several methods. For this comparison analysis, 69 pairs of COS proband–healthy full sibling were included. There was no significant difference in gender distribution and age between patient group and healthy oldest sibling group (Supplementary Table S3).
We compared the rates of disease-related CNVs for COS probands and their oldest healthy siblings. Among 69 COS proband–healthy sibling pairs, disease-related CNVs were more common among patients (13.0%, n = 9) than healthy siblings (1.5%, n = 1) (two-tailed P = 0.017) (Figure 1). Excluding 22q11.2 deletion from the comparison, the COS group tended to have higher rate than healthy sibling group (10.1% vs 1.5%, two-tailed P = 0.062).
Figure 1.
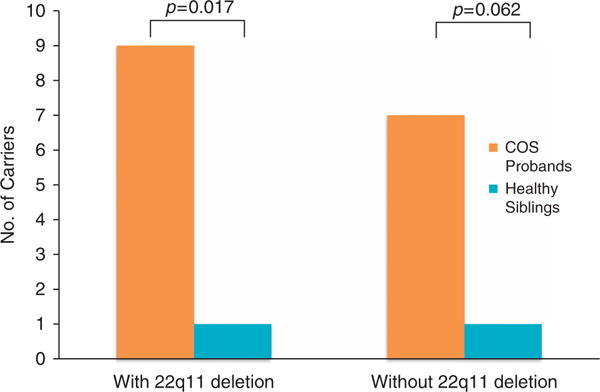
Comparisons of childhood onset schizophrenia (COS) proband–Healthy Sibling Pairs (n = 69).
As 30 probands have more than 1 healthy sibling, we conducted two additional probands–sibling comparison methods. We included all available healthy siblings and used the weighted scores depending on the number of healthy siblings in each family; we also selected one sibling randomly and computed permutations. All three methods exhibited a significant increase in rate for the COS group compared with their healthy sibling.
Finally, we explored the paternal age at birth with the rates of CNV in COS probands and healthy siblings. The significant difference between siblings and probands of CNVs remained even after adjusting the observed rate for paternal age at birth.
Comparison with AOS
Two AOS cohorts were available for comparison using our 46 candidate CNV regions; 977 caucasian AOS samples from the Genetic Association Information Network (GAIN) and the University of Pennsylvania (rate = 4.9%, P<0.0001)31 and another published data set for 649 Ashkenazi AOS patients from Israel (rate = 1.4%, P<0.0001).32 Our COS sample had a higher rate of CNVs than that found in either of these samples (Figure 2).
Figure 2.
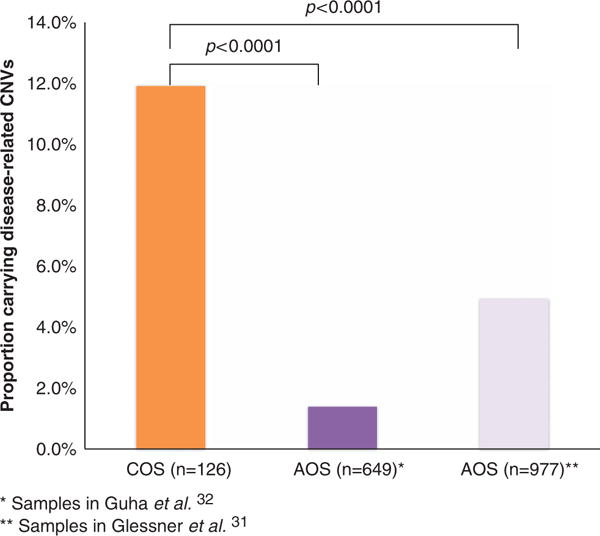
Rates of neurodevelopmental disease-related copy number variants (CNVs) in childhood onset schizophrenia (COS) and adult onset schizophrenia (AOS).
DISCUSSION
Overall, we identified a high rate (11.9%) of disease-related CNVs for COS, which was certainly greater than that expected in the general population and exceeding, in this sample, the rates seen in other neurodevelopmental disorders. Interestingly, 4 (26.7%) of the15 subjects with a CNV had an additional CNV associated with neuropsychiatric disorder. This observation is consistent with a ‘two-hit’ hypothesis in these cases, as recently stressed by Girirajan et al.50 As the rarer variant was considered to be the primary-site variant,50,51 2p25.3, 8q11.2, 17q21.3 and 22q13.3 are assumed to be the primary site for subjects 1358, 534, 1546 and 676, respectively.
Five of the COS carried CNVs that, to date, have been associated with only ID or with autism; for three of these subjects, these were ‘second hits’, but for two cases, these were the only disease-related CNV identified. This finding is not surprising given that many of the CNVs associated with ID are noted to have a variety of accompanying behavioral disturbances.51 However, our findings do extend the phenotypic spectrum for these particular structural variants.
Of considerable interest is the finding that 4% of these COS patients carried a 2.5–3 Mb 22q11.2 deletion. This is not only higher than the rate of 0.2% in the general population but also higher than the 0.3–1% rate seen in adult onset cases (P<0.001). This marked association with early onset schizophrenia is not accounted for by unusually severe prepsychotic developmental patterns. We also note that the five COS probands (4 females and 1 male) with 22q11.2 deletions had a mean age of onset of 10.2 years (range 7–12). In all cases, onset was insidious with withdrawal, deterioration and symptoms not seen earlier, which included bizarre behavior, hallucinations and delusions. The mean full-scale IQ was 86 (range 70–96). Earlier development included a variety of speech (articulation), language and motor delays; three had repeated an early grade. In three cases, there was obsessive ideation but none met the criteria for OCD. No cases had received a diagnosis of mood disorder. None had cardiac abnormalities.
Although comparisons with two sets of adult onset cases showed that our COS sample carried more disease-related CNVs than either of the adult samples, neither is an ideal comparison for our ethnically heterogeneous COS sample. Moreover, our sample is necessarily small, given the rarity of true COS, and unknown ascertain biases could enhance certain findings in the studies of rare disorders. We do note, however, that for 22q11.2 2p25.3, 15q13.3 and 16p11.2, the rate we observed in our cohort is higher than we would expect for AOS.37–39,42,43
Finally, the nonspecificity of these risk factors needs to be further explored. Ideally a pediatric population study could be used to examine phenotypic variance in a group identified by these CNVs. Such a study would give a more precise indication of the true clinical burden associated with these intriguing but still poorly understood chromosomal abnormalities.
Supplementary Material
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
References
- 1.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders: DSM-IV. 4th. American Psychiatric Association; Washington, DC, USA: 1994. p. 866. [Google Scholar]
- 2.Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, Huey B, et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250:1684–1689. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- 3.Murrell J, Farlow M, Ghetti B, Benson MD. A mutation in the amyloid precursor protein associated with hereditary Alzheimer’s disease. Science. 1991;254:97–99. doi: 10.1126/science.1925564. [DOI] [PubMed] [Google Scholar]
- 4.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 5.Ingason A, Rujescu D, Cichon S, Sigurdsson E, Sigmundsson T, Pietilainen OP, et al. Copy number variations of chromosome 16p13.1 region associated with schizophrenia. Mol Psychiatry. 2011;16:17–25. doi: 10.1038/mp.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.ISC. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P, et al. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rujescu D, Ingason A, Cichon S, Pietilainen OP, Barnes MR, Toulopoulou T, et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009;18:988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Consortium IS. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingason A, Rujescu D, Cichon S, Sigurdsson E, Sigmundsson T, Pietilainen OP, et al. Copy number variations of chromosome 16p13.1 region associated with schizophrenia. Mol Psychiatry. 2009;16:17–25. doi: 10.1038/mp.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mefford HC, Eichler EE. Duplication hotspots, rare genomic disorders, and common disease. Curr Opin Genet Dev. 2009;19:196–204. doi: 10.1016/j.gde.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shinawi M, Liu P, Kang SH, Shen J, Belmont JW, Scott DA, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet. 2010;47:332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 17.O’Donovan MC, Kirov G, Owen MJ. Phenotypic variations on the theme of CNVs. Nat Genet. 2008;40:1392–1393. doi: 10.1038/ng1208-1392. [DOI] [PubMed] [Google Scholar]
- 18.van Bon BW, Mefford HC, Menten B, Koolen DA, Sharp AJ, Nillesen WM, et al. Further delineation of the 15q13 microdeletion and duplication syndromes: a clinical spectrum varying from non-pathogenic to a severe outcome. J Med Genet. 2009;46:511–523. doi: 10.1136/jmg.2008.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helbig I, Mefford HC, Sharp AJ, Guipponi M, Fichera M, Franke A, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009;41:160–162. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Kovel CG, Trucks H, Helbig I, Mefford HC, Baker C, Leu C, et al. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 2010;133(Part 1):23–32. doi: 10.1093/brain/awp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levinson DF, Duan J, Oh S, Wang K, Sanders AR, Shi J, et al. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry. 2011;168:302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller DT, Shen Y, Weiss LA, Korn J, Anselm I, Bridgemohan C, et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J Med Genet. 2009;46:242–248. doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharp AJ, Mefford HC, Li K, Baker C, Skinner C, Stevenson RE, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Girirajan S, Brkanac Z, Coe BP, Baker C, Vives L, Vu TH, et al. Relative burden of large CNVs on a range of neurodevelopmental phenotypes. PLoS Genet. 2011;7:e1002334. doi: 10.1371/journal.pgen.1002334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumra S, Frazier JA, Jacobsen LK, McKenna K, Gordon CT, Lenane MC, et al. Childhood-onset schizophrenia. A double-blind clozapine-haloperidol comparison. Arch Gen Psychiatry. 1996;53:1090–1097. doi: 10.1001/archpsyc.1996.01830120020005. [DOI] [PubMed] [Google Scholar]
- 27.Shaw P, Sporn A, Gogtay N, Overman GP, Greenstein D, Gochman P, et al. Childhood-onset schizophrenia: a double-blind, randomized clozapine–olanzapine comparison. Arch Gen Psychiatry. 2006;63:721–730. doi: 10.1001/archpsyc.63.7.721. [DOI] [PubMed] [Google Scholar]
- 28.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colella S, Yau C, Taylor JM, Mirza G, Butler H, Clouston P, et al. QuantiSNP: an objective Bayes hidden-Markov model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–2025. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glessner JT, Reilly MP, Kim CE, Takahashi N, Albano A, Hou C, et al. Strong synaptic transmission impact by copy number variations in schizophrenia. Proc Natl Acad Sci USA. 2010;107:10584–10589. doi: 10.1073/pnas.1000274107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guha S, Rees E, Darvasi A, Ivanov D, Ikeda M, Bergen SE, et al. Implication of a rare deletion at distal 16p11.2 in schizophrenia. JAMA Psychiatry. 2013;70:253–260. doi: 10.1001/2013.jamapsychiatry.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bassett AS, Chow EW. Schizophrenia and 22q11.2 deletion syndrome. Curr Psychiatry Rep. 2008;10:148–157. doi: 10.1007/s11920-008-0026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niklasson L, Rasmussen P, Oskarsdottir S, Gillberg C. Autism, ADHD, mental retardation and behavior problems in 100 individuals with 22q11 deletion syndrome. Res Dev Disabil. 2009;30:763–773. doi: 10.1016/j.ridd.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 35.Vorstman JA, Morcus ME, Duijff SN, Klaassen PW, Heineman-de Boer JA, Beemer FA, et al. The 22q11.2 deletion in children: high rate of autistic disorders and early onset of psychotic symptoms. J Am Acad Child Adolesc Psychiatry. 2006;45:1104–1113. doi: 10.1097/01.chi.0000228131.56956.c1. [DOI] [PubMed] [Google Scholar]
- 36.Goodship J, Cross I, LiLing J, Wren C. A population study of chromosome 22q11 deletions in infancy. Arch Dis Child. 1998;79:348–351. doi: 10.1136/adc.79.4.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Need AC, Ge D, Weale ME, Maia J, Feng S, Heinzen EL, et al. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5:e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grozeva D, Conrad DF, Barnes CP, Hurles M, Owen MJ, O’Donovan MC, et al. Independent estimation of the frequency of rare CNVs in the UK population confirms their role in schizophrenia. Schizophr Res. 2012;135:1–7. doi: 10.1016/j.schres.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoogendoorn ML, Vorstman JA, Jalali GR, Selten JP, Sinke RJ, Emanuel BS, et al. Prevalence of 22q11.2 deletions in 311 Dutch patients with schizophrenia. Schizophr Res. 2008;98:84–88. doi: 10.1016/j.schres.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rapoport J, Chavez A, Greenstein D, Addington A, Gogtay N. Autism spectrum disorders and childhood-onset schizophrenia: clinical and biological contributions to a relation revisited. J Am Acad Child Adolesc Psychiatry. 2009;48:10–18. doi: 10.1097/CHI.0b013e31818b1c63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vorstman JA, Breetvelt EJ, Thode KI, Chow EW, Bassett AS. Expression of autism spectrum and schizophrenia in patients with a 22q11.2 deletion. Schizophr Res. 2012;143:55–59. doi: 10.1016/j.schres.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 42.Lee Y, Mattai A, Long R, Rapoport JL, Gogtay N, Addington AM. Microduplications disrupting the MYT1L gene (2p25.3) are associated with schizophrenia. Psychiatr Genet. 2012;22:206–209. doi: 10.1097/YPG.0b013e328353ae3d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vrijenhoek T, Buizer-Voskamp JE, van der Stelt I, Strengman E, Sabatti C, Geurts van Kessel A, et al. Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am J Hum Genet. 2008;83:504–510. doi: 10.1016/j.ajhg.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balciuniene J, Feng N, Iyadurai K, Hirsch B, Charnas L, Bill BR, et al. Recurrent 10q22–q23 deletions: a genomic disorder on 10q associated with cognitive and behavioral abnormalities. Am J Hum Genet. 2007;80:938–947. doi: 10.1086/513607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Bon BW, Balciuniene J, Fruhman G, Nagamani SC, Broome DL, Cameron E, et al. The phenotype of recurrent 10q22q23 deletions and duplications. Eur J Hum Genet. 2011;19:400–408. doi: 10.1038/ejhg.2010.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bachmann-Gagescu R, Mefford HC, Cowan C, Glew GM, Hing AV, Wallace S, et al. Recurrent 200-kb deletions of 16p11.2 that include the SH2B1 gene are associated with developmental delay and obesity. Genet Med. 2010;12:641–647. doi: 10.1097/GIM.0b013e3181ef4286. [DOI] [PubMed] [Google Scholar]
- 47.Grisart B, Willatt L, Destree A, Fryns JP, Rack K, de Ravel T, et al. 17q21.31 microduplication patients are characterised by behavioural problems and poor social interaction. J Med Genet. 2009;46:524–530. doi: 10.1136/jmg.2008.065367. [DOI] [PubMed] [Google Scholar]
- 48.Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rapoport J, Chavez A, Greenstein D, Addington A, Gogtay N. Autism spectrum disorders and childhood-onset schizophrenia: clinical and biological contributions to a relation revisited. J Am Acad Child Adolesc Psychiatry. 2009;48:10–18. doi: 10.1097/CHI.0b013e31818b1c63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, Goldstein A, et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N Engl J Med. 2012;367:1321–1331. doi: 10.1056/NEJMoa1200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


