Summary
Tumor suppressor protein 53 (p53) plays a central role in the control of genome stability, acting primarily through the transcriptional activation of stress-response genes. However, many p53 binding sites are located at genomic locations with no obvious regulatory-link to known stress-response genes. We recently discovered p53 binding sites within retrotransposon-derived elements in human and mouse subtelomeres. These retrotransposon-derived p53 binding sites protected chromosome ends through transcription activation of telomere repeat RNA, as well as through the direct modification of local chromatin structure in response to DNA damage. Based on these findings, I hypothesize that a class of p53 binding sites, including the retrotransposon-derived p53-sites found in subtlomeres, provide a primary function in genome stability by mounting a direct and local protective chromatin-response to DNA damage. I speculate that retrotransposon-derived p53 binding sites share features with telomere-repeats through an evolutionary drive to monitor and maintain genome integrity.
Keywords: enhancers, epigenetics, p53, retrotransposons, telomeres
Introduction
Repetitive DNA elements constitute a large proportion of eukaryotic genomes, and are thought to be the evolutionary building blocks of genes and genetic regulatory elements. Telomeres are a unique type of repetitive DNA element that bind sequence-specific factors to protect the ends of linear chromosomes. Telomeres may be evolutionarily derived from type II introns, retrotransposons, and related mobile genetic elements [1]. In most eukaryotes, telomere repeats can be propagated by telomerase, a sequence-specific reverse-transcriptase [2]. Telomere repeats are bound by a collection of telomere-specific factors, termed shelterin, that regulate the local DNA damage response and prevent the chromosome ends from being mistaken for double strand breaks. A critical loss of telomere repeat number destabilizes shelterin and elicits a DNA damage signal that can lead to telomere elongation, or alternatively, cell cycle arrest and senescence. Thus, there is a dynamic homeostasis between telomere repeat number, telomere maintenance, cell cycle control, and cellular replicative capacity.
Telomere damage signaling is processed by the p53 pathway [3, 4]. Phosphorylation of p53 by ATM or ATR leads to increased DNA binding and transcriptional activation of p53 target genes important for cell cycle arrest, while additional post-translational modifications of p53, including lysine acetylation, further activate genes important for apoptosis and autophagy [5]. Although p53 binds to promoter elements in several p53 responsive genes, recent genome-wide studies have revealed that a majority of p53 binding sites are found in distal enhancer elements or in a variety of proto-enhancers, that often include repetitive LTR-like elements [6]. These proto-enhancers show an increase in p53 binding and alteration in histone modifications in response to DNA damage, but it is not known how they may regulate any downstream target genes. Here, I consider a potential function of these distal, proto-enhancer p53 sites in genome protection and how they may function similar to telomere repeats.
Telomeres and subtelomeres are actively transcribed
Telomeres are known to assemble into distinct chromatin structures characterized by condensed heterochromatin and transcriptional silence [7, 8]. More recent studies reveal a much more dynamic behavior at telomeres, and other sites of heterochromatin, involving the generation of non-coding RNA transcripts that can alter local chromatin conformation and interact with various target proteins [9]. Telomeres can switch from closed to open conformations that allow transcription, replication, and telomere repeat addition by telomerase, or in some less frequent cases, by homologous recombination [10, 11]. Most of the regulation at telomere repeats can be attributed to the shelterin components. However, the genomic regions surrounding telomere repeats, referred to as subtelomeres, have complex DNA structures that can also influence telomere function and chromosome stability.
Subtelomeres typically consist of tandem and degenerate repeat elements. In S. cerevisiae, subtelomeres include a number of functionally important regulatory elements, including latent origins of DNA replication and binding sites for scTBF1, an orthologue of human telomere-repeat binding factors. scTBF1 can interact with telomere-specific factor scRAP1 to regulate the DNA damage response, indicating that there are regulatory interactions between telomeres and subtelomeres [12]. Human subtelomeres consist of various length tandem duplications and repeat elements, including portions of retrotransposon-like repeats [13, 14]. Most of these human retrotransposons are vestigial remnants and no longer retain coding sequences for active transposition. In contrast, Drosophila and other dipteran telomeres are unique among metazoans in their utilization of active retrotransposons for telomere length regulation [15, 16]. Drosophila telomeres lack terminal repeats, and their genomes do not encode telomerase. Rather, Drosophila telomeres consist of tandem duplications of specialized retrotransposons that are maintained by reverse transcription and site-specific integration. The evolutionary relationship between telomerase and reverse transcriptases suggests that these two telomere maintenance mechanisms share a common ancestor. And while telomerase protein component (TERT) has been reported to function as an RNA-dependent RNA polymerase, it is not known if this function contributes to telomere maintenance [17].
Telomere retrotransposition is not frequently observed in metazoans other than diptera. However, telomere-repeat encoding RNA (TERRA) that could serve as templates for retrotransposition are frequently observed in diverse organisms, ranging from yeast [18, 19] to mouse [20] and human [21], (reviewed in [9, 22, 23]). And like retrotransposed RNA, TERRA is retained at telomere repeat DNA by mechanisms involving both protein-interaction and RNA-DNA hybrid formation [24, 25]. In human, telomere-repeat encoded RNA (TERRA) is induced in response to genomic stress, including DNA damage, and arises from GC-rich promoter-like elements in the subtelomeres. Some of these GC-rich elements overlap with CTCF binding sites, and are enriched with RNA polymerase II and cohesin, suggesting that they are sites of RNA polymerase II initiation, pausing, or DNA-DNA loop interactions [14, 26]. TERRA transcripts maintain telomeric heterochromatin and regulate telomere DNA accessibility [9, 27]. Although numerous stress responses can activate TERRA transcription [28], the specific mechanisms of TERRA transcript regulation remain poorly understood. The discovery of p53 binding sites within subtelomeres provided a potential mechanism for transcription activation of TERRA in response to DNA damage signaling.
p53 mounts a stress response at telomeres
P53 is well known for its many functions in genome integrity [29]. In response to genotoxic and other stress-responses, p53 is subject to a variety of post-translational modifications that increase its protein stability, induce its DNA binding at various chromosomal sites, and enhance transcription activation of numerous target genes. Telomere uncapping is one type of genotoxic stress that activates p53 through ATM and ATR signaling [3, 4]. Shelterin proteins, TRF2 and Pot1, prevent ATM and ATR from recognizing chromosome DNA ends as double strand breaks [30]. Telomere uncapping induced by loss of shelterin binding leads to ATM and ATR activation, and ultimately p53 phosphorylation and stabilization. Changes in TRF2 levels and stability, as well as normal changes in telomere conformation associated with telomere DNA replication, and repeat elongation, may also involve sub-threshold ATM and p53 signaling pathways [11, 31]. While telomere uncapping can lead to p53 activation, telomeres are themselves subject to p53 signaling. For example, p53 is required for the induction of TERRA transcription following TRF2 loss and telomere uncapping [10, 28]. p53 can also function upstream of telomere uncapping by ubiquitin-mediated degradation of TRF2 [32]. This positive feedback loop promotes ATM phosphorylation of p53 and the further uncapping of telomeres [33]. This may be important for fine tuning telomere length regulation and damage signaling. However, a direct role for p53 in TERRA transcription and telomere regulation had not been reported.
Subtelomeric p53 binding sites are derived from retrotransposons
Genome-wide studies reveal that p53 can bind to a large number of chromosomal positions, including human and mouse subtelomeres [34]. Epigenetic analysis of p53 ChIP-Seq data revealed that only 23% of sites localized at transcription start sites (promoter) or at enhancers (colocalization with H3K4me1 and H3K27ac), and an equal number of p53 sites were at positions not associated with any obvious target genes [35, 36]. Many of the p53 sites also correspond with retrotransposon-like elements. A computational search for p53 binding sites identified ~400,000 possible sites within Alu elements [37]. These p53 sites occur within the internal RNA polymerase III binding sites, designated Boxes A/A’ and B within Alu elements. The strongest p53 sites were found in the most recently evolved Alu repeat subfamilies. P53 binding sites consist of half sites that are separated by a variable length spacer. Sites with shorter spacers tend to function in transcription activation, while sites with longer spacers tend to function in transcription repression [38]. However, more recent metadata analyses suggest that p53 transcriptional repression is an indirect effect of the transcriptional activation of CDKN1A/p21 and the repressor proteins E2F4 and E2F7 [39, 40]. Sites with no spacer have been referred to as unsplit and tend to be more differentially bound upon p53 activation [36]. Many of the unsplit P53 binding sites are found within SINE/Alu, SINE/MIR and LTR/ERV repeats. Experimentally determined p53 binding sites from genome-wide ChIP-Seq showed that ~1,500 p53 sites were within ERV/LTR [41]. Relative to all p53 binding sites, the transposon-associated p53 sites were found to have stronger binding and low phylogenetic conservation, suggesting that they are recently evolved high-affinity p53 binding sites [42].
The p53 binding sites that we identified in human subtelomeres [34] fall within several categories, including retroviral-associated transposon elements LTR10, MER61, and Alu-associated p53 response elements. The LTR10b/MER61 family are among the most conserved of these mobile elements (oldest), but also among the most occupied by p53 [42]. The p53 binding sites within the LTR10b/MER61 family, which are most proximal to terminal telomere repeats in human subtelomeres, tend to be unsplit and contain imperfect tandem duplications. This is consistent with these sites arising from a recent fusion of MIR and LTR transposon elements in human subtelomeres.
With respect to heterochromatin structure, these p53 binding sites are predicted to fall on the external surface of nucleosomes [37, 43], which is consistent with genome-wide studies showing a correlation between nucleosome occupancy and p53 binding sites [42]. Many of these unassigned sites acquire H3acK27 and H3acK16 upon p53 binding [6]. These sites also correspond to regions of the genome that are nucleosome dense (as measured by ATAC-seq and MNase-seq studies), suggesting that p53 has ‘pioneer’ activity by virtue of its ability to invade and modify previously condensed chromatin [44]. Further epigenetic analyses suggest that these sites are commonly elevated in H4K16 acetylation after p53 binding [6]. H4K16ac is most commonly elevated at p53 binding sites, especially the distal types. The lysine acetyl transferases KAT5 (TIP60) and KAT 8 (P/CAF) are mostly responsible for H4K16Ac, and also directly modify p53 [45]. H4K16 acetylation is also enriched at telomeres. In yeast, SAS (Something About Silencing) complex acetylates H4K16 to control telomeric silencing. This raises the possibility that p53-associated KATs may alter local chromatin structure to increase DNA accessibility for transcription, replication, or repair.
p53 inhibits retrotransposition
P53 binding to transposable elements has been proposed to have a function in their transcriptional regulation [46]. On the one hand, p53 binding to transposable elements may be responsible for their transcriptional activation and mobilization during DNA damage and tumor progression. p53 has been shown to bind and activate transcription of human retrotransposon L1 LTR elements [47]. The authors propose that retrotransposition of p53 binding sites increases genome stability by increasing the overall number of p53 response elements throughout the genome.
On the other hand, loss of p53 tends to correlate with an increase in retrotransposon activity and consequent genomic instability [46]. In several animal models, including Drosophila, p53 has been shown to repress retrotransposon expression [48]. The loss of p53 corresponded to a loss of H3K9me3 and heterochromatin at the LTRs [48]. This includes the telomere-specific TAHRE elements that regulate telomere length control [49].
A similar finding was made in mouse and human, where p53 was required to block the massive transcription of many short interspersed elements ( SINEs) and tandem repeats [46]. SINE transcription was induced by demethylating agent 5’ azacytidine, suggesting that p53 disruption, by itself, is not sufficient to activate retrotransposon transcription. Activation of mouse retrotransposons induces the interferon response and causes cell cycle arrest, a phenomenon referred to as TRAIN (transcription of repeats activates interferon). The TRAIN response is hypothesized to prevent the propagation of genetically unstable cells, thus acting as an alternative tumor suppressor if p53 fails [46] In human, p53 binding to Alu repeats was found to inhibit the RNA polymerase III internal promoter utilization and transcription [50–52]. Thus, p53 may directly inhibit Alu transcription in human. It may be possible that p53 inhibits pol III transcription, but activates pol II transcription from alternative start sites that ultimately restrict retrotransposition.
p53 binding sites induce eRNA expression
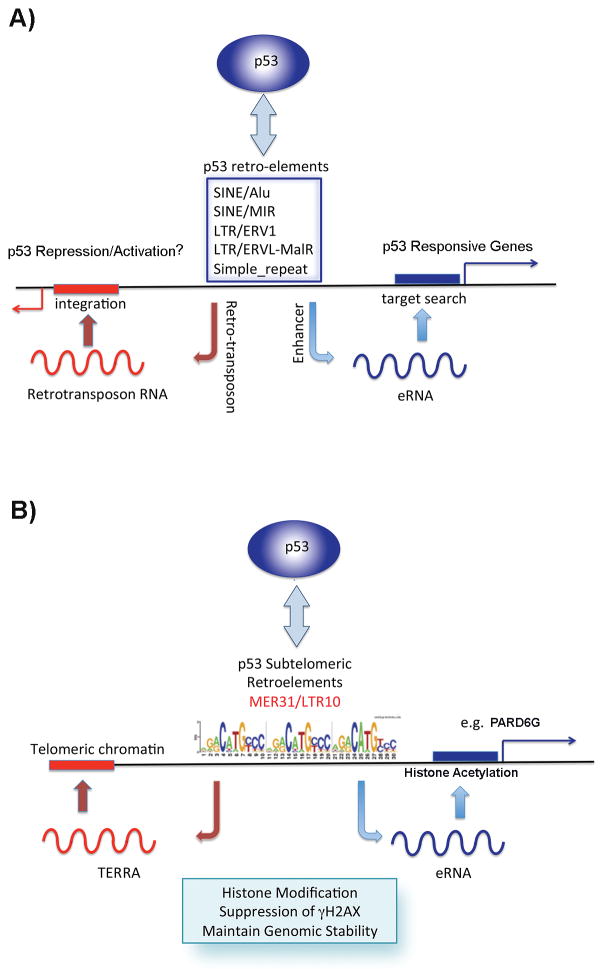
In addition to binding to retrotransposons, p53 binds to many enhancer and proto-enhancer elements. Most enhancers generate bi-directional non-coding RNAs, termed enhancer-RNAs (eRNAs) in response to enhancer activation [53]. This is also observed for p53-bound enhancer like elements [54–56]. Enhancer RNAs are low abundance, non-coding RNAs that are thought to direct enhancers to find promoters. Whether they target promoters through base-pair complementarity or RNA-polymerase tracking or structural scaffold for protein complexes is not yet known. However, the enhancer search for promoters is reminiscent of retrotransposon re-integration at specific sites, such as that observed in Drosophila telomeres. We have found that p53 induces enhancer-like RNA at the subtelomeric retrotransposon-like sites, and that these sites function as transcriptional enhancer elements for TERRA as well as some subtelomeric genes, like PARD6G (Fig. 1). Thus, retrotransposon-derived p53 binding sites found in subtlomeres function as proto-enhancers [6]. We also found that subtelomeric p53 maintains local telomere repeat stability. Whether other p53 proto-enhancer elements function to maintain local genetic stability remains an important unanswered question.
Figure 1.
Model of p53 binding to retrotransposon-like elements throughout the genome (A) and in human subtelomeres (B). A: High-affinity p53 binding sites exist in many retrotransposon-like elements, including Alu and LTR repeats. Some of these sites are implicated in the repression of retrotransposons, while others have been implicated as transcriptional enhancers that can target and activate distal promoters. B: Similar properties have been found for a MER31/LTR10 repeat found in human subtelomeres that binds p53 and activates telomere-encoded repeat RNA (TERRA) and distal subtelomeric genes (e.g. PARD6G).
p53 regulates the alternative lengthening of telomeres (ALT)
P53 has also been implicated in the suppression of telomere recombination. Alternative lengthening of telomeres (ALT) is a telomerase-independent mechanism of telomere repeat expansion. Most ALT can be attributed to homologous recombination-based mechanisms. At least two lines of evidence suggest that p53 plays a role in preventing ALT. First, most ALT tumors carry p53 mutations [57–59]. Secondly, most telomeres in ALT cells colocalizes with PML nuclear bodies to form ALT-associated PML bodies (APBs) [60]. p53 is also found at these APBs, and this colocalization correlates with unrepaired DNA double strand breaks [60]. The role of p53 at these sites may be complicated since p53 may facilitate HR-repair, but suppress other forms of repeat-recombination associated with ALT .
Telomeres in ALT cells frequently colocalize with γH2AX foci. In our study, we observed that p53 expression and p53 binding sites in subtelomeres prevented the accumulation of γH2AX at sites proximal to the p53 binding sites. The suppression of γH2AX also correlated with the formation of p53-dependent histone H3K9 acetylation and eRNA transcription. Based on this, we proposed a model where p53 binding prevents the accumulation of persistent DNA damage signaling, by inducing local transcription and histone modifications conducive to DNA repair and resolution of γH2AX signaling.
Inherited mutations in p53 have telomere deficiencies
Genetic evidence also suggests that there is a close connection between p53 and telomere regulation. Specific p53 inactivating mutations (e.g. R175H) in LoVo colon cancer cells altered telomere nuclear architecture and individual telomere lengths [61]. Mutant p53 telomeres tended to cluster in the periphery, while wildtype p53 telomeres clustered in the center of the nucleus. P53 mutants led to more telomere aggregates and tetraploidization, potentially due to telomere uncapping. In mice, a germ-line deletion of p53 carboxy-terminus (P53 Δ31/Δ31) produced a dyskeratosis congenita phenotype with severe telomere deficiencies [62, 63]. The p53Δ31 is known to increases p53 transcription activity, but the authors attribute the telomere deficiency to the decreased expression of telomere regulatory proteins, dyskerin, Tinf2, and RTEL1 [62, 63]. While these telomere defects may be attributed to indirect changes in p53 transcription targets, the potential direct effects on telomeres through subtelomeric binding has not been examined.
Li-Fraumeni patients have inherited mutations in p53 and have a high incidence of cancer. Telomere length is shorter and telomere attrition is faster in Li-Fraumeni patients relative to genetically matched control parents or siblings [64]. Li Fraumeni-like syndromes that have normal p53 were found to have inherited mutations in Pot1, suggesting that loss of telomere capping function phenocopies loss of p53 [65]. In mouse, POT1 deficiency in the context of p53 mutation leads to a prolonged DNA damage signal and mitotic failure [66]. Furthermore, a prolonged telomere damage signaling in mitosis relies on the loss of p53 [67] even though p53 is not required for the M phase checkpoint signal. This raises the possibility that p53 has a direct role in regulating the mitotic accumulation of γH2AX at telomeres.
p53 orthologues function in telomere regulation
Evolutionarily related orthologues of p53, such as C. elegans CEP1 and Drosophila DM_p53, have strong conservation in the DNA binding domains, but lack detectable or measurable transcription activation function, suggesting that DNA binding functions distinct from transcription control are important evolutionary functions of p53 [68]. p53 paralogues, p63, and p73, bind to the same recognition site [69], but have different functions. p53 paralog delta ( )Np73 localizes directly to sites of DNA damage, interacts with p53BP1 and inhibits ATM activation and p53 phosphorylation [70]. Earlier studies found evidence that p53 could also localize to sites of DNA damage, where it can have a direct effect on local chromatin and DNA repair processes [71]. Both p63 and p73 have more specialized functions in cellular differentiation and environmental responses, but it is not yet known whether these factors can localize at p53 consensus sites in retrotransposon-like elements, including those found at subtelomeres to regulate telomere functions and genetic stability.
Conclusions, remaining questions, and future experiments
P53 binding sites have been identified in retrotransposon-like elements in human subtelomeres. These sites are induced by DNA damage, alter local chromatin structure, and promote transcription of local eRNAs. These sites activate TERRA and confer DNA-damage protection at the adjacent telomere repeat. Loss of p53 or deletion of the p53 binding site leads to a loss of TERRA transcription and telomere instability. Our understanding of the function of p53 binding sites in subtelomeres remains incomplete. I propose that these p53 binding sites induce changes in local chromatin structure in response to DNA damage stress. These changes, including histone acetylation and activation of local eRNA-like transcription increase the efficiency of local DNA repair and protection. In the absence of p53, DNA damage signals, such as γH2AX persist. Thus, I propose that these p53 binding sites provide direct chromatin modifying activity to prevent the accumulation of persistent DNA damage signaling.
The functional significance of p53 binding sites at subtelomeres and other retrotransposon-like elements throughout the genome requires further experimental investigation. At least one paradoxical observation needs to be resolved. How does p53 repress retrotransposon activation and the TRAIN phenomenon, yet activate transcription at subtelomeric retrotransposons and enhancer eRNAs?. Is the p53-depdenent repression of retrotransposons an indirect effect of p53 activating transcriptional repressors, like E2F7? Many addditional questions arise. What, if any, is the relationship between eRNAs and retrotransposon transcripts? Are these transcripts processed similarly, and do they share common mechanisms in binding proteins that target DNA sequence through base complementarity or strand invasion? Perhaps more relevant to cancer biology is whether p53 mutations alter retrotransposon activity and subtelomeric transcription? How do p53 mutation alter telomeric chromatin and length regulation? These and many other questions (see Box 1) need to be addressed to further validate any hypothesis connecting retrotransposon-derived p53 binding sites in subtelomeres with telomere regulation and genomic stability.
Box 1. Outstanding questions.
Are p53 binding sites found in retrotransposons functionally significant throughout the genome?
If so, are these p53 sites important in the regulation of retrotransposon repression or activation?
How might p53 binding at subtelomeres regulate telomere length and genomic stability?
What other factors are recruited to p53 subtelomeric sites in response to DNA damage, and how may these factors influence telomere regulation.
Do p53 mutations directly alter telomere maintenance and genomic stability?
Some future experiments that can test these hypotheses include determining if p53 mutations, like p53Δ31, have direct affects on subtelomeric chromatin and TERRA transcription. Another important experiment will be to directly inhibit subtelomeric eRNA by anti-sense or morpholino to guage their role in telomere length regulation and DNA damage protection. Finally, it will be valuable to determine how mutation of p53 binding sites in an active retrotransposon alters transcription and retrotransposition.
Acknowledgments
This work was supported by grants from NIH (RO1CA140652) to PML and from the NCI Cancer Center Core Grant (P30 CA10815) and the Commonwealth Universal Research Enhancement Program, PA Department of Health.
Abbreviations
- eRNA
enhancer RNA
- HR
homologous recombination
- LTR
long terminal repeat
- TERRA
telomere-repeat encoded RNA
References
- 1.de Lange T. A loopy view of telomere evolution. Front Genet. 2015;6:321. doi: 10.3389/fgene.2015.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackburn EH, Collins K. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol. 2011;3:a003558. doi: 10.1101/cshperspect.a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doksani Y, de Lange T. The role of double-strand break repair pathways at functional and dysfunctional telomeres. Cold Spring Harb Perspect Biol. 2014;6:a016576. doi: 10.1101/cshperspect.a016576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnoult N, Karlseder J. Complex interactions between the DNA-damage response and mammalian telomeres. Nat Struct Mol Biol. 2015;22:859–66. doi: 10.1038/nsmb.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meek DW. Regulation of the p53 response and its relationship to cancer. Biochem J. 2015;469:325–46. doi: 10.1042/BJ20150517. [DOI] [PubMed] [Google Scholar]
- 6.Sammons MA, Zhu J, Drake AM, Berger SL. TP53 engagement with the genome occurs in distinct local chromatin environments via pioneer factor activity. Genome Res. 2015;25:179–88. doi: 10.1101/gr.181883.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shore D. Telomeric chromatin: replicating and wrapping up chromosome ends. Curr Opin Genet Dev. 2001;11:189–98. doi: 10.1016/s0959-437x(00)00178-7. [DOI] [PubMed] [Google Scholar]
- 8.Schoeftner S, Blasco MA. Chromatin regulation and non-coding RNAs at mammalian telomeres. Semin Cell Dev Biol. 2010;21:186–93. doi: 10.1016/j.semcdb.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Azzalin CM, Lingner J. Telomere functions grounding on TERRA firma. Trends Cell Biol. 2015;25:29–36. doi: 10.1016/j.tcb.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Porro A, Feuerhahn S, Delafontaine J, Riethman H, et al. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat Commun. 2014;5:5379. doi: 10.1038/ncomms6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cesare AJ, Karlseder J. A three-state model of telomere control over human proliferative boundaries. Curr Opin Cell Biol. 2012;24:731–8. doi: 10.1016/j.ceb.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukunaga K, Hirano Y, Sugimoto K. Subtelomere-binding protein Tbf1 and telomere-binding protein Rap1 collaborate to inhibit localization of the Mre11 complex to DNA ends in budding yeast. Mol Biol Cell. 2012;23:347–59. doi: 10.1091/mbc.E11-06-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ambrosini A, Paul S, Hu S, Riethman H. Human subtelomeric duplicon structure and organization. Genome Biol. 2007;8:R151. doi: 10.1186/gb-2007-8-7-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stong N, Deng Z, Gupta R, Hu S, et al. Subtelomeric CTCF and cohesin binding site organization using improved subtelomere assemblies and a novel annotation pipeline. Genome Res. 2014;24:1039–50. doi: 10.1101/gr.166983.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abad JP, De Pablos B, Osoegawa K, De Jong PJ, et al. TAHRE, a novel telomeric retrotransposon from Drosophila melanogaster, reveals the origin of Drosophila telomeres. Mol Biol Evol. 2004;21:1620–4. doi: 10.1093/molbev/msh180. [DOI] [PubMed] [Google Scholar]
- 16.Silva-Sousa R, Lopez-Panads E, Casacuberta E. Drosophila telomeres: an example of co-evolution with transposable elements. Genome Dyn. 2012;7:46–67. doi: 10.1159/000337127. [DOI] [PubMed] [Google Scholar]
- 17.Maida Y, Yasukawa M, Furuuchi M, Lassmann T, et al. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461:230–5. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bah A, Wischnewski H, Shchepachev V, Azzalin CM. The telomeric transcriptome of Schizosaccharomyces pombe. Nucleic Acids Res. 2011;40:2995–3005. doi: 10.1093/nar/gkr1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenwood J, Cooper JP. Non-coding telomeric and subtelomeric transcripts are differentially regulated by telomeric and heterochromatin assembly factors in fission yeast. Nucleic Acids Res. 2012;40:2956–63. doi: 10.1093/nar/gkr1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10:228–36. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 21.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, et al. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 22.Azzalin CM, Lingner J. Telomere functions grounding on TERRA firma. Trends Cell Biol. 2015;25:29–36. doi: 10.1016/j.tcb.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Cusanelli E, Chartrand P. Telomeric repeat-containing RNA TERRA: a noncoding RNA connecting telomere biology to genome integrity. Front Genet. 2015;6:143. doi: 10.3389/fgene.2015.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luke B, Panza A, Redon S, Iglesias N, et al. The Rat1p 5' to 3' exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell. 2008;32:465–77. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 25.Balk B, Maicher A, Dees M, Klermund J, et al. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat Struct Mol Biol. 2013;20:1199–205. doi: 10.1038/nsmb.2662. [DOI] [PubMed] [Google Scholar]
- 26.Deng Z, Wang Z, Stong N, Plasschaert R, et al. A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. EMBO J. 2012;31:4165–78. doi: 10.1038/emboj.2012.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng Z, Norseen J, Wiedmer A, Riethman H, et al. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell. 2009;35:403–13. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caslini C, Connelly JA, Serna A, Broccoli D, et al. MLL associates with telomeres and regulates telomeric repeat-containing RNA transcription. Mol Cell Biol. 2009;29:4519–26. doi: 10.1128/MCB.00195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lane D, Levine A. p53 Research: the past thirty years and the next thirty years. Cold Spring Harb Perspect Biol. 2010;2:a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karlseder J, Hoke K, Mirzoeva OK, Bakkenist C, et al. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2004;2:E240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cesare AJ, Hayashi MT, Crabbe L, Karlseder J. The telomere deprotection response is functionally distinct from the genomic DNA damage response. Mol Cell. 2013;51:141–55. doi: 10.1016/j.molcel.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujita K, Horikawa I, Mondal AM, Jenkins LM, et al. Positive feedback between p53 and TRF2 during telomere-damage signalling and cellular senescence. Nat Cell Biol. 2010;12:1205–12. doi: 10.1038/ncb2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horikawa I, Fujita K, Harris CC. p53 governs telomere regulation feedback too, via TRF2. Aging. 2011;3:26–32. doi: 10.18632/aging.100271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tutton S, Azzam GA, Stong N, Vladimirova O, et al. Subtelomeric p53 binding prevents accumulation of DNA damage at human telomeres. EMBO J. 2016;35:193–207. doi: 10.15252/embj.201490880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tonelli C, Amati B, Morelli MJ. p53 transcriptional programs in B cells upon exposure to genotoxic stress in vivo: Computational analysis of next-generation sequencing data. Genom Data. 2016;7:29–31. doi: 10.1016/j.gdata.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tonelli C, Morelli MJ, Bianchi S, Rotta L, et al. Genome-wide analysis of p53 transcriptional programs in B cells upon exposure to genotoxic stress in vivo. Oncotarget. 2015;6:24611–26. doi: 10.18632/oncotarget.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui F, Sirotin MV, Zhurkin VB. Impact of Alu repeats on the evolution of human p53 binding sites. Biol Direct. 2011;6:2. doi: 10.1186/1745-6150-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffman WH, Biade S, Zilfou JT, Chen J, et al. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem. 2002;277:3247–57. doi: 10.1074/jbc.M106643200. [DOI] [PubMed] [Google Scholar]
- 39.Schlereth K, Heyl C, Krampitz AM, Mernberger M, et al. Characterization of the p53 cistrome--DNA binding cooperativity dissects p53's tumor suppressor functions. PLoS Genet. 2013;9:e1003726. doi: 10.1371/journal.pgen.1003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischer M, Steiner L, Engeland K. The transcription factor p53: not a repressor, solely an activator. Cell Cycle. 2014;13:3037–58. doi: 10.4161/15384101.2014.949083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei CL, Wu Q, Vega VB, Chiu KP, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–19. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 42.Su D, Wang X, Campbell MR, Song L, et al. Interactions of chromatin context, binding site sequence content, and sequence evolution in stress-induced p53 occupancy and transactivation. PLoS Genet. 2015;11:e1004885. doi: 10.1371/journal.pgen.1004885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cui F, Zhurkin VB. Rotational positioning of nucleosomes facilitates selective binding of p53 to response elements associated with cell cycle arrest. Nucleic Acids Res. 2014;42:836–47. doi: 10.1093/nar/gkt943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lidor Nili E, Field Y, Lubling Y, Widom J, et al. p53 binds preferentially to genomic regions with high DNA-encoded nucleosome occupancy. Genome Res. 2010;20:1361–8. doi: 10.1101/gr.103945.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sykes SM, Mellert HS, Holbert MA, Li K, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–51. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leonova KI, Brodsky L, Lipchick B, Pal M, et al. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc Natl Acad Sci USA. 2013;110:E89–98. doi: 10.1073/pnas.1216922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harris CR, Dewan A, Zupnick A, Normart R, et al. p53 responsive elements in human retrotransposons. Oncogene. 2009;28:3857–65. doi: 10.1038/onc.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wylie A, Jones AE, D'Brot A, Lu WJ, et al. p53 genes function to restrain mobile elements. Genes Dev. 2016;30:64–77. doi: 10.1101/gad.266098.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pardue ML, DeBaryshe PG. Retrotransposons that maintain chromosome ends. Proc Natl Acad Sci USA. 2011;108:20317–24. doi: 10.1073/pnas.1100278108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crighton D, Woiwode A, Zhang C, Mandavia N, et al. p53 represses RNA polymerase III transcription by targeting TBP and inhibiting promoter occupancy by TFIIIB. EMBO J. 2003;22:2810–20. doi: 10.1093/emboj/cdg265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White RJ. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet. 2008;24:622–9. doi: 10.1016/j.tig.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 52.Cairns CA, White RJ. p53 is a general repressor of RNA polymerase III transcription. EMBO J. 1998;17:3112–23. doi: 10.1093/emboj/17.11.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 2016;17:207–23. doi: 10.1038/nrg.2016.4. [DOI] [PubMed] [Google Scholar]
- 54.Leveille N, Melo CA, Rooijers K, Diaz-Lagares A, et al. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat Commun. 2015;6:6520. doi: 10.1038/ncomms7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Allen MA, Andrysik Z, Dengler VL, Mellert HS, et al. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. eLife. 2014;3:e02200. doi: 10.7554/eLife.02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Melo CA, Drost J, Wijchers PJ, van de Werken H, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49:524–35. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 57.Mangerel J, Price A, Castelo-Branco P, Brzezinski J, et al. Alternative lengthening of telomeres is enriched in, and impacts survival of TP53 mutant pediatric malignant brain tumors. Acta Neuropathol. 2014;128:853–62. doi: 10.1007/s00401-014-1348-1. [DOI] [PubMed] [Google Scholar]
- 58.Farooqi AS, Dagg RA, Choi LM, Shay JW, et al. Alternative lengthening of telomeres in neuroblastoma cell lines is associated with a lack of MYCN genomic amplification and with p53 pathway aberrations. J Neurooncol. 2014;119:17–26. doi: 10.1007/s11060-014-1456-8. [DOI] [PubMed] [Google Scholar]
- 59.Makinen N, Aavikko M, Heikkinen T, Taipale M, et al. Exome sequencing of uterine leiomyosarcomas identifies frequent mutations in TP53, ATRX, and MED12. PLoS Genet. 2016;12:e1005850. doi: 10.1371/journal.pgen.1005850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shen H, Maki CG. p53 and p21(Waf1) are recruited to distinct PML-containing nuclear foci in irradiated and Nutlin-3a-treated U2OS cells. J Cell Biochem. 2010;111:1280–90. doi: 10.1002/jcb.22852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Samassekou O, Bastien N, Lichtensztejn D, Yan J, et al. Different TP53 mutations are associated with specific chromosomal rearrangements, telomere length changes, and remodeling of the nuclear architecture of telomeres. Genes Chromosomes Cancer. 2014;53:934–50. doi: 10.1002/gcc.22205. [DOI] [PubMed] [Google Scholar]
- 62.Simeonova I, Jaber S, Draskovic I, Bardot B, et al. Mutant mice lacking the p53 C-terminal domain model telomere syndromes. Cell Rep. 2013;3:2046–58. doi: 10.1016/j.celrep.2013.05.028. [DOI] [PubMed] [Google Scholar]
- 63.Jaber S, Toufektchan E, Lejour V, Bardot B, et al. p53 downregulates the Fanconi anaemia DNA repair pathway. Nat Commun. 2016;7:11091. doi: 10.1038/ncomms11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tabori U, Nanda S, Druker H, Lees J, et al. Younger age of cancer initiation is associated with shorter telomere length in Li-Fraumeni syndrome. Cancer Res. 2007;67:1415–8. doi: 10.1158/0008-5472.CAN-06-3682. [DOI] [PubMed] [Google Scholar]
- 65.Calvete O, Martinez P, Garcia-Pavia P, Benitez-Buelga C, et al. A mutation in the POT1 gene is responsible for cardiac angiosarcoma in TP53-negative Li-Fraumeni-like families. Nat Commun. 2015;6:8383. doi: 10.1038/ncomms9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Palm W, Hockemeyer D, Kibe T, de Lange T. Functional dissection of human and mouse POT1 proteins. Mol Cell Biol. 2009;29:471–82. doi: 10.1128/MCB.01352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hayashi MT, Cesare AJ, Fitzpatrick JA, Lazzerini-Denchi E, et al. A telomere-dependent DNA damage checkpoint induced by prolonged mitotic arrest. Nat Struct Mol Biol. 2012;19:387–94. doi: 10.1038/nsmb.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lion M, Raimondi I, Donati S, Jousson O, et al. Evolution of p53 transactivation specificity through the lens of a yeast-based functional assay. PloS One. 2015;10:e0116177. doi: 10.1371/journal.pone.0116177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nicolai S, Rossi A, Di Daniele N, Melino G, et al. DNA repair and aging: the impact of the p53 family. Aging. 2015;7:1050–65. doi: 10.18632/aging.100858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilhelm MT, Rufini A, Wetzel MK, Tsuchihara K, et al. Isoform-specific p73 knockout mice reveal a novel role for delta Np73 in the DNA damage response pathway. Genes Dev. 2010;24:549–60. doi: 10.1101/gad.1873910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu Y, Kulesz-Martin M. p53 protein at the hub of cellular DNA damage response pathways through sequence-specific and non-sequence-specific DNA binding. Carcinogenesis. 2001;22:851–60. doi: 10.1093/carcin/22.6.851. [DOI] [PubMed] [Google Scholar]