Abstract
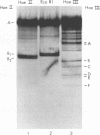
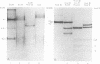
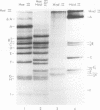
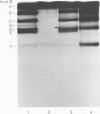
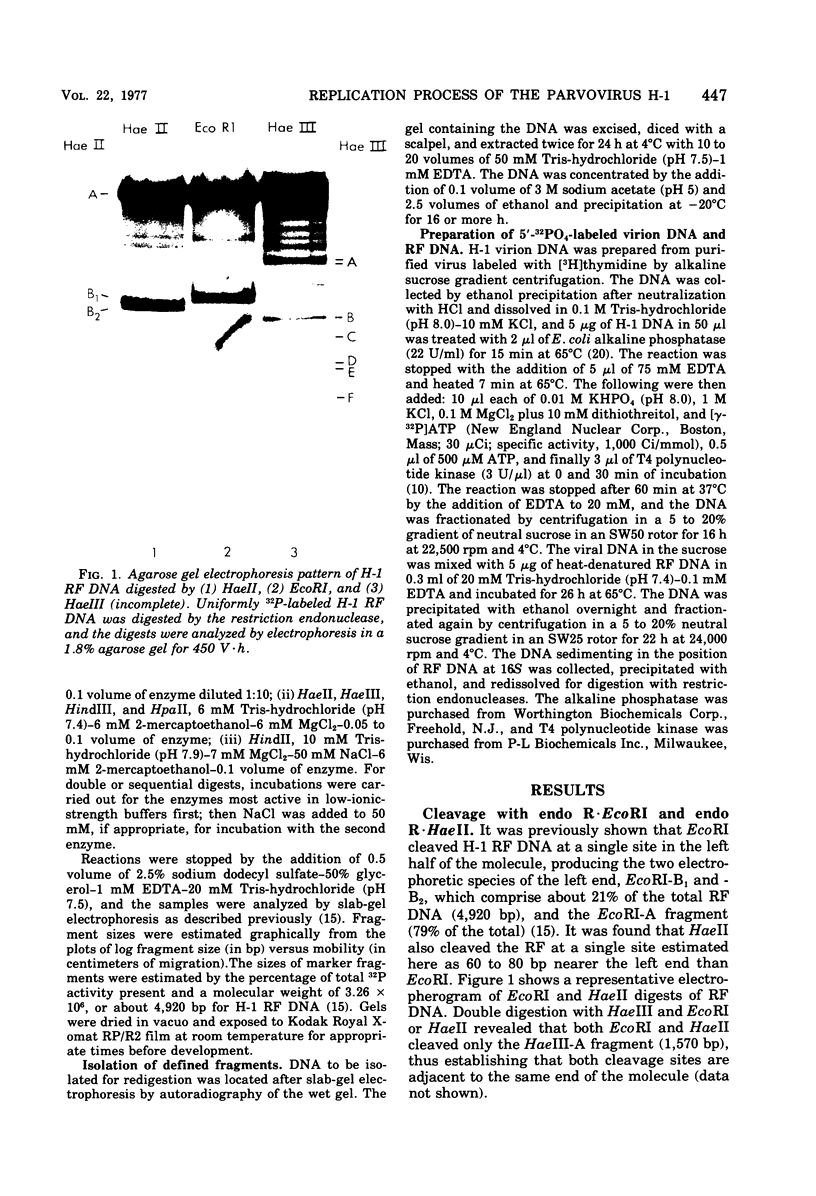
The cleavage map of H-1 replicative-form DNA to the bacterial restriction endonuclease EcoRI, HaeII, HaeIII, HindII, HindIII, and HpaII has been determined. The 5'-phosphoryl end of the viral strand is on the right end of the molecule at or near the replication origin. Evidence is presented for the presence of inverted self-complementary sequences at the right end that differ from those at the left end. These sequences allow a foldback of the DNA after denaturation, and a minority of the native replicative-form DNA has the foldback configuration. The possible role of these structures in H-1 DNA synthesis is discussed.
Full text
PDF



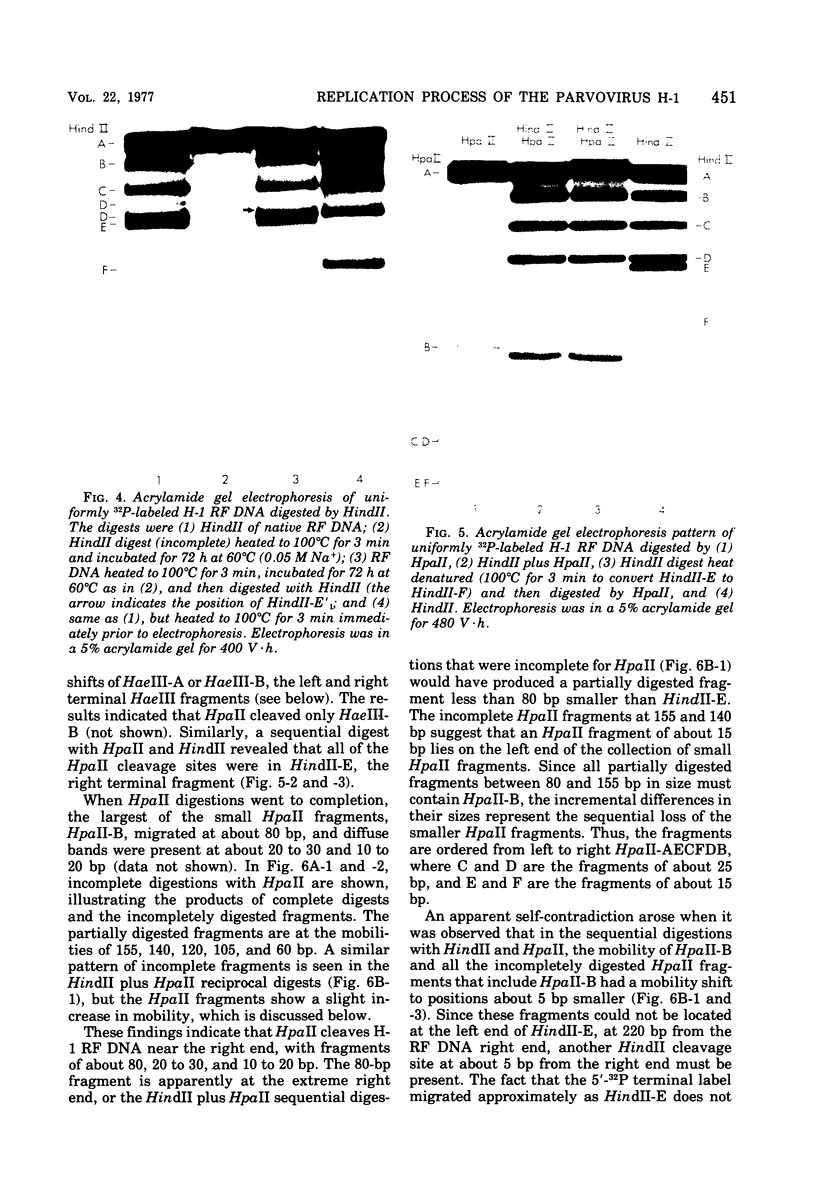



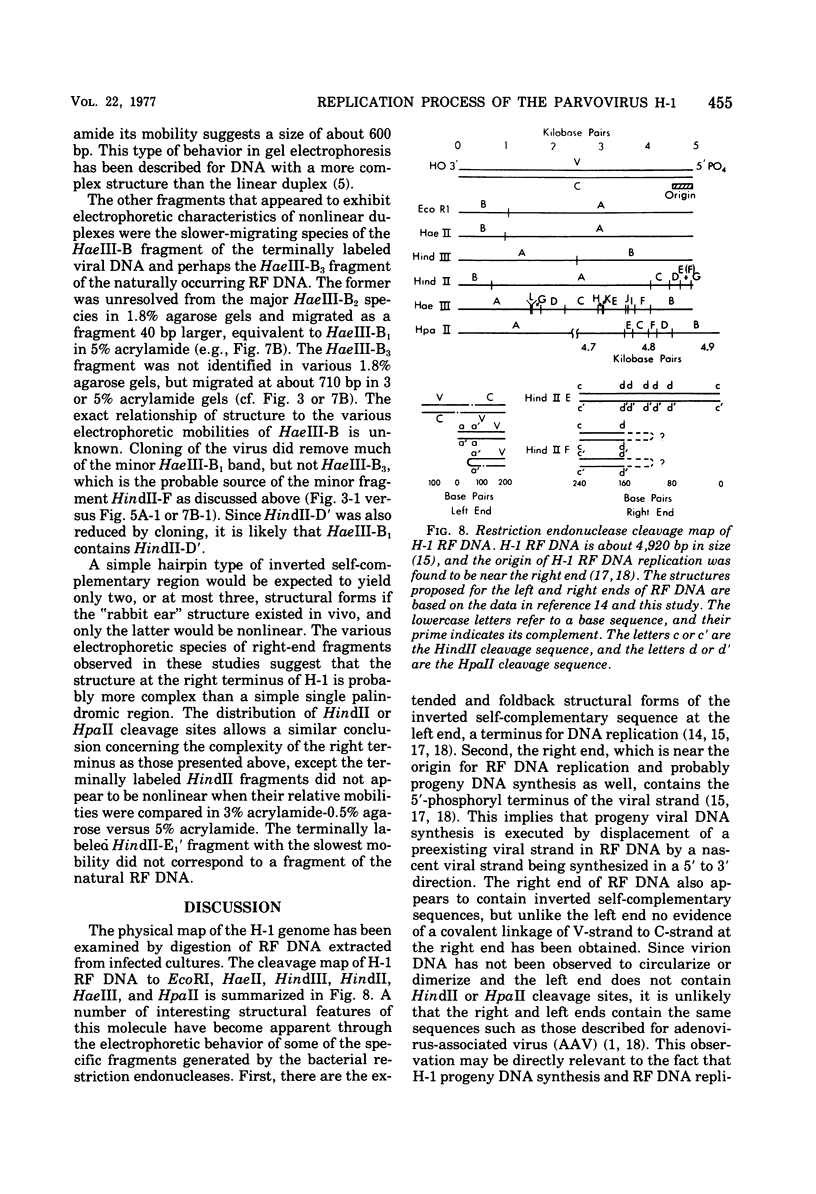



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berns K. I., Kelly T. J., Jr Letter: Visualization of the inverted terminal repetition in adeno-associated virus DNA. J Mol Biol. 1974 Jan 15;82(2):267–271. doi: 10.1016/0022-2836(74)90344-1. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Palindromic base sequences and replication of eukaryote chromosome ends. Nature. 1974 Aug 9;250(5466):467–470. doi: 10.1038/250467a0. [DOI] [PubMed] [Google Scholar]
- Davoli D., Fareed G. C. Amplification of a circular segment of SV40 DNA. Nature. 1974 Sep 13;251(5471):153–155. doi: 10.1038/251153a0. [DOI] [PubMed] [Google Scholar]
- Delius H., Clements J. B. A partial denaturation map of herpes simplex virus type 1 DNA: evidence for inversions of the unique DNA regions. J Gen Virol. 1976 Oct;33(1):125–133. doi: 10.1099/0022-1317-33-1-125. [DOI] [PubMed] [Google Scholar]
- Dingman C. W., Fisher M. P., Kakefuda T. Role of molecular conformation in determining the electrophoretic properties of polynucleotides in agarose-acrylamide gels. II. Biochemistry. 1972 Mar 28;11(7):1242–1250. doi: 10.1021/bi00757a020. [DOI] [PubMed] [Google Scholar]
- Garfin D. E., Goodman H. M. Nucleotide sequences at the cleavage sites of two restriction endonucleases from Hemophilus parainfluenzae. Biochem Biophys Res Commun. 1974 Jul 10;59(1):108–116. doi: 10.1016/s0006-291x(74)80181-6. [DOI] [PubMed] [Google Scholar]
- KISSANE J. M., ROBINS E. The fluorometric measurement of deoxyribonucleic acid in animal tissues with special reference to the central nervous system. J Biol Chem. 1958 Jul;233(1):184–188. [PubMed] [Google Scholar]
- Kongsvik J. R., Gierthy J. F., Rhode S. L., 3rd Replication process of the parvovirus H-1. IV. H-1-specific proteins synthesized in synchronized human NB kidney cells. J Virol. 1974 Dec;14(6):1600–1603. doi: 10.1128/jvi.14.6.1600-1603.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledinko N. Plaque assay of the effects of cytosine arabinoside and 5-iodo-2'-deoxyuridine on the synthesis of H-I virus particles. Nature. 1967 Jun 24;214(5095):1346–1347. doi: 10.1038/2141346a0. [DOI] [PubMed] [Google Scholar]
- Lillehaug J. R., Kleppe K. Effect of salts and polyamines on T4 polynucleotide kinase. Biochemistry. 1975 Mar 25;14(6):1225–1229. doi: 10.1021/bi00677a021. [DOI] [PubMed] [Google Scholar]
- Rhode S. L., 3rd Replication process of the parvovirus H-1 V. Isolation and characterization of temperature-sensitive H-1 mutants. J Virol. 1976 Feb;17(2):659–667. doi: 10.1128/jvi.17.2.659-667.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode S. L., 3rd Replication process of the parvovirus H-1. I. Kinetics in a parasynchronous cell system. J Virol. 1973 Jun;11(6):856–861. doi: 10.1128/jvi.11.6.856-861.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode S. L., 3rd Replication process of the parvovirus H-1. II. Isolation and characterization of H-1 replicative form DNA. J Virol. 1974 Feb;13(2):400–410. doi: 10.1128/jvi.13.2.400-410.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode S. L., 3rd Replication process of the parvovirus H-1. III. Factors affecting H-1 RF DNA synthesis. J Virol. 1974 Oct;14(4):791–801. doi: 10.1128/jvi.14.4.791-801.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode S. L., 3rd Replication process of the parvovirus H-1. VI. Characterization of a replication terminus of H-1 replicative-form DNA. J Virol. 1977 Feb;21(2):694–712. doi: 10.1128/jvi.21.2.694-712.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C. K., Hearst J. E. Psoralen-crosslinked secondary structure map of single-stranded virus DNA. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2649–2653. doi: 10.1073/pnas.73.8.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer I. I., Rhode S. L., 3rd Replication process of the parvovirus H-1. VII. Electron microscopy of replicative-form DNA synthesis. J Virol. 1977 Feb;21(2):713–723. doi: 10.1128/jvi.21.2.713-723.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer I. I., Rhode S. L., 3rd Replication process of the parvovirus H-1. VIII. Partial denaturation mapping and localization of the replication origin of H-1 replicative-form DNA with electron microscopy. J Virol. 1977 Feb;21(2):724–731. doi: 10.1128/jvi.21.2.724-731.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattersall P., Ward D. C. Rolling hairpin model for replication of parvovirus and linear chromosomal DNA. Nature. 1976 Sep 9;263(5573):106–109. doi: 10.1038/263106a0. [DOI] [PubMed] [Google Scholar]
- Weiss B., Live T. R., Richardson C. C. Enzymatic breakage and joining of deoxyribonucleic acid. V. End group labeling and analysis of deoxyribonucleic acid containing single straned breaks. J Biol Chem. 1968 Sep 10;243(17):4530–4542. [PubMed] [Google Scholar]
- Wilkie N. M., Cortini R. Sequence arrangement in herpes simplex virus type 1 DNA: identification of terminal fragments in restriction endonuclease digests and evidence for inversions in redundant and unique sequences. J Virol. 1976 Oct;20(1):211–221. doi: 10.1128/jvi.20.1.211-221.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie N. M. Physical maps for Herpes simplex virus type 1 DNA for restriction endonucleases Hind III, Hpa-1, and X. bad. J Virol. 1976 Oct;20(1):222–233. doi: 10.1128/jvi.20.1.222-233.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]