Abstract
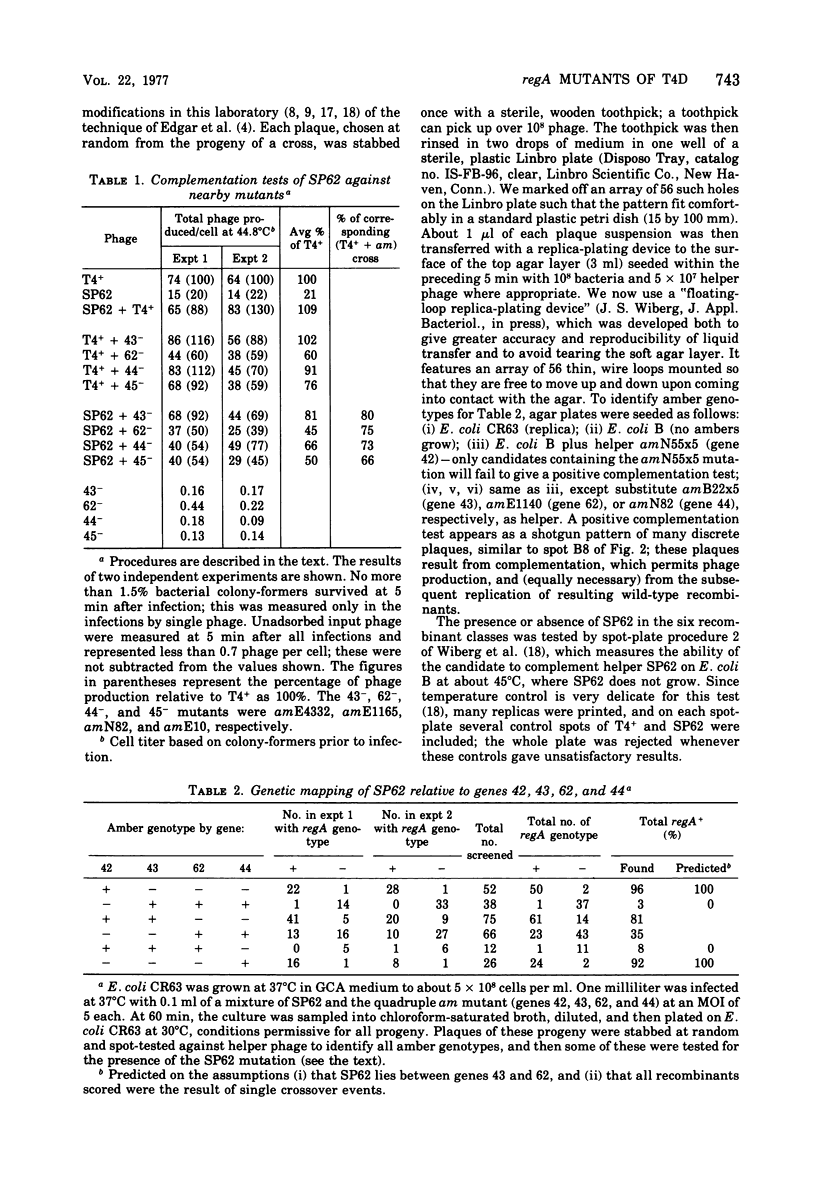
SP62, a mutant of bacteriophage T4 shown by Wiberg et al. (1973) to be defective in regulation of T4 protein synthesis, was shown by complementation tests to define a new gene, regA, and by intergenic mapping to lie between genes 43 and 62. The mapping involved crossing SP62 with a quadruple amber mutant defective in genes 42, 43, 62, and 44, selecting all six classes of amber-containing recombinants caused by single crossover events, and then scoring the presence or absence of SP62 in these recombinants. In addition, 15 new, spontaneous regA mutants were isolated, and 13 of these were mapped against each other; a total of eight different mutation sites were thus defined. Most of the new mutants were isolated as pseudorevertants of a leaky amber mutant in gene 62, according to Karam and Bowles (1974), whereas one was identified by virtue of the "white ring" around its plaque, a phenotype possessed by all the regA mutants at high temperature, SP62 was renamed regA1, and the new mutants were named regA2, regA3, etc.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berger H., Warren A. J. Effects of deletion mutations on high negative interference in T4D bacteriophage. Genetics. 1969 Sep;63(1):1–5. doi: 10.1093/genetics/63.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broker T. R., Lehman I. R. Branched DNA molecules: intermediates in T4 recombination. J Mol Biol. 1971 Aug 28;60(1):131–149. doi: 10.1016/0022-2836(71)90453-0. [DOI] [PubMed] [Google Scholar]
- Chase M, Doermann A H. High Negative Interference over Short Segments of the Genetic Structure of Bacteriophage T4. Genetics. 1958 May;43(3):332–353. doi: 10.1093/genetics/43.3.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EDGAR R. S., DENHARDT G. H., EPSTEIN R. H. A COMPARATIVE GENETIC STUDY OF CONDITIONAL LETHAL MUTATIONS OF BACTERIOPHAGE T4D. Genetics. 1964 Apr;49:635–648. doi: 10.1093/genetics/49.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRASER D., JERREL E. A. The amino acid composition of T3 bacteriophage. J Biol Chem. 1953 Nov;205(1):291–295. [PubMed] [Google Scholar]
- Hall D. H. Mutants of bacteriophage T4 unable to induce dihydrofolate reductase activity. Proc Natl Acad Sci U S A. 1967 Aug;58(2):584–591. doi: 10.1073/pnas.58.2.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D. H., Tessman I., Karlström O. Linkage of T4 genes controlling a series of steps in pyrimidine biosynthesis. Virology. 1967 Mar;31(3):442–448. doi: 10.1016/0042-6822(67)90224-3. [DOI] [PubMed] [Google Scholar]
- Hercules K., Munro J. L., Mendelsohn S., Wiberg J. S. Mutants in a nonessential gene of bacteriophage T4 which are defective in the degradation of Escherichia coli deoxyribonucleic acid. J Virol. 1971 Jan;7(1):95–105. doi: 10.1128/jvi.7.1.95-105.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hercules K., Wiberg J. S. Specific suppression of mutations in genes 46 and 47 by das, a new class of mutations in bacteriophage T4D. J Virol. 1971 Nov;8(5):603–612. doi: 10.1128/jvi.8.5.603-612.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. R., Hall D. H. Isolation and characterization of mutants of bacteriophage T4 resistant to folate analogs. Virology. 1973 Jun;53(2):413–426. doi: 10.1016/0042-6822(73)90221-3. [DOI] [PubMed] [Google Scholar]
- Karam J. D., Bowles M. G. Mutation to overproduction of bacteriophage T4 gene products. J Virol. 1974 Feb;13(2):428–438. doi: 10.1128/jvi.13.2.428-438.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris C. F., Sinha N. K., Alberts B. M. Reconstruction of bacteriophage T4 DNA replication apparatus from purified components: rolling circle replication following de novo chain initiation on a single-stranded circular DNA template. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4800–4804. doi: 10.1073/pnas.72.12.4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIMON E. H., TESSMAN I. THYMIDINE-REQUIRING MUTANTS OF PHAGE T4. Proc Natl Acad Sci U S A. 1963 Sep;50:526–532. doi: 10.1073/pnas.50.3.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessman I., Greenberg D. B. Ribonucleotide reductase genes of phage T4: map location of the thioredoxin gene nrdC. Virology. 1972 Jul;49(1):337–338. doi: 10.1016/s0042-6822(72)80040-0. [DOI] [PubMed] [Google Scholar]
- Warner H. R., Snustad D. P., Koerner J. F., Childs J. D. Identification and genetic characterization of mutants of bacteriophage T4 defective in the ability to induce exonuclease A. J Virol. 1972 Mar;9(3):399–407. doi: 10.1128/jvi.9.3.399-407.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiberg J. S., Mendelsohn S., Warner V., Hercules K., Aldrich C., Munro J. L. SP62, a viable mutant of bacteriophage T4D defective in regulation of phage enzyme synthesis. J Virol. 1973 Oct;12(4):775–792. doi: 10.1128/jvi.12.4.775-792.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiberg J. S. Mutants of bacteriophage T4 unable to cause breakdown of host DNA. Proc Natl Acad Sci U S A. 1966 Mar;55(3):614–621. doi: 10.1073/pnas.55.3.614. [DOI] [PMC free article] [PubMed] [Google Scholar]




