Abstract
Rhabdomyosarcomas (RMS) in children and adolescents are heterogeneous sarcomas broadly defined by skeletal muscle features and the presence/absence of PAX3/7-FOXO1 fusion genes. MicroRNAs are small non-coding RNAs that regulate gene expression in a cell context specific manner. Sequencing analyses of microRNAs in 64 RMS revealed expression patterns separating skeletal muscle, fusion gene positive and negative RMS. Integration with parallel gene expression data assigned biological functions to 12 co-expression networks/modules that reassuringly included myogenic roles strongly correlated with microRNAs known in myogenesis and RMS development. Modules also correlated with clinical outcome and fusion status. Regulation of microRNAs by the fusion protein was demonstrated after PAX3-FOXO1 reduction, exemplified by miR-9-5p. MiR-9-5p levels correlated with poor outcome, even within fusion gene positive RMS, and were higher in metastatic versus non-metastatic disease. MiR-9-5p reduction inhibited RMS cell migration. Our findings reveal microRNAs in a regulatory framework of biological and clinical significance in RMS.
Keywords: MicroRNAs, Next generation sequencing, Co-expression modules, Rhabdomyosarcoma, Fusion protein
Abbreviations: RMS, rhabdomyosarcoma; MiRNAs, microRNAs
Highlights
-
•
RNAseq profiled miRNA expression in 64 rhabdomyosarcomas (RMS).
-
•
MiRNA expression distinguished muscle and RMS on the basis of fusion gene status.
-
•
Co-expression networks linked to function, clinical data and fusion gene status.
-
•
Identified miRNAs, including miR-9-5p, altered by the PAX3-FOXO1 fusion protein.
-
•
Demonstrated clinical and functional role for miR-9-5p in RMS.
Introduction
Rhabdomyosarcomas (RMS) are the most frequent soft tissue sarcoma in children and RMS cells resemble those in the early stages of myogenic differentiation [1], [2]. Histologically there are two major subtypes, embryonal (ERMS) accounting for 60–70% of cases and alveolar (ARMS). Approximately 70% of ARMS harbor fusion genes that join the 5′ sequence of PAX3 or PAX7 to the 3′ sequence of FOXO1 gene [3], [4]. The encoded fusion proteins play an oncogenic role in the development of ARMS in association with co-operating events, notably overexpression of MYCN that is a transcriptional target of the fusion protein and genomically amplified in approximately 20% of cases [5], [6], [7], [8]. The presence of PAX3-FOXO1 has been correlated with aggressive clinical behavior in several studies [9], [10], [11], [12] and gene expression profiles and genomic imbalances are similar in fusion gene negative ARMS and ERMS [10], [13].
MicroRNAs (miRNAs) are small (20–22 nt) RNA molecules that can alter cellular processes including proliferation, differentiation and apoptosis [14]. MiRNAs inhibit protein synthesis of specific genes by degrading specific mRNA species or hindering translation. Specific mRNA degradation involves base-pairing to partially complementary sequences within the 3′ UTR [15] or other less frequently studied mechanisms including their binding to the 5′ UTR [16] and gene promoters regions [17]. Each miRNA potentially regulates hundreds of target gene products and it is suggested that the entire protein coding genome is regulated by miRNAs [14]. Individual miRNAs have been ascribed oncogenic or tumor suppressor roles in cancer and their roles can clearly be cell context specific [18]. Roles for individual miRNAs in RMS have been described such as miR-1, miR-206, miR-133a/b and miR-378 that maintain an undifferentiated myogenic state [19], [20], [21], [22], [23], [24], [25] and miR-29 that is described with a tumor suppressor role [26]. However, a comprehensive view of how miRNAs shape the gene expression and biological features of a large number of RMS has not been described but could contribute to identification of prognostic markers/signatures and ultimately therapeutic targets, as indicated by previous studies of other tumor types [27], [28], [29].
Here we derived miRNA expression profiling data from high-throughput sequencing analyses of 64 primary RMS samples and took a systems biology approach to integrate this with parallel gene expression profiling data. This allowed us to identify co-expression mRNA networks (modules) with assigned biological functions that correlated with both patient outcome data and the presence of PAX3/7-FOXO1. This led us to demonstrate that PAX3-FOXO1 regulates the expression of many miRNAs, including miR-9-5p that was indirectly altered by PAX3-FOXO1 and shown to be of clinical and functional relevance.
Materials and methods
Primary tumor samples and cell lines
Sixty-four samples (36 ARMS and 28 ERMS) were available from RMS patients as part of a previously described cohort [30]. Clinico-pathological features of the cases used in this study for miRNA profiling are summarized in Supplementary Table S1A. Three normal skeletal muscle samples were also obtained. Samples from a similarly treated cohort of 154 RMS patients were used for miRNA validation analyses and their clinico-pathological details are summarized in Supplementary Table S1B. RMS from U.K. centres were collected through the Children's Cancer and Leukaemia Group (Local Research Ethics Committee protocol Nos. 1836 and 2015 and Multi-Regional Research Ethics Committee 06/4/71 with consent, where required). Human cell lines derived from embryonal (RH2, RH36, RD, RMSYM, RUCH2, RUCH3, CT10) and alveolar (RH3, RH4, RH5, RH18, RH41, RMS, SCMCRMZ, CW9019) were cultured and validated by DNA fingerprinted as previously described [7], [31].
miRNA library preparation, sequencing and analyses
RNA extraction was performed using Trizol reagent following manufacturer's instructions (Invitrogen – Carlsbad, CA). Size selection and library preparation was performed by Fasteris, Plan-les Ouates, Switzerland including duplicates for quality control using 5 μg of RNA in a maximum volume of 10–15 μl that was sent to the company. Briefly, small RNA fragments were purified by acrylamide gel purification and 3′ and 5′ adapters were ligated to the ends of single stranded small RNA molecules. Reverse transcription and PCR amplification was performed to generate the DNA colonies template library, which was diluted to 10 nM. The miRNA expression profiling was performed by high-throughput sequencing analysis using a Solexa/Illumina Genome Analyzer II. The samples were analyzed by grouping in pools of four per channel. Each sample specific sequence was then retrieved using a bar-coding system made of 4 nucleotides. The Solexa fasta-Q format sequence of each sample was aligned to 2 databases: mature miRNA sequence (miRBase release 21) and EST (NCBI). The mature miRNA sequence was extended with Ns at each end to allow the alignment with segments of RNA one or two nucleotides longer than the wild type sequence. All the alignments were performed using novoalign 2.06.09 with default parameters, after removing the 3′ adapter and cutting the sequence to 30 nt. Following alignment, the output files were imported into R for further analysis. First of all the sequence with low Solexa quality score (which did not passed the cut-off in the novoalign software) and those having less than 5 reads in at least 2 samples were filtered out from the analysis. When multiple alignments with the same score were detected, the sequence was randomly assigned to one of them. After the alignment each sequence was assigned to an RNA class based on Entrez Gene type. The variance of the counts data were stabilized using a power transformation function and normalized by applying a non-linear function (lowess). Differentially expressed miRNAs were identified by fitting a linear model (limma) and adjusting for multiple testing by computing FDR values from p-values using the Benjamini-Hochberg procedure (limma package). Clinical correlations between levels of miRNA expression and patient outcome data were performed by fitting a Cox Proportional Hazards regression model and adjusting for multiple testing, as described above. A classifier was generated using pamr package [32].
Gene network analysis
RNA gene expression profiling data for 64 RMS samples parallel to those profiled for miRNAs were obtained and pre-analyzed as previously described [30]. Probe sets were filtered in three steps: 1) probe sets with log intensity below 6 across all the sample were removed; 2) if multiple probe sets per gene were present, only the one with the highest coefficient of variation was retained; 3) the first 8000 most variable probe sets associated to a unique entrez ID were selected for further analysis.
The samples underwent Weighted Gene Co-expression Network Analysis (WGCNA) using the R package version 0.96 [33]. Briefly, a co-expression similarity measure was calculated by raising the absolute correlation coefficient between genes to a selected power. Then using a soft thresh-holding procedure an adjacency matrix was obtained. This was used as distance to generate a hierarchical cluster of genes. Modules were defined by cutting the breaches using a high cut-off of 0.99 and a minimum number of genes within the resulting dendrogram equal or above 30. Enrichment for Gene Ontology terms was evaluated using a hypergeometric test. The expression of all the genes within each module was summarized as eigengene, representation of the first component was obtained by singular value decomposition of standardized values. Association between modules expression and clinico-pathological parameters were evaluated using the Kruskal-Wallis rank sum test applied to both the eigengene and the gene expression levels. In particular, for the gene expression, the statistical values were obtained by testing the association of each gene with a specific parameter and averaged within each module. Therefore, high mean values of the module were interpreted as a larger number of genes within that module associated to the parameter tested. Cox's regression model was used to correlation module expression with patient overall survival (OS) or progression free survival (PFS). Finally, correlation of miRNA expression levels with the modules was assessed calculating Pearson correlation considering module eigengenes. MiRNAs were associated to the modules if they showed an absolute correlation coefficient above 0.4 and p value < 0.001.
Modulating and quantifying miRNA and gene expression
Mir-9-5p knockdown in the RH30 cell line was achieved by transfecting the cells with miScript miRNA inhibitor using HiPerfect reagent (Qiagen GmbH, Hilden, Germany). Levels of expression during experiments and analyses of levels in primary RMS samples were assayed where appropriate using TaqMan® MicroRNA Assays according to the manufacturer's instructions (Applied Biosystems, CA, USA). Briefly, RNA was reverse transcribed using TaqMan® MicroRNA Reverse Transcription (RT) Kit (Applied Biosystems, CA, USA) with specific-miRNA primers, then the product used in the real-time PCR reaction in the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, CA, USA). Expression data were analyzed by the comparative threshold cycle (Ct) method accordingly to User Bulletin #2 (Applied Biosystems). Results were expressed in term of the ΔCt value and obtained as follows: ΔCt miRNA = Average (Ct U6; CtRUN48) – Ct miRNA, where Ct miRNA, Ct U6 and CtRUN48 represents the comparative threshold cycle for miRNA and the two endogenous controls U6 and RUN48, respectively. All experiments were performed in triplicate and differences between groups were evaluated using the Mann-Whitney test. MYCN expression was modulated using siRNA as previously described [7]. SiRNA against PAX3-FOXO1 [34] was synthesized by Thermo Fisher Scientific (MA, USA) and RH4 cells were transfected with this using Lipofectamine RNAiMAX (Invitrogen, CA, USA) according to the manufacturer's instructions. Cells were cultured for 48 h before total RNA and miRNA was extracted using miRNeasy Mini Kit (Qiagen, Hilden, Germany). To analyze PAX3-FOXO1 levels after RNA interference, cDNA was synthesized using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, CA, USA) and quantitative RT-PCR was performed using ViiA™ 7 Real-Time PCR System (Applied Biosystems, CA, USA). Primers and probes designed for analysis of PAX3-FOXO1 were as follows: Forward 5′-GAACCCACCATTGGCAAT-3′, Probe 5′-CCTCTCACCTCAGAATTCAATTCGTCATAATCTG-3′, Reverse 5′-TCTGCACACGAATGAACTTGCT-3′. To analyze miRNA expression after PAX3-FOXO1 knock-down in RH4 cells, cDNA was synthesized using TaqMan® MicroRNA Reverse Transcription Kit and Megaplex™ Primer Pools (Applied Biosystems, CA, USA). Quantification of miRNAs was performed on TaqMan® Array Human MicroRNA A+B Cards Set v3.0 using ViiA™ 7 Real-Time PCR System according to the manufacturer's instructions and analyzed with ViiA™ 7 Software (Applied Biosystems, CA, USA).
Migration and viability assays
At 72 h post transfection, cells were re-suspended in DMEM plus 1% FCS and placed in triplicate into cell culture inserts (BD, NJ, USA) before being submerged into specially adapted 24 well plates (BD, NJ, USA) containing DMEM plus 10% FCS. 24 h later, cells that had not migrated were removed using a cotton bud, and cells that had migrated to the base of the inserts were fixed overnight in 100% methanol at −20 °C and then stained for 2 min with 2% crystal violet solution (Sigma-Aldrich, MO, USA). Cells were photographed at 10× magnification using a Leica DM IRB microscope with a Leica 420C camera attached (Leica Microsystems, Milton Keynes, UK) (4 fields of view) and counted manually in photoshop CS3. Viability was assessed at 72 h post transfection using Cell Titer Aqueous ONE reagent (Promega, WI, USA) as previously described [7].
Results
miRNA profiles in skeletal muscle and RMS
MiRNA expression profiling was performed by high-throughput sequencing analysis of 68 samples (3 skeletal muscle, 1 myoblast and 64 RMS patient samples (Supplementary Table S1)). On average, around 2,069,412 tags were sequenced for each sample (range: 753,614–7,390,288). After sequence alignment and filtering, each tag was assigned to a RNA class based on Entrez Gene type and 78% and 73% were shown to be miRNAs in RMS and skeletal muscle, respectively, confirming their successful enrichment (Supplementary Fig. S1A). Since miRNAs are often included in transcription clusters, their expression is more coordinated when located in close proximity compare to those further apart [35]. This feature was clearly observed in our dataset (Supplementary Fig. 1B).
Direct comparisons of levels of expression across molecular (fusion positive versus negative RMS), histological (skeletal muscle versus RMS), and clinical (metastatic M1 versus non metastatic M0) entities produced lists of miRNAs significantly over or under-expressed in these groups (Supplementary Tables S2-5). Correlation between miRNA expression and overall survival (OS) and progression free survival (PFS) was also assessed by Cox proportional hazard regression model (Supplementary Tables S6-9).
miRNA profile distinguishes RMS according to fusion gene status
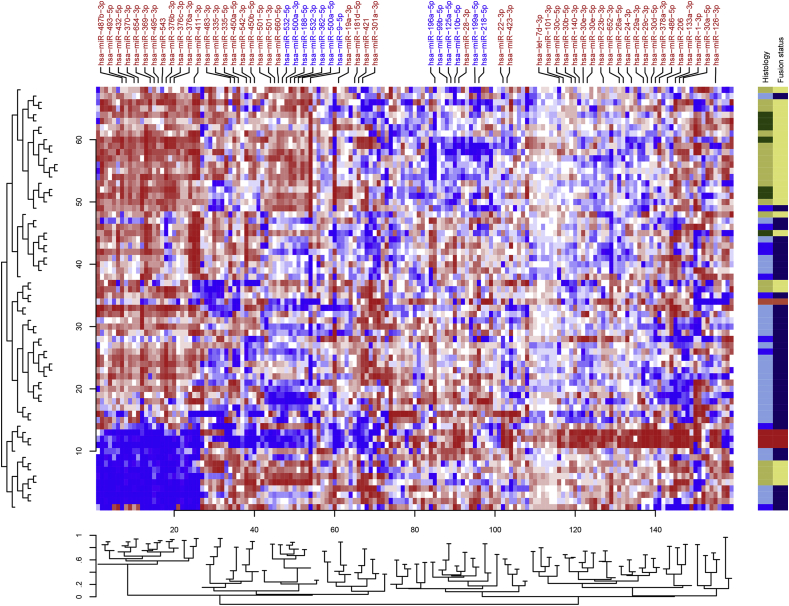
Unsupervised hierarchical clustering analysis performed using miRNAs with at least 100 tags in 50% of the samples showed the presence of distinct patterns of expression across RMS samples (Fig. 1). Notably, miRNA expression patterns mostly distinguished RMS histological subtypes showing sets of miRNAs positively and negatively associated with fusion gene status. The well-known muscle specific miRNAs including miR-1, miR-133 and miR-206 showed higher expressed in skeletal muscle versus RMS subtypes.
Fig. 1.
Heatmap based on the expression profile of 161 miRNAs in RMS. Skeletal muscle samples (red) cluster together showing high expression of a group of miRNA which contains “muscle specific” miR-1, miR-133 and miR-206. Myoblast sample is in separate (tomato red). The histology is color-coded as follows: ARMS_PAX3-FOXO1 (khaki), ARMS_PAX7-FOXO1 (dark green), ERMS (sky blue) and ARMS negative (blue). Fusion gene positive (light khaki) and negative (navy) patients mainly cluster into two separate groups. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Application of class prediction analysis based on nearest shrunken centroid classifier [32] showed that miRNA expression can distinguish fusion positive and negative patients with around 89% accuracy (Supplementary Fig. S2) using the expression level of 11 miRNAs. These data suggest a strong interplay between fusion protein and miRNA expression that potentially contributes to the malignant phenotype of RMS.
RMS gene co-expression-modules are assigned biological functions and correlate with clinicopathological features
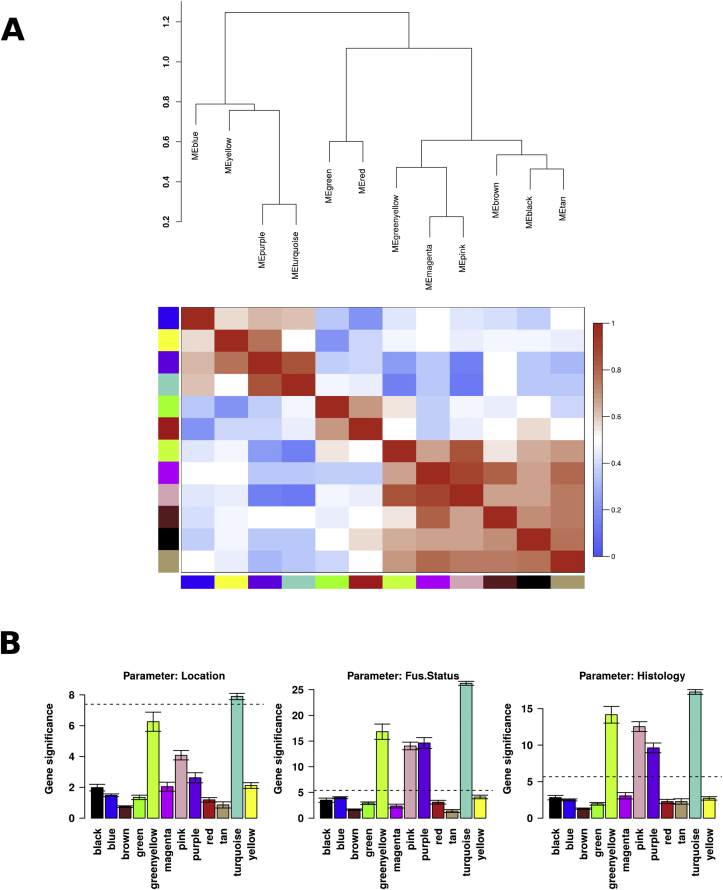
Based on the principle that co-expressed genes are commonly associated with similar biological function/pathways, we employed WGCNA using parallel gene expression data from the 64 RMS patients [30] and identified 12 groups of co-expressing genes/modules. Each module was color coded and tested for GO term enrichment in order to associate each module with biological functions (Table 1). Consistent modular gene structure was also seen using a larger population of 132 publicly available RMS samples [13] suggesting that these modules are a good representation of the biological characteristic of RMS tumors (data not shown). In addition, an eigengene per module was generated for each patient to summarize the expression of all the genes contained in each module. These values were used to correlate modules to each other as well as to clinico-pathological features (Fig. 2). Modules did not show significant correlation with IRS stage, metastasis, tumor size or age of patient at diagnosis (data not shown). However notably, the turquoise, green/yellow, pink and purple modules showed a significant correlation fusion status and histological subtype (Fig. 2B). The association between modules and overall survival (OS) or progression free survival (PFS) was also tested. The pink and turquoise modules showed a significant correlation with the OS and PFS (Supplementary Table S10).
Table 1.
Functional annotation, predicted by GeneGo software, of the miR/gene co-expression modules for RMS identified by WGCNA.
| Module | No. of genes | Go processes | GeneGo networks |
|---|---|---|---|
| Black | 116 | Epidermis development | Immune response_Th17-derived cytokines |
| Ectoderm development | Cytoskeleton_Intermediate filaments | ||
| Epithelial cell differentiation | Inflammation_Amphoterin signaling | ||
| Epithelium development | Chemotaxis | ||
| Blue | 516 | Cellular macromolecule metabolic process | Signal Transduction_TGF-beta, GDF and Activin signaling |
| Regulation of metabolic process | Signal Transduction_BMP and GDF signaling | ||
| Nucleic acid metabolic process | Cell cycle_Mitosis | ||
| Regulation of gene expression | Cytoskeleton_Cytoplasmic microtubules | ||
| Brown | 460 | Immune system process | Inflammation _Interferon signaling |
| Immune response | Chemotaxis | ||
| Defense response | Cell adhesion_Leucocyte chemotaxis | ||
| Response to stimulus | Immune response Phagocytosis | ||
| Green | 206 | M phase | Cell cycle_Core |
| Cell cycle phase | Cell cycle_Mitosis | ||
| Mitosis | Cell cycle_G2-M | ||
| Nuclear division | Cytoskeleton_Spindle microtubules | ||
| Greenyellow | 41 | Regulation of atrial cardiomyocyte membrane depolarization | Neurophysiological process_Long-term potentiation |
| Keratan sulfate biosynthetic process | Signal transduction_Neuropeptide signaling pathways | ||
| response to nickel ion | Reproduction_FSH-beta signaling pathway | ||
| multicellular organismal process | Reproduction_Feeding and Neurohormone signaling | ||
| Magenta | 79 | Tissue development | Cell adhesion_Cell-matrix interactions |
| Wound healing | Cell adhesion_Platelet-endothelium-leucocyte interactions | ||
| Extracellular matrix organization | Development_EMT_Regulation of epithelial-to-mesenchymal transition | ||
| Skeletal system development | Development_Skeletal muscle development | ||
| Pink | 103 | Extracellular matrix organization | Cell adhesion_Cell-matrix interactions |
| Positive regulation of dendritic spine development | Proteolysis_ECM remodeling | ||
| positive regulation of smooth muscle cell chemotaxis | Development_Cartilage development | ||
| Anatomical structure morphogenesis | Signal Transduction_Cholecystokinin signaling | ||
| Purple | 64 | Cardiac muscle contraction | Muscle contraction |
| Heart contraction | Cytoskeleton_Actin filaments | ||
| Striated muscle contraction | Development_Skeletal muscle | ||
| Muscle contraction | Development_Neuromuscular junction | ||
| Red | 156 | Protein localization | Immune response_Phagosome in antigen presentation |
| Cellular process | Transcription_mRNA processing | ||
| Macromolecule localization | Cell cycle_Mitosis | ||
| Protein transport | Development_Hedgehog signaling | ||
| Tan | 31 | Positive regulation of gene expression | Reproduction_Gonadotropin regulation |
| Response to external stimulus | Immune response_Th17-derived cytokines | ||
| Positive regulation of transcription, DNA-dependent | Inflammation_IL-6 signaling | ||
| Positive regulation of RNA metabolic process | Cell cycle_G1-S Interleukin regulation | ||
| Turquoise | 529 | Nervous system development | Development_Neurogenesis in general |
| Developmental process | Development_Hedgehog signaling | ||
| Anatomical structure development | Development_Blood vessel morphogenesis | ||
| Multicellular organismal development | Development_Ossification and bone remodeling | ||
| Yellow | 209 | Muscle system process | Muslce contraction |
| Muscle contraction | Development_Skeletal muscle development | ||
| Striated muscle contraction | Cytoskeleton_Actin filaments | ||
| Myofibril assembly | Cytoskeleton_Regulation of cytoskeleton rearrangement |
Fig. 2.
Relationship between modules and with other clinic-molecular variables. WGCNA analysis identified 12 modules based on gene co-expression patterns. Each module was uniquely color labeled. The expression profile of the genes included in each module was summarized by a module eigengene (first principal component of the expression matrix). These eigengenes were used to assess relationships between modules as well as with other variables A. Hierarchical clustering of module eigengenes and a heatmap showing the relatedness of these to each other. In the heatmap, red indicates modules are highly related and blue that they are not related. Modules that cluster closer together tend to be more related. In B. we averaged the statistics computed using the Kruskal-Wallis rank sum test of the genes within each module – higher values indicate the presence of a larger number of genes contained in that module which are associated to a specific patient-specific parameter. The horizontal bars mark the threshold level for significance. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
RMS gene-expression-modules associated with skeletal muscle development correlate with known myomiRs
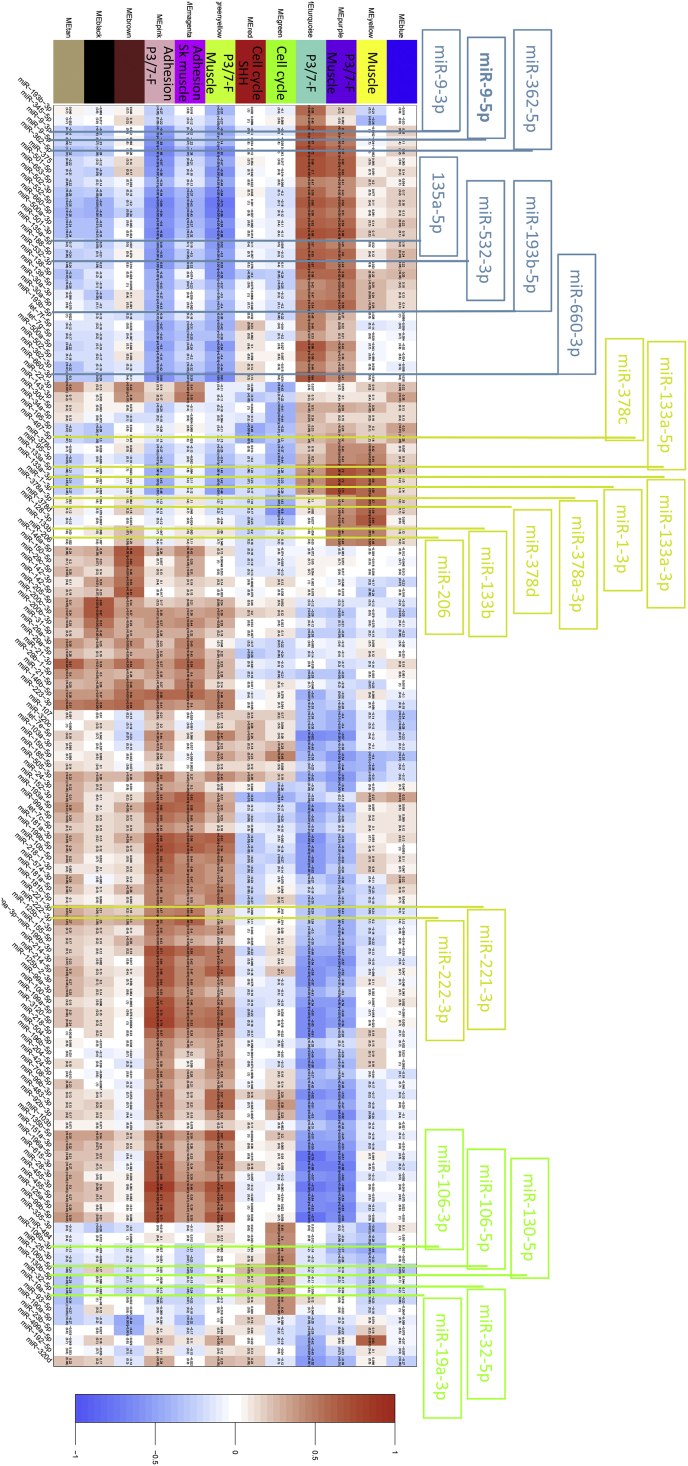
As miRNAs are known to modulate many of their targets through mRNA transcript degradation [14], [36], we correlated the expression of miRNAs to the module eigengenes in order to evaluate their level of association. Fig. 3 represents a heatmap of the correlation between module eigengenes and a subset of miRNAs selected based on their coefficient values (absolute value above 0.4) and significance (p value < 0.001). The power of this approach is demonstrated by well-known myogenic miRs (myomiRs) -1, -206 and -133a that are most highly expressed in more differentiated skeletal muscle. These myomiRs correlated positively with yellow (miR-1, direct pearson (DP) 0.59; miR-206, DP 0.49; miR-133a, DP 0.58), and purple modules (miR-1, DP 0.73; miR-206, DP 0.46; miR-133a, DP 0.70) (Fig. 4). In an inverse manner, miR-221 and -222 are myoD-associated myomiRs most highly expressed early in myogenic differentiation [37] and show a negative correlations with the yellow and purple modules (miR-221, DP-0.24, -0.043; miR-222, DP-0.2, -0.041, respectively). Both modules had functional annotations linked to skeletal muscle development. Additionally miR-1 and -133a showed inverse correlation with green module (miR-1, DP-0.31; miR-133a, DP-0.37) that was linked to cell cycle, indicating that low levels of these myomiRs correlated with genes involved in cell cycle progression. These myomiRs/gene expression correlations associated with differentiation processes strongly support the ability of the approach to identify key roles for miRNAs in RMS.
Fig. 3.
Heatmap showing the correlation between module eigengenes in a subset of miRNAs. The colored modules have been labeled with some features (see Table 1) as well as those that correlated with fusion gene positivity (P3/7-F). MiRNAs are indicated in black at the base and positive and negative correlations with modules indicated in red and blue, respectively. These were selected based on an absolute Pearson correlation coefficient above 0.4 and a p value < 0.001. MiRNAs highlighted above the heatmap are those positively associated with fusion gene positivity (blue), myoMiRs associated with muscle differentiation (yellow) and miRNAs linked to regulating the cell cycle (green) (see discussion for further details). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
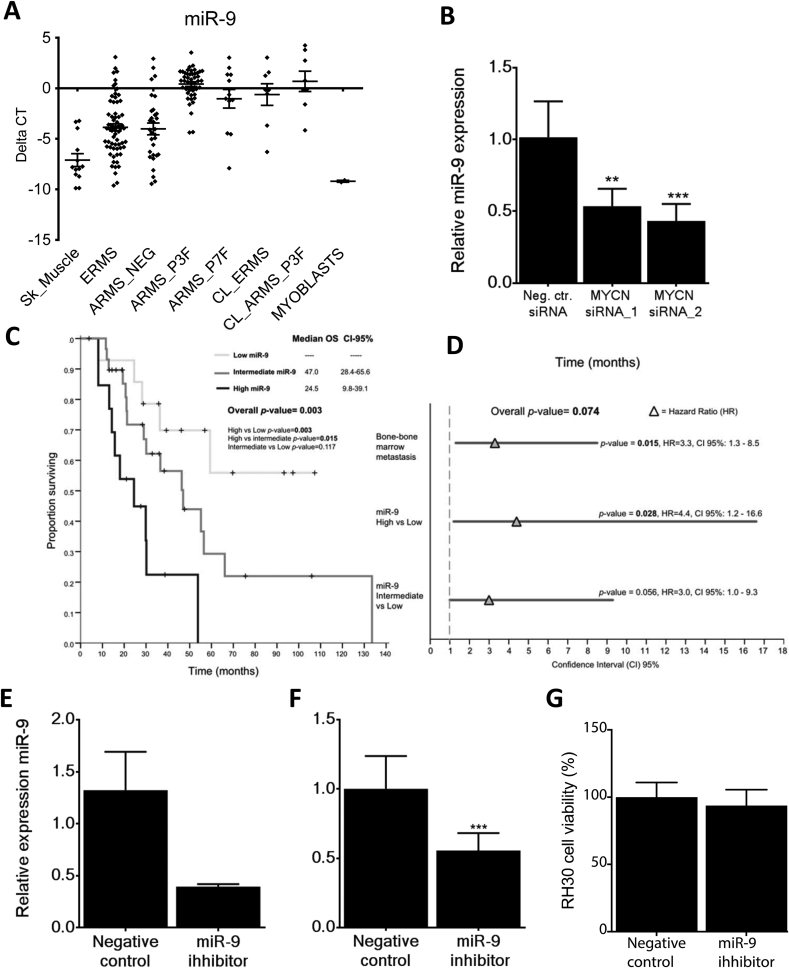
Fig. 4.
Validation of clinical correlations. A. Validation of NGS results on larger cohort of RMS. Box and whisker plots representing the expression levels (QRT-PCR) of miR-9 in 13 normal skeletal muscles, 66 ERMS, 31 ARMS fusion negative (ARMS_NEG), 45 ARMS PAX3-FOXO1A (ARMS_P3F), 12 ARMS PAX7-FOXO1A (ARMS_P7F), 8 ERMS cell lines (CL_ERMS), 8 ARMS cell lines (CL_ARMS) and 2 myoblast samples (Myoblasts). B. Silencing of MYCN using siRNAs resulted in a reduction in the expression level of miR-9 in RH30 cells when compared to control cells. Data represent a triplicate measurement for each sample at 72 h post addition of negative control and MYCN siRNAs. **p < 0.01; ***p < 0.001. C. Kaplan-Meier plots for miR-9 expression and overall survival (OS) Expression levels within the first quartile was considered ‘Low’ (<Q25), ‘Intermediate’ when expression was between the second and third quartile (>Q25 and < Q75) and ‘High’ when within the top quartile (>Q75). p values were obtained using Log-Rank test (Mantel-Cox). Median survival for each patient group, the CI-95% values and pairwise comparison p values are shown on each graph. D. A forest plot showing the multivariate analysis (MVA) results of miR-9 expression in overall survival (OS). Bone-marrow metastasis at diagnosis (predictive factor in univariate analysis) was incorporated in to the Cox regression model for OS. An HR value greater than 1 indicates a predictive factor. HR, Hazard Ratio; CI, Confidence Interval. E. Synthetic miR-9 inhibitors reduce miR-9 expression levels in the cell line RH30. F. This significantly reduces the ability of the cells to migrate in a transwell assay (***p < 0.001) but G. had no detectable effect on cell viability, as assessed by the MTS assay.
PAX3-FOXO1 alters the profile of miR expression
Consistent with miRNA expression accurately distinguishing fusion positive from negative patients, four of the 12 modules correlated with fusion gene positivity (turquoise, purple, green/yellow, and pink) with turquoise correlating most strongly (Fig. 2B and indicated in Fig. 3). MiRNAs were also overexpressed in PAX3/7-FOXO1 positive cases compared to RMS without the fusion gene (Supplementary Table S3). Based on these results, we hypothesized that the fusion protein could regulate miRNA expression. To identify miRNAs altered by PAX3-FOXO1, RNA interference (RNAi) was used to reduce expression levels of the fusion gene in the cell line RH4 (Supplementary Fig. S3A), and corresponding miRNAs levels screened for in duplicate (Supplementary Table S11). To prioritize miRNAs of likely relevance in RMS patients for further investigation we identified those that were reduced by knockdown of the fusion protein, were most strongly positively correlated with the turquoise and purple modules and that were also differentially overexpressed comparing fusion gene positive with negative RMS. This identified miRNAs as candidates for further investigation (Table 2), including miR-9-5p that was additionally correlated with metastatic behavior and poor outcome (Supplementary Tables S4-S9).
Table 2.
MiRNAs in modules associated with fusion genes, differential expressed between fusion positive versus negative RMS and fold change reduction by decreasing PAX3-FOXO1.
| MiRNA | Turquoise module (Pearson correlation coefficient) | Purple module (Pearson correlation coefficient) | log2 fold change expression fusion positive vs negative RMS | RH4 set 1 fold expression relative to non targeting control | RH4 set 2 fold expression relative to non targeting control |
|---|---|---|---|---|---|
| miR-9-5p | 0.67 | 0.51 | 1.14 | −2.49 | −1.15 |
| miR-660-3p | 0.64 | 0.41 | 0.53 | −3.54 | −1.22 |
| miR-9-3p | 0.70 | 0.53 | 1.32 | −1.42 | −1.32 |
| miR-532-3p | 0.67 | 0.49 | 0.46 | −1.40 | −1.37 |
| miR-135a-5p | 0.57 | 0.45 | 1.45 | −13.01 | −8.85 |
| miR-193b-5p | 0.58 | 0.19 | 0.43 | −3.18 | −2.14 |
| miR-362-5p | 0.72 | 0.51 | 0.28 | −4.98 | −1.28 |
Validation of MiR-9-5p levels in RMS driven by PAX3-FOXO1 via MYCN
miR-9-5p expression levels were analyzed by quantitative RT-PCR in a larger cohort of RMS patients (n = 154, Supplementary Table S1B) and, consistent with the sequencing data (Supplementary Table S3), results showed that levels of miR-9-5p were significantly higher in fusion gene positive versus negative RMS (Wilcoxon test p < 0.0001), skeletal muscle and myoblasts (Wilcoxon test p < 0.0001) (Fig. 4A). In addition, fusion gene positive RMS cell lines had significantly higher miR-9-5p levels than ERMS cell lines (Wilcoxon test p = 0.04) (Supplementary Fig. S3B).
As miR-9-5p levels decreased by reducing PAX3-FOXO1 expression (Table 2, Supplementary Table S11), we examined available data for PAX3-FOXO1 [38] and MYCN (Walters et al. unpublished) DNA binding sites near the miR-9-5p locus in RMS. No sites were identified for PAX3-FOXO1 but MYCN binding sites were found consistent with those reported for MYC and MYCN in neuroblastoma [39]. Based on MYCN being a downstream transcriptional target of PAX3-FOXO1 [5], [38], we also tested the effects of decreasing MYCN levels on miR-9-5p levels in a PAX3-FOXO1 positive cell line. MiR-9-5p levels were significantly decreased by MYCN reduction (Fig. 4B). Therefore, PAX3-FOXO1 regulates miR-9-5p levels via MYCN in RMS.
MiR-9-5p levels correlate with poor patient outcome, metastases and cell migration in fusion positive ARMS
Modules associated with the PAX3-FOXO1 fusion genes (turquoise and purple) positively correlated with miR-9-5p expression. Significantly higher levels of miR-9-5p expression were noted in metastatic versus non-metastatic considering both all RMS and fusion positive RMS (Supplementary Tables S4 and S5). Significantly lower OS and PFS in all RMS was associated with high miR-9-5p expression (Supplementary Tables S6 and S8) and also within fusion positive RMS cases (Supplementary Tables S7 and S9). Univariate analyses, including only PAX3-FOXO1 cases, also revealed significant correlations with outcome (Supplementary Table S12), although the sample size was very limited.
Univariate analyses of the quantitative RT-PCR data for the second validation cohort (n = 154, Supplementary Table S1B) using a Cox regression model with miR-9-5p as a continuous variable revealed significant correlations in fusion gene positive RMS with OS (p = 0.037, HR = 1.242, CI 95% 1.013–1.522) and PFS (p = 0.034, HR = 1.239, CI 95% 1.016–1.511). In PAX3-FOXO1 cases (n = 45), a significant correlation with OS was also observed (p = 0.012, HR = 1.445, CI 95% 1.084–1.925) and a trend with PFS (p = 0.055, HR = 1.29, CI 95% 0.995–1.676). No correlations with miR-9-5p levels and survival were observed in fusion gene negative patients (OS; p = 0.65, HR = 0.971, CI 95% 0.853–1.104. PFS; p = 0.65, HR = 0.994, CI 95% 0.853–1.104). Kaplan-Meier analyses were consistent with these results: log-rank test identified significant correlations with both OS (p = 0.003) (Fig. 4C) and PFS (p = 0.004) (Supplementary Fig. 3C) when RMS fusion positive patients (n = 57) were split into three groups based on their levels of miR-9-5p expression (percentiles 0–33%, 34–66%, 67–100%). When the highest miR-9-5p expressers were compared to the lowest in multivariate analysis (MVA), miR-9-5p retained its independent predictive value for OS (p = 0.028, HR = 4.4, CI 95% 1.2–16.6) (Fig. 4D) and PFS (p = 0.012, HR = 3.74, CI 95% 1.34–10.44) (Supplementary Fig. S3D). When MVA analyses was repeated considering only the 45 PAX3-FOXO1, high miR-9-5p expression remained an independent predictive variable correlating with outcome (p = 0.004, HR = 4.55, CI 95% 1.63–12.76) correlating with outcome.
Due to the significant correlations of high miR-9-5p levels with metastatic disease and poor survival as well as strong negative correlations with a module associated with processes in cell adhesion (pink module) (Fig. 3), we tested whether miR-9-5p functionally influenced the migratory behavior of RMS cells. Cell migration in the RH30 cell line was assessed 72 h after transfection with a synthetic inhibitor against miR-9-5p by placing cells into a transwell migratory assay for 24 h. At the end point of experiments, levels of miR-9-5p were decreased on average by 70% compared to control treated cells (Fig. 4E). There was significant reduction in levels of migration in cells with reduced miR-9-5p levels versus the controls (p < 0.001, Fig. 4F) but no effect was on cell viability, as assessed by the MTS metabolic assay (Fig. 4G).
Discussion
Here we describe the sequencing analyses of miRNAs in a large series (n = 64) of well-characterized RMS samples. We were broadly able to distinguish tumor from skeletal muscle and also fusion gene positive from fusion gene negative RMS. This is consistent with a previous study of sarcomas that included only 7 RMS cases in which cluster analyses of miRNA data separated skeletal muscle from RMS and other types of sarcoma and placed 3 alveolar RMS in a different group from one embryonal case and 3 adult pleomorphic cases [40]. Furthermore, our integrated analyses of miRNA expression data with parallel gene expression levels using WCGNA identified co-expression modules that we were able to assign with biological functions and also correlate with clinical outcome data and fusion gene status. Given that miRNAs regulate the expression of many genes in a dynamic and cell context specific manner [14], these results provide a valuable framework and resource for further investigations of RMS.
Reassuringly, modules identified as associated with skeletal muscle development (Fig. 3) included correlations with miRNAs that have been previously described as miRNAs involved in myogenesis (myomiRs) and also shown to functionally impact on the phenotype of RMS cell lines. Specifically these included miRs -1, -206, -133a and miR-378 that correlated positively with muscle associated modules (yellow and purple, Table 1, Fig. 3) [19], [20], [21], [22], [23], [24], [25]. In addition, we previously showed that lower levels of miR-206 expression correlated with poorer outcome in samples from RMS patients that was linked to their undifferentiated myogenic phenotype, indicating relevance to the clinical behavior of patients [22]. MiR-221 and miR-222 are also described as myomiRs and their overexpression and reduction is reported to enhance and decrease myoD protein levels, respectively [37]. As predicted from this, our analyses of miRs-221-3p and -222-3p inversely correlated their expression with the skeletal muscle associated modules (yellow and purple) in contrast to miRs -1, -133 and -378 which were positively correlated to these functional modules. MiR-128-3p shows a similar expression pattern in these modules to miRs -1, -133 and -378 and is described with a clear role in muscle development [41]. A definitive role for miRs -221, -222 and 128-3p in RMS remains to be determined, as well as other miRNAs correlated with the modules defined with myogenic functions, such as miR-96-3p, miR-152-3p and miR-156-5p.
Interestingly, miR-1, 133, 206, 378 and 128-3p negatively, and miR-221 and -222 positively, correlated with gene expression processes for cell adhesion, chemotaxis and extracellular organisation (pink module). This is consistent with the differentiation status of RMS cells shown in oncogenic KRAS (G12D) induced zebrafish models of RMS where embryonal RMS cells recapitulate normal myogenesis and their associated migratory and proliferative capacity [42]. Also, targets of these miRNAs in these networks are likely to contribute to these invasive and migratory phenotypes in RMS, for example miR-206 targets the MET tyrosine-kinase receptor which is highly expressed in RMS and enhances RMS cell migration [24]. Similarly, miR-221 and -222 have strong evidence to support that they target TIMP3 and MMP1 proteins known to influence the extracellular matrix (http://mirtarbase.mbc.nctu.edu.tw/index.php).
The myomiRs -1, -133a and miR-378 also inversely correlated with modules linked to cell cycle progression (green and red), consistent with their role in cell cycle arrest when overexpressed in RMS [21], [23] and the requirement of this prior to normal myogenic differentiation [43]. These cell cycle related modules were positively correlated with other miRNAs that may play a role in RMS, including miR-106b, that is overexpressed and associated with proliferation in medulloblastoma [44], miR-32 that when down-regulated in mesenchymal stem cells inhibits cell cycle progression [45] and miR-130b that enhances profileration through inactivating HIPPO signaling in gliomas [46]. Mir-19a-3p, as part of the polycistronic miRNA cluster miR-17-92, also positively correlated with these cell cycle modules as well as a module associated with fusion genes (turquoise). The latter and red modules were both linked to Sonic hedgehog (SHH) signaling. Together this is consistent with high expression of the miR17-92 cluster being genomically amplified in around 20% of RMS and MYCN driven, mostly in those with PAX7-FOXO1 fusion genes, and possible involvement of the cluster in proliferation through SHH signaling [47], [48].
The strongest positive and negative correlations between genes and miRNA expression levels were associated with fusion gene positive RMS cases (Fig. 2, Fig. 3). In addition, the turquoise and pink modules linked to fusion gene positivity were also most significantly associated with poor outcome (Supplementary Table S10). This is consistent with several studies, including our own, demonstrating the prognostic value of the presence of a fusion gene in RMS [9], [10], [11], [12]. Our analyses indicates that the fusion protein impacts on the expression of miRNAs and through screening changes to miRNA levels following reduction of PAX3-FOXO1 expression in a cell line, we were able to show that the fusion protein alters the expression levels of microRNAs. Interestingly, miR-9-5p was one of the top hits in the screen, was highly correlated with the modules associated with the presence of the fusion gene and it was more highly expressed in fusion gene positive RMS compared to fusion gene negative RMS (Table 2). Although no PAX3-FOXO1 binding sites were identified near miR-9-5p, we showed miR-9-5p to be indirectly driven by PAX3-FOXO1 via MYCN. MYC/MYCN have previously been shown to bind to the miR-9 locus and activate expression in other cell types [39]. MYCN is known to be a direct transcriptional target of the fusion protein [5] and to operate in a positive feedback loop mechanism [7]. This overlap in miRNA targets for PAX3-FOXO1 and MYCN is to the gene expression changes identified through altering MYCN levels [7] and PAX3-FOXO1 expression in RMS that show considerable overlap [13]. Taken together, this indicates that the fusion protein together with MYCN, and other transcription factors downstream of PAX3-FOXO1, such as MYOD1, regulate the expression of miRNAs and their many target genes, to massively re-programme gene expression in RMS cells towards a tumorigenic phenotype [49]. The contribution of these altered levels of miRNAs is predicted to impact on the phenotype of fusion positive RMS in ways that include the developmental, myogenic, immunological and cell adhesion processes indicated in the co-expression modules (Fig. 3).
As representative of a miRNA associated with the presence of PAX3-FOXO1, miR-9-5p levels are shown to contribute to the clinical and biological behavior of RMS. MiR-9-5p levels were higher in metastatic versus non-metastatic cases and correlated with a poor outcome of patients, including within fusion gene positive group. Through reducing miR-9-5p we deduce a role for miR-9-5p in enhancing RMS cell migration and this cellular phenotype potentially facilitates metastastic behavior. This is similar to the roles for miR-9-5p in the proliferation, differentiation and migration of neural progenitor cells but miR-9-5p may alter different gene targets dependent on cell context [50]. Also, aberrant miR-9-5p levels have been indicated in several cancer types where it may be either overexpressed or underexpressed. Overexpression has been described in Hodgkin's lymphoma [51], brain tumors [52], neuroblastoma [39] and gastric cancers, in the latter affecting proliferation via the miR-9 target CDX2 [53]. Reduced levels of miR-9-5p play a role in ovarian cancer cells where this, in part, enhances cell proliferation through NFkappaB1 [54]. Furthermore, in normal epithelial and breast cancer cells, miR-9-5p induces epithelial to mesenchymal transition by targeting E cadherin (CDH1) [39]. Recently, reducing miR-9-5p in an osteosarcoma cell line and identification of high levels of expression in primary tumors suggests a role for this miRNA in regulating the progression of osteosarcoma [55]. This role in osteosarcomas in addition to RMS described here, indicates a role for miR-9-5p in mesenchymal malignancies further to that previously described epithelial and neural cancers.
MiRNAs altered by the fusion protein, including indirectly miR-9-5p (Table 2, Supplementary Tables S3 and S11), are likely to play an important role in RMS. Furthermore, the expression levels and the functional framework presented have assigned likely functions for miRNAs in RMS for further investigation. These could identify additional tumor biomarkers and, depending on their expression in bodily fluids, useful markers for disease monitoring, as exemplified by the detection of the myomiR miR-206 in serum from RMS patients [56]. Achieving good in vivo delivery for therapeutically targeting or introducing miRNAs has potential but is still challenging [29]. However, further understanding of miRNAs in RMS may ultimately enable targeting their key roles via regulation of multiple genes and pathways in ways that could improve outcome for patients.
Acknowledgments
This work was supported by Cancer Research UK [grant references C5066/A9541, A10399], the Chris Lucas Trust, the “Carte d’Identitié” and Equipe Labellisé programs of La Ligue Nationale Contre le Cancer, INCA. We also acknowledge NHS funding to the ICR/RMH NIHR Biomedical Research Centre for Cancer. We thank the Children's Cancer and Leukaemia Group for their role in tumor and data collection and are grateful to Rebecca Allen for her technical support.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.canlet.2016.10.011.
Conflict of interest
None.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Anderson J., Gordon A., Pritchard-Jones K., Shipley J. Genes, chromosomes, and rhabdomyosarcoma. Genes Chromosom. Cancer. 1999;26:275–285. [PubMed] [Google Scholar]
- 2.Pappo A.S., Shapiro D.N., Crist W.M., Maurer H.M. Biology and therapy of pediatric rhabdomyosarcoma. J. Clin. Oncol. 1995;13:2123–2139. doi: 10.1200/JCO.1995.13.8.2123. [DOI] [PubMed] [Google Scholar]
- 3.Barr F.G., Galili N., Holick J., Biegel J.A., Rovera G., Emanuel B.S. Rearrangement of the PAX3 paired box gene in the paediatric solid tumour alveolar rhabdomyosarcoma. Nat. Genet. 1993;3:113–117. doi: 10.1038/ng0293-113. [DOI] [PubMed] [Google Scholar]
- 4.Davis R.J., D'Cruz C.M., Lovell M.A., Biegel J.A., Barr F.G. Fusion of PAX7 to FKHR by the variant t(1;13) (p36; q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994;54:2869–2872. [PubMed] [Google Scholar]
- 5.Mercado G.E., Xia S.J., Zhang C., Ahn E.H., Gustafson D.M., Lae M. Identification of PAX3-FKHR-regulated genes differentially expressed between alveolar and embryonal rhabdomyosarcoma: focus on MYCN as a biologically relevant target. Genes Chromosom. Cancer. 2008;47:510–520. doi: 10.1002/gcc.20554. [DOI] [PubMed] [Google Scholar]
- 6.Naini S., Etheridge K.T., Adam S.J., Qualman S.J., Bentley R.C., Counter C.M. Defining the cooperative genetic changes that temporally drive alveolar rhabdomyosarcoma. Cancer Res. 2008;68:9583–9588. doi: 10.1158/0008-5472.CAN-07-6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tonelli R., McIntyre A., Camerin C., Walters Z.S., Di Leo K., Selfe J. Antitumor activity of sustained N-myc reduction in rhabdomyosarcomas and transcriptional block by antigene therapy. Clin. Cancer Res. 2012;18:796–807. doi: 10.1158/1078-0432.CCR-11-1981. [DOI] [PubMed] [Google Scholar]
- 8.Williamson D., Lu Y.J., Gordon T., Sciot R., Kelsey A., Fisher C. Relationship between MYCN copy number and expression in rhabdomyosarcomas and correlation with adverse prognosis in the alveolar subtype. J. Clin. Oncol. 2005;23:880–888. doi: 10.1200/JCO.2005.11.078. [DOI] [PubMed] [Google Scholar]
- 9.Anderson J., Gordon T., McManus A., Mapp T., Gould S., Kelsey A. U.K.C.s.C.S. Group, U.K.C.C.G. the, Detection of the PAX3-FKHR fusion gene in paediatric rhabdomyosarcoma: a reproducible predictor of outcome? Br. J. Cancer. 2001;85:831–835. doi: 10.1054/bjoc.2001.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Missiaglia E., Williamson D., Chisholm J., Wirapati P., Pierron G., Petel F. PAX3/FOXO1 fusion gene status is the key prognostic molecular marker in rhabdomyosarcoma and significantly improves current risk stratification. J. Clin. Oncol. 2012;30:1670–1677. doi: 10.1200/JCO.2011.38.5591. [DOI] [PubMed] [Google Scholar]
- 11.Rosenberg A.R., Skapek S.X., Hawkins D.S. The inconvenience of convenience cohorts: rhabdomyosarcoma and the PAX-FOXO1 biomarker. Cancer Epidemiol. Biomark. Prev. 2012;21:1012–1018. doi: 10.1158/1055-9965.EPI-12-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skapek S.X., Anderson J., Barr F.G., Bridge J.A., Gastier-Foster J.M., Parham D.M. PAX-FOXO1 fusion status drives unfavorable outcome for children with rhabdomyosarcoma: a children's oncology group report. Pediatr. Blood Cancer. 2013;60:1411–1417. doi: 10.1002/pbc.24532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davicioni E., Finckenstein F.G., Shahbazian V., Buckley J.D., Triche T.J., Anderson M.J. Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res. 2006;66:6936–6946. doi: 10.1158/0008-5472.CAN-05-4578. [DOI] [PubMed] [Google Scholar]
- 14.Giza D.E., Vasilescu C., Calin G.A. Key principles of miRNA involvement in human diseases. Discov. (Craiova) 2014;2:e34. doi: 10.15190/d.2014.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brodersen P., Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat. Rev. Mol. Cell Biol. 2009;10:141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 16.Lytle J.R., Yario T.A., Steitz J.A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3' UTR. Proc. Natl. Acad. Sci. U. S. A. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim D.H., Saetrom P., Snove O., Jr., Rossi J.J. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esquela-Kerscher A., Slack F.J. Oncomirs – microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 19.Ciesla M., Dulak J., Jozkowicz A. MicroRNAs and epigenetic mechanisms of rhabdomyosarcoma development. Int. J. Biochem. Cell Biol. 2014;53:482–492. doi: 10.1016/j.biocel.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Li L., Sarver A.L., Alamgir S., Subramanian S. Downregulation of microRNAs miR-1, -206 and -29 stabilizes PAX3 and CCND2 expression in rhabdomyosarcoma. Lab. Invest. 2012;92:571–583. doi: 10.1038/labinvest.2012.10. [DOI] [PubMed] [Google Scholar]
- 21.Megiorni F., Cialfi S., McDowell H.P., Felsani A., Camero S., Guffanti A. Deep Sequencing the microRNA profile in rhabdomyosarcoma reveals down-regulation of miR-378 family members. BMC Cancer. 2014;14:880. doi: 10.1186/1471-2407-14-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Missiaglia E., Shepherd C.J., Patel S., Thway K., Pierron G., Pritchard-Jones K. MicroRNA-206 expression levels correlate with clinical behaviour of rhabdomyosarcomas. Br. J. Cancer. 2010;102:1769–1777. doi: 10.1038/sj.bjc.6605684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao P.K., Missiaglia E., Shields L., Hyde G., Yuan B., Shepherd C.J. Distinct roles for miR-1 and miR-133a in the proliferation and differentiation of rhabdomyosarcoma cells. FASEB J. 2010;24:3427–3437. doi: 10.1096/fj.09-150698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taulli R., Bersani F., Foglizzo V., Linari A., Vigna E., Ladanyi M. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J. Clin. Invest. 2009;119:2366–2378. doi: 10.1172/JCI38075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taulli R., Foglizzo V., Morena D., Coda D.M., Ala U., Bersani F. Failure to downregulate the BAF53a subunit of the SWI/SNF chromatin remodeling complex contributes to the differentiation block in rhabdomyosarcoma. Oncogene. 2014;33:2354–2362. doi: 10.1038/onc.2013.188. [DOI] [PubMed] [Google Scholar]
- 26.Wang H., Garzon R., Sun H., Ladner K.J., Singh R., Dahlman J. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garzon R., Garofalo M., Martelli M.P., Briesewitz R., Wang L., Fernandez-Cymering C. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc. Natl. Acad. Sci. U. S. A. 2008;105:3945–3950. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu M., Zhang Q., Deng M., Miao J., Guo Y., Gao W. An analysis of human microRNA and disease associations. PLoS One. 2008;3:e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monroig-Bosque Pdel C., Rivera C.A., Calin G.A. MicroRNAs in cancer therapeutics: “from the bench to the bedside. Expert Opin. Biol. Ther. 2015;15:1381–1385. doi: 10.1517/14712598.2015.1074999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williamson D., Missiaglia E., de Reynies A., Pierron G., Thuille B., Palenzuela G. Fusion gene-negative alveolar rhabdomyosarcoma is clinically and molecularly indistinguishable from embryonal rhabdomyosarcoma. J. Clin. Oncol. 2010;28:2151–2158. doi: 10.1200/JCO.2009.26.3814. [DOI] [PubMed] [Google Scholar]
- 31.Missiaglia E., Selfe J., Hamdi M., Williamson D., Schaaf G., Fang C. Genomic imbalances in rhabdomyosarcoma cell lines affect expression of genes frequently altered in primary tumors: an approach to identify candidate genes involved in tumor development. Genes Chromosom. Cancer. 2009;48:455–467. doi: 10.1002/gcc.20655. [DOI] [PubMed] [Google Scholar]
- 32.Tibshirani R., Hastie T., Narasimhan B., Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. U. S. A. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinforma. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kikuchi K., Tsuchiya K., Otabe O., Gotoh T., Tamura S., Katsumi Y. Effects of PAX3-FKHR on malignant phenotypes in alveolar rhabdomyosarcoma. Biochem. Biophys. Res. Commun. 2008;365:568–574. doi: 10.1016/j.bbrc.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 35.Wang L., Oberg A.L., Asmann Y.W., Sicotte H., McDonnell S.K., Riska S.M. Genome-wide transcriptional profiling reveals microRNA-correlated genes and biological processes in human lymphoblastoid cell lines. PLoS One. 2009;4:e5878. doi: 10.1371/journal.pone.0005878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pasquinelli A.E. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 37.Tan S.B., Li J., Chen X., Zhang W., Zhang D., Zhang C. Small molecule inhibitor of myogenic microRNAs leads to a discovery of miR-221/222-myoD-myomiRs regulatory pathway. Chem. Biol. 2014;21:1265–1270. doi: 10.1016/j.chembiol.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 38.Cao L., Yu Y., Bilke S., Walker R.L., Mayeenuddin L.H., Azorsa D.O. Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer. Cancer Res. 2010;70:6497–6508. doi: 10.1158/0008-5472.CAN-10-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma L., Young J., Prabhala H., Pan E., Mestdagh P., Muth D. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramanian S., Lui W.O., Lee C.H., Espinosa I., Nielsen T.O., Heinrich M.C. MicroRNA expression signature of human sarcomas. Oncogene. 2008;27:2015–2026. doi: 10.1038/sj.onc.1210836. [DOI] [PubMed] [Google Scholar]
- 41.Shi L., Zhou B., Li P., Schinckel A.P., Liang T., Wang H. MicroRNA-128 targets myostatin at coding domain sequence to regulate myoblasts in skeletal muscle development. Cell Signal. 2015;27:1895–1904. doi: 10.1016/j.cellsig.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 42.Storer N.Y., White R.M., Uong A., Price E., Nielsen G.P., Langenau D.M. Zebrafish rhabdomyosarcoma reflects the developmental stage of oncogene expression during myogenesis. Development. 2013;140:3040–3050. doi: 10.1242/dev.087858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saab R., Spunt S.L., Skapek S.X. Myogenesis and rhabdomyosarcoma the Jekyll and Hyde of skeletal muscle. Curr. Top. Dev. Biol. 2011;94:197–234. doi: 10.1016/B978-0-12-380916-2.00007-3. [DOI] [PubMed] [Google Scholar]
- 44.Li K.K., Xia T., Ma F.M., Zhang R., Mao Y., Wang Y. miR-106b is overexpressed in medulloblastomas and interacts directly with PTEN. Neuropathol. Appl. Neurobiol. 2015;41:145–164. doi: 10.1111/nan.12169. [DOI] [PubMed] [Google Scholar]
- 45.Zhu G., Chai J., Ma L., Duan H., Zhang H. Downregulated microRNA-32 expression induced by high glucose inhibits cell cycle progression via PTEN upregulation and Akt inactivation in bone marrow-derived mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2013;433:526–531. doi: 10.1016/j.bbrc.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 46.Zhu G., Wang Y., Mijiti M., Wang Z., Wu P.F., Jiafu D. Upregulation of miR-130b enhances stem cell-like phenotype in glioblastoma by inactivating the Hippo signaling pathway. Biochem. Biophys. Res. Commun. 2015;465:194–199. doi: 10.1016/j.bbrc.2015.07.149. [DOI] [PubMed] [Google Scholar]
- 47.Northcott P.A., Fernandez L.A., Hagan J.P., Ellison D.W., Grajkowska W., Gillespie Y. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009;69:3249–3255. doi: 10.1158/0008-5472.CAN-08-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reichek J.L., Duan F., Smith L.M., Gustafson D.M., O'Connor R.S., Zhang C. Genomic and clinical analysis of amplification of the 13q31 chromosomal region in alveolar rhabdomyosarcoma: a report from the Children's Oncology Group. Clin. Cancer Res. 2011;17:1463–1473. doi: 10.1158/1078-0432.CCR-10-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahn E.H., Mercado G.E., Lae M., Ladanyi M. Identification of target genes of PAX3-FOXO1 in alveolar rhabdomyosarcoma. Oncol. Rep. 2013;30:968–978. doi: 10.3892/or.2013.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuva-Aydemir Y., Simkin A., Gascon E., Gao F.B. MicroRNA-9: functional evolution of a conserved small regulatory RNA. RNA Biol. 2011;8:557–564. doi: 10.4161/rna.8.4.16019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nie K., Gomez M., Landgraf P., Garcia J.F., Liu Y., Tan L.H. MicroRNA-mediated down-regulation of PRDM1/Blimp-1 in Hodgkin/Reed-Sternberg cells: a potential pathogenetic lesion in Hodgkin lymphomas. Am. J. Pathol. 2008;173:242–252. doi: 10.2353/ajpath.2008.080009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nass D., Rosenwald S., Meiri E., Gilad S., Tabibian-Keissar H., Schlosberg A. MiR-92b and miR-9/9* are specifically expressed in brain primary tumors and can be used to differentiate primary from metastatic brain tumors. Brain Pathol. 2009;19:375–383. doi: 10.1111/j.1750-3639.2008.00184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rotkrua P., Akiyama Y., Hashimoto Y., Otsubo T., Yuasa Y. MiR-9 downregulates CDX2 expression in gastric cancer cells. Int. J. Cancer. 2011;129:2611–2620. doi: 10.1002/ijc.25923. [DOI] [PubMed] [Google Scholar]
- 54.Guo L.M., Pu Y., Han Z., Liu T., Li Y.X., Liu M. MicroRNA-9 inhibits ovarian cancer cell growth through regulation of NF-kappaB1. FEBS J. 2009;276:5537–5546. doi: 10.1111/j.1742-4658.2009.07237.x. [DOI] [PubMed] [Google Scholar]
- 55.Qi X.J., Wang J.F., Wang G.D., Xu Q., Sun H.L. Pivotal role of microRNA-9 in osteosarcoma tumorigenesis and tumor progression. Genet. Mol. Res. 2016;15 doi: 10.4238/gmr.15017318. [DOI] [PubMed] [Google Scholar]
- 56.Miyachi M., Tsuchiya K., Yoshida H., Yagyu S., Kikuchi K., Misawa A. Circulating muscle-specific microRNA, miR-206, as a potential diagnostic marker for rhabdomyosarcoma. Biochem. Biophys. Res. Commun. 2010;400:89–93. doi: 10.1016/j.bbrc.2010.08.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.