ABSTRACT
During seed imbibition at supra-optimal temperature, an increase in the abscisic acid (ABA)/gibberellin (GA) ratio imposes secondary dormancy to prevent germination (thermoinhibition). FUSCA3 (FUS3), a positive regulator of seed dormancy, accumulates in seeds imbibed at high temperature and increases ABA levels to inhibit germination. Recently, we showed that ABA inhibits FUS3 degradation at high temperature, and that ABA and high temperature also inhibit the ubiquitin-proteasome system, by dampening both proteasome activity and protein polyubiquitination. Here, we investigated the role of ABA signaling components and the ABA antagonizing hormone, GA, in the regulation of FUS3 levels. We show that the ABA receptor mutant, pyl1-1, is less sensitive to ABA and thermoinhibition. In this mutant background, FUS3 degradation in vitro is faster. Similarly, GA alleviates thermoinhibition and also increases FUS3 degradation. These results indicate that inhibition of FUS3 degradation at high temperature is dependent on a high ABA/GA ratio and a functional ABA signaling pathway. Thus, FUS3 constitutes an important node in ABA-GA crosstalk during germination at supra-optimal temperature.
KEYWORDS: ABA, dormancy, FUSCA3, GA, germination, high temperature, protein degradation, proteasome, thermoinhibition, ubiquitin proteasome system, UPS
Seed dormancy is an important adaptive trait that prevents seedling establishment under unfavourable conditions to maximize plant survival. Dormancy prevents pre-harvest sprouting of mature seeds and precocious germination or vivipary of immature seeds when still attached to the mother plant. These processes can lead to severe yield loss in agricultural crops. While primary dormancy is induced during seed maturation, secondary dormancy can be imposed in mature seeds with non-deep dormancy to prevent their germination under unfavourable growth conditions such as high temperature.1-2
In Arabidopsis, the transcription factor FUSCA3 (FUS3) is essential for seed maturation and regulates several processes including the establishment of primary dormancy during embryogenesis. Loss-of-function fus3 embryos skip dormancy and can germinate precociously, while mutants ectopically expressing FUS3 post-embryonically display increased dormancy and show delayed germination, growth and flowering among several other phenotypes.3-6 FUS3 represses developmental phase transitions by increasing the levels of the dormancy-promoting hormone, abscisic acid (ABA), while repressing the synthesis of the germination/growth-promoting hormone, gibberellin A (GA).5,7,8 FUS3 is itself regulated by these hormones: the protein is more abundant in the presence of ABA and less abundant in the presence of GA. Exogenous GA application or reduction of endogenous ABA both partially rescue FUS3 overexpression phenotypes, such as the delayed growth, flowering and reduced cell cycling.5 While FUS3 was shown to directly bind and repress GA biosynthetic genes,4,8 the mechanisms behind the regulation of FUS3 protein levels by these hormones are unknown.
FUS3 promoter activity, mRNA and protein levels show little correlation throughout embryogenesis and germination, indicative of post-transcriptional regulation.5,9,10 FUS3 is degraded by the 26S proteasome and regulated post-translationally by a C-terminal PEST degron.10 This region contains negatively charged amino acids and is intrinsically unstructured, making it prone to degradation by proteases. The PEST degron confers FUS3 a very short half-life of 10–15 minutes, based on in-vitro degradation assays. Interestingly, ABA or GA treatments do not affect the stability of FUS3 truncation mutants lacking the PEST degron, suggesting that this region modulates sensitivity to ABA and GA.10
At optimal temperatures, the FUS3 protein accumulates during embryogenesis, but is not detected post-embryonically.5,10 At elevated temperatures, ABA levels increase and this leads to a delay or inhibition of germination (thermoinhibition) and FUS3 accumulation.11,12,13 Overexpression of FUS3 at 32°C completely inhibit germination through de novo ABA synthesis, this allows higher seedling survival when seeds are transferred to optimal temperature.13 To elucidate the mechanism behind FUS3 protein accumulation under heat stress, we recently characterized FUS3 degradation rates in extracts of seed imbibed at optimal temperature (21°C) in the presence of ABA and at supra-optimal (32°C) temperatures.14 By using ABA biosynthetic mutants and fluridone, an inhibitor of ABA biosynthesis, we showed that FUS3 degradation in vitro is strongly inhibited at 32°C in an-ABA-dependent manner, and is also inhibited by ABA at 21°C. Thus, the accumulation of FUS3 at 32°C is due to the increased ABA level.
We then characterized the overall activity of the ubiquitin-proteasome system (UPS) under these conditions. We reported a strong correlation between UPS activity and seed germination.14 During germination at 21°C, the activity of the 26S proteasome and protein polyubiquitination increase rapidly. Inhibition of proteasome activity with MG132 is sufficient to prevent non-dormant mature embryos from transitioning to vegetative development under dormancy-releasing conditions. Elongation of the radicle, cotyledon expansion, cotyledon greening, and development of true leaves were inhibited in MG132-treated embryos, indicating that an active proteasome is required for these germination processes. Accordingly, protein polyubiquitination and 26S proteasome activity are strongly reduced when germination is inhibited at 32°C or 21°C + ABA. Inhibition of proteasome activity and protein polyubiquitination at 32°C are also ABA-dependent. Interestingly, the inhibitory effect of ABA on UPS activity occurs only within the first 2 days of germination, prior to reaching seedling establishment, a timeframe that defines seed sensitivity to ABA. We propose that dampening of proteasome activity may be one of the mechanisms put in place to prevent germination at supra-optimal temperature, by reducing the degradation of FUS3 and likely of other germination inhibitors, such as ABI3/ABI5/DELLA.14
In this study, we used a mutant affected in one of the ABA receptor family genes, the PYRABACTIN-RESISTANT 1 (PYR1), to understand if FUS3 accumulation during heat stress is dependent on ABA signaling.15 Members of the PYR/PYL family, including PYL1, contribute to ABA sensitivity in seeds.15,16 Therefore, we first characterized the response of the pyl1-1 mutant (Salk_054640c) to high temperature (32°C) and concentrations of ABA that mimic the inhibitory effect of high temperature during germination. When imbibed at 21°C + 0.5µM ABA, pyl1-1 seeds germinated faster than WT (Fig. 1A). A similar pattern was observed at 32°C, with pyl1-1 seeds germinating quicker than WT (Fig. 1A). Thus, the ABA receptor mutant pyl1-1 was less sensitive to germination inhibition caused by ABA or high temperature.
Figure 1.
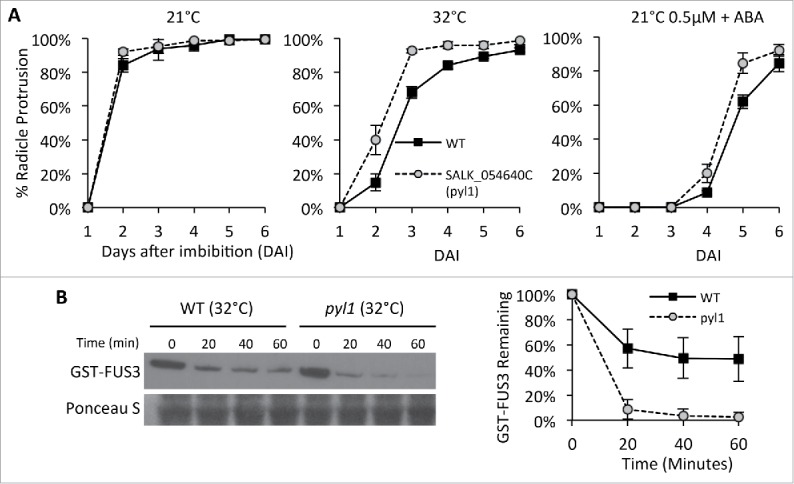
PYL1 is required for the increased FUS3 stability at high temperature (A) Germination rates of wild type (WT) and pyl1-1 T-DNA insertion mutant (SALK_054640; Guzman et al. 2012) imbibed at control (21°C), supraoptimal (32°C) temperatures, or 21°C + 0.5 micromolar ABA. 3 months-old seeds were sterilized and imbibed on half strength MS plates as previously described (Chiu et al. 2016). Averages from triplicates ± standard deviation (SD) are shown. (B) Immunoblots showing degradation of GST-FUS3 in cell extracts of 3 DAI WT and pyl1-1 seeds imbibed at 32°C. Cell free degradation assays were performed as previously described (Chiu et al. 2016). Immunoblots were probed with 1:1000 anti-GST antibody (one representative is shown). Ponceau S stain is shown as a loading control. A plot showing the quantification of GST-FUS3 levels in 3 biological replicates ± SD is shown.
Next, we measured the degradation rates of FUS3-GST in cell extracts of WT and pyl1-1 seeds imbibed at 32°C. Degradation of FUS3 was faster in the cell lysate of pyl1-1 than WT, with FUS3 being almost completely degraded within 20 minutes. The short 15 minute half-life of FUS3 in pyl1-1 extracts at 32°C is similar to that measured in WT extracts at 21°C, or when ABA accumulation at 32°C is prevented by the ABA biosynthesis inhibitor, fluridone.14 This indicates that ABA signaling is required to delay FUS3 degradation at high temperature.
Repression of GA synthesis is necessary to inhibit germination at high temperature and is induced by the rise in ABA level.12 Since addition of GA alleviates thermoinhibition,4 here we examined if GA does so by increasing FUS3 turnover. As expected, GA displayed a dormancy-releasing effect on WT seeds imbibed at 32°C (Fig. 2A), and also accelerated FUS3 degradation (Fig. 2B). When GA was supplied during imbibition at 32°C, the half-life of FUS3 was shortened from 50 minutes to 15 minutes (Fig. 2B), similarly to FUS3 degradation rates at 21°C.14 These results, together with those presented in our recent study,14 show that inhibition of FUS3 degradation at high temperature is dependent on ABA synthesis and signaling. They also show that GA negatively regulates FUS3 levels by accelerating FUS3 degradation. We conclude that during germination at high temperature, an increase in ABA level activates ABA signaling to antagonize GA action, and the increased ABA/GA ratio prevents degradation of FUS3. These results corroborate previous finding showing that the FUS3-GFP fusion protein is more stable (stronger fluorescence) in the presence of ABA and uniconazole, an inhibitor of ABA catabolism, while unstable (weaker fluorescence) in the presence of GA, and show that these hormones regulate FUS3 post-translationally.5 Thus, FUS3 is an important node of crosstalk between ABA and GA signaling during germination at high temperature, with ABA and GA having antagonistic roles in the regulation of FUS3 levels.17
Figure 2.
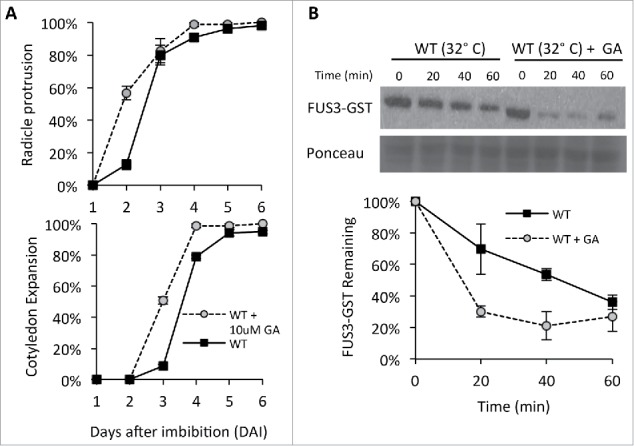
GA accelerates FUS3 degradation at 32°C (A) Germination rates (% radíele protrusion) and seedling growth (% cotyledon expansion) of wild type (WT) seeds imbibed at 32°C in the presence or absence of 10μM GA. Six-months-old dry seeds were sterilized and ¡mbibed on MS/Agar plates as previously described (Chiu et al., 2016). Averages from triplicates ± standard deviation (SD) are shown. (B) Immunoblots showing GST-FUS3 degradation rates in cell extraets of 3 DAI WT seeds imbibed at 32°C in the presence or absence of 10μM GA. Cell free degradation assays were performed as previously described (Chiu et al., 2016). Immunoblots were probed with 1:1000 anti-GST antibody and Ponceau S stain is shown as a loading control. A plot showing the quantification of GST-FUS3 levels in 3 biological replicates ± SD is shown.
Together with ABI3/ABI5/DELLA complex, FUS3 regulates the expression of heat-induced genes to promote thermoinhibition.13,18,19 It will be important to determine if the accumulation of the ABI3/ABI5/DELLA complex at high temperature is also due to inhibition of proteasome activity by ABA. Given that GA partially rescues thermoinhibition and increases FUS3 degradation, it is expected that GA would have a stimulatory effect on proteasome activity and protein ubiquitination during germination, by antagonizing ABA action.
Besides ABA, also auxin has been shown to regulate the activity of the proteasome. In addition to targeting IAA repressors for degradation, auxin has been recently shown to inhibit the activity of the proteasome through PI31 as a mechanism to prevent excessive degradation of IAAs.20 Thus, both ABA and auxin negatively regulate proteasome activity. Understanding the mechanism through which hormones regulate the proteasome will further our understanding of protein homeostasis during high temperature and other abiotic stresses.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Gazzarrini S is supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC).
References
- 1.Holdsworth M, Bentsink L, Soppe W. Molecular networks regulating Arabidopsis seed maturation, after ripening, dormancy and germination. New Phytologist 2008; 74:767-780; PMID:18422904; http://dx.doi.org/12244252 10.1111/j.1469-8137.2008.02437.x [DOI] [PubMed] [Google Scholar]
- 2.Nambara E, Okamoto M, Tatematsu K, Yano R, Seo M, Kamiya Y. Abscisic acid and the control of seed dormancy and germination. Seed Science Research 2010; 20:55-67; http://dx.doi.org/ 10.1017/S0960258510000012 [DOI] [Google Scholar]
- 3.Keith K, Kraml M, Dengler NG, McCourt P. fusca3: A Heterochronic Mutation Affecting Late Embryo Development in Arabidopsis. Plant Cell 1994; 6:589-600; PMID:12244252; http://dx.doi.org/ 10.1105/tpc.6.5.589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meinke DW, Franzmann LH, Nickle TC, Yeung EC. Leafy Cotyledon Mutants of Arabidopsis. Plant Cell 1994; 6:1049-64; PMID:12244265; http://dx.doi.org/ 10.1105/tpc.6.8.1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P. The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev Cell 2004; 7:373-85; PMID:15363412; http://dx.doi.org/ 10.1016/j.devcel.2004.06.017 [DOI] [PubMed] [Google Scholar]
- 6.Tsai AY, Gazzarrini S. Overlapping and distinct roles of AKIN10 and FUSCA3 in ABA and sugar signaling during seed germination. Plant Signal Behav 2012; 7:1238-42; PMID:22902692; http://dx.doi.org/ 10.4161/psb.21549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nambara E, Hayama R, Tsuchiya Y, Nishimura M, Kawaide H, Kamiya Y, Naito S. The role of ABI3 and FUS3 loci in Arabidopsis thaliana on phase transition from late embryo development to germination. Dev Biol 2000; 220:412-23; PMID:10753527; http://dx.doi.org/ 10.1006/dbio.2000.9632 [DOI] [PubMed] [Google Scholar]
- 8.Curaba J, Moritz T, Blervaque R. AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol 2004; 136:3660-69; PMID:15516508; http://dx.doi.org/ 10.1104/pp.104.047266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsuchiya Y, Nambara E, Naito S, McCourt P. The FUS3 transcription factor functions through the epidermal regulator TTG1 during embryogenesis in Arabidopsis. Plant J 2004; 37:73-81; PMID:14675433; http://dx.doi.org/ 10.1046/j.1365-313X.2003.01939.x [DOI] [PubMed] [Google Scholar]
- 10.Lu QS, Paz JD, Pathmanathan A, Chiu RS, Tsai AY, Gazzarrini S. The C-terminal domain of FUSCA3 negatively regulates mRNA and protein levels, and mediates sensitivity to the hormones abscisic acid and gibberellic acid in Arabidopsis. Plant J 2010; 64:100-13; PMID:20663088; http://dx.doi.org/ 10.1111/j.1365-313X.2010.04307.x [DOI] [PubMed] [Google Scholar]
- 11.Tamura N, Yoshida T, Tanaka A, Sasaki R, Bando A, Toh S, Lepiniec L, Kawakami N. Isolation and characterization of high temperature-resistant germination mutants of Arabidopsis thaliana. Plant Cell Physiol 2006; 47:1081-94; PMID:16816409; http://dx.doi.org/ 10.1093/pcp/pcj078 [DOI] [PubMed] [Google Scholar]
- 12.Toh S, Imamura A, Watanabe A, Nakabayashi K, Okamoto M, Jikumaru Y, Hanada A, Aso Y, Ishiyama K, Tamura N, Iuchi S, Kobayashi M, Yamaguchi S, Kamiya Y, Nambara E, Kawakami N. High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol 2008; 146:1368-85; PMID:18162586; http://dx.doi.org/ 10.1104/pp.107.113738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiu RS, Nahal H, Provart NJ, Gazzarrini S. The role of the Arabidopsis FUSCA3 transcription factor during inhibition of seed germination at high temperature. BMC Plant Biol 2012; 12:15; PMID:22279962; http://dx.doi.org/ 10.1186/1471-2229-12-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiu RS, Pan S, Zhao R, Gazzarrini S. ABA-dependent inhibition of the ubiquitin proteasome system during germination at high temperature in Arabidopsis. Plant J 2016; PMID:27496613; http://dx.doi.org/ 10.1111/tpj.13293 [DOI] [PubMed] [Google Scholar]
- 15.Park S, Fung P, Nishimura N, Jensen DR, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow T, Alfred S, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu JK, Schroeder JI, Volkman BF, Cutler SR. Abscisic acid inhibits PP2Cs via the PYR/PYL family of ABA-binding START proteins. Science 2009; 324:1068-71; PMID: 19407142; http://dx.doi.org/22739828 10.1126/science.1173041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez-Guzman M, Pizzio GA, Antoni R, Vera-Sirera F, Merilo E, Bassel GW, Fernandez MA, Holdsworth MJ, Perez-Amador MA, Kollist H, Rodriguez PL. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 2012; 24:2483-96; PMID:22739828; http://dx.doi.org/ 10.1105/tpc.112.098574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gazzarrini S, Tsai AY. Hormone cross-talk during seed germination. Essays Biochem. 2015; 58:151-64; PMID:26374893; http://dx.doi.org/ 10.1042/bse0580151 [DOI] [PubMed] [Google Scholar]
- 18.Lim S, Park J, Lee N, Jeong J, Toh S, Watanabe A, Kim J, Kang H, Kim DH, Kawakami N, Choi G. ABA-insensitive3, ABA-insensitive5, and DELLAs interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 2013; 25:4863-78; PMID:24326588; http://dx.doi.org/ 10.1105/tpc.113.118604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang F, Perry SE. Identification of Direct Targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol 2013; 161:1251-1264; PMID:23314941; http://dx.doi.org/ 10.1104/pp.112.212282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang BJ, Han XX, Yin LL, Xing MQ, Xu ZH, Xue HW. Arabidopsis PROTEASOME REGULATOR1 is required for auxin-mediated suppression of proteasome activity and regulates auxin signaling. Nat Commun 2016; 7:11388; PMID:27109828; http://dx.doi.org/ 10.1038/ncomms11388 [DOI] [PMC free article] [PubMed] [Google Scholar]


