Abstract
It is well established that antiretroviral therapy (ART), while highly effective in controlling HIV replication, cannot eliminate virus from the body. Therefore, the majority of HIV-1-infected individuals remain at risk for developing AIDS due to persistence of infected reservoir cells serving as a source of virus re-emergence. Several reservoirs containing replication competent HIV-1 have been identified, most notably CD4+ T cells. Cells of the myeloid lineage, which are the first line of defense against pathogens and participate in HIV dissemination into sanctuary organs, also serve as cellular reservoirs of HIV-1. In latently infected resting CD4+ T cells, the integrated copies of proviral DNA remain in a dormant state, yet possess the ability to produce replication competent virus after cellular activation. Studies have demonstrated that modification of chromatin structure plays a role in establishing persistence, in part suggesting that latency is, controlled epigenetically. Current efforts to eradicate HIV-1 from this cell population focus primarily on a “shock and kill” approach through cellular reactivation to trigger elimination of virus producing cells by cytolysis or host immune responses. However, studies revealed several limitations to this approach that require more investigation to assess its clinical application. Recent advances in gene editing technology prompted use of this approach for inactivating integrated proviral DNA in the genome of latently infected cells. This technology, which requires a detailed understanding of the viral genetics and robust delivery, may serve as a powerful strategy to eliminate the latent reservoir in the host leading to a sterile cure of AIDS.
Introduction
Approximately thirty-seven million people are living with HIV-1 infection, and two million new infections were reported in 2014 worldwide [1]. With the introduction of anti-retroviral therapy (ART) that is highly effective in suppressing HIV replication in vivo, there has been a significant reduction in morbidity and mortality associated with this infection [2]. However, to date, only fifteen million people living with HIV have access to ART [1]. Unfortunately, long-term ART also has significant adverse effects such as drug toxicities, incomplete immune reconstitution, and residual immune activation/inflammation that enhance the risk of selected co-morbidities [3–5]. Recent studies have shown a substantial shift in the subtypes of lymphoma observed in HIV-infected patients treated with ART [6]. Also, long-term treatment with ART of HIV-1 infected patients worldwide is unsustainable. Therefore, there is an urgent need for discovering a cure for HIV-1 infection.
The resurgence of HIV-1 in patients, relatively soon after the discontinuation of ART, suggests the presence of long-lived viral reservoir(s) that are resistant to ART [7]. Persistence of HIV-1 in patients under suppressive ART is due to the latent cellular reservoirs in circulation as well as anatomical sanctuaries such as the gut-associated lymphoid tissue (GALT), central nervous system (CNS) and bone marrow hematopoietic progenitor cells [8–14]. Studies thus far have demonstrated that the most important cellular reservoir of latent HIV-1 provirus is the resting CD4+ T cells. Estimates suggest that the latently infected CD4+ T cell reservoir may take as long as 60 years to decay naturally [15]. Furthermore, the doses of ART effective in suppressing peripheral viremia may not be sufficient to target HIV-1 that crosses the blood-brain barrier (BBB), and, therefore, may not prevent the establishment of latency. Thus, the existence of a CNS viral reservoir is envisioned, wherein, HIV-1 can exist in perivascular macrophages, microglia, and astrocytes [9, 10 16]. In addition, recent studies have shown HIV-1 in alveolar macrophages in patients on ART with undetectable plasma viral loads [17]. Thus, the lung is a potential reservoir for the virus. The establishment and maintenance of HIV-1 latency are a combination of multifactorial mechanisms [see reviews, 18–21]. Therefore, purging the viral reservoir will likely require a combination of pharmacological agents with unique mechanisms of action with no adverse associated toxicity to effectively target all latent virus, both in circulation and at various anatomical sites. In summary, the primary challenge in the field is how to purge and kill the latent reservoirs from resting CD4+ T cells and other sanctuaries and thus effectively eliminate infected cells. At the same time, new approaches can be developed using gene editing strategies to eradicate completely the HIV-1 genome from infected cells in whole animal models and then in the clinical setting. Success in this approach, indeed, requires an understanding of the viral genetic variations and robust delivery of the editing materials to latently infected cells to these circulating cells and anatomical reservoirs.
Host factors involved in HIV-1 latency maintenance
Histone deacetylation is one of the predominant mechanisms involved in repression of HIV-1 transcription in the maintenance of HIV-1 latency. Elucidation of the mechanisms involved in histone deacetylases (HDACs) mediated repression of HIV-1 latency demonstrates recruitment of HDAC1, 2 and 3 by cellular transcription factors. Binding of cellular factors such as, c-promoter-binding factor (CBF-1), NF-κB p50 homodimer, Ying-Yang 1 (YY1), late SV40 factor (LSF), COUP-TF-interacting protein (CTIP2), c-myc and Sp1 are among some of the potent transcription factors that recruit HDAC1 to the HIV-1 promoter [22–27]. CTIP2 and Sp1 have also been shown to promote recruitment of HDAC2 [23]. In addition, RBF-2 (USF1/2-TFII-I) promotes HDAC3 binding to HIV-1 LTR [28].
Reversible histone methylation also contributes to the establishment of HIV-1 latency in CD4+ T cells and cells of myeloid lineage. Studies in several cell models and peripheral blood mononuclear cells (PBMC) isolated from HIV-1-infected patients have shown the involvement of histone lysine methyltransferases (HKMT) Suv39H1 and HP1 gamma in tri-methylation of histone H3 lysine (H3K9me3) in silencing of HIV-1 [29]. A similar phenomenon is also seen in HIV-1 infected microglia cells that result in the formation of heterochromatin, and ultimately HIV-1 silencing [30]. G9a, a histone methyltransferase (HMT), has been shown to promote HIV-1 latency in ACH2 and OM-10.1 cells by governing H3K9me2 and formation of repressive chromatin [31]. Earlier studies have demonstrated that HKMT enhancer of zeste homolog 2 (EZH2) is also recruited at the silenced 5’LTR in latently infected Jurkat T-cell lines to increase local H3K9me3 level that is rapidly displaced following proviral reactivation [32].
Current strategies for elimination of latent reservoir “Shock and Kill”
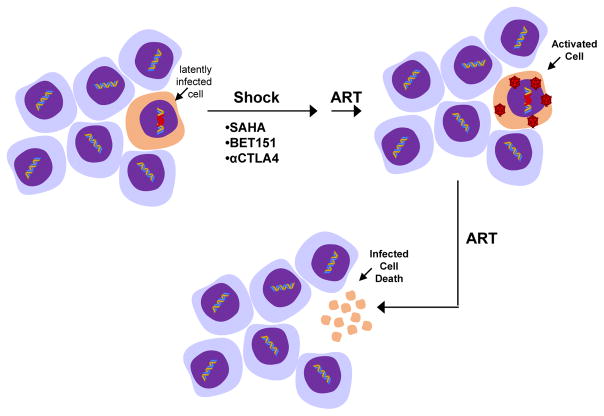
The major barrier to elimination of latent reservoirs of HIV-1 is multifactorial. Latently infected cells are long-lived, immunologically invisible, may undergo homeostatic proliferation, and are refractory to combinatorial antiretroviral therapy (cART). Current pharmacological efforts are designed towards a ‘shock and kill’ [33] approach to eradicate viral reservoirs (Fig. 1). The strategy, in a simplistic view, is to reactivate viral transcription with various latency-reversing agents (LRA) that will result in the death of the productively infected cells by the virus itself, or by the host immune system, and circumvention of new infection by cART. There are several LRA that have been extensively investigated, most notably HDAC inhibitors. In vitro and ex vivo studies have shown that there is significant variability in the potency of HDAC inhibitors (HDACis) in activation of latent reservoirs. Furthermore, there are limited studies on the efficacy of these HDACis in purging the latent HIV-1 reservoir in individuals on cART. Here, we highlight the findings observed with HDAC that belong to four major structural families.
Figure 1.
Shock and kill approach aiming to reactivate latently infected cells by HDAC inhibition including suberoylanilide hydroxamic acid (SAHA), the BET bromodomain protein inhibitor (BET151), and anti-CTLA4 antibody and induce subsequent cell death due to viral toxicity and/or host immune defense while HIV-1 replication is inhibited by ART.
Short-chain aliphatic acids, e.g. Valproic acid (VPA). VPA is an FDA-approved anti-epileptic agent that inhibits class I and II HDACs [34]. It reactivates latent HIV in cell line models of latency that include U1 and J-LAT [35]. VPA also reactivates latent HIV in primary CD4+ T-cell model (ex vivo) of HIV [35]. In a proof-of-concept study, VPA was effective in reducing the frequency of latent infection in three out of four patients on ART [36]. However, VPA failed in reducing latent reservoir size in subsequent clinical trials in HIV-1 infected individuals [37, 38].
Hydroxamic acids, e.g. Trichostatin A (TSA), Suberoylanilide hydroxamic acid (SAHA), and Panobinostat (LBH-589). TSA has been shown to reactivate latent HIV-1 in cell line models U1, ACH2, J49 and OM 10.1, and in CD4 T+ cells isolated from patients on ART [39, 40]. SAHA, an FDA approved HDACi named Vorinostat as been shown to be effective in HIV-1 reactivation in J89, ACH-2, U1 and J-LAT [37, 41, 42] and reactivation of latent HIV-1 in primary CD4+ T cell and CD4 T+ cells isolated from patients on ART [39, 42]. Panobinostat has been shown to induce HIV-1 expression in latently infected cell lines (ACH2 and U1), and in a latent primary CD4+ T-cell infection model with greater potency than other HDAC inhibitors, including givinostat, belinostat, SAHA, and VPA. [39]. Subsequently, in a recent clinical trial Panobinostat was shown to effectively disrupted HIV latency in vivo [43].
Benzamides, e.g. Entinostat. Entinostat is effective in inducing HIV expression in the latently infected cell lines ACH2 and J-Lat as well as in latently infected chemokine ligand 19 (CCL19)-treated primary CD4+T cells. [44].
Cyclic tetrapeptides and depsipeptide, e.g. romidepsin. Romidepsin (RMD) has been shown to be a potent inducer of HIV-1 in an in vitro T-cell model of HIV latency compared with vorinostat and other HDACis in clinical development including panobinostat [45]. Also, RMD induced HIV-1 in both resting and memory CD4+ T cells isolated from HIV-infected patients on suppressive cART [46]. In a recent proof-of-concept phase Ib/IIa trial, RMD significantly reversed HIV-1 latency in vivo without blunting T cell-mediated immune responses [47].
In addition to the HDAC inhibitors, several other agents have been shown to have the ability to reactivate the HIV-1 genome. For example, histone methyltransferase (HKMT) inhibitors such as the Suv39H1 inhibitor chaetocin [48–50], G9a inhibitor BIX01294 [31], and EZH2 inhibitor DZNep [32] have been effective in stimulating latent HIV-1 expression and replication in cell line models or primary CD4+ T-cell model. Another potential reactivation mechanism is through induction of NF-κB, which binds to specific sites on integrated HIV-1 LTR [51] in latently infected cells and thereby promotes transcription. Prostratin, a PKC agonist, is effective in activating latent virus in patient-derived CD4+ T cells (ex vivo), and J-Lat cell lines by inducing NFkB [52, 53]. Similarly, Bryostatin-2, a PKC agonist also reactivates latency in cell line models and CD4+T-cells [54–57]. Indeed, one can predict the universal impact of these LRAs on cellular genes and their toxicity to HIV-1 negative cells. More recently, to achieve HIV-1 targeted specificity for reactivating latent HIV-1, several gene-editing strategies have been modified, by recruiting target specific transcriptional activators such as VP64 [58, 59]
HIV-1 latency and eradication in humanized mouse models
Studies have shown that HIV-1-infected BLT, hu-Rag2−/−γ(c)−/− and NSG mice can develop latent infection as a consequence of virus integration, and is inducible and replication competent [60–66]. These mice models have been used to investigate HIV latency and persistence during therapy, and in the evaluation of pharmacologic agents intended to eliminate the latent reservoir of HIV. Studies have shown that prostatin, a non-tumor-promoting phorbol ester efficiently reactivated HIV expression from latently infected cells generated in the SCID-hu mouse [67]. Using the SCID-hu model, it has been shown that IL-7 induces substantial expression of latent HIV while having minimal effects on the cell phenotype [68]. In a recent study using HIV-1 infected NSG humanized mice with undetected basal levels of viral replication, a dramatic increase in HIV-1 RNA in plasma, lung and brain tissues were observed following ionizing radiation-induced cellular stress [69].
Limitations of LRA and strategies to improve efficacy
SAHA has been used to purge HIV-1 latency both in vitro and in vivo. However, SAHA was shown to increase significantly the susceptibility of CD4+ T cells to infection by HIV-1 in a dose- and time-dependent manner [70]. This was found to be dependent on the efficiency of post entry viral events [70]. A recent study using transcriptomic and proteomic profiling has shown that SAHA modulates the expression of a number of genes and proteins involved in HIV-1 transcriptional regulation, however, some of these genes and proteins appear to be inhibitory with respect to HIV reactivation [71]. In another independent study, SAHA modulated, in a dose-responsive manner, a number of genes in CD4 T cells that could negatively influence HIV reactivation from latency [72].
Among the PKC activators, prostratin and bryostatin are not suitable for use in vivo due to limited availability and major side effects. However, SAHA has been used as a synergistic agent in vitro with prostratin and bryostatin [43, 73]. Therefore, it is essential to identify the counter-regulatory effects of other potential LRAs such as Romidepsin [46] and Panobinostat [42] that have better potencies than SAHA in induction of HIV-1 before advancement into clinical trials specifically for reactivation of latent HIV-1 in CNS and GALT where the availability of therapeutic levels of drugs is a major concern for successful eradication of the latent reservoir.
HIV-1 latency and Cytotoxic T lymphocyte (CTL)
Cytotoxic T lymphocyte (CTL) response contributes to the control of HIV-1 infection in vivo [74, 75]. However, it is unknown whether virus-specific immune mechanisms, including CTLs, can eliminate infected cells in ART-treated patients after reactivation of HIV-1 in latently infected cells. A recent study demonstrated that there is a predominance of CTL-resistant viruses in the latent reservoir that poses a significant challenge in the ‘shock and kill’ approach for viral eradication [76]. A recent study also showed that treatment with HDACis, such as romidepsin and panobinostat, to reactivate the latent reservoir had an adverse impact on CTL-mediated IFN-γ production, and elimination of HIV-infected or peptide-pulsed target cells [77]. More recently, a new strategy that activates and lyses latently infected CD4+ T-cells with HIV-1 has been developed by two independent research teams [78, 79]. This strategy includes the development of a novel bi-specific immunomodulatory protein that combines the broad recognition of HIV-1 Env with binding to the T-cell activation glycoprotein, CD3. Known as Dual-Affinity- Re- Targeting (DARTs) molecules, this therapy seems to be effective at targeting latent and activated cells for killing in ex vivo experiments [79, 80].
Therapeutic approaches for efficient delivery of LRA and ART to anatomical sanctuaries
One of the major hurdles that limit the efficacy of LRAs and ART in the “shock and kill” approach is the penetration and availability of these drugs at therapeutic levels in viral sanctuaries of latency. To circumvent these limitations, micro- to nanoformulated ART referred to as “NanoArt” has been developed [81, 82]. The strategies involved the use of macrophage-based nanoparticle for ART delivery [83, 84], magnetic nanoparticles [85], and folic acid receptor-based nanoparticle [86]. A recent study has shown that a polymer-based pluronic nanocarrier containing anti-HIV drug called efavirenz can efficiently target microfold cells of GALT and inhibit HIV-1 infection [87]. Furthermore, it was also shown that SAHA could be packaged into nanoparticles in conjunction with an antiretroviral drug, tenofovir. The therapeutic potential of this type of nanoparticle has been shown to reactivate and kill HIV-1 across the BBB [82].
Role of genetic variation and quasispecies
While initial infection with HIV has been described as occurring from a small number of transmitter or founder viral strains, genetic diversity of the viral genome occurs rapidly as a consequence of continued replication in multiple cell types with an error prone polymerase (reverse transcriptase), producing numerous and distinct viral quasispecies. These quasispecies accumulate within the patient as the disease progresses, and this accumulation is thought to continue at low-levels even in the presence of suppressive ART. Continued variation has been confirmed in the Drexel Medicine CARES cohort by longitudinal sampling of the LTR (Fig. 2) and by others [88].
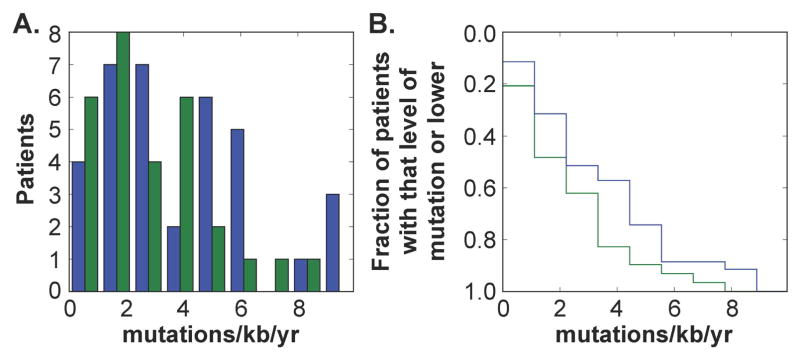
Figure 2.
ART therapy reduces mutation rate of the LTR by an average of 1.1 mutations per Kb per year. A) A histogram of the average mutation rate per patient while naive to therapy (blue) and after virologic control on cART (green). B) A cumulative normalized histogram of the same data in A) showing the fraction of patients with a mutation rate. Fourty two non-drug using patients that currently have undetectable viral load and at least 5 years in the study were selected. from the Drexel Medicine CNS AIDS Research and Eradication Study (CARES) Cohort. The LTR region was PCR amplified from genomic DNA isolated from PBMCs and phylogenetic trees were constructed to estimate the time varying mutation rate of the virus.
Furthermore, the variations within the HIV genome can vary substantially from one tissue or cell type to another, thereby creating compartments. Compartmentalization of HIV has been well established in the literature with compartments described in the brain, cerebrospinal fluid, gut (especially gut associated lymphoid tissue or GALT), lung, liver, and genital tract. The mechanism(s) behind compartmentalization have not been fully elucidated, however, differential pressure from the immune system, cell type differences within these different compartments, accessibility of the compartment to ART, as well as co-infections that may preferentially affect particular compartments may all play a role [89].
Early in HIV infection, the brain is seeded with virus due to increased permeability of the BBB because of a number of factors including increased release of proinflammatory cytokines, exogenous viral proteins including Tat and gp120, infection of perivascular macrophages and microglial cells, and possible infection of brain microvascular endothelial cells and astrocytes. The brain as a viral reservoir with distinct viral genetics is an area that has been heavily researched especially with relation to neurocognitive impairment [reviewed in 90]. Studies have shown that both the brain and the CSF contain distinctly different viral quasispecies as compared to peripheral blood and are therefore distinct compartments [91–97]. Not only were brain sequences found to be distinctly different from those found in the peripheral blood, but also different from those found in other compartments, such as lymphoid tissue [91, 98–100]. In fact, there is even evidence for regional compartmentalization of the virus within the brain of infected individuals based on variable viral replication as measured by viral loads as well as looking at the distinct viral variants [101–104]. Even though it has been shown that the brain contains replication competent, integrated virus and autonomous replication can occur [105], it is unclear what role the brain plays in systemic viral reseeding, although studies utilizing viral sequencing have shown that the meninges harbors virus from both the brain and peripheral tissues suggesting that HIV is capable of migrating out of the brain [78, 90].
In addition to the brain, CSF, and lymphoid tissue discussed above, the GALT, lung [106], genital tract [107–109], and liver [110] have also been shown to act as compartments that differ from what is seen in the peripheral blood. The role that each of these compartments plays in disease course, as well as potential reseeding of the periphery, is variable and results can vary between studies. Gut, for example, has been shown to be an important reservoir and compartment in HIV infection. Initial studies showed that the GALT contained distinct quasispecies in different parts of the gut that differed from what was seen in the peripheral blood [13]. Additional studies also suggested that these reservoirs were maintained even in the presence of long-term ART [111], and the levels of T cell restoration and activation differed between different parts of the gut [112]. However, other studies have suggested that if ART is initiated during primary infection there is an absence of HIV-1 evolution [113]. Although it has been suggested that there is active viral replication to a low level in the presence of active, suppressive ART [114], each of these sites requires further study to determine what role exactly these sites play in HIV compartmentalization and reseeding of the periphery as the disease progresses.
The peripheral blood can also be viewed as its own compartment, and is the most widely studied compartment. Within the peripheral blood, resting memory CD4+ T cells constitute the most abundant latently infected cells [115–117]. The abundance of this reservoir as well as the long half-life of these cells is a major barrier to eradication [118, 119]. It has been assumed that the size of this reservoir can be directly assessed by the ability to reactive the resting cells to again produce infectious virus. However, cellular based assays to assess the size of this reservoir indicate the frequency to be 300-fold lower than the frequency of resting CD4+ T cells that harbor proviruses as detectable by PCR [120]. It has been assumed that the non-induced virus is defective, however, it is unclear if it is truly defective or if it is in fact replication competent [121] and simply not inducible through the systems used. Further molecular characterization of these integrated proviruses is necessary to fully understand the nature of this reservoir to be able to effectively develop an eradication strategy. This has implications for infected cells in both the peripheral blood compartment as well as cells in other anatomical sanctuaries.
While a number of studies have been completed looking at the differences and similarities between different potential anatomical sanctuaries or compartments and the peripheral blood, little work has been completed looking at the differences in genetic variability/quasispecies production, susceptibility to long-term ART, ability to reseed the periphery all within the context of the same model (Fig. 2). It is important to understand the potential variability between not only individual sites and the blood, but also between the sites themselves. Each site represents a different compartment and understanding the differences and similarities in the quasispecies, the effects of ART on these quasispecies, as well as the activity of these species is important in the context of developing a sterilizing or functional cure. To date, a sterilizing cure has only been realized in one patient coined the “Berlin” patient, wherein the patient received an allogeneic hematopoietic stem cell transplant in 2007 and to date all tests indicate that he is HIV-free in the absence of ART [122, 123]. This can be compared to the two “Boston” patients who also received allogeneic hematopoietic stem cell transplants. In these patients, they maintained undetectable viral loads in the presence of ART for 4.3 years and 2.6 years, however when ART was removed both patients experienced viral rebounds at 12 weeks and 32 weeks post therapy removal [124–126]. This suggests that long-lived reservoirs remained in both patients and in the absence of therapy began to reseed the patient. However from the viewpoint of the virus, genetics studies have not been reported that have made any comments as to where the virus may have come from. While it may be very difficult to perform these types of studies in a human, this could be accomplished in the SIV system that was recently published [114] to obtain clues to what reservoirs may reseed the peripheral blood. Therefore, it is important to understand these reservoirs so that an appropriate sterilizing strategy can target these compartments.
Gene editing strategies for eliminating HIV-1
Similar to other retroviruses, after infection, HIV-1 proviral DNA permanently integrates into the genome of its target cells and presents a strong challenge for curing AIDS. Currently, combination ART is effective at reducing viral loads but it doesn’t reduce the source of the infection, which is the stable, integrated provirus in the cellular genome. While the shock and kill strategies discussed above can reactivate and cause eradication of a portion of this virus, largely the inducible, replication competent virus, it has been proven over many studies that it can not reactivate all proviral DNA, including hypermutated and defective deleted forms which are two among many that exists in the latent reservoir, and this in turn is potentially its Achilles heal. In recent years, several novel systems for targeting endogenous genes have been developed including homing endonucleases (HE) or meganucleases, zinc finger nucleases (ZFN), transcription activator-like effector nucleases (TALEN) and most recently clustered regularly interspaced short palindromic repeats (CRISPR)-associated system 9 (Cas9) proteins [127–132], which utilize site-specific, double-strand DNA break (DSB)-mediated DNA repair mechanisms. These DSB-mediated genome editing techniques have enabled genetic studies including targeted gene deletion, insertion, or modification that were previously difficult to perform, and are currently being explored as novel therapeutic approaches. While all three are still being developed, the ZFN and TALEN approaches have limitations with respect to complicated design and size of the molecules necessary to carry out the editing. However, the CRISPR/Cas9 technology, in particular, has captured the attention of many investigators due to its simplicity, precision, and versatility. In addition, the CRISPR/cas9 technology is the only one of the three that has been shown to successfully excise the entire HIV-1 genome from cells. We recently modified this system to enable recognition of specific HIV-1 LTR sequences while excluding sequences that might trigger host off-target effects (Fig. 3). Using this modified system, for the first time, we were able to demonstrate complete excision of a 9.7 kb DNA fragment corresponding to HIV-1 proviral DNA incorporated in various chromosomes of the latently infected cells with no off-target effects [133, 134]. Furthermore, these studies demonstrated that CRISPR/Cas9 could protect uninfected cells against HIV-1 infection, suggesting that this technology can be refined to provide a specific and efficacious prophylactic and therapeutic approach against HIV/AIDS. Several other studies describe utilization of CRISPR and ZFN to introduce InDels and large deletions of the HIV-1 provirus from the host genome [132, 135–137]. TALEN, ZFN and Cas9 have also been used experimentally for efficient disruption of cellular genes that are important for HIV-1 infection including CCR5 and CXCR4 [138–144]. The gene editing strategy for eliminating HIV-1 co-receptors has also entered clinical phase. By using ZFN strategy for targeting CCR5 [141, 145, 146], it was shown that in autologous CD4+ T cells from a small cohort of patients, the viral load remained undetectable at the time of treatment [147]. Given the ease and rapidity of Cas9/gRNA development, it is expected that CRISPR/Cas9 will soon enter clinical trials for the treatment of AIDS. However, there are several important issues that deserve close attention. For example, the CRISPR/Cas9 gene editing strategy utilizes multiplexes of gRNAs that can be designed to be broadly recognized by all HIV-1 isolates that have been characterized to date. However, due to the presence of HIV-1 quasispecies, it is important to determine the most conserved regions of the LTR quasispecies and use this information to develop a personalized therapeutic strategy that effectively eliminates HIV-1 DNA from the patient’s genome. While CD4+ T cells are recognized as a site of latency and serve as a critical reservoir for virus during ART, there are several studies that have ascribed a role for other cell types, including macrophages, astrocytes, brain-derived microglia, as well as lymph nodes of gastric origin, in hosting HIV-1 in a latent state and may therefore serve as additional reservoirs. Another important issue related to the delivery of the gene editing apparatus that is comprised of Cas9 endonuclease and the gRNAs. This can be accomplished by gene therapy approach utilizing a suitable and efficient vector that robustly delivers the genes expressing both Cas9 and gRNAs. In this respect, several viral vectors including lentivirus, adenovirus, and adeno-associated virus have been employed for delivery of Cas9 and gRNAs to various human cell lines [148–150]. However, its employment in the clinic with respect to efficiency, specificity and safety remains to be seen. On the other hand, a broad range of non-viral vehicles including nanomolecules, lipid-based or polymer-based such as polyethylamine and polylysine, which have the ability to carry its cargo at the protein, RNA and DNA levels [for review see 151]. Further, by decorating the surface of the nanoparticles with specific targeting ligands, targeted nanoparticles can be developed for cell type specific delivery of the payload. These issues are of critical importance to precisely target all of the HIV-1 viral quasispecies within an individual.
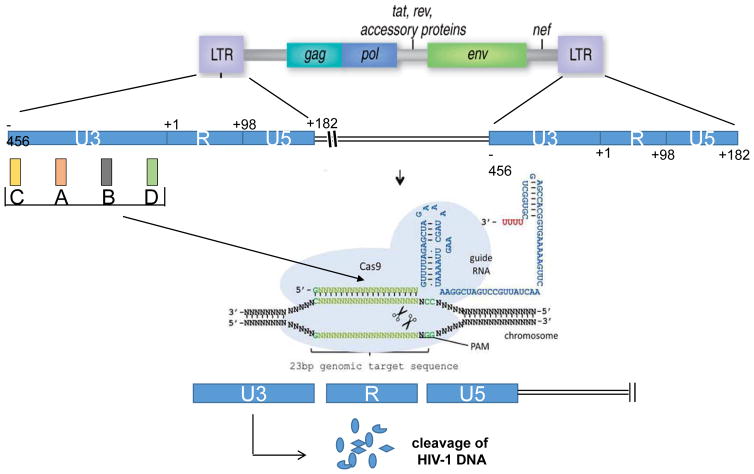
Figure 3.
Strategy for CRISPR/Cas9 mediated cleavage of the HIV-1 genome. Guide RNAs (gRNAs) targeting the U3 region of the LTR can recruit Cas9 to the viral DNA sequence integrated in the host chromosome and results in cleavage of the viral DNA at the designated sites and introduces InDel mutations. Cleavage at both the 5′ and 3′ LTRs can lead to removal of the complete coding region of HIV-1 from the host genome and lead to eradication of HIV-1 from the host [133, 134].
Future considerations
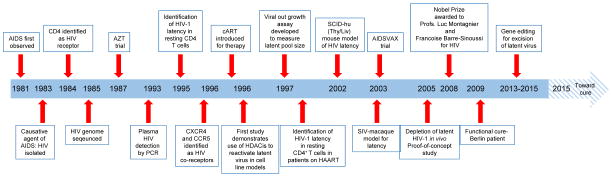
Figure 4 highlights several notable discoveries and events that have occurred since the first cases of AIDS appeared in 1981. After 34 years we have reached the point of being able to identify, characterize and manipulate the integrated proviral DNA in the latently infected cells in the laboratory setting. Clearly, gene editing strategy with its ability to completely eliminate the HIV-1 genome from the latently infected human CD4+ T cells as well as the other cell types harboring HIV-1 proviral DNA has brought new hope that AIDS may be a curable disease. However, there are several challenges that need to be met to assess its efficacy in eliminating HIV-1 prior to implementation in the clinic. While at this stage, it is difficult to predict the extent of gene editing delivery and high efficiency of the viral DNA excision by CRISPR/Cas9 in an in vivo system, one may speculate that under ART protection of a fraction of the cells from HIV-1 infection and death could have dramatic functional consequences in the infected subject, once the ART is interrupted after a certain period of time post repeated application of gene editing molecules. Under these circumstances, it is possible that the protected uninfected cells would be free to become activated, promote CD8+ T-cell and B-cell responses, yet remain immune to virus-induced cellular death and/or dysregulation. In such a scenario, even an incomplete transduction could result in complete eradication assuming the promotion of a significant immunologic effect. Such an effect would be analogous to the Berlin patient’s donor derived cells (CCR5Δ32) being able to clear infected cells within the transplant recipient. Thus, one may envision that eliminating HIV-1 DNA and diminishing the toxicity associated with the presence of the viral genome and expression of the viral proteins, even at very low levels, by gene editing strategy can empower the fragile immune system and collaboratively rid the virus from the remaining infected cells in the patient.
Figure 4.
HIV-1, thirty years of latency: from discovery to excision.
Acknowledgments
The authors wish to thank past and present members of the Department of Neuroscience/Center for Neurovirology at Temple University and the Department of Microbiology and Immunology, Center for Molecular Virology and Translational Neuroscience, Institute for Molecular Medicine and Infectious Disease at Drexel University College of Medicine.
Funding Information
This work was partially supported by P30 MH092177 Comprehensive NeuroAIDS Center (Program Director: Kamel Khalili, Brian Wigdahl, PI of the Drexel subcontract). These studies were also supported by the Public Health Service, National Institutes of Health, through grants from the National Institute of Neurological Disorders and Stroke, NS32092 and NS46263, the National Institute of Drug Abuse, DA19807 (Dr. Brian Wigdahl, Principal Investigator), and the National Institute of Mental Health under the Ruth L. Kirschstein National Research Service Award 5T32MH079785 (Jay Rappaport, PI, Brian Wigdahl, PI of the Drexel subcontract). Dr. Prasun K. Datta was supported in part by the Public Health Service, National Institutes of Health, through grants from National Institute of Drug Abuse, 5R01DA033213 and 1P01DA037830-01A1. Dr. Michael Nonnemacher was supported in part by the Public Health Service, National Institutes of Health, through grants from the National Institute of Neurological Disorders and Stroke, NS089435 and faculty development funds provided by the Department of Microbiology and Immunology and the Institute for Molecular Medicine and Infectious Disease. The contents of the paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Conflict of Interests
The authors have no conflict of interest to declare
References
- 1.UNAIDS. 2015 http://www.unaids.org/sites/default/files/media_asset/AIDS_by_the_numbers_2015_en.pdf/
- 2.Lewden C, Chene G, Morlat P, et al. HIV-infected adults with a CD4 cell count greater than 500 cells/mm3 on long-term combination antiretroviral therapy reach same mortality rates as the general population. J Acquir Immune Defic Syndr. 2007;46:72–7. doi: 10.1097/QAI.0b013e318134257a. [DOI] [PubMed] [Google Scholar]
- 3.Anzinger JJ, Butterfield TR, Angelovich TA, Crowe SM, Palmer CS. Monocytes as regulators of inflammation and HIV-related comorbidities during cART. J Immunol Res. 2014;2014:569819. doi: 10.1155/2014/569819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hearps AC, Martin GE, Rajasuriar R, et al. Inflammatory co-morbidities in HIV+ individuals: learning lessons from healthy ageing. Curr HIV/AIDS Rep. 2014;11:20–34. doi: 10.1007/s11904-013-0190-8. [DOI] [PubMed] [Google Scholar]
- 5.Samaras K. Prevalence and pathogenesis of diabetes mellitus in HIV-1 infection treated with combined antiretroviral therapy. J Acquir Immune Defic Syndr. 2009 Apr 15;50(5):499–505. doi: 10.1097/QAI.0b013e31819c291b. [DOI] [PubMed] [Google Scholar]
- 6.Carroll V, Garzino-Demo A. HIV-associated lymphoma in the era of combination antiretroviral therapy: shifting the immunological landscape. Pathog Dis. 2015;73 doi: 10.1093/femspd/ftv044. pii: ftv044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim SH, Gerver SM, Fidler S, et al. Adherence to antiretroviral therapy in adolescents living with HIV: systematic review and meta-analysis. AIDS. 2014;28:1945–1956. doi: 10.1097/QAD.0000000000000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter CC, Onafuwa-Nuga A, McNamara LA, et al. HIV-1 infects multipotent progenitor cells causing cell death and establishing latent cellular reservoirs. Nat Med. 2010;16:446–451. doi: 10.1038/nm.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Churchill MJ, Wesselingh SL, Cowley D, et al. Extensive astrocyte infection is prominent in human immunodeficiency virus-associated dementia. Ann Neurol. 2009;66:253–258. doi: 10.1002/ana.21697. [DOI] [PubMed] [Google Scholar]
- 10.Clements JE, Gama L, Graham DR, et al. A simian immunodeficiency virus macaque model of highly active antiretroviral treatment: viral latency in the periphery and the central nervous system. Curr Opin HIV AIDS. 2011;6:37–42. doi: 10.1097/COH.0b013e3283412413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisele E, Siliciano RF. Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity. 2012;37:377–388. doi: 10.1016/j.immuni.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothenberger MK, Keele BF, Wietgrefe SW, et al. Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proc Natl Acad Sci U S A. 2015;112:E1126–34. doi: 10.1073/pnas.1414926112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Marle G, Gill MJ, Kolodka D, et al. Compartmentalization of the gut viral reservoir in HIV-1 infected patients. Retrovirology. 2007;4(4):87. doi: 10.1186/1742-4690-4-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wightman F, Solomon A, Khoury G, et al. Both CD31(+) and CD31− naive CD4(+) T cells are persistent HIV type 1-infected reservoirs in individuals receiving antiretroviral therapy. J Infect Dis. 2010;202:1738–1748. doi: 10.1086/656721. [DOI] [PubMed] [Google Scholar]
- 15.Finzi D, Blankson J, Siliciano JD, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 16.Alexaki A, Liu Y, Wigdahl B. Cellular reservoirs of HIV-1 and their role in viral persistence. Curr HIV Res. 2008;6:388–400. doi: 10.2174/157016208785861195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cribbs SK, Lennox J, Caliendo AM, et al. Healthy HIV-1-infected individuals on highly active antiretroviral therapy harbor HIV-1 in their alveolar macrophages. AIDS Res Hum Retroviruses. 2015;31:64–70. doi: 10.1089/aid.2014.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bruner KM, Hosmane NN, Siliciano RF. Towards an HIV-1 cure: measuring the latent reservoir. Trends Microbiol. 2015;23:192–203. doi: 10.1016/j.tim.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dahabieh MS, Battivelli E, Verdin E. Understanding HIV latency: the road to an HIV cure. Annu Rev Med. 2015;66:407–21. doi: 10.1146/annurev-med-092112-152941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar A, Darcis G, Van Lint C, et al. Epigenetic control of HIV-1 post integration latency: implications for therapy. Clin Epigenetics. 2015;7:103. doi: 10.1186/s13148-015-0137-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mbonye U, Karn J. Transcriptional control of HIV latency: cellular signaling pathways, epigenetics, happenstance and the hope for a cure. Virology. 2014 Apr;454–455:328–39. doi: 10.1016/j.virol.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang G, Espeseth A, Hazuda DJ, et al. c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase 1 to the human immunodeficiency virus type 1 promoter. J Virol. 2007;81:10914–10923. doi: 10.1128/JVI.01208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marban C, Suzanne S, Dequiedt F, et al. Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J. 2007;26:412–423. doi: 10.1038/sj.emboj.7601516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margolis DM, Somasundaran M, Green MR. Human transcription factor YY1 represses human immunodeficiency virus type 1 transcription and virion production. J Virol. 1994;68:905–910. doi: 10.1128/jvi.68.2.905-910.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romerio F, Gabriel MN, Margolis DM. Repression of human immunodeficiency virus type 1 through the novel cooperation of human factors YY1 and LSF. J Virol. 1997;71:9375–9382. doi: 10.1128/jvi.71.12.9375-9382.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tyagi M, Karn J. CBF-1 promotes transcriptional silencing during the establishment of HIV-1. EMBO J. 2007;26:4985–4995. doi: 10.1038/sj.emboj.7601928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams SA, Chen LF, Kwon H, et al. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 2006;25:139–149. doi: 10.1038/sj.emboj.7600900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malcolm T, Chen J, Chang C, et al. Induction of chromosomally integrated HIV-1 LTR requires RBF-2 (USF/TFII-I) and Ras/MAPK signaling. Virus Genes. 2007;35:215–223. doi: 10.1007/s11262-007-0109-9. [DOI] [PubMed] [Google Scholar]
- 29.du Chéné I, Basyuk E, Lin YL, et al. Suv39H1 and HP1 gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J. 2007;26:424–435. doi: 10.1038/sj.emboj.7601517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marban C, Redel L, Suzanne S, et al. COUP-TF interacting protein 2 represses the initial phase of HIV-1 gene transcription in human microglial cells. Nucl Acids Res. 2005;33:2318–2331. doi: 10.1093/nar/gki529. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Imai K, Togami H, Okamoto T. Involvement of histone H3 lysine 9 (H3K9) methyltransferase G9a in the maintenance of HIV-1 latency and its reactivation by BIX01294. J Biol Chem. 2010;285:16538–16545. doi: 10.1074/jbc.M110.103531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friedman J, Cho WK, Chu CK, et al. Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of Zeste 2. J Virol. 2011;85:9078–9089. doi: 10.1128/JVI.00836-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamer DH. Can HIV be Cured? Mechanisms of HIV persistence and strategies to combat it. Curr HIV Res. 2004;2:99–111. doi: 10.2174/1570162043484915. [DOI] [PubMed] [Google Scholar]
- 34.Gottlicher M, Minucci S, Zhu P, et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20:6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reuse S, Calao M, Kabeya K, et al. Synergistic activation of HIV-1 expression by deacetylase inhibitors and prostratin: implications for treatment of latent infection. PLoS One. 2009;4:e6093. doi: 10.1371/journal.pone.0006093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehrman G, Hogue IB, Palmer S, et al. Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet. 2005;366:549–555. doi: 10.1016/S0140-6736(05)67098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Archin NM, Eron JJ, Palmer S, et al. Valproic acid without intensified antiviral therapy has limited impact on persistent HIV infection of resting CD4+ T cells. AIDS. 2008;22:1131–1135. doi: 10.1097/QAD.0b013e3282fd6df4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Routy JP, Tremblay CL, Angel JB, et al. Valproic acid in association with highly active antiretroviral therapy for reducing systemic HIV-1 reservoirs: results from a multicentre randomized clinical study. HIV Med. 2012;13:291–296. doi: 10.1111/j.1468-1293.2011.00975.x. [DOI] [PubMed] [Google Scholar]
- 39.Lu HK, Gray LR, Wightman F, et al. Ex vivo response to histone deacetylase (HDAC) inhibitors of the HIV long terminal repeat (LTR) derived from HIV-infected patients on antiretroviral therapy. PLoS One. 2014;9:e113341. doi: 10.1371/journal.pone.0113341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Lint C, Emiliani S, Ott M, Verdin E. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 1996;15:1112–1120. [PMC free article] [PubMed] [Google Scholar]
- 41.Contreras X, Schweneker M, Chen CS, et al. Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J Biol Chem. 2009;284:6782–6789. doi: 10.1074/jbc.M807898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rasmussen TA, Schmeltz Søgaard O, et al. Comparison of HDAC inhibitors in clinical development: effect on HIV production in latently infected cells and T-cell activation. Hum Vaccin Immunother. 2013;9:993–1001. doi: 10.4161/hv.23800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rasmussen TA, Tolstrup M, Brinkmann CR, et al. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV. 2014;1:e13–21. doi: 10.1016/S2352-3018(14)70014-1. [DOI] [PubMed] [Google Scholar]
- 44.Wightman F, Lu HK, Solomon AE, et al. Entinostat is a histone deacetylase inhibitor selective for class 1 histone deacetylases and activates HIV production from latently infected primary T cells. AIDS. 2013;27:2853–2862. doi: 10.1097/QAD.0000000000000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mejia EJ, Loveridge ST, Stepan G, et al. Study of marine natural products including resorcyclic acid lactones from Humicola fuscoatra that reactivate latent HIV-1 expression in an in vitro model of central memory CD4+ T cells. J Nat Prod. 2014;77:618–624. doi: 10.1021/np400889x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei DG, Chiang V, Fyne E, et al. Histone deacetylase inhibitor romidepsin induces HIV expression in CD4 T cells from patients on suppressive antiretroviral therapy at concentrations achieved by clinical dosing. PLoS Pathog. 2014;10:e1004071. doi: 10.1371/journal.ppat.1004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Søgaard OS, Graversen ME, Leth S, et al. The depsipeptide romidepsin reverses HIV-1 latency in vivo. PLoS Pathog. 2015;11:e1005142. doi: 10.1371/journal.ppat.1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bernhard W, Barreto K, Saunders A, Dahabieh MS, Johnson P, Sadowski I. The Suv39H1 methyltransferase inhibitor chaetocin causes induction of integrated HIV-1 without producing a T cell response. FEBS Lett. 2011;585:3549–3554. doi: 10.1016/j.febslet.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 49.Bouchat S, Gatot JS, Kabeya K, et al. Histone methyltransferase inhibitors induce HIV-1 recovery in resting CD4(+) T cells from HIV-1-infected HAART-treated patients. AIDS. 2012;26:1473–1482. doi: 10.1097/QAD.0b013e32835535f5. [DOI] [PubMed] [Google Scholar]
- 50.Narasipura SD, Kim S, Al-Harthi L. Epigenetic regulation of HIV-1 latency in astrocytes. J Virol. 2014;88:3031–3038. doi: 10.1128/JVI.03333-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karn J, Stoltzfus CM. Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb Perspect Med. 2012;2:a006916. doi: 10.1101/cshperspect.a006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kulkosky J, Sullivan J, Xu Y, et al. Expression of latent HAART-persistent HIV type 1 induced by novel cellular activating agents. AIDS Res Hum Retroviruses. 2004;20:497–505. doi: 10.1089/088922204323087741. [DOI] [PubMed] [Google Scholar]
- 53.Williams SA, Chen LF, Kwon H, et al. Prostratin antagonizes HIV latency by activating NF-kappaB. J Biol Chem. 2004;279:42008–420017. doi: 10.1074/jbc.M402124200. [DOI] [PubMed] [Google Scholar]
- 54.Darcis G, Kula A, Bouchat S, et al. An in-depth comparison of latency-reversing agent combinations in various in vitro and Ex vivo HIV-1 latency models identified bryostatin-1 + JQ1 and ingenol-B + JQ1 to potently reactivate viral gene expression. PLoS Pathog. 2015;11:e1005063. doi: 10.1371/journal.ppat.1005063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Díaz L, Martínez-Bonet M, Sánchez J, et al. Bryostatin activates HIV-1 latent expression in human astrocytes through a PKC and NF-κB-dependent mechanism. Sci Rep. 2015;5:12442. doi: 10.1038/srep12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kovochich M, Marsden MD, Zack JA. Activation of latent HIV using drug loaded nanoparticles. PLoS One. 2011;6:e18270. doi: 10.1371/journal.pone.0018270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pérez M, de Vinuesa AG, Sanchez-Duffhues G, et al. Bryostatin-1 synergizes with histone deacetylase inhibitors to reactivate HIV-1 from latency. Curr HIV Res. 2010;8:418–429. doi: 10.2174/157016210793499312. [DOI] [PubMed] [Google Scholar]
- 58.Wang X, Wang P, Fu Z, et al. Designed transcription activator-like effector proteins efficiently induced the expression of latent HIV-1 in latently infected cells. AIDS Res Hum Retroviruses. 2015;31:98–106. doi: 10.1089/aid.2014.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, Chaoran Y, Zhang T, et al. CRISPR/gRNA directed synergistic activaiton mediator (SAM) induces specific, persistent and robust reactivation of the HIV-1 latent reservoir. Sci Rep. 2015 doi: 10.1038/srep16277. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berges BK, Akkina SR, Remling L, Akkina R. Humanized Rag2(−/−)gammac(−/−) (RAG-hu) mice can sustain long-term chronic HIV-1 infection lasting more than a year. Virology. 2010;397:100–103. doi: 10.1016/j.virol.2009.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Choudhary SK, Archin NM, Cheema M, et al. Latent HIV-1 infection of resting CD4+ T cells in the humanized Rag2−/− γc−/− mouse. J Virol. 2012;86:114–120. doi: 10.1128/JVI.05590-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Denton PW, Olesen R, Choudhary SK, et al. Generation of HIV latency in humanized BLT mice. J Virol. 2012;86:630–634. doi: 10.1128/JVI.06120-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duyne RV, Narayanan A, K-Hall K, Saifuddin M, Shultz L, Kashanchi F. Humanized mouse models of HIV-1 latency. Curr HIV Res. 2011;9:595–605. doi: 10.2174/157016211798998781. Review. [DOI] [PubMed] [Google Scholar]
- 64.Honeycutt JB, Wahl A, Archin N, et al. HIV-1 infection, response to treatment and establishment of viral latency in a novel humanized T cell-only mouse (TOM) model. Retrovirology. 2013;10:121. doi: 10.1186/1742-4690-10-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marsden MD, Kovochich M, Suree N, et al. HIV latency in the humanized BLT mouse. J Virol. 2012;86:339–347. doi: 10.1128/JVI.06366-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nischang M, Sutmuller R, Gers-Huber G, et al. Humanized mice recapitulate key features of HIV-1 infection: a novel concept using long-acting anti-retroviral drugs for treating HIV-1. PLoS One. 2012;7:e38853. doi: 10.1371/journal.pone.0038853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Korin YD, Brooks DG, Brown S, et al. Effects of prostratin on T-cell activation and human immunodeficiency virus latency. J Virol. 2002;76:8118–8123. doi: 10.1128/JVI.76.16.8118-8123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scripture-Adams DD, Brooks DG, Korin YD, et al. Interleukin-7 induces expression of latent human immunodeficiency virus type 1 with minimal effects on T-cell phenotype. J Virol. 2002;76:13077–13082. doi: 10.1128/JVI.76.24.13077-13082.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iordanskiy S, Van Duyne R, Sampey GC, et al. Therapeutic doses of irradiation activate viral transcription and induce apoptosis in HIV-1 infected cells. Virology. 2015;485:1–15. doi: 10.1016/j.virol.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lucera MB, Tilton CA, Mao H, et al. The histone deacetylase inhibitor vorinostat (SAHA) increases the susceptibility of uninfected CD4+ T cells to HIV by increasing the kinetics and efficiency of postentry viral events. J Virol. 2014;88:10803–10812. doi: 10.1128/JVI.00320-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.White CH, Johnston HE, Moesker B, et al. Mixed effects of suberoylanilide hydroxamic acid (SAHA) on the host transcriptome and proteome and their implications for HIV reactivation from latency. Antiviral Res. 2015;123:78–85. doi: 10.1016/j.antiviral.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reardon B, Beliakova-Bethell N, Spina CA, et al. Dose-responsive gene expression in suberoylanilide hydroxamic acid-treated resting CD4+ T cells. AIDS. 2015;29:2235–2244. doi: 10.1097/QAD.0000000000000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laird GM, Bullen CK, Rosenbloom DI, et al. Ex vivo analysis identifies effective HIV-1 latency-reversing drug combinations. J Clin Invest. 2015;1(25):1901–1912. doi: 10.1172/JCI80142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jin X, Bauer DE, Tuttleton SE, et al. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kalams SA, Goulder PJ, Shea AK, et al. Levels of human immunodeficiency virus type 1-specific cytotoxic T-lymphocyte effector and memory responses decline after suppression of viremia with highly active antiretroviral therapy. J Virol. 1999;73:6721–6728. doi: 10.1128/jvi.73.8.6721-6728.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Deng K, Pertea M, Rongvaux A, et al. Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature. 2015;517:381–385. doi: 10.1038/nature14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jones RB, O’Connor R, Mueller S, et al. Histone deacetylase inhibitors impair the elimination of HIV-infected cells by cytotoxic T-lymphocytes. PLoS Pathog. 2014;10:e1004287. doi: 10.1371/journal.ppat.1004287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pegu A, Mangaiarkarasi A, Lan W, et al. Activation and lysis of human CD4 cells latently infected with HIV-1. Nat Commun 2015; Nat Commun. 2015;6:8447. doi: 10.1038/ncomms9447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sung JA, Pickeral J, Liu L, et al. Dual-Affinity Re-Targeting proteins direct T cell-mediated cytolysis of latently HIV-infected cells. J Clin Invest. 2015;125:4077–4090. doi: 10.1172/JCI82314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sloan DD, Lam CY, Irrinki A, et al. Targeting HIV Reservoir in Infected CD4 T Cells by Dual-Affinity Re-targeting Molecules (DARTs) that Bind HIV Envelope and Recruit Cytotoxic T Cells. PLoS Pathog. 2015;11:e1005233. doi: 10.1371/journal.ppat.1005233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nowacek A, Gendelman HE. NanoART, neuroAIDS and CNS drug delivery. Nanomedicine (Lond) 2009;4:557–574. doi: 10.2217/nnm.09.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jayant RD, Atluri VS, Agudelo M, et al. Sustained-release nanoART formulation for the treatment of neuroAIDS. Int J Nanomedicine. 2015;10:1077–1093. doi: 10.2147/IJN.S76517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dou H, Destache CJ, Morehead JR, et al. Development of a macrophage-based nanoparticle platform for antiretroviral drug delivery. Blood. 2006;108:2827–2835. doi: 10.1182/blood-2006-03-012534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nowacek AS, Miller RL, McMillan J, et al. NanoART synthesis, characterization, uptake, release and toxicology for human monocyte-macrophage drug delivery. Nanomedicine (Lond) 2009;4:903–917. doi: 10.2217/nnm.09.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saiyed ZM, Gandhi NH, Nair MP. AZT 5′-triphosphate nanoformulation suppresses human immunodeficiency virus type 1 replication in peripheral blood mononuclear cells. J Neurovirol. 2009;15:343–347. doi: 10.1080/13550280903062813. [DOI] [PubMed] [Google Scholar]
- 86.Puligujja P, Araínga M, Dash P, et al. Pharmacodynamics of folic acid receptor targeted antiretroviral nanotherapy in HIV-1-infected humanized mice. Antiviral Res. 2015;120:85–88. doi: 10.1016/j.antiviral.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Roy U, Ding H, Pilakka-Kanthikeel S, et al. Preparation and characterization of anti-HIV nanodrug targeted to microfold cell of gut-associated lymphoid tissue. Int J Nanomedicine. 2015;10:5819–5835. doi: 10.2147/IJN.S68348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Josefsson L, von Stockenstrom S, Faria NR, et al. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc Natl Acad Sci USA. 2013;110:E4987–4996. doi: 10.1073/pnas.1308313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Blackard JT. HIV compartmentalization: a review on a clinically important phenomenon. Curr HIV Res. 2012;10:133–142. doi: 10.2174/157016212799937245. [DOI] [PubMed] [Google Scholar]
- 90.Nath A. Eradication of human immunodeficiency virus from brain reservoirs. J Neurovirol. 2015;21:227–234. doi: 10.1007/s13365-014-0291-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Agopian K, Wei BL, Garcia JV, et al. CD4 and MHC-I downregulation are conserved in primary HIV-1 and Nef alleles from brain and lymphoid tissues, but Pak2 activation is highly variable. Virology. 2007;358:119–135. doi: 10.1016/j.virol.2006.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dunfee RL, Thomas ER, Gorry PR, et al. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc Natl Acad Sci USA. 2006;103:15160–15165. doi: 10.1073/pnas.0605513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gray LR, Gabuzda D, Cowley D, et al. CD4 and MHC class 1 down-modulation activities of nef alleles from brain- and lymphoid tissue-derived primary HIV-1 isolates. J Neurovirol. 2011;17:82–91. doi: 10.1007/s13365-010-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stam AJ, Nihhuis M, van den Bergh WM, et al. Differential genotypic evolution of HIV-1 quasispecies in cerebrospinal fluid and plasma: a systematic review. AIDS Rev. 2013;15:152–161. [PubMed] [Google Scholar]
- 95.Strain MC, Letendre S, Pillai SK, et al. Genetic composition of human immunodeficiency virus type 1 in cerebrospinal fluid and blood without treatment and during failing antiretroviral therapy. J Virol. 2005;79:1772–1788. doi: 10.1128/JVI.79.3.1772-1788.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sturdevant CB, Joseph SB, Schnell G, et al. Comparmentalized replication of R5 T cell-tropic HIV-1 in the central nervous system early in the course of infection. PLoS Pathog. 2015;11:e1004720. doi: 10.1371/journal.ppat.1004720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Thompson KA, Churchill MJ, Gorry PR, et al. Astrocyte specific viral strains in HIV dementia. Ann Neurol. 2004;56:873–877. doi: 10.1002/ana.20304. [DOI] [PubMed] [Google Scholar]
- 98.Chang J, Jozwiak R, Wang B, et al. Unique HIV type 1 V3 region sequences derived from six different regions of brain: region-specific evolution within host-determined quasispecies. AIDS Res Hum Retroviruses. 1998;14:25–30. doi: 10.1089/aid.1998.14.25. [DOI] [PubMed] [Google Scholar]
- 99.Olivieri KC, Agopian KA, Mukerji J, et al. Evidence for adaptive evolution at the divergence between lymphoid and brain HIV-1 nef genes. AIDS Res Hum Retroviruses. 2010;26:495–500. doi: 10.1089/aid.2009.0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thomas ER, Dunfee RL, Stanton J, et al. High frequency of defective vpu compared with tat and rev gene in brain from patients with HIV type 1-associated dementia. AIDS Res Hum Retroviruses. 23:575–580. doi: 10.1089/aid.2006.0246. [DOI] [PubMed] [Google Scholar]
- 101.Fujimura RK, Goodkin K, Petito CK, et al. HIV-1 proviral DNA load across neuranatomic regions of individuals with evidence for HIV-1 associated dementia. J Acquir Immune Defic Syndr Hum. 1997;16:146–152. doi: 10.1097/00042560-199711010-00002. [DOI] [PubMed] [Google Scholar]
- 102.Lamers SL, Gray RR, Salemi M, et al. HIV-1 phylogenetic analysis shows HIV-1 transits through meninges to brain and peripheral tissues. Infect Genet Evol. 2011;11:31–37. doi: 10.1016/j.meegid.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shapshak P, Segal DM, Crandall KA, et al. Independent evolution of HIV type 1 in different brain regions. AIDS Res Hum Retroviruses. 1999;15:811–820. doi: 10.1089/088922299310719. [DOI] [PubMed] [Google Scholar]
- 104.Wiley CA, Soontornniyomkij V, Radhakrishnan L, et al. Distribution of brain HIV load in AIDS. Brain Pathol. 1998;8:277–284. doi: 10.1111/j.1750-3639.1998.tb00153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hellmuth J, Valcour V, Spudich S. CNS reservoirs for HIV: implications for eradication. J Virus Erad. 2015;1:67–71. [PMC free article] [PubMed] [Google Scholar]
- 106.Heath L, Fox A, McClure J, et al. Evidence for limited genetic comparmentalization of HIV-1 between lung and blood. PloS One. 2009;4:e6949. doi: 10.1371/journal.pone.0006949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bull M, Learn G, Genowati I, et al. Compartmentalization of HIV-1 within the female genital tract is due to monotypic and low-diverisity variants not distinct viral populations. PLoS One. 2009;4:e7122. doi: 10.1371/journal.pone.0007122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Miller CJ. Localization of Simian immunodeficiency virus-infected cells in the genital tract of male and female Rhesus macaques. J Reprod Immunol. 1998;41:331–339. doi: 10.1016/s0165-0378(98)00069-2. [DOI] [PubMed] [Google Scholar]
- 109.Philpott S, Burger H, Tsoukas C, et al. Human immunodeficiency virus type 1 genomic RNA sequences in the female genital tract and blood: compartmentalization and intrapatient recombination. J Virol. 2005;79:353–363. doi: 10.1128/JVI.79.1.353-363.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Penton PK, Blackard JT. Analysis of HIV quasispecies suggests comparmentalization in the liver. AIDS Res Hum Retroviruses. 2014;30:394–402. doi: 10.1089/aid.2013.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chun TW, Nickle DC, Justement JS, et al. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J Infect Dis. 2008;197:714–720. doi: 10.1086/527324. [DOI] [PubMed] [Google Scholar]
- 112.Yukl SA, Gianella S, Sinclair E, et al. Differences in HIV burden and immune activation within the gut of HIV-positive patients receiving suppressive antiretroviral therapy. J Infect Dis. 2010;202:1553–1561. doi: 10.1086/656722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Evering TH, Mehandru S, Racz P, et al. Absence of HIV-1 evolution in the gut-associated lympohid tissue from patients on combination antiviral therapy initiated during primary infection. PLoS Pathog. 2012;8:e1002506. doi: 10.1371/journal.ppat.1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Santangelo PJ, Rogers KA, Zurla C, et al. Whole-body immunoPET reveals active SIV dynamics in viremic and antiretroviral therapy-treated macaques. Nat Methods. 2015;12:427–432. doi: 10.1038/nmeth.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chun TW, Carruth L, Finzi D, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 116.Finzi D, Hermankova M, Pierson T, et al. Identification of a reservoir for HIV-1 in patients in highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 117.Piatek M, Jr, Saag MS, Yang LC, et al. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science. 1993;259:1749–1754. doi: 10.1126/science.8096089. [DOI] [PubMed] [Google Scholar]
- 118.Ramratnam B, Mittler JE, Zhang L, et al. The decay of latent reservoir of replication-competent HIV-1 is inversely correlated with the extent of residual viral replication during prolonged anti-retroviral therapy. Nat Med. 2000;6:82–85. doi: 10.1038/71577. [DOI] [PubMed] [Google Scholar]
- 119.Siliciano JD, Kajdas J, Finzi D, et al. Long-term follow up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 120.Eriksson S, Graf EH, Dahl V, et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 2013;9:e1003174. doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ho YC, Shan L, Hosmane NN, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Allers K, Hutter G, Hofmann J, et al. Evidence for the cure of HIV infection by CCRΔ32/Δ32 stem cell transplantation. Blood. 2011;117:2791–2799. doi: 10.1182/blood-2010-09-309591. [DOI] [PubMed] [Google Scholar]
- 123.Hütter G, Nowak D, Mossner M, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 124.Henrich TJ, Gandhi RT. Early treatment and HIV-1 reservoirs: a stitch in time? J Infect Dis. 2013a;208:1189–1193. doi: 10.1093/infdis/jit307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Henrich TJ, Hu Z, Li JZ, et al. Long-term reduction in peripheral blood HIV type 1 reservoirs following reduced-intensity conditioning allogeneic stem cell transplantation. J Infect Dis. 2013b;207:1694–1702. doi: 10.1093/infdis/jit086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Henrich TJ, Hanhauser E, Marty FM, et al. Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann Intern Med. 2014;161:319–327. doi: 10.7326/M14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bedell VM, Wang Y, Campbell JM, et al. In vivo genome editing usign a high efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mali P, Aach J, Stranges PB, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013a;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013b;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Qu X, Wang P, Ding D, et al. Zinc finger-nucleases mediate specific and efficient excision of HIV-1 proviral DNA from infected and latently infected human T cells. Nucleic Acids Res. 2013;41:7771–7782. doi: 10.1093/nar/gkt571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hu W, Kaminski R, Yang F, et al. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc Natl Acad Sci USA. 2014;111:11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Khalili K, Kaminski R, Gordon J, et al. Genome editing strategies: potential tools for eradicating HIV-1/AIDS. J Neurovirol. 2015;21:310–321. doi: 10.1007/s13365-014-0308-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ebina H, Misawa N, Kanemura Y, et al. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liao H-K, Gu Y, Diaz A, et al. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nature Commun. 2015;6:6413. doi: 10.1038/ncomms7413. [DOI] [PubMed] [Google Scholar]
- 137.Wayengera M. Proviral HIV-genome-wide and pol-gene specific zinc finger nucleases: usability for targeted HIV gene therapy. Theor Biol Med Model. 2011;8:26. doi: 10.1186/1742-4682-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nature Biotech. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 139.Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucl Acids Res. 2013;41:9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liu J, Gaj T, Patterson JT, et al. 3rd Cell-penetrating peptide-mediated delivery of TALEN proteins via bioconjugation for genome engineering. PLoS One. 2014;9:e85755. doi: 10.1371/journal.pone.0085755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Manjunath N, Yi G, Dang Y, et al. Newer gene editing technologies toward HIV gene therapy. Viruses. 2013;5:2748–2766. doi: 10.3390/v5112748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Manotham K, Chattong S, Setpakdee A. Generation of CCR5-defective CD34 cells from ZFN-driven stop codon-integrated mesenchymal stem cell clones. J Biomed Sci. 2015;22:25. doi: 10.1186/s12929-015-0130-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ye L, Wang J, Beyer AI, et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers resistance to HIV infection. Proc Natl Acad Sci U S A. 2014;111:9591–9596. doi: 10.1073/pnas.1407473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Didigu CA, Wilen CB, Wang J, et al. Simultaneous zinc-finger nuclease editing of the HIV coreceptors ccr5 and cxcr4 protects CD4+ T cells from HIV-1 infection. Blood. 2014;123:61–69. doi: 10.1182/blood-2013-08-521229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hofer U, Henley JE, Exline CM, et al. Pre-clinical modeling of CCR5 knockout in human hematopoietic stem cells by zinc finger nucleases using humanized mice. J Infect Dis. 2013;208(Suppl 2):S160–164. doi: 10.1093/infdis/jit382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Stone D, Kiem HP, Jerome KR. Targeted gene disruption to cure HIV. Curr Op HIV AIDS. 2013;8:217–223. doi: 10.1097/COH.0b013e32835f736c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tebas P, Stein D, Tang WW, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. New Engl J Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Koike-Yusa H, Li Y, Tan EP, et al. Genome wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 149.Maggio I, Holkers M, Liu J, et al. Adenoviral vector delivery of RNA-guided CRISPR/Cas9 nuclease complexes induces targeted mutagenesis in a divese array of human cells. Sci Rep. 2014;4:5105. doi: 10.1038/srep05105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Senis E, Fatouros C, Groβe S, et al. CRISPR/Cas9 mediated genome engineering: an adeno-associated viral (AAV) vector toolbox. Biotechnol J. 2014;9:1402–1412. doi: 10.1002/biot.201400046. [DOI] [PubMed] [Google Scholar]
- 151.Li L, Wei Y, Gong C. Polymeric Nanocarriers for Non-Viral Gene Delivery. J Biomed Nanotechnol. 2015;11:739–770. doi: 10.1166/jbn.2015.2069. [DOI] [PubMed] [Google Scholar]