Abstract
The M2 proton channel of the Influenza A virus has been the subject of extensive studies because of its critical role in viral replication. As such, we now know a great deal about its mechanism of action, especially how it selects and conducts protons in an asymmetric fashion. The conductance of this channel is tuned to conduct protons at a relatively low biologically useful rate, which allows acidification of the viral interior of a virus entrapped within an endosome, but not so great as to cause toxicity to the infected host cell prior to packaging of the virus. The dynamic, structural and chemical features that give rise to this tuning are not fully understood. Herein, we use a tryptophan (Trp) analog, 5-cyanotryptophan, and various methods, including linear and nonlinear infrared spectroscopies, static and time-resolved fluorescence techniques, and molecular dynamics simulations, to site-specifically interrogate the structure and hydration dynamics of the Trp41 gate in the transmembrane domain of the M2 proton channel. Our results suggest that the Trp41 sidechain adopts the t90 rotamer whose χ2 dihedral angle undergoes an increase of approximately 35° upon changing the pH from 7.4 to 5.0. Furthermore, we find that Trp41 is situated in an environment lacking bulk-like water, and somewhat surprisingly, the water density and dynamics do not show a measurable difference between the high (7.4) and low (5.0) pH states. Since previous studies have shown that upon channel opening water flows into the cavity above the histidine tetrad (His37), the present finding thus provides evidence indicating that the lack of sufficient water molecules near Trp41 needed to establish a continuous hydrogen bonding network poses an additional energetic bottleneck for proton conduction.
Graphical Abstract

INTRODUCTION
The M2 protein of the influenza A virus forms an α-helical homotetrameric channel in the viral envelope, allowing selective proton conduction across the viral membrane upon endosomal acidification on virus entry.1,2 Because it is vital to the viral life-cycle, the M2 proton channel has been the subject of extensive studies and also targeted for development of anti-influenza drugs.3–7 It is well recognized that, lining the channel pore, His37 and Trp41 (shown in Figure 1), are the two most important residues that are responsible for channel opening and unidirectional or asymmetric proton conduction from the exterior into the interior of the virus.8 In particular, the role of His37 is to select protons for conduction via its imidazole ring and to trigger the opening of the channel at low pH through protonation of approximately three His37 residues, whereas the Trp41 tetrad functions as a gate to allow asymmetric proton conductance only when the pH is low on the outside of the virus.9–11 In addition, recent studies3,12–14 indicate that Asp44 plays an essential role in stabilizing the Trp41 gate in the closed state via hydrogen bonding through a structured water network and perhaps more importantly, this stabilization helps the channel achieve its asymmetric proton conduction property.
Figure 1.
X-ray crystallographic structure of the transmembrane domain of the M2 proton channel (Protein Data Bank ID: 3LBW), showing the two key sidechains, His37 (blue) and Trp41 (magenta), as well as three channel water clusters (red) labeled as A–C.
Over the past decade, several models have been proposed to explain the mechanism of proton transport through the M2 channel. An early model suggests that in the open state the channel hosts a water wire with which protons conduct via the Grotthuss mechanism.15–17 A second model, the “shuttle mechanism”, proposes that His37 is directly involved in proton conduction by cycling through a protonation-deprotonation process.18–22 Similarly, a third model, involving the formation of a His37 dimer via hydrogen bonding in the closed state, suggests that protonation of the third His37 disrupts this dimer and promotes a His37-Trp41 cation-π interaction, which occasionally is disrupted by conformational motions of the helix backbone and/or Trp41 causing exposure of His37 to C-terminal water and hence proton release.8 Finally, a transporter model,13,23 which involves two distinct conformational ensembles with only one capable of proton conduction, proposes that acidification of the virus interior is achieved by oscillating between these two conformations, triggered by protonation and deprotonation of His37.
One particular property of the M2 channel that makes it unique is that it has a low proton conductance rate. Both whole-cell patch clamp and liposome experiments have put the proton conducting rate in the range of 101 – 104 protons per second,24–26 which is one to three orders of magnitude slower than expected for transmission of a proton through a pore of similar dimension at the same pH.27 In particular, existing evidence indicates that, when scaled for changes in the permeant ion concentration, the proton conduction rate is in fact attenuated as the pH approaches 5.0.27 This observation suggests that additional kinetic barriers arise for proton conduction at low pH due to structural fluctuations and/or protonation/deprotonation of His37 necessary for conduction.24,28 For example, the recent electrophysiological study of DiFrancesco et al.,29 suggested a recycling step, where after proton release the channel resets its conformation by closing the Trp41 gate and opening the Val27 gate to prepare for proton intake from the exterior. On the other hand, others have proposed that a rate-limiting step arises from the necessity of tautomerization or ring reorientation of the imidazoliums in the low pH, conducting state.18,22 This type of pre-equilibration is consistent with electrophysiological data showing that His37 equilibrates with protons from the outside of the virus prior to a rate-determining step, which allows protons to diffuse away from the His37 on the cytoplasmic side of the channel.24 Indeed, Hong and coworkers30 have determined that the His37-water proton exchange rate (~105 s−1) is significantly higher than the time-averaged unitary proton flux of M2, indicating that not every proton that is exchanged with His37 exits the channel. Williams et al.31 proposed that such futile exchanges are caused by the His37-Trp41 cation-π interaction, which periodically forms and breaks at low pH. Yet, another possibility is that the channel resides in a predominantly closed state even at low pH and a conducting state is only transiently populated to allow proton release. In a theoretical treatment of M2 proton conductance Zhou found that in order to use a proton transfer model that takes into account the primary and secondary gating of Trp41 and Val27 as well as backbone dynamics to quantitatively describe the proton conductance data of M2, the C-terminal open conformation (i.e., the conformation that allows transfer of protons from His37 to the interior of the virus) needs to be a minor population (<10%).32 To provide further insight into the proton conducting mechanism of M2, herein we approach the problem from the perspective of channel water.
It is well known that water plays a pivotal role in maintaining a viable pathway for proton conduction in biological systems.33 While water is an integral part of the M2 channel, as revealed by high-resolution crystal structures of the transmembrane (TM) domain of the M2 proton channel,13,34 only a few previous studies13,35–42 have focused on elucidating the structure and dynamics of the channel water. The current understanding is that the region above His37 is a water-filled pore; thus protons can diffuse up to His37 through this water pore via a Grotthuss mechanism. However, the structure and dynamics of the water in this region have been shown to depend on pH (and the binding of pore-blocking drugs): at high pH (>7), the channel water is more structured and less mobile in comparison to bulk water, whereas at low pH (e.g., 5.0), the water density near Gly34 is increased37 and the water cluster exhibits bulk-like dynamics.43 While these previous studies provide invaluable insight into the hydration environment of the region immediately above the key His37 tetrad, less is known about the structure and dynamics of the water clusters below the His37 gate, especially those surrounding the Trp41 tetrad. Recently, Voth and coworkers40 have calculated the potential of mean force (PMF) of proton conduction across the transmembrane (TM) domain of M2 and identified two energy barriers, with one (~10 kcal/mol) associated with the proton passing through the His37-Trp41 section of the channel. PMF calculations conducted in a second study and restricted to this region also show a similar barrier.42 These studies suggest that deprotonation of the His37 tetrad is the rate-determining step; however, the intrinsic proton release rate is modulated by the structure and dynamics of water below the His37 gate and/or conformational fluctuations of the channel. In addition to the role of water, previous resonance Raman44 and electrophysiological measurements point to a tight coupling between His37 and Trp41 at low pH for the most common variants of M2 that contain Asp at position 44.14 However, other spectroscopic and crystallographic studies suggest that the His37/Trp41 interaction might be largely broken at low pH.31,45 Thus, considering these previous studies, it would be quite useful to design an experiment that would allow one to directly assess the dynamics of water surrounding the Trp41 tetrad under different conditions.
In order to use infrared (IR) spectroscopy to interrogate the water environment and local electrostatics directly surrounding His37 and Trp41, a site-specific IR probe is required. For this reason, we employed a nitrile-derivatized Trp, 5-cyanotryptophan (TrpCN) in the current study. Previously studies have shown that the frequency and linewidth of the C≡N stretching vibration of TrpCN is highly sensitive to local solvation environment, due to the fact that even solvent interactions that do not directly involve the nitrile moiety, such as H-bonding with the indole amine (N-H) group, can be propagated through the aromatic ring to affect the C≡N stretching frequency.46–49 In addition, the well defined direction of the C≡N stretching vibrational transition dipole moment can be exploited to determine the structural transition of Trp41 in response to channel closing or opening, akin to the strategy of Hong and coworkers31 that used 5-19F-Trp and NMR spectroscopy to probe the pH-dependent rotameric states of Trp41. An added advantage of using TrpCN is that its fluorescence exhibits a strong dependence on the presence of water and hence can be used as a complementary method to assess the local hydration status of the Trp41 region of the channel.50 Specifically, we employed the TM domain of M2 (sequence: 22SSDPLVVAASIIGILH37LILW41ILDRL46) as our model system and incorporated the TrpCN spectroscopic probe via a Trp41/TrpCN mutation. We chose this M2 construct (hereafter referred to as M2TM) because it has been shown that: 1) it fully recapitulates the tetrameric assembly of the full length M2; 2) there is little, if any, difference in its rate of conduction or amantadine inhibition versus the full-length protein when both are expressed in cellular membranes;51 3) the conductance measured for this peptide, when reconstituted in an appropriate lipid composition, is in close agreement with the magnitude of flux expected from whole-cell channel recordings;51 and 4) it is easily synthesized in high yield and purity and has been used in many structural studies.13,14,45,52–55 Despite this convenience, it is worth noting that this truncated version of M2 does not capture other biological functions of the full length protein, such as facilitating budding of the virus in an ESCRT-independent manner and interaction with the M1 protein. Therefore, the current study does not shed light on these aspects of M2’s function.
Results obtained with all spectroscopic techniques, including FTIR, two-dimensional (2D) IR, and steady-state and time-resolved fluorescence spectroscopies, indicate that the water is well-structured around the Trp41 gate at both high and low pH states and does not show evidence of a significant increase in local hydration upon channel acidification. Furthermore, we find that the Trp41 sidechain adopts the t90 rotamer that rotates approximately 35° along its χ2 dihedral angle upon lowering the pH from 7.4 to 5.0. Taken together, these results suggest that there is a hydration bottleneck within the M2TM channel, particularly in the vicinity of Trp41, which may cause a significant barrier for the proton to diffuse across this region of the channel.
EXPERIMENTAL DETAILS
Materials and Peptide Synthesis
The peptide (M2TM-WCN) used in this work corresponds to the transmembrane domain of the M2 proton channel with a single mutation at the 41 position, i.e., Trp41 to TrpCN, which has the following sequence: 22SSDPLVVAASIIGILHLIL-TrpCN-ILDRL46. All peptides were synthesized using standard 9-fluorenylmethoxy-carbonyl (Fmoc) methods on a PS3 peptide synthesizer (Protein Technologies, Woburn, MA) and cleaved from resin with trifluoroacetic acid (TFA). Fmoc-5-cyanotryptophan was purchased from RSP amino acids (Shirley, MA), and all other amino acids were purchased from Advanced ChemTech (Louisville, KY). 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG), cholesterol, and dodecylphosphocholine (DPC) were purchased from Avanti Polar Lipids Inc. (Alabaster, Alabama). The crude peptides were purified by reverse-phase HPLC (Agilent Technologies 1260 Infinity) with a C18 preparative column (Vydac). For purification of the M2TM peptides, a linear gradient of buffer A (Millipore water) and buffer B (6:3:1 2-propanol:acetonitrile:water), both containing 0.1% TFA, was used, starting from 30% buffer B. For purification of the tripeptide Gly-TrpCN-Gly, buffer B was replaced with acetonitrile and 0.1% TFA. The mass of every peptide was verified by either liquid-chromatography mass spectrometry (LC-MS) or matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) where appropriate.
LUV Preparation
Large unilamellar vesicles (LUVs) with a 100 nm diameter containing the M2TM-WCN peptide were prepared by first cosolubilizing the peptide with a ternary lipid mixture consisting of POPC, POPG, and cholesterol at 4:1:2 ratios in a mixture of chloroform and ethanol using the desired peptide to lipid ratio. Then, the organic solvent was removed using a stream of nitrogen gas and the resulting film was lyophilized for at least 2 hours to remove any remaining solvent. The film was rehydrated with 1 mL of either a pH 7.4 buffer (50 mM phosphate and 100 mM NaCl) or a pH 5.0 buffer (50 mM cacodylate and 100 mM NaCl) and the resultant solution was then subjected to 8 freeze-thaw cycles, which include slow vortexing, freezing, and thawing. Further extrusion of this lipid-peptide solution using an extruder (Avanti Polar Lipids Inc.) equipped with a 100 nm membrane yielded the desired LUV sample. All LUV solutions were stored at 4 °C and used within 5 days of preparation.
Circular Dichroism, Fluorescence, and FTIR Measurements
Circular dichroism (CD) spectra were measured on an Aviv 62A DS spectropolarimeter (Aviv Associates, NJ) with a 1 mm cuvette. Fluorescence spectra were collected on a Jobin Yvon Horiba Fluorolog 3.10 spectrofluorometer using a 1 cm quartz cuvette. FTIR spectra were collected on a Thermo Nicolet 6700 FTIR spectrometer at a resolution of 1 cm−1 using a home-built CaF2 sample cell with an optical pathlength of 25 μm. For all the FTIR spectra shown, a background arising from solvent absorption has been subtracted. All the static spectroscopic measurements were conducted at room temperature.
Attenuated Total Reflectance (ATR)-FTIR Spectroscopy
Polarized ATR-FTIR spectra were recorded on a Nicolet 6700 FT-IR spectrometer (Thermo Fisher Scientific Inc., Madison, WI) with a Harrick’s Horizon multiple-reflection attachment equipped with a Ge crystal (50 mm × 10 mm × 2 mm SPT45). The instrument was purged with nitrogen gas during data acquisition. Each spectrum corresponds to an average of 128 scans with a resolution of 2 cm−1. A solution of M2TM-WCN peptide in TFE was first mixed with a stock solution of POPC/POPG/cholesterol (4:1:2) in chloroform and ethanol in a 1:25 peptide to lipid ratio. A multilayer lipid film was prepared by introducing 100 μL of the mixture onto the surface of the Ge crystal, which was previously plasma cleaned, and left to dry. After a dry film was obtained, 100 μL of either a pH 7.4 buffer (10 mM phosphate buffer and 10 mM NaCl) or a pH 5.0 buffer (10 mM cacodylate buffer and 10 mM NaCl) was uniformly added to the surface and allowed to dry under nitrogen. Then, the crystal was placed back in a hydration chamber and incubated for at least 24 hours before measurement.
The C≡N orientation and helix tilt angles, all with respect to the membrane normal, were calculated from the dichroic ratio (R) of the respective C≡N and amide I bands following the published protocols.56,57 In the current study, R was calculated using the integrated areas of the bands measured with parallel and perpendicular polarizations. The detail of Trp41 rotamer analysis based on the ATR results is described in the ESI.
2D IR Measurements
The samples used in the 2D IR measurements were prepared by first solubilizing the M2TM-WCN peptide and DPC in trifluoroethanol (TFE) at a 1:35 peptide to lipid ratio. A stream of nitrogen gas was used to remove the organic solvent and the resultant film was lyophilized for at least 4 hours to eliminate the residual solvent. Then, this peptide-detergent film was re-dissolved in either a pH 7.4 buffer (50 mM phosphate and 100 mM NaCl) or a pH 5.0 buffer (50 mM cacodylate and 100 mM NaCl). The final concentration of the peptide was approximately 15 mM. The primary reason to use DPC micelles, instead of the lipid membrane used in the static spectroscopic measurements, in the 2D IR experiments was that they significantly decrease the noise arising from the scattered light. DPC micelles have been used in many previous 2D IR studies of similar nature.41,43 As shown (Figure S2, ESI), the C≡N stretching band of M2TM-WCN in DPC micelles was almost identical to that obtained in membranes composed of POPC/POPG/cholesterol (4:1:2), suggesting that the use of DPC micelles in the current case has not significantly changed the assembly and function of the M2TM-WCN peptide.
The details of the 2D IR setup have been described elsewhere.58 In short, a train of 800 nm, 65 fs, and 300 μJ pulses were generated by a Libra Ti:sapphire regenerative amplifier (Coherent, Santa Clara, CA) laser system at 1 kHz, which were used to pump a home-built optical parametric amplifier with different frequency generation capability to generate mid-IR pulses with frequency centered at 2220 cm−1. Then, these mid-IR pulses were split into three groups and focused on to the sample in a boxcar-geometry. The resultant photon echo signal was collected and overlapped with a local oscillator pulse for heterodyne detection. A grating in the spectrometer was used to disperse the heterodyned signal onto a 64 element mercury-cadmium-telluride (MCT) array detector (Infrared Associates, Stuart, FL). The data were collected at different waiting times (T) and the resulting 2D IR spectra were obtained from three Fourier transforms of the raw data. In addition, a tapered cosine window function was applied to all 2D IR time domain data before applying the Fourier transform.
Time-Resolved Fluorescence Measurements
Time-resolved fluorescence measurements were collected on a home-built time-correlated single photon counting (TCSPC) apparatus with a 0.4 cm quartz cuvette at 25 °C. The details of the TCSPC setup have been described elsewhere.50,59 Fluorescence decays were deconvoluted with the instrument response function (IRF) and fit to a multi-exponential function to minimize χ2 below an acceptable value (i.e., 1.2) using FLUOFIT (Picoquant GmbH).
RESULTS AND DISCUSSION
As indicated above, the site-specific spectroscopic probe, TrpCN, was introduced into M2TM via a Trp41/TrpCN mutation (this mutant is hereafter referred to as M2TM-WCN). As shown below, this mutation does not impede the folding and proton conduction activity of M2TM in any significant manner, but does allow us to monitor the local electrostatic and hydration environment of Trp41 through the C≡N stretching vibration via FTIR, 2D IR, static and time-resolved fluorescence techniques.
CD and Proton Conductance Measurements of M2TM-WCN Channel
As shown (Figure 2), the far-UV CD spectra of M2TM-WCN in vesicles consisting of a ternary mixture of POPC, POPG, and cholesterol at both pH 5.0 and 7.4 (also referred to in the following text as the low pH and high pH state, respectively) show characteristic features (i.e., minima at 208 and 222 nm) of α-helical secondary structures and are similar to those of the wild-type (Figure S3, ESI).60 Interestingly, unlike that of M2TM, the CD spectrum of M2TM-WCN in a membrane environment contains an additional weak band located at about 245 nm. The fact that this band disappears when M2TM-WCN is solubilized in a mixture of water and isopropanol (Figure S4, ESI), wherein the peptide is expected to remain monomeric, suggests that it arises from a M2TM assembly that promotes electronic exciton coupling between nearby TrpCN residues. A recent study has shown that such electronic interaction between two nearby TrpCN residues would produce an exciton CD couplet in this wavelength range of 235–255 nm.61 Thus, taken together, these CD results suggest that the chosen Trp41/TrpCN mutation does not disrupt the secondary structure and presumably the tetrameric assembly of M2TM in lipid membranes.
Figure 2.
CD spectra of M2TM-WCN variant in 100 nm vesicles composed of POPC:POPG:cholesterol (4:1:2) at pH 5.0 and 7.4, as indicated. The peptide concentration was 30 μM and the peptide to lipid ratio was 1:100.
To ensure that the TrpCN mutation does not impede the proton conduction function of the tetramer, we further performed proton flux measurements on M2TM and M2TM-WCN channels reconstituted in large unilamellar vesicles (LUVs) following a protocol previously published by Ma et al.51 (see details in ESI). Briefly, a M2TM or M2TM-WCN vesicle solution was first prepared in potassium phosphate buffer (pH 7.4), and then was diluted by a sodium phosphate buffer solution (pH 7.4) that also contained a potassium ion selective carrier, valinomycin.25 This mixing caused an electrochemical gradient between the interior and exterior of the liposome, which, when a proton-selective channel is embedded in the lipid bilayer, enabled transfer of potassium ions from the inside to the outside of the liposome through valinomycin and, in the meantime, a proton flow in the opposite direction. This proton flux was detected and quantified by the fluorescence time-profile of a pH-sensitive dye, pyranine, which was preloaded inside the vesicles (see details in ESI). As indicated (Figure S5, ESI), the cumulative proton flux per M2TM tetramer at this pH was determined to be 6.0 ± 0.5 H+/s, which is almost identical to that reported previously,51 and 4.2 ± 0.6 H+/s for M2TM-WCN. As expected, when the pH of the exterior solution is decreased to 5.0, the cumulative proton flux increases by approximately a factor of 2 for both the wild-type M2TM and M2TM-WCN channels (Figure S5, ESI). Thus, these results provide direct evidence that the proton conduction activity of the M2TM channel tolerates the Trp41 to TrpCN mutation. In addition, the proton conduction rates obtained here are within an order of magnitude of previously reported values for the M2 channel.4,26,51,62
Structural Transition of Trp41
Hong and coworkers have investigated the structure and structural transition of the Trp41 gate in M2TM at acidic pH using ssNMR.10,31 By replacing Trp41 with 5-19F-Trp in M2TM, they were able to use 19F spin diffusion NMR measurements to characterize the distance between His37 and Trp41 as well as the conformational dynamics of the indole sidechain. Their results indicated that the Trp41 sidechain adopts a t90 rotamer at both high and low pH; however, the two dihedral angles of Trp41, χ1 and χ2, are increased by ~20° upon channel activation. Furthermore, they found that at low pH, Trp41 undergoes Gaussian fluctuations in both χ1 and χ2, suggesting that the cation-π interaction between His37 and Trp41 is sporadic, and may serve as the trigger to release a proton into the C-terminal region of the channel.
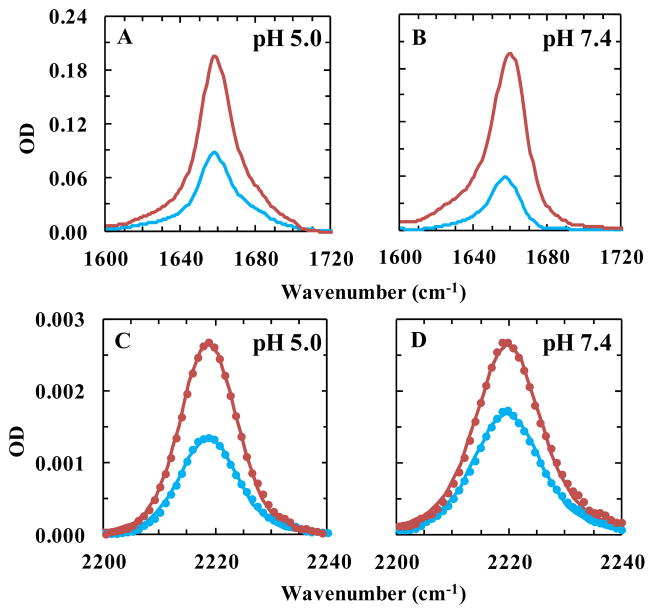
To provide further validation of these previous findings and also to confirm the functionality of the M2TM-WCN channel, herein we employed polarized attenuated total reflectance IR (ATR-IR) spectroscopy to assess the orientation of the Trp41 sidechain and also the helix tilt angles. This assessment is based on the notion that the C≡N stretching vibration is a local vibrational mode and its transition dipole moment is along the molecular axis of the C≡N moiety, which lies in the same plane of the indole ring. Therefore, the orientation angle of TrpCN in a well-oriented M2TM-WCN tetramer, such as when embedded in a surface-supported lipid bilayer, can be determined by measuring the dependence of the absorbance the C≡N stretching vibration on the polarization of the IR light.57 As shown (Figure 3), the dichroic ratio of the C≡N stretching vibration, defined as the ratio between the integrated areas of the absorption spectra obtained with parallel and perpendicular polarizations, is changed from 1.55 ± 0.24 at pH 7.4 to 1.95 ± 0.18 at pH 5.0. This indicates that TrpCN undergoes a distinct conformational change in response to a protonation state change of the channel. Based on these dichroic ratios, we further calculated the average angle of the C≡N axis with respect to the membrane normal56,63 or z-axis (θCN) to be 60.2 ± 4.7° for pH 7.4 and 53.1 ± 3.7° for pH 5 (Table 1).
Figure 3.
ATR-FTIR spectra of M2TM-WCN in a hydrated lipid bilayer composed of POPC:POPG:cholesterol (4:1:2) at pH 5.0 and 7.4, obtained with parallel (red) and perpendicular (blue) polarizations. For comparison, the spectrum measured at pH 7.4 was scaled by a factor of 1.2. (A) and (B): amide I mode of the peptide. (C) and (D): C≡N stretching vibrational band of TrpCN, where the solid line is the best fit of the corresponding spectrum to a Voigt profile and the resultant orientational parameters were given in Table 1.
Table 1.
Dichroic ratio (R), order parameter (S), helix tilt angle (β), and C≡N orientation angle (θCN) of M2TM-WCN obtained based on the ATR-FTIR measurements at pH 5.0 and 7.4.
| Parameter | pH 5.0 | pH 7.4 |
|---|---|---|
| RCN | 1.95 ± 0.18 | 1.55 ± 0.24 |
| SCN | 0.04 ± 0.07 | −0.13 ± 0.11 |
| RHelix | 2.31 ± 0.26 | 2.94 ± 0.37 |
| SHelix | 0.16 ± 0.07 | 0.30 ± 0.07 |
| β (°) | 39.7 ± 7.4 | 24.2 ± 9.0 |
| θCN (°) | 53.1 ± 3.7 | 60.2 ± 4.7 |
We have also performed classical molecular dynamics (MD) simulations (see details in ESI) to examine the pH-induced orientational changes of the C≡N group in the M2TM-WCN channel. The protocols employed were consistent with previous studies.13,14 Specifically, these simulations were carried out using the high-resolution X-ray structure of M2TM (PDB: 3LBW) as the starting point, where Trp41 was replaced with TrpCN. To represent different pH states, four His37 protonation states (i.e., His1+, His2+, His3+, His4+) were used in the simulations. We found that the overall structure and water density profiles mirror previously published results up to the +3 state,13,14 which corresponds to the lowest pH state in the experiment. The distribution of the angle between the C≡N vector and the pore axis (i.e., membrane normal), θCN-MD, determined from a 120-ns simulation trajectory, is shown in Figure S6. In support of our experimental results, it is evident that as the protonation state of the His37 tetrad increases, the θCN-MD angle decreases. Furthermore, the mean values of θCN-MD obtained at His2+ and His3+ states, which correspond to the experimental pH conditions of 7.4 and 5.0, respectively, are on the same order of those determined from the ATR measurements (Table 1 and Figure S6). A similar comparative trend is also observed in the MD simulations of the wild-type M2TM (Figure S7) published by Acharya et al.13
Similarly, we used the amide I band of the peptide and polarized ATR-IR measurements to estimate the helix tilt angle of M2TM-WCN in supported lipid bilayers at different pH values. As shown (Figure 3A and 3B), in both cases the absorbance of the amide I band measured with polarization parallel to the membrane normal is significantly higher than that measured with IR light whose polarization is perpendicular to membrane normal, indicating, as expected, that the helix is oriented perpendicular to the membrane plane. Further calculations using the experimentally determined dichroic ratios indicate that the average helix tilt angle (β) increases from 24.2 ± 9.0° to 39.7 ± 7.4° with respect to the z-axis upon changing the pH from 7.4 to 5.0 (Table 1). This pH-induced global structural change is consistent with several other previously published M2TM structures.45,64,65 In addition, the angle determined herein at high pH is in agreement with the ATR-IR study of Manor et al.,36 which reported a helix tilt angle of 27° at pH 7.0 obtained via a protein model constructed from backbone carbonyl angles calculated from the dichroic ratios of backbone isotope labels.
Because θCN depends on the torsion angles (i.e., χ1 and χ2) of the TrpCN sidechain and also on the helix tilt angles of individual helices, the rotameric state of Trp41 cannot be determined in an absolute sense using the θCN alone. However, similar to the strategy used by Hong and coworkers,31 this information can be used as constraints to exclude rotameric states that do not meet the angle criteria obtained from experiment, especially when the backbone conformation is known. In the current case, we began by identifying PDB structures of M2TM that could be used as a structural template for the subsequent rotameric analysis of Trp41. Based on the similarity of their helix tilt angle with the β angle determined from experiment, two PDB structures5,13 obtained at neutral pH and pH 6.5, 2KQT and 3LBW, were chosen for the high pH case. Similarly, a pH 5.3 crystal structure (PDB: 3C9J)45 and a ssNMR structure (PDB: 2KAD)54 obtained at pH 7 were chosen for the low pH state, because the helix tilt angle in both is between 35–38° and their C-terminal pore is relatively more expanded compared to that of the structure models of the high pH state. For each PDB structure, we then allowed the Trp41 sidechain to sample all the possible rotameric conformers by varying its χ1 and χ2 dihedral in 1° increments and the value of θCN was calculated for each and every possible combination. As shown (Figure S8, ESI) and expected, there are a large number of combinations of χ1 and χ2 that can satisfy the experimentally determined C≡N angle constraints. However, out of all the possible rotameric states that tryptophan can adopt, the majority of them can be ruled out due to steric clashes either with the protein backbone or with the His37 sidechain. As indicated (Figure S8A, ESI), the two allowable rotamers are t90 and t-105. To further determine which of these two rotamers is the most probable, one needs additional information. As discussed in detail below, the C≡N stretching frequency allowed us to further conclude that Trp41 adopts the t90 rotamer in both the high and low pH states. Also in support of this picture, the torsion angles (Figure S9, ESI) determined from the MD simulations of both M2TM-WCN and wild-type M2TM channels show a distribution that is centered primarily in the regime of the t90 rotamer, where χ1, χ2 = ~180º, 90º.
To further refine the possible χ1 and χ2 values in the t90 rotamer regime, we only considered combinations of χ1 and χ2 that satisfy the θCN angle within ± 7.0° of the reported value to account for the experimental error within the ATR measurement. As indicated (Figure 4), this analysis allowed us to determine the minimum RMSD values within this range and hence better resolved rotamer angles (Table S1). For the PDB structures 2KAD (low pH) and 2KQT (high pH), a single minimum was found. On the other hand, structures 3C9J (low pH) and 3LBW (high pH) yielded two minima within the t90 rotamer region in each case and both are sterically allowed. However, the χ1 and χ2 angles corresponding to one of the minima are comparable to those obtained with 2KAD and 2KQT structures. Therefore, we chose this set of χ1 and χ2 angles (i.e., 162°, 93° and 190°, 70°) to describe the pH-induced structural transition of Trp41. Interestingly, the χ1 angle determined herein is similar to that reported by Williams et al.,31 although our χ2 angle is significantly different from theirs. In particular, their χ2 angle suggests that the benzene ring is pointed downward and hence closer to the C-terminus, whereas our result suggests that the benzene ring is more buried within the pore. In addition, the dihedral angles determined from the IR measurements indicate that upon lowering the pH, χ2 undergoes a significant increase (i.e., from ~65° at pH 7.4 to ~100° at pH 5.0). This increase shifts the benzene ring of Trp41, specifically atoms Cη2, Cζ3, and Cε3, away from the center and into the lipid region, only exposing the N-H side of the pyrrole to the inner pore. Accompanied by a larger helix tilt angle, the His37 imidazole rings come into closer contact with Trp41 (Figure S8E, ESI), potentially allowing an optimal proximity for a cation-π interaction between adjacent helices as was observed in UV resonance Raman44 and ssNMR31 experiments. Therefore, these structural data on Trp41 and the helix backbone provide further validation that the mutation strategy used, i.e., the Trp41 to TrpCN mutation, does not disrupt the conformational transitions that are critical for the proton conducting function of the M2TM channel.
Figure 4.
Contour plot of the RMSD between the measured θCN from the ATR-FTIR spectra (Table 1) and the calculated θCN in the t90 rotamer range, where only values within ± 7.0° of the experimental value were plotted, as a function of Trp41 (χ1, χ2) torsion angles for high pH (A) and low pH (B) cases. The minimum values are depicted in Table S1.
Infrared Measurement of Local Electrostatic and Hydration Environment of Trp41
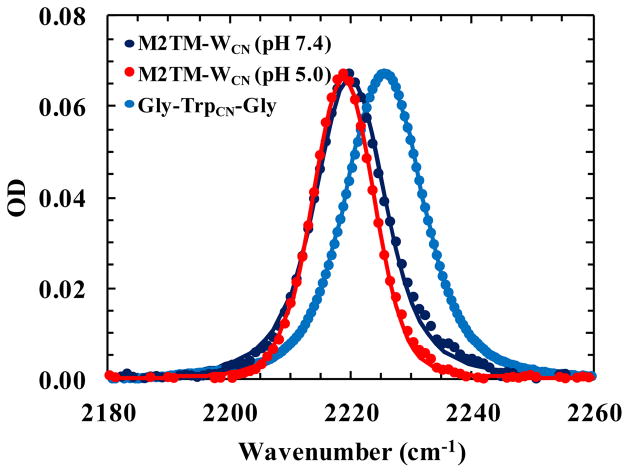
It has been shown that the C≡N stretching vibrational band of TrpCN is a sensitive IR reporter of the hydration status of its indole sidechain.47 For example, in water its C≡N band is centered at 2224 cm−1 with a broad bandwidth of 18 cm−1, whereas in tetrahydrofuran (THF) the band is shifted to 2220 cm−1 and narrowed to 8 cm−1.47 As shown (Figure 5 and Table 2), the C≡N band of M2TM-WCN in a hydrated lipid bilayer composed of POPC, POPG, and cholesterol is centered at 2219.8 cm−1 at pH 7.4, which becomes slightly red-shifted (~1 cm−1) and narrower when the pH is decreased to 5.0. In comparison, the C≡N stretching vibration of a fully hydrated tripeptide, Gly-TrpCN-Gly, has a peak frequency of 2225.5 cm−1 and also a broader bandwidth. Taken together, these results suggest that the TrpCN sidechain in the M2TM-WCN channel is mostly dehydrated and situated in a THF-like environment even when the channel is in its low pH state. This is a significant finding as previous studies have shown that more mobile water will be accumulated in the Gly34 region under more acidic conditions.23,37,40,43,53 In addition, the observed frequency shift toward lower wavenumbers upon changing the pH from 7.4 to 5.0 is consistent with the notion that the His37 tetrad is transitioned from a +2 state to a +3 state. This is because the associated increase in electric field at the nitrile site is predicted to decrease its stretching frequency, due to the vibrational Stark effect. Boxer and coworkers66 have shown that the C≡N stretching vibrational frequency of 5-cyanoindole decreases with increasing electric field with a Stark tuning rate of 0.86 cm−1/(MV cm−1). Thus, a simple calculation of the electric field change (ΔE) experienced by the C≡N bond allowed us to estimate the frequency shift of the C≡N stretching vibration of TrpCN. Assuming that a single point charge is added to the center of the His37 tetrad and using the PDB structure 3C9J and the Trp41 rotamer obtained above for distance and angle determination, ΔE was calculated to be 4.9 MV cm−1 when vacuum permittivity was used, which gives rise to a frequency shift of −4.2 cm−1. Comparing to the 1 cm−1 red-shift observed in the experiment, this value suggests that the effective dielectric constant in this region of the channel is on the order of 4. This finding thus supports the notion that the Trp41 tetrad is immersed in a THF-like environment, which has a dielectric constant of 7.6. Moreover, as discussed above, this red-shift would argue against the idea that the Trp41 sidechain adopts the t-105 rotamer, as this conformation would point the C5 position, where the nitrile moiety is located in TrpCN, directly toward the center of the channel, resulting in a blue-shift47,67 due to direct H-bonding between the C≡N group and the few water molecules in this region (see below). Finally, the fact that at pH 5.0 the C≡N stretching band becomes narrower is consistent with the MD simulation results (Table S2, ESI), which show that for both the M2TM-WCN and wild-type M2TM channels the percentage of H-bonds formed between the indole N-H and pore water, an interaction that is known to broaden the linewidth of the C≡N stretching vibration of TrpCN,48 decreases as the protonation state increases (e.g., from His2+ to His3+).
Figure 5.
Comparison of the C≡N stretching vibrational bands of M2TM-WCN in a lipid bilayer at pH 5.0 and 7.4 with that of Gly-TrpCN-Gly in H2O. The M2TM-WCN spectra are identical to those used in Figure 4. For comparison, the spectra of M2TM-WCN was scaled by a factor of 25. The smooth lines are fits to a Voigt profile and the resultant parameters are listed in Table 2.
Table 2.
Center frequency (ω0) and width (FWHM) of the C≡N stretching vibration determined from linear IR experiments and parameters obtained from 2D IR measurements.
| Peptide | ω0, cm−1 | FWHM, cm−1 | A | τ, ps | B |
|---|---|---|---|---|---|
| M2TM-WCN pH 5.0 | 2218.8 ± 0.2 | 11.9 ± 0.2 | 0.48 ± 0.04 | 2.92 ± 0.52 | 0.11 ± 0.04 |
| M2TM-WCN pH 7.4 | 2219.8 ± 0.3 | 13.7 ± 0.3 | 0.60 ± 0.08 | 2.84 ± 0.82 | 0.15 ± 0.09 |
| Gly-TrpCN-Gly | 2225.5 ± 0.1 | 14.9 ± 0.4 | 0.42 ± 0.05 | 1.63 ± 0.33 | 0.04 ± 0.05 |
Within the M2TM channel, the Trp41 cluster forms the narrowest region of the channel and is sequestered in a hydrophobic environment with the exception of the confined waters, especially in the closed state.68 Therefore, the characteristics of the C≡N stretching vibration obtained at pH 7.4, indicative of a more ordered and less hydrated environment for the TrpCN sidechain, is not surprising. At pH 5.0, which leads to the C-terminal region to expand, it is expected, as observed in simulations,16,23,40 that more water will come to the vicinity of Trp41, leading to an increase in the degree of hydration of the indole ring. However, the IR results suggest otherwise. One concern was that the M2TM-WCN channel may be still closed at pH 5.0 due to the TrpCN mutation. Thus, we also measured the FTIR spectrum of M2TM-WCN in the aforementioned lipid environment at pH 3.0, where all four His37 sidechains are protonated. The result (Figure S10, ESI) shows that the C≡N stretching frequency is identical to that measured at pH 5.0 and, therefore, we rule out this possibility. Moreover, our findings are in line with a previous UV resonance Raman study,44 which showed that the W17 Raman band of M2, which is a marker of the H-bonding status of the indole N-H, did not show any appreciable change upon lowering the pH from 7.4 to 5.4 and that the relative intensity of the W7 doublet, which measures the overall hydrophobicity of the indole ring environment, also remained unchanged.
Hydration Dynamics of Trp41
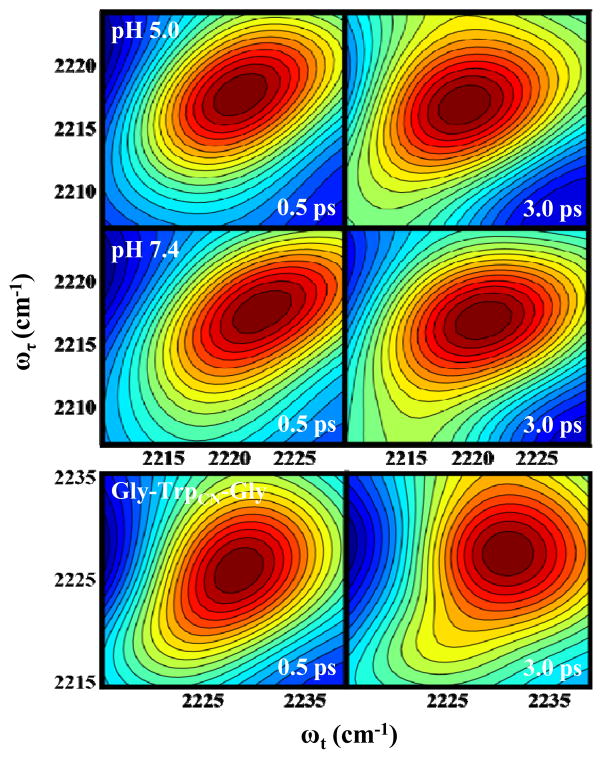
The motions of water molecules surrounding a vibrator can induce fluctuations in its vibrational frequency, causing its frequency-frequency correlation function (FFCF) to decay on a timescale that characterizes the dynamics of these motions. Two-dimensional IR (2D IR) spectroscopy is capable of assessing such dynamic events and has been widely used to study water dynamics.69,70 Thus, to further validate the aforementioned findings obtained via static spectroscopic methods, we carried out 2D IR experiments on the C≡N stretching vibration of the tripeptide Gly-TrpCN-Gly in water and the M2TM-WCN channel in a membrane environment. As shown (Figure 6), at the same waiting time (T) the 2D IR spectrum of the tripeptide is more circular than that of the M2TM-WCN channel, indicating that the spectral evolution or diffusion rates of these systems are different. To quantify these spectral diffusion dynamics, we used the center line slope (CLS) method developed by Fayer and coworkers71 to determine their respective rate constants. Specifically, the center line represents the maxima of the v=0 → v=1 peak of a series of cuts through the 2D IR spectrum. A plot of the inverse CLS as a function of waiting time (i.e., T) provides a measure of the underlying FFCF, and is described here by the following exponential function:
| (1) |
where A, B, and τ are the amplitude, offset, and spectral diffusion decay time constant. As shown (Figure 7 and Table 2), the spectral diffusion dynamics of the C≡N vibrational mode in the tripeptide occurs on a timescale of 1.6 ± 0.3 ps, which is characteristic of the H-bonding dynamics of bulk water.72,73 Thus, this result is consistent with the expectation that the TrpCN sidechain in this case is fully hydrated. On the other hand, as indicated (Figure 7), the CLS of M2TM-WCN decays on a slower timescale and with a time constant of approximately 3.0 ± 0.8 ps for both pH 7.4 and 5.0. Taken together, these results indicate that the water in the immediate vicinity of Trp41 in the M2TM channel does not exhibit bulk-water-like dynamics. The slower dynamics suggest that the water molecules in this critical region of the channel form a more structured and hence more strongly hydrogen-bonded network. This is entirely consistent with the notion put forward by Acharya et al.13 as well as by Schnell and Chou3 that a strongly hydrogen bonded bridging water cluster resides between Trp41 and Asp44 when the channel is closed (see below). However, our finding that this water cluster remains intact even in the low pH condition has strong implication on the rate of proton conduction.
Figure 6.
Representative 2D IR spectra of the C≡N vibrational mode of M2TM-WCN (15 mM) in DPC micelles (1:35 ratio) at pH 5.0 (A) and pH 7.0 (B). Shown in (C) are the 2D IR spectra of Gly-TrpCN-Gly (80 mM) in H2O. In each case, the respective waiting time (T) is indicated.
Figure 7.
Center line slope (CLS) vs. waiting time (T) plots obtained from the 2D IR spectra of M2TM-WCN in DPC micelles at pH 7.4 (red), pH 5.0 (green), and Gly-TrpCN-Gly in H2O (blue). The smooth lines are fit of this data to the following equation: CLS(T) = A·e−T/t + B, and the resulting parameters are listed in Table 2.
Fluorescence Assessment of the Hydration Status of Trp41
Another concern was that the C≡N stretching vibration may lack the necessary sensitivity to reveal a distinct change in the hydration status of Trp41 between the high and low pH scenarios. Accordingly, to provide further evidence to support the conclusions reached through IR experiments, we carried out steady-state and time-resolved fluorescence measurements on M2TM-WCN. Recently, we have shown that the fluorescence quantum yield and lifetime of TrpCN are highly sensitive to the degree of hydration as water acts as an efficient quencher of its fluorescence.50 For example, in water the florescence lifetime of TrpCN is ~0.5 ns, which is increased to ~6.0 ns in 1,4-dioxane. Because of this sensitivity, we believe that the TrpCN fluorescence will be able to detect any significant changes in the hydration status near the Trp41 gate.
As shown (Figure 8), the TrpCN fluorescence spectra of M2TM-WCN in vesicles composed of POPC, POPG, and cholesterol in a 4:1:2 ratio at both pH 7.4 and 5.0 closely resemble that of Gly-TrpCN-Gly obtained in THF,50 indicating that the Trp41 sidechain is situated in a relatively hydrophobic environment in both high and low pH states of the channel. Consistent with our results, a previous study on the wild-type M2TM in DHPC/POPC bicelles showed that the native Trp41 fluorescence spectrum is peaked at about 334 nm at both pH 8.0 and 4.5.74 Interestingly, a closer inspection indicates that the TrpCN fluorescence obtained at pH 5.0 is approximately 1.2 times more intense than that obtained at pH 7.4 and also shows a small but measurable blue-shift (i.e., from 371 to 368 nm), suggesting that the TrpCN sidechain becomes slightly less hydrated at pH 5.0. As indicated (Figure 9), time-resolved fluorescence measurements provide additional support of the notion that Trp41 is situated in a relatively dry environment, even at low pH, as the intensity weighted average lifetimes (τave) of TrpCN in the M2TM-WCN channel are 6.9 ns at pH 5.0 and 6.7 ns at pH 7.4. Thus, taken together, these fluorescence results corroborate the conclusions we have reached above based on the IR measurements.
Figure 8.
TrpCN fluorescence spectra of M2TM-WCN in 100 nm vesicles composed of POPC:POPG:cholesterol (4:1:2) at pH 5.0 and 7.4, as indicated. The peptide concentration was 30 μM and the peptide to lipid ratio was 1:100.
Figure 9.
TrpCN fluorescence decay kinetics of M2TM-WCN in 100 nm vesicles composed of POPC, POPG, and cholesterol (4:1:2) at pH 5.0 and 7.4, as indicated. In each case, the smooth line corresponds to a fit of the kinetics to a double-exponential function with the following time constants (relative amplitudes): 7.8 ns (0.62) and 2.5 ns (0.38) for pH 5.0; 7.5 ns (0.62) and 2.4 ns (0.38) for pH 7.4. Top panel shows the residuals of the fits.
Implication on Proton Conductance
As an integral part of the M2 proton channel, water must be considered in order to achieve a quantitative understanding of the underlying proton conducting mechanism and kinetics. As indicated by a recent high-resolution crystal structure of M2TM,13 which represents an intermediate state (pH 6.5) between the open and closed states, three well-ordered channel water clusters exist. As shown (Figure 1), the first cluster is situated above His37 and consists of six water molecules where four are hydrogen bonded (H-bonded) to the His37 tetrad. The second cluster, consisting of two water molecules with each H-bonded to a His37, is located between His37 and Trp41. Perhaps more importantly, the third cluster, which consists of five well-ordered waters that form H-bonds between the indole N-H of Trp41 and the carboxylate oxygen of Asp44, is located near the C-terminal region of the channel and serves to stabilize the Trp41 gate. It should, however, be appreciated that these clusters were observed in the structure determined at cryogenic temperatures. MD simulations indicate that a similar distribution is also present near room temperature, although the waters have more dynamic character on the nano- to microsecond time scale. Thus, the crystallographically observed water clusters reflect the approximate positions of more dynamic waters at various positions in the channel.13,75 The water distribution pattern clearly indicates that in order for a proton to diffuse from the first cluster to the third, or vice versa, it has to overcome two barriers imposed by His37 and Trp41, respectively. Thus, this picture justifies, from the perspective of water, why when the channel is closed protons are unable to diffuse across the channel beginning from either direction.
How does this water distribution pattern change upon viral acidification to facilitate proton conduction? Several recent experimental and simulation studies8,23,37,40,43,53 showed that both the density and mobility of the first water cluster are significantly increased under acidic conditions, indicating that in the low pH state water can readily flow into this region of the channel, hence allowing protons to access His37 from the N-terminal end of the channel. On the other hand, our results indicate that the degree of hydration surrounding the Trp41 tetrad does not show any appreciable change upon decreasing the pH from 7.4 to 5.0, suggesting that the second and third water clusters present at high pH are preserved at low pH. In other words, our results argue against the idea that, even under acidic conditions where the channel has a wider C-terminal pore, more water will gain access to the Trp41 gate region to establish a continuous water wire below His37. This notion is consistent with other experimental findings: (1) it is consistent with the study of Pinto and coworkers,9 which showed that replacing Trp41 with a smaller residue, such as Ala, Cys, or Phe, results in an increase in both the forward and backward proton flux. This can be rationalized by considering that a smaller sidechain in this region will allow more water to be accumulated and, as a result, either decreasing or eliminating the proton conduction barrier caused by Trp41 in the wild type; (2) it is consistent with the study of Chizhmakov et al. and Ma et al.,12,14 which demonstrated that in the absence of Asp44, the rate of proton conductance increases at high pH and also at pH 5.0, and was attributed to the weakening of the H-bonding network of the water cluster below Trp41.14 This once again points to the fact that the water network surrounding Trp41 plays an important role in controlling the forward and backward proton flows; and (3) it is consistent with the QM/MM MD simulations of Voth and coworkers40 and Klein and coworkers,42 which showed that there is a significant energy barrier arising from deprotonation of His37 and ion passage through Trp41, and also with the MD simulation of Acharya et al.,13 which showed that when the channel is transitioned from the +2 to +3 state the water cluster between His37 and Trp41 only accumulates one additional water molecule. Finally, and perhaps more importantly, the current study provides new insight into the low proton conductance of the M2 proton channel; it suggests, besides the previously well-recognized barrier imposed by His37,22,28,30 that a discontinued water wire in the Trp41 region produces a second barrier for proton conduction. A phenomenological kinetic scheme summarizing these points is give below:
where in the first step the third His37 uptakes a proton from water cluster A ((H2O)A), leading to formation of the +3 state of the channel, which is followed by a tautomerization and/or ring reorientation step (i.e., conformational change from (His37+)A to (His37+)B) required to orient His37 in a position primed to release the proton to water cluster B ((H2O)B), in the third step. Previously, the proton exchange rate between water and the imidazole of His37, assessed by a ssNMR study,30 is at least one to two orders of magnitude faster than the overall proton flux of M2 measured with whole-cell patch-clamp and liposome assays24–26 and is synchronized with ring reorientations that produce a barrier of ~60 kJ/mol.22 In the last step, the rate of which is not diffusion-limited (kBC < kD), the proton is transferred from water cluster B to C ((H2O)C). While the current study does not provide any direct information about the structural nature of this step, it likely suggests that an additional conformational change, either at the sidechain (e.g., Trp41) or the backbone level,8,22,29,31,40 or a transiently occupied proton diffusive state32 is required to successfully relay a proton from His37 to the water at the C-terminal end of the channel, instead of a purely two-state equilibrium process proposed by the transporter model.23 Interestingly, a recent single-molecule fluorescence study55 on the M2TM channel showed that the Trp41 residue exhibits a slow conformational motion, on the timescale of ~500 μs. This slow motion, whose rate is similar to that of proton conduction, could potentially be the aforementioned conformational change required to deliver a proton from B to C.
CONCLUSIONS
The tryptophan residue (Trp41) in the transmembrane domain of the M2 protein of influenza A virus serves as a gate to allow asymmetric proton conductance into virion interior under external acid conditions. Herein, we used an analog of tryptophan, 5-cyanotrptophan (TrpCN), and various spectroscopic methods to interrogate the structure and hydration dynamics of Trp41 of a model M2 proton channel (M2TM) in a membrane environment. Specifically, we mutated the Trp41 residue in the transmembrane domain of the M2 protein (M2TM) with TrpCN and used the C≡N stretching vibration of the resulting peptide (M2TM-WCN) as a site-specific infrared probe. The proton conductivity of the M2TM-WCN channel was found to be comparable to the wild-type M2TM channel, indicating that this mutation does not impede, in any significant manner, the native function. Consistent with many previous studies on M2TM, polarized ATR-FTIR measurements showed that both the backbone and TrpCN sidechain of the M2TM-WCN channel undergo conformational changes in response to a pH change from 7.4 to 5.0. Furthermore, a rotamer analysis based on the ATR-FTIR results suggested that Trp41 adopts the t90 rotamer conformation, and upon channel opening, the Trp41 sidechain rotates by +35° along the χ2 axis, loosening the initially tightly bound Trp41 cluster. As expected, the C≡N stretching frequency indicated that the Trp41 tetrad is sequestered in a THF-like environment at pH 7.4, and the spectral diffusion dynamics of this vibrational mode indicated that the surrounding water is not bulk-like. What is not expected, however, is that these characteristics are preserved at pH 5.0, or in the open state of the channel. In addition, measurements on the fluorescence spectra and decay kinetics of TrpCN in the M2TM-WCN channel at different pH values yielded consistent results. Taken together, these findings suggest that (1) there are a small number of water molecules in the Trp41 region of the channel, as observed in a high-resolution crystal structure of M2TM; (2) the water molecules in this critical region of the channel are less mobile compared to bulk water; and (3) unlike the cavity above His37, where more water molecules will accumulate upon channel acidification, the water density surrounding Trp41 does not show any significant increase when the pH is decreased from 7.4 to 5.0. We believe that this invariability of water density between the high and low pH states in this gating region is closely linked to the low rate of proton conduction in the M2 channel, as it suggests that a continuous water wire passing through this region has a low probability to be established. In other words, besides the energetic barrier inflicted by the His37 reorientation and protonation-deprotonation steps, the inward proton flux encounters a second barrier imposed by the Trp41 tetrad.
Supplementary Material
Highlight.
The Trp41 tetrad of the M2 proton channel is found to be relatively dehydrated even at pH 5, indicative of the existence of an additional barrier for proton conduction.
Acknowledgments
We gratefully acknowledge financial support from the National Institutes of Health (P41-GM104605). B.N.M. is an NIH Ruth Kirschstein Predoctoral Fellow (AG-046010). G.F. acknowledges support from the National Science Foundation (CHE-1212416).
Footnotes
Electronic Supplementary Information (ESI) available: Details of the proton flux assay, Trp41 rotamer analysis, and molecular dynamics simulations and Tables S1–S2 and Figures S1–S10.
NOTES AND REFERENCES
- 1.Shimbo K, Brassard DL, Lamb RA, Pinto LH. Biophys J. 1996;70:1335–1346. doi: 10.1016/S0006-3495(96)79690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pinto LH, Lamb RA. J Biol Chem. 2006;281:8997–9000. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- 3.Schnell JR, Chou JJ. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pielak RM, Schnell JR, Chou JJ. Proc Natl Acad Sci U S A. 2009;106:7379–7384. doi: 10.1073/pnas.0902548106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cady SD, Schmidt-Rohr K, Wang J, Soto CS, Degrado WF, Hong M. Nature. 2010;463:689–692. doi: 10.1038/nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balannik V, Carnevale V, Fiorin G, Levine BG, Lamb RA, Klein ML, DeGrado WF, Pinto LH. Biochemistry. 2010;49:696–708. doi: 10.1021/bi901799k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu YB, Canturk B, Jo H, Ma CL, Gianti E, Klein ML, Pinto LH, Lamb RA, Fiorin G, Wang J, DeGrado WF. J Am Chem Soc. 2014;136:17987–17995. doi: 10.1021/ja508461m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma M, Yi MG, Dong H, Qin HJ, Peterson E, Busath DD, Zhou HX, Cross TA. Science. 2010;330:509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang Y, Zaitseva F, Lamb RA, Pinto LH. J Biol Chem. 2002;277:39880–39886. doi: 10.1074/jbc.M206582200. [DOI] [PubMed] [Google Scholar]
- 10.Luo W, Mani R, Hong M. J Phys Chem B. 2007;111:10825–10832. doi: 10.1021/jp073823k. [DOI] [PubMed] [Google Scholar]
- 11.Witter R, Nozirov F, Sternberg U, Cross TA, Ulrich AS, Fu R. J Am Chem Soc. 2008;130:918–924. doi: 10.1021/ja0754305. [DOI] [PubMed] [Google Scholar]
- 12.Chizhmakov IV, Ogden DC, Geraghty FM, Hayhurst A, Skinner A, Betakova T, Hay AJ. J Physiol. 2003;546:427–438. doi: 10.1113/jphysiol.2002.028910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acharya R, Carnevale V, Fiorin G, Levine BG, Polishchuk AL, Balannik V, Samish I, Lamb RA, Pinto LH, DeGrado WF, Klein ML. Proc Natl Acad Sci U S A. 2010;107:15075–15080. doi: 10.1073/pnas.1007071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma CL, Fiorin G, Carnevale V, Wang J, Lamb RA, Klein ML, Wu YB, Pinto LH, DeGrado WF. Structure. 2013;21:2033–2041. doi: 10.1016/j.str.2013.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sansom MSP, Kerr ID, Smith GR, Son HS. Virology. 1997;233:163–173. doi: 10.1006/viro.1997.8578. [DOI] [PubMed] [Google Scholar]
- 16.Smondyrev AM, Voth GA. Biophys J. 2002;83:1987–1996. doi: 10.1016/S0006-3495(02)73960-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kass I, Arkin IT. Structure. 2005;13:1789–1798. doi: 10.1016/j.str.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 18.Pinto LH, Dieckmann GR, Gandhi CS, Papworth CG, Braman J, Shaughnessy MA, Lear JD, Lamb RA, DeGrado WF. Proc Natl Acad Sci U S A. 1997;94:11301–11306. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gandhi CS, Shuck K, Lear JD, Dieckmann GR, DeGrado WF, Lamb RA, Pinto LH. J Biol Chem. 1999;274:5474–5482. doi: 10.1074/jbc.274.9.5474. [DOI] [PubMed] [Google Scholar]
- 20.Schweighofer KJ, Pohorille A. Biophys J. 2000;78:150–163. doi: 10.1016/S0006-3495(00)76581-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiorin G, Carnevale V, DeGrado WF. Science. 2010;330:456–458. doi: 10.1126/science.1197748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu FH, Luo WB, Hong M. Science. 2010;330:505–508. doi: 10.1126/science.1191714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khurana E, Peraro MD, DeVane R, Vemparala S, DeGrado WF, Klein ML. Proc Natl Acad Sci U S A. 2009;106:1069–1074. doi: 10.1073/pnas.0811720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mould JA, Li HC, Dudlak CS, Lear JD, Pekosz A, Lamb RA, Pinto LH. J Biol Chem. 2000;275:8592–8599. doi: 10.1074/jbc.275.12.8592. [DOI] [PubMed] [Google Scholar]
- 25.Lin TI, Schroeder C. J Virol. 2001;75:3647–3656. doi: 10.1128/JVI.75.8.3647-3656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moffat JC, Vijayvergiya V, Gao PF, Cross TA, Woodbury DJ, Busath DD. Biophys J. 2008;94:434–445. doi: 10.1529/biophysj.107.109082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeCoursey TE. Physiol Rev. 2003;83:1067–1067. doi: 10.1152/physrev.00028.2002. [DOI] [PubMed] [Google Scholar]
- 28.Chen H, Wu Y, Voth GA. Biophys J. 2007;93:3470–3479. doi: 10.1529/biophysj.107.105742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DiFrancesco ML, Hansen UP, Thiel G, Moroni A, Schroeder I. PLoS One. 2014;9:e107406. doi: 10.1371/journal.pone.0107406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu F, Schmidt-Rohr K, Hong M. J Am Chem Soc. 2012;134:3703–3713. doi: 10.1021/ja2081185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams JK, Zhang Y, Schmidt-Rohr K, Hong M. Biophys J. 2013;104:1698–1708. doi: 10.1016/j.bpj.2013.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou HX. Biophys J. 2011;100:912–921. doi: 10.1016/j.bpj.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasaiah JC, Garde S, Hummer G. Annu Rev Phys Chem. 2008;59:713–740. doi: 10.1146/annurev.physchem.59.032607.093815. [DOI] [PubMed] [Google Scholar]
- 34.Thomaston JL, Alfonso-Prieto M, Woldeyes RA, Fraser JS, Klein ML, Fiorin G, DeGrado WF. Proc Natl Acad Sci U S A. 2015;112:14260–14265. doi: 10.1073/pnas.1518493112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voth GA. Acc Chem Res. 2006;39:143–150. doi: 10.1021/ar0402098. [DOI] [PubMed] [Google Scholar]
- 36.Manor J, Mukherjee P, Lin Y-S, Leonov H, Skinner JL, Zanni MT, Arkin IT. Structure. 2009;17:247–254. doi: 10.1016/j.str.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo W, Hong M. J Am Chem Soc. 2010;132:2378–2384. doi: 10.1021/ja9096219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manor J, Feldblum ES, Zanni MT, Arkin IT. J Phys Chem Lett. 2012;3:939–944. doi: 10.1021/jz300150v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hong M, Fritzsching KJ, Williams JK. J Am Chem Soc. 2012;134:14753–14755. doi: 10.1021/ja307453v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang RB, Li H, Swanson JMJ, Voth GA. Proc Natl Acad Sci U S A. 2014;111:9396–9401. doi: 10.1073/pnas.1401997111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghosh A, Wang J, Moroz YS, Korendovych IV, Zanni M, DeGrado WF, Gai F, Hochstrasser RM. J Chem Phys. 2014;140:235105. doi: 10.1063/1.4881188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong H, Fiorin G, DeGrado WF, Klein ML. J Phys Chem B. 2014;118:12644–12651. doi: 10.1021/jp5102225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghosh A, Qiu J, DeGrado WF, Hochstrasser RM. Proc Natl Acad Sci U S A. 2011;108:6115–6120. doi: 10.1073/pnas.1103027108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okada A, Miura T, Takeuchi H. Biochemistry. 2001;40:6053–6060. doi: 10.1021/bi0028441. [DOI] [PubMed] [Google Scholar]
- 45.Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, Soto CS, Tereshko V, Nanda V, Stayrook S, DeGrado WF. Nature. 2008;451:596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waegele MM, Gai F. J Phys Chem Lett. 2010;1:781–786. doi: 10.1021/jz900429z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waegele MM, Tucker MJ, Gai F. Chem Phys Lett. 2009;478:249–253. doi: 10.1016/j.cplett.2009.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang W, Markiewicz BN, Doerksen RS, Smith AB, III, Gai F. Phys Chem Chem Phys. 2015;18:7027–7034. doi: 10.1039/c5cp04413h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding B, Hilaire MR, Gai F. J Phys Chem B. 2016;120:5103–5113. doi: 10.1021/acs.jpcb.6b03199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Markiewicz BN, Mukherjee D, Troxler T, Gai F. J Phys Chem B. 2016;120:936–944. doi: 10.1021/acs.jpcb.5b12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma C, Polishchuk AL, Ohigashi Y, Stouffer AL, Schön A, Magavern E, Jing X, Lear JD, Freire E, Lamb RA, DeGrado WF, Pinto LH. Proc Natl Acad Sci U S A. 2009;106:12283–12288. doi: 10.1073/pnas.0905726106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stouffer AL, Nanda V, Lear JD, DeGrado WF. J Mol Biol. 2005;347:169–179. doi: 10.1016/j.jmb.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 53.Yi MG, Cross TA, Zhou HX. Proc Natl Acad Sci U S A. 2009;106:13311–13316. doi: 10.1073/pnas.0906553106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cady SD, Mishanina TV, Hong M. J Mol Biol. 2009;385:1127–1141. doi: 10.1016/j.jmb.2008.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rogers JMG, Polishchuk AL, Guo L, Wang J, DeGrado WF, Gai F. Langmuir. 2011;27:3815–3821. doi: 10.1021/la200480d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goormaghtigh E, Raussens V, Ruysschaert JM. Biochim Biophys Acta, Rev Biomembr. 1999;1422:105–185. doi: 10.1016/s0304-4157(99)00004-0. [DOI] [PubMed] [Google Scholar]
- 57.Tucker MJ, Getahun Z, Nanda V, DeGrado WF, Gai F. J Am Chem Soc. 2004;126:5078–5079. doi: 10.1021/ja032015d. [DOI] [PubMed] [Google Scholar]
- 58.Asplund MC, Zanni MT, Hochstrasser RM. Proc Natl Acad Sci U S A. 2000;97:8219–8224. doi: 10.1073/pnas.140227997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hill PA, Wei Q, Troxler T, Dmochowski IJ. J Am Chem Soc. 2009;131:3069–3077. doi: 10.1021/ja8100566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kochendoerfer GG, Salom D, Lear JD, Wilk-Orescan R, Kent SBH, DeGrado WF. Biochemistry. 1999;38:11905–11913. doi: 10.1021/bi990720m. [DOI] [PubMed] [Google Scholar]
- 61.Mukherjee D, Gai F. Anal Biochem. 2016;507:74–78. doi: 10.1016/j.ab.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pielak RM, Chou JJ. J Am Chem Soc. 2010;132:17695–17697. doi: 10.1021/ja108458u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Frey S, Tamm LK. Biophys J. 1991;60:922–930. doi: 10.1016/S0006-3495(91)82126-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kukol A, Adams PD, Rice LM, Brunger AT, Arkin IT. J Mol Biol. 1999;286:951–962. doi: 10.1006/jmbi.1998.2512. [DOI] [PubMed] [Google Scholar]
- 65.Cady SD, Luo W, Hu F, Hong M. Biochemistry. 2009;48:7356–7364. doi: 10.1021/bi9008837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suydam IT, Boxer SG. Biochemistry. 2003;42:12050–12055. doi: 10.1021/bi0352926. [DOI] [PubMed] [Google Scholar]
- 67.Bagchi S, Fried SD, Boxer SG. J Am Chem Soc. 2012;134:10373–10376. doi: 10.1021/ja303895k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Phongphanphanee S, Rungrotmongkol T, Yoshida N, Hannongbua S, Hirata F. J Am Chem Soc. 2010;132:9782–9788. doi: 10.1021/ja1027293. [DOI] [PubMed] [Google Scholar]
- 69.Piletic IR, Moilanen DE, Spry DB, Levinger NE, Fayer MD. J Phys Chem A. 2006;110:4985–4999. doi: 10.1021/jp061065c. [DOI] [PubMed] [Google Scholar]
- 70.Kim YS, Hochstrasser RM. J Phys Chem B. 2009;113:8231–8251. doi: 10.1021/jp8113978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kwak K, Park S, Finkelstein IJ, Fayer MD. J Chem Phys. 2007;127:124503. doi: 10.1063/1.2772269. [DOI] [PubMed] [Google Scholar]
- 72.Ma JQ, Pazos IM, Gai F. Proc Natl Acad Sci U S A. 2014;111:8476–8481. doi: 10.1073/pnas.1403224111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chung JK, Thielges MC, Lynch SR, Fayer MD. J Phys Chem B. 2012;116:11024–11031. doi: 10.1021/jp304058x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nanda V, Cristian L, Toptygin D, Brand L, DeGrado WF. Chem Phys. 2013;422:73–79. doi: 10.1016/j.chemphys.2012.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang J, Ma C, Fiorin G, Carnevale V, Wang T, Hu F, Lamb RA, Pinto LH, Hong M, Klein ML, DeGrado WF. J Am Chem Soc. 2011;133:12834–12841. doi: 10.1021/ja204969m. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.