Abstract
Rafflesia is a biologically enigmatic species that is very rare in occurrence and possesses an extraordinary morphology. This parasitic plant produces a gigantic flower up to one metre in diameter with no leaves, stem or roots. However, little is known about the floral biology of this species especially at the molecular level. In an effort to address this issue, we have generated and characterised the transcriptome of the Rafflesia cantleyi flower, and performed a comparison with the transcriptome of its floral bud to predict genes that are expressed and regulated during flower development. Approximately 40 million sequencing reads were generated and assembled de novo into 18,053 transcripts with an average length of 641 bp. Of these, more than 79% of the transcripts had significant matches to annotated sequences in the public protein database. A total of 11,756 and 7,891 transcripts were assigned to Gene Ontology categories and clusters of orthologous groups respectively. In addition, 6,019 transcripts could be mapped to 129 pathways in Kyoto Encyclopaedia of Genes and Genomes Pathway database. Digital abundance analysis identified 52 transcripts with very high expression in the flower transcriptome of R. cantleyi. Subsequently, analysis of differential expression between developing flower and the floral bud revealed a set of 105 transcripts with potential role in flower development. Our work presents a deep transcriptome resource analysis for the developing flower of R. cantleyi. Genes potentially involved in the growth and development of the R. cantleyi flower were identified and provide insights into biological processes that occur during flower development.
Introduction
Rafflesia is a genus of holoparasitic flowering plants found only in the tropical rainforest of Southeast Asia. The genus is well known for members with extraordinary flower size and is deemed to represent the world’s largest single flower [1]. In contrast with conventional flowering plants, Rafflesia species are highly reduced in vegetative parts and the only visible component being the mycelium-like structure which penetrates their host plants [2]. Previous studies have reported on the absence of a plastid/chloroplast genome in Rafflesia [3]. This suggests that Rafflesia has lost its plastidial organelle, and it is therefore not surprising that this plant does not have the capability to carry out photosynthesis and thus depends completely on its host, the vines of the Tetrastigma tree species for nutrients and water supply. More interestingly, a recent study demonstrated that Rafflesia acquires benefits from its host through horizontal gene transfer mechanism from host to flower [4].
To date, more than 30 species of Rafflesia have been reported. Despite rapidly increasing discovery of new species, the biology of Rafflesia remains poorly understood. This may be mainly due to the limited resources available for research. The occurrence of Rafflesia is very rare as it takes a long time to complete its lifecycle [5]. The bud takes up to nine months to develop into a flower, but the flower lifespan is very short, as it blooms for only five to seven days before decomposing.
Molecular sequence information on Rafflesia is scarce. As a result, genome-driven elucidation of biological processes, particularly those that govern the growth and development of this giant flower plant remains under explored. In an effort to address this matter, we utilised next generation sequencing (NGS) technology to characterise the transcriptome of the perigone lobe (petal-like structure) of Rafflesia cantleyi. Sequence data were processed, assembled, annotated and classified into putative functional categories according to the Gene Ontology (GO) framework. The transcripts were further grouped into pathways based on the Kyoto Encyclopaedia of Genes and Genomes (KEGG) mapping. Transcripts from the developing flower were then compared to transcripts from a floral bud generated in a previous study [4] to identify genes involved in flower development. This is the deepest transcriptomic study to date of the developing flower of R. cantleyi and the results will serve as a reference for further gene expression and functional genomics studies.
Materials and Methods
Plant material and RNA isolation
The R. cantleyi flower (Fig 1) was collected from Lata Jarum, Pahang, Malaysia (Permission from Pahang State Forestry Department—reference no. PHN. PHG. (PEM) 118/146 Bhg. 3 (123)). The perigone lobe tissue was collected from the flower three days after it began blooming. One of the five perigone lobes present in the flower was dissected and transported back to the laboratory. Once in the laboratory, samples were fragmented into small pieces and stored at -80°C. Total RNA was extracted according to the method described by López-Gomez et al [6]. RNA integrity and quantity were quantified by Agilent BioAnalyzer (Agilent Technologies, USA) and Spectrophotometer ND-1000 (NanoDrop, USA).
Fig 1. The Rafflesia cantleyi flower collected from Lata Jarum, Pahang, Malaysia.
Sequencing and de novo assembly
A cDNA library was constructed with the TruSeq RNA Sample Prep kit (Illumina, USA) and sequenced on a HiSeq 2000 (Illumina, USA) with paired-end 90 bp read lengths. The reads were trimmed and filtered with FASTX-Toolkit to eliminate low quality reads. Reads were considered as high quality if more than 70% of the bases had phred values of more than Q20. Reads shorter than 30 bp after trimming were discarded from further analysis. Reads produced in this study have been deposited in the NCBI Sequence Read Archive (SRA) (accession number SRP075857). Three assembly tools, namely Oases [7], Trinity [8] and CLC Genomics Workbench (Version 4.9, CLC Bio), were evaluated for the de novo assembly of the R. cantleyi perigone lobe transcriptome. Preliminary assembled transcripts from each assembly tool were compared based on the number of transcripts generated, and average and total length of transcripts. The generated transcripts from each tool were then compared against the Arabidopsis thaliana proteome using BLASTX to determine the transcript set with the highest database hits by applying an E-value cut-off of 1E-06. Final assembly was carried out using Oases v0.2.08 (http://www.ebi.ac.uk/~/zerbino/oases/) [7]. The appropriate k-mer and coverage cut-off value used in the assembly was determined by a perl script (VelvetOptimiser-2.2.0.pl). K-mer size range from 25 to 79 were tested for the best N50 value while the coverage cut-off was automatically determined by the script. All other parameters were on default settings. Transcripts with a minimum length of 200bp obtained from final assembly were used for annotation and further analysis. This Transcriptome Shotgun Assembly project has been deposited at DDBJ/EMBL/GenBank under the accession GFBU00000000. The version described in this paper is the first version, GFBU01000000.
Similarity search and functional annotation
To deduce the putative function, all transcripts were subjected to BLASTX analysis against the GenBank non-redundant protein database. Hits were considered as significant if E-value ≤ 1E-06. Gene ontology analysis was conducted on the annotated sequences through Blast2GO [9] followed by functional classification based on plant-specific GO slim ontology [10]. Putative metabolic pathway assignments were carried out according to KEGG mapping with the E-value cut-off of 1E-06. KOBAS (version 2.0.2) [11] was carried out in a stand-alone environment to identify enrichment in the KEGG pathway. To further characterise the transcriptome, we searched the annotated transcripts for possible functions involved in KOG classification. In addition, transcription factors represented in our samples were searched by performing BLASTX with an E-value cut-off of 1E-06 against transcription factor protein sequences obtained from the Plant Transcription Factor Database (PlantTFDB: http://planttfdb.cbi.pku.edu.cn/download.php) [12].
Transcript abundance estimation
To analyse transcript abundance levels in the flower transcriptomes, Tophat [13] incorporating the Bowtie algorithm was used to align RNA-seq reads to the transcriptome. The aligned read files were processed by Cufflinks [14] to estimate the relative abundances of transcripts. The abundance of transcripts were upper-quartile normalised and corrected for sequence bias internally in Cufflinks. The unit of measurement for transcript abundance used is fragments per kilobase of exon per million fragments mapped (FPKM).
Differential expression analysis
To analyse the difference of transcript abundance between the two floral development stages, which is bud and flower of R. cantleyi, the Cuffdiff algorithm was used. Cuffdiff allowed for the discovery of transcripts that are common, differentially expressed or present/absent between bud and flower tissues of R. cantleyi. Transcriptome data of floral bud used in this analysis was downloaded from NCBI SRA (http://www.ncbi.nlm.nih.gov/sra) (accession number: SRA052224) [4]. A reference transcriptome was built by performing a combine assembly of bud and flower RNA-seq reads using Oases. Tophat was used to align reads to the reference transcriptome while Cufflinks processed the alignment files produced by TopHat. Cuffmerge produces a combined GTF file, which is passed to Cuffdiff to re-estimate the abundance of transcripts listed in the GTF file and concurrently tests for differential expression. Genes with FDR < 0.05 were considered significant. A heatmap of selected differentially expressed genes was generated by CummeRbund (http://compbio.mit.edu/cummeRbund/).
Reverse transcription quantitative real-time PCR (RT-qPCR)
For RT-qPCR analysis, eight candidate genes were selected from various levels of abundance. Actin was selected as the reference gene for relative quantification of gene expression. Gene-specific primers (S1 Table) were designed using Primer-Blast (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). The primers pairs specificity and efficiency were examined using standard real-time PCR, and were further validated by gel electrophoresis to ensure correct amplicons. RT-qPCR was performed on DNase-treated RNA (RIN number 7.5; 20 ng/μl) in three technical replicates using QuantiNova® SYBR® Green RT-PCR Kit (Qiagen) following the manufacturer’s instructions. The amplification protocol: 10 min at 50°C and 2 min at 95°C for the reverse transcription, followed by 40 cycles of 5 s at 95°C and 10 s at 60°C using the CFX96™ Real-Time Detection System (Bio-Rad). The relative expression of each gene to actin was calculated from mean Ct ± standard error mean using the 2-ΔCt method. Relative expression values from RNA-seq data were calculated from the ratio of FPKM value of individual genes relative to that of actin. Correlation analysis of relative expression values from RNA-seq and RT-qPCR was performed using Microsoft EXCEL 2016.
Results and Discussion
Sequence generation and de novo assembly
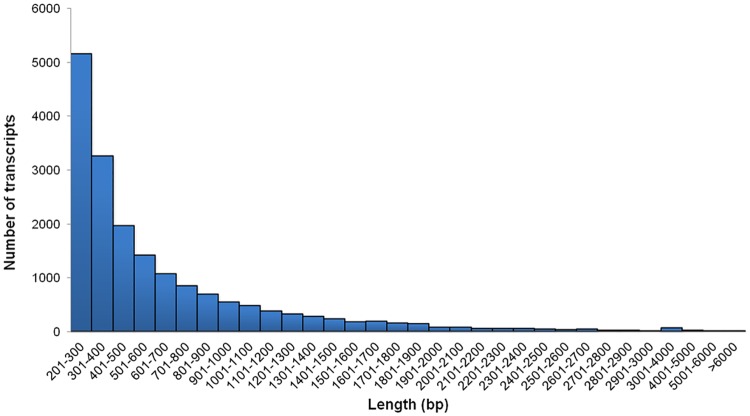
Deep sequencing of transcriptome derived from the perigone lobe of Rafflesia was performed using the Illumina RNA-seq platform. More than 20 million paired-end reads with a length of 90 bp were obtained and subjected to filtering and trimming to remove low quality reads. After pre-processing, a total number of 40,768,187 high quality reads, corresponding to an average length of 87 bp were obtained. More than 99% of the raw reads were of quality more than Q20 (a sequencing error probability of 0.01), indicating the high quality of cDNA library and reads obtained. The summary of sequencing data is shown in Table 1. After cleaning and quality checks, the high quality reads were subjected to sequence assembly. In order to generate better assemblies, several assembly tools were evaluated for their performance, namely Oases, Trinity and CLC Genomics Workbench. Based on the number of transcripts, and average and total transcript length, Oases stood out as the best performer. Oases produced 38,783 transcripts with an average length of 1,172 bp, followed by Trinity (31,363 transcripts, average length of 865 bp) and CLC Genomics Workbench (26,294 transcripts, average length of 632 bp). Oases assembly also showed the largest significant similarity with Arabidopsis proteins (68.8%) compared to the transcripts generated by Trinity (55.6%) and CLC Genomics Workbench (44.9%) [Table 2]. Taken together, the results suggested Oases as the most suitable assembly tool for our dataset. As this study emphasises more on downstream analyses, the assembly output is crucial in determining the quality of the results. Thus, more stringent and conservative parameters were utilised during the assembly process to increase the confidence level of downstream analyses. A k-mer distribution generated by the Velvet perl script (S1 Fig) predicted that a k-mer size of 69 will produce the best N50 value, and was used in the final assembly process. In addition, a coverage cut-off of 4.86 was set to discard low-coverage transcripts. Subsequently, the high quality reads were assembled into 18,053 transcripts with a mean length of 641 bp. The size distribution of these transcripts is shown in Fig 2. Although most transcripts were between 200 and 300 bp in length, the assembly did produce a substantial number of longer contigs where 7,666 transcripts were more than 500 bp in length.
Table 1. Statistical summary of Rafflesia cantleyi sequence data.
| Total number of paired-end reads (before trimming) | 40,993,980 |
| Total number of read base pairs (bp) | 3,689,458,200 |
| Average read length (before trimming; bp) | 90 |
| Total number of read paired-end (after trimming) | 40,768,187 |
| Total number of read singletons (after trimming) | 306,421 |
| Average read length (after trimming; bp) | 87 |
| Total number of reads assembled | 33,002,956 |
| Total number of transcripts produced | 18,053 |
| Average length of transcripts (bp) | 641 |
| Total number of transcript base pairs (bp) | 11,572,001 |
Table 2. Comparison of de novo assembly data using Oases, Trinity and CLC Genomics Workbench.
| Oases | Trinity | CLC Genomics Workbench | |
|---|---|---|---|
| Number of transcripts | 38,783 | 31,363 | 26,294 |
| Average transcript length (bp) | 1172 | 865 | 632 |
| Total transcript length (bp) | 45,471,333 | 27,056,569 | 16,624,957 |
| Annotated transcripts | 26,672 (68.8%) | 17,452 (55.6%) | 11,807 (44.9%) |
Fig 2. Length distribution of Rafflesia cantleyi flower transcripts obtained from de novo assembly.
Functional annotation
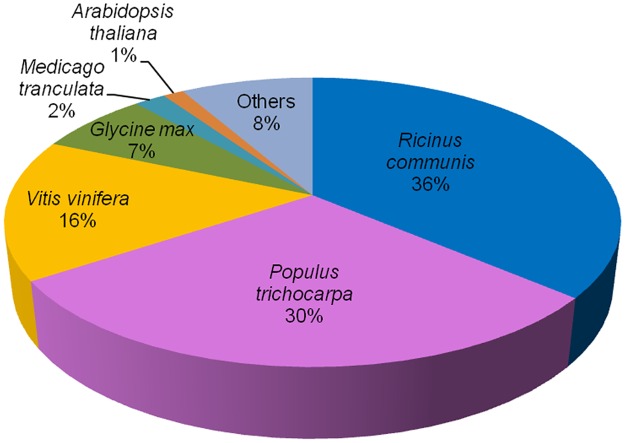
A comparison of the transcript sequences against the GenBank non-redundant protein database found a total of 14,336 (79.41%) transcripts with significant matches (S2 Table). The majority of these annotated transcripts displayed the highest similarity to genes from plants, with more than a third of the transcripts matched to Ricinus communis, partly due to the over representation of sequences from this species in the database and its close phylogenetic relationship with R. cantleyi (Fig 3).
Fig 3. Species distribution of annotated Rafflesia cantleyi flower transcripts.
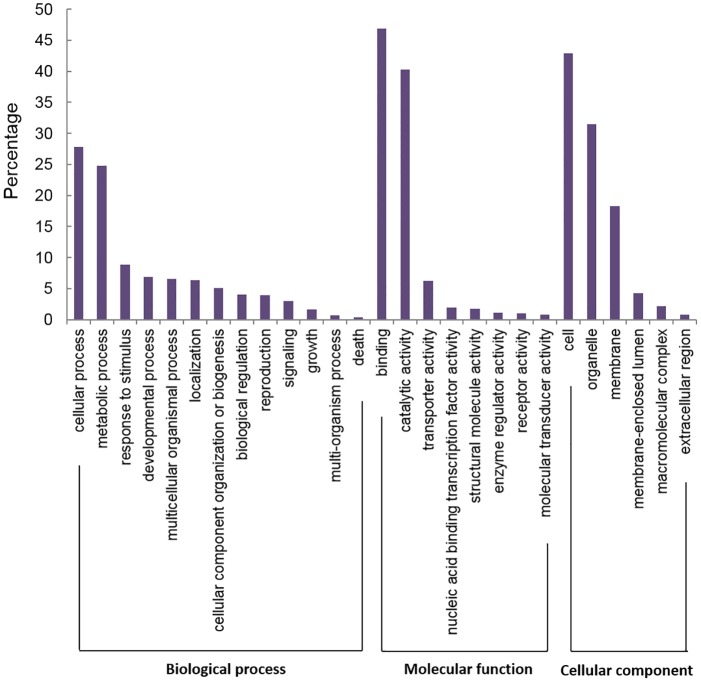
All transcripts with BLAST hits were further classified using a set of plant specific GO slim terms. Information on protein domains were retrieved by InterProScan via Blast2GO and the corresponding annotations were merged with the annotated GOSlim terms. A total of 11,756 transcripts (65.1%) were annotated and classified to the three main categories: biological processes, molecular functions and cellular components (Fig 4). The biological process category was dominated by cellular (27.80%) and metabolic (24.74%) processes, which may be related to the developmental activities taking place during flowering. Besides that, a total of 930 transcripts were annotated with GOSlim terms related to post-embryonic development (GO: 0009791), 268 transcripts with flower development (GO: 0009908) and another 167 transcripts with pollination (GO: 0009856). The molecular function category was most highly represented by binding (46.89%) and catalytic activity (40.29%), while the cellular component category was dominated by cell (42.91%) and organelle (31.51%).
Fig 4. GO annotation of Rafflesia cantleyi flower transcripts.
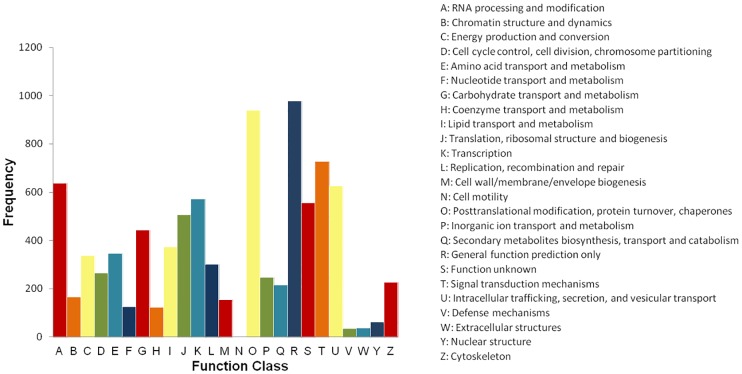
Overall, 7,981 transcripts had KOG classification, which were distributed into 25 categories (Fig 5). Among these, the most represented group was “general function prediction only” (10.87%), followed by “post-translational modification, protein turnover, chaperone” (10.43%), “signal transduction mechanisms” (8.07%) and “RNA processing and modification” (7.24%). A total of 627 transcripts (6.96%) were in the intracellular trafficking, secretion, and vesicular transport category and 572 transcripts (6.35%) were categorised as having a role in transcription.
Fig 5. KOG functional classification of Rafflesia cantleyi flower transcripts.
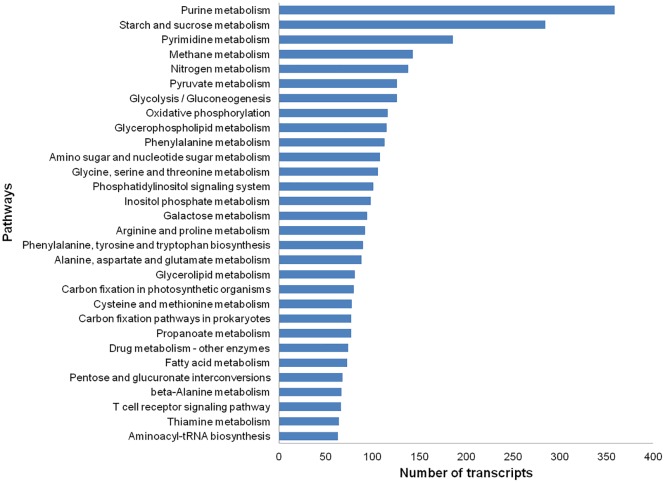
We also retrieved KEGG pathways in which the transcripts were mapped by performing Enzyme Commission (EC) annotation via Blast2GO. In total, 2,537 transcripts were annotated with 627 ECs and were assigned to 129 different pathways (Fig 6; S3 Table). KOBAS analysis showed that the top five enriched pathways (FDR<0.05) were “metabolic pathways”, “biosynthesis of secondary metabolites”, “biosynthesis of amino acids”, “carbon metabolism” and “pyruvate metabolism” (S4 Table). A large assortment of pathways derived from this analysis showed that genes sampled from the R. cantleyi developing flower transcriptome represent a range of highly diverse functions. Notable dominant categories of metabolisms included carbohydrate metabolism (1322 transcripts), amino acid metabolism (860), lipid metabolism (620), and purine metabolism (545). Interestingly, a number of transcripts mapped to plant hormone signal transduction pathways that are known to regulate a variety of developmental processes such as senescence, stress response, and plant vegetative and reproductive growth. As shown in Table 3, such transcripts included those related to the signalling pathways mediated by ethylene, gibberellin, indole acetic acid, jasmonic acid, salicylic acid, auxin and cytokinin.
Fig 6. Top KEGG pathways represented in the Rafflesia cantleyi flower transcriptome.
Table 3. The pathways and products involved in plant hormone signal transduction.
| Pathway | Product | Pathway ID | Number of transcripts |
|---|---|---|---|
| Phenylalanine metabolism | Salicylic acid | Ko00360 | 113 |
| Cysteine and methionine metabolism | Ethylene | Ko00270 | 78 |
| Tryptophan metabolism | Auxin | Ko00380 | 57 |
| α-Linolenic acid metabolism | Jasmonic acid | Ko00591 | 55 |
| Indole alkaloid biosynthesis | Indole acetic acid | Ko00901 | 3 |
| Diterpenoid biosynthesis | Gibberellin | Ko00904 | 2 |
| Zeatin biosynthesis | Cytokinin | Ko00908 | 2 |
Distribution of transcription factors
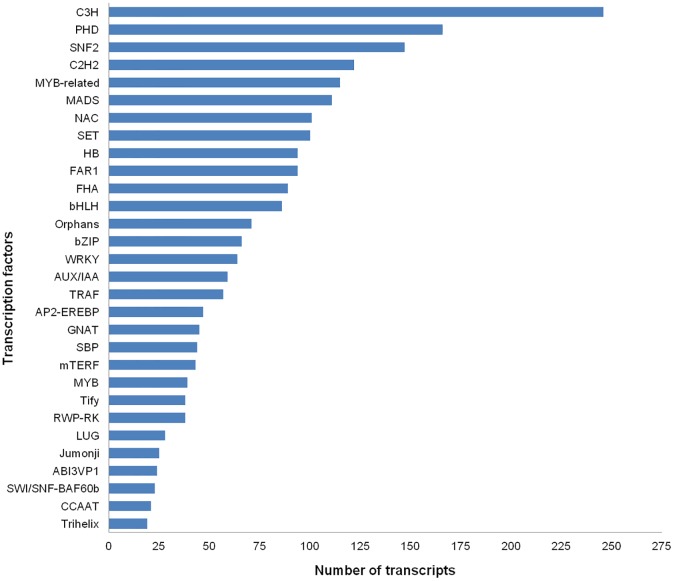
Transcription factors are proteins consisting of various multigene families that bind to specific DNA sequences. In plants, the function of transcription factors are closely linked to gene expression and plant development. A total of 2,504 putative genes distributed in 75 transcription factor families were identified in the R. cantleyi developing flower transcriptome, representing 13.87% of the total transcripts (Fig 7; S5 Table). The majority of these annotated transcripts displayed the highest homology to the C3H family (246 transcripts, 9.82%). A total of 166 transcripts (6.63%) fell into the PHD family, followed by the SNF2 family (147 transcripts, 5.87%) and the C2H2 family (122 transcripts, 4.87%).
Fig 7. Transcription factor families identified in the Rafflesia cantleyi flower transcriptome.
Previous studies have shown that one of the members of the C2H2 family, JAGGED (JAG) is expressed in the distal petal blade during late phase of development. It is believed to control petal growth and to promote cell-cycle progression and cell expansion [15–17]. In addition, quite a number of transcripts matched with transcription factors of the MADS family. MADS-box genes in plants have been intensively studied and many of the MADS family members have been shown to be involved in floral organ specification and development [18]. Several other transcription factor families associated with flower development were also found, including basic helix-loop-helix (bHLH) [19] and basic leucine zipper (bZIP) [20]. Previous work has demonstrated that mutations in the bHLH transcription factor, BIG PETAL (BPE) caused the failure of post-mitotic cell expansion and resulted in petal size change [21]. In short, the results obtained indicate that a diverse range of transcription factors is active in the R. cantleyi developing flower, consistent with what has been reported in floral development biology. Profiling of transcription factors will be a crucial starting point in elucidating Rafflesia-specific floral development.
Transcript abundance analysis
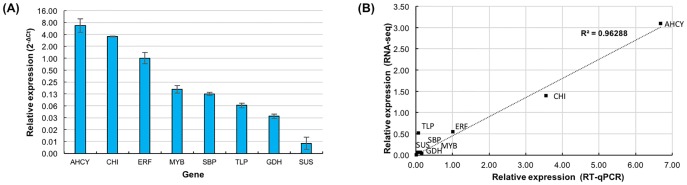
To analyse the abundance of R. cantleyi transcripts within the sample, quality-trimmed RNA-seq reads were mapped to the transcripts of R. cantleyi. The relative abundance of transcripts was calculated in fragments per kilobase of exon per million fragments mapped (FPKM) value. FPKM values (>0) for more than 76% of the transcripts were determined. The highest FPKM value is 17634.90 while the lowest FPKM value is 2.57, indicating a wide range of expression levels of R. cantleyi transcripts. Based on these FPKM values, transcripts were arbitrarily classified into five categories (very low, low, moderate, high and very high) (Table 4). The category with the largest fraction of transcripts (97.4%) showed low abundance (FPKM >10–500), followed by moderate (FPKM >500–1000) and very low (FPKM <10) abundance categories. Categories described as very low and very high each constituted less than 1% of the transcripts analysed (Table 4). To validate the transcript abundance, RT-qPCR was performed to quantify the relative expression of eight selected genes with different expression levels, i.e. two genes with a very high level of abundance, two genes with a moderate level of abundance, three genes with a low level of abundance and one gene with a very low level of abundance. The results showed a high correlation (R2>0.96) between the RNA-seq data and RT-qPCR (Fig 8).
Table 4. Distribution of transcripts according to the levels of abundance.
| Levels of abundance | FPKM value | Number of transcripts | Percentage (%) |
|---|---|---|---|
| Very low | <10 | 98 | 0.71 |
| Low | >10–500 | 13,421 | 97.40 |
| Moderate | >500–1000 | 164 | 1.19 |
| High | >1000–1500 | 50 | 0.36 |
| Very high | >1500 | 52 | 0.38 |
Fig 8. RT-qPCR validation of transcript abundance from RNA-seq analysis.
(A) Relative expression of eight genes based on actin as the reference housekeeping gene. Y-axis is shown in log base 2 scale for clearer representation of genes with low expression values. (B) Correlation analysis of relative gene expression values from RT-qPCR and RNA-seq analysis. Genes studied include adenosylhomocysteinase (AHCY), chitinase (CHI), ethylene responsive transcription factor (ERF), MYB transcription factor (MYB), selenium binding protein (SBP), thaumatin-like protein (TLP), glutamate dehydrogenase (GDH) and sucrose synthase (SUS). The relative expression for RT-qPCR was calculated by using the 2-ΔCt method based on the reference gene, actin; whereas the relative expression values for RNA-seq are FPKM ratios of individual genes relative to that of actin.
For further analysis, we focused our analysis on selected genes in the highest abundance category (52 transcripts, 0.38%), which display functions that could provide insightful leads into Rafflesia floral development. Thirty-five out of the 52 transcripts that showed a very high abundance level had significant matches with protein sequences in the GenBank non-redundant database (S6 Table). The transcript with the highest FPKM value (17634.90) matched with the sufE-like protein of Cucumis sativa. Previous studies have suggested that the sufE-like protein in Arabidopsis (AtSufE) acts as an inter-organellar coordinator of Fe-S cluster biogenesis by interacting with the cysteine desulfurases (AtSufS) in plastids and AtNifS1 in mitochondria [22]. This ensures a balance of Fe-S cluster biogenesis between plastids and mitochondria, which is vital during embryogenesis. Three different SufE proteins, termed SufE1, SufE2 and SufE3 can be found in Arabidopsis. SufE1 is expressed in all tissues of Arabidopsis and its knockout is lethal [23, 24]. SufE2 was found to be abundantly expressed in the Arabidopsis flower, while SufE3 is ubiquitously expressed in all Arabidopsis organs [24]. In contrast to SufE1 and SufE2, SufE3 contains both a SufE-like domain and a domain similar to the bacterial quinolinate synthase NadA. SufE3 is considered as the quinolinate synthase enzyme of Arabidopsis, which is involved in a critical step during NAD biosynthesis. Our transcriptome showed the presence of both the SufE-like and quinolinate synthase domains. This may imply that the SufE-like protein found in our transcriptome displays a similar role with SufE3 in Arabidopsis, which also possesses both SufE and quinolinate synthase activities.
A putative homolog of fructose biphosphate aldolase (FBA) also showed a similar overrepresentation (FPKM value of 10157.30) in the flower transcriptome of Rafflesia. In plants, FBA is a key metabolic enzyme for glycolysis and gluconeogenesis [25]. It catalyses the condensation of fructose-1,6-biphosphate and sedoheptulose-1,7-biphosphate in the Calvin cycle [26]. It also plays important roles in sugar production, as well as abscisic acid (ABA) and stress signalling in plant. Previous study of eight FBA genes in Arabidopsis showed that the AtFBA gene family is greatly diversified in terms of subcellular and tissue localisation, and abiotic stress responsive expression patterns [27]. Studies conducted on these AtFBAs suggested that AtFBA1, AtFBA2, AtFBA5 and AtFBA7 were highly expressed in shoots, while AtFBA4 and AtFBA8 were found to have a higher expression in flowers. AtFBA3 and AtFBA6 have lower expression than the other AtFBAs but were shown to be specifically expressed in roots and flowers respectively. Moreover, AtFBA genes have been suggested to not only play important roles in development, but also in sugar, light and abiotic stress responses [27]. Given the significantly high levels of FBA transcripts in the R. cantleyi flower, we speculate that these genes may be involved in the flower development process, as in Arabidopsis.
Another transcript identified to be highly expressed in the R. cantleyi flower transcriptome is a putative homolog of the late embryogenesis abundant (LEA) gene (FPKM value of 4093.94). LEA proteins were first characterised in cotton and were abundantly found in the embryos at the late stage of seed development [28]. However, recent studies showed that LEA proteins in Arabidopsis are of a wide range of sequence diversity, intracellular localisation and expression patterns [29]. Besides that, LEA protein has been hypothesised to play a protective role in plant cells that might be essential for the survival of plant under various stress conditions [28, 30–32]. Many LEA genes can also be induced by ABA in both reproductive and vegetative tissues [29, 33, 34]. In general, expression of LEA proteins has been largely associated with water deficiency in plant tissues. The perigone lobes of Rafflesia have a high water content and water losses are at the highest level when the surface area is fully exposed during blooming. Thus, we suggest that the abundant LEA genes found in the R. cantleyi flower transcriptome confer a protective function towards water lost in perigone tissues. Accumulation of LEA proteins is believed to serve as a defence mechanism and inhibit excessive fluid loss.
Detection of differentially expressed genes (DEGs)
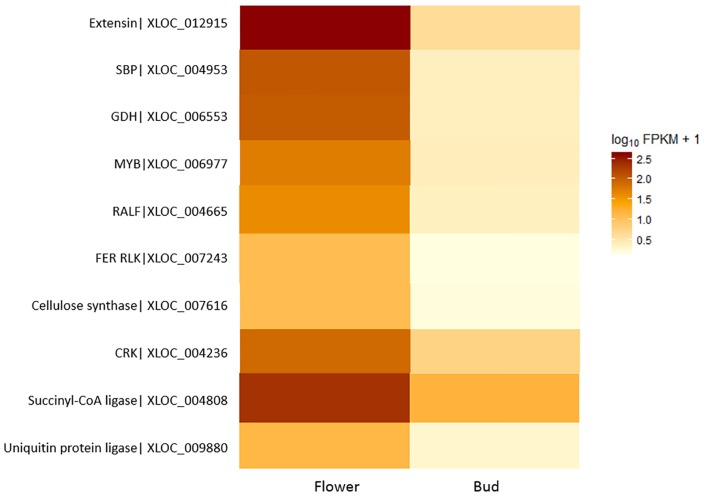
Due to difficulty in obtaining Rafflesia material growing in the wild, this study was limited to transcriptome analysis of the perigone lobe. However, an existing floral bud transcriptome data [4] presented an opportunity to perform a preliminary comparative gene expression analysis of two floral development stages. One of the difficulties we had faced during this work was the absence of a suitable reference genome. Due to this, our strategy to quantify gene expression was to align RNA-seq reads back to the contigs that were reconstructed from the floral bud and developing flower transcriptome data sets. We had used Cuffdiff because it is more optimally designed to detect differential expression at the transcript level compared to the other DEG analysis tools. While edgeR has been reported to be able to detect more DEGs than Cuffdiff in many of previous comparative studies, it is more prone to introduce false positives. With the lack of reference data, we felt that it was more prudent to limit the introduction of false positives that we would not be able to filter or address appropriately at a downstream stage of the analysis. Furthermore, due to the limitation in getting biological replicates for DEG analysis, we had applied a more stringent gating to define DEGs. As Cuffdiff is more conservative in making differential expression calls, we think that it is more suitable for our study. Comparison of transcript abundance between the two transcriptome data sets revealed a total of 840 transcripts that were differentially expressed between the developing flower and the floral bud (S7 Table). Of these, 105 genes displayed higher expression in the perigone tissue of the flower compared to the floral bud, whereby 91 genes had GenBank matches (S7 and S8 Tables). To improve our understanding of Rafflesia floral development, we decided to examine several genes with functions related to stress response, cellular signalling, cell wall formation and transcriptional regulation. A heat map of these selected genes is shown in Fig 9.
Fig 9. Heat map of selected genes that are differentially expressed between the developing flower and the floral bud of Rafflesia cantleyi.
Perigone lobes are petal-like structures of the Rafflesia flower. In angiosperms, petal development typically involves two stages, which is cell division and differentiation, followed by rapid growth through cell expansion [35, 36]. Differential analysis revealed that a number of genes associated with stress response were differentially expressed between the developing flower and the floral bud. This is not surprising as studies have shown that a large proportion of stress-related genes were also highly expressed in developing flowers of Arabidopsis, Antirrhinum and rose [37–39]. Such accumulation of stress-related proteins is believed to be induced by the rapid growth of petals and sudden increase in the rate of cellular metabolism [38]. Also, it has been suggested that petals are extremely sensitive toward stress conditions and therefore, abundant expression of stress response genes protects cells against intracellular or external stresses during flower development [39].
One of the transcripts encoding the stress-related gene, glutamate dehydrogenase (GDH) (XLOC_006553) showed more than 6-fold higher expression in the R. cantleyi flower in relation to the floral bud (S7 Table). GDH is known to catalyse the reversible reaction of glutamate and 2-oxyglutarate, which is a key reaction in carbon and nitrogen metabolisms in plants [40]. Previous studies of GDH in the tobacco plant showed that the gene was up-regulated by elevated ammonium levels, suggesting that it functions in ammonium detoxification by assimilating some of the ammonium ions [41]. Several other studies also reported the function of GDH in responding to abiotic and physiological stress conditions [42, 43]. Furthermore, GDH was found to localised in the cytosol of senescing organs, implying that it plays a role in controlling plant growth and productivity [44]. Taken together, we suggest that GDH plays an important role in enabling R. cantleyi developing flower cells to adapt or tolerate stress conditions during the rapid growth of perigone tissues.
Another differently expressed stress-related gene identified is the selenium binding protein (SBP) (XLOC_004953), which showed more than 6-fold higher expression in the R. cantleyi flower compared to the floral bud. High expression of SBP in Oryza sativa has been reported to enhance tolerance against pathogens [45]. In Arabidopsis, SBP was suggested to counter metal-related stresses and take part in the cadmium detoxification process [46, 47]. In addition, SBP has been reported to express ubiquitously in non-stress conditions especially in actively growing tissues and during seed development, highlighting its possible role in plant development [46]. Besides that, gene encoding succinyl-CoA ligase (XLOC_004808) and ubiquitin protein ligase (XLOC_009880) were also identified as up-regulated stress-related genes in the R. cantleyi flower.
Several genes involved in cellular signalling were differentially expressed between the R. cantleyi flower and floral bud. For example, XLOC_004665 that codes for a putative rapid alkalinisation factor (RALF) precursor showed a 4-fold increase of expression in the flower compared to the bud. RALF was first isolated from tobacco leaves and previous studies have shown that it arrested the growth of roots and pollen tubes in Nicotiana attenuate, Arabidopsis and tomato (Solanum lycopersicum) [48–50]. Besides that, there is evidence showing that RALF is a regulator of nodulation in M. truncatula [51]. Characterisation of five RALF-like genes in Solanum chacoense has also revealed the role of RALF proteins in plant development [52]. Increased expression of the RALF precursor gene in the R. cantleyi flower indicates that it plays an important role in regulating flower development.
Receptor-like kinase (RLK) proteins are known to play important roles in signal transduction pathways [53]. Several genes encoding RLKs were also identified to be differentially expressed in R. cantleyi, including XLOC_007243 (4.35-fold increase) and XLOC_004236 (3.97-fold increase), which are homologs of FERONIA (FER) receptor-like kinase and cysteine-rich receptor-like protein kinase, respectively. FER was previously revealed to act as a regulator for female fertility and mediating pollen tube rupture in the female gametophyte of Arabidopsis [54, 55]. It was also found to regulate RHO GTPase signalling of root hair development. Taken together, these findings indicate that FER plays specific growth and development roles in R. cantleyi.
Morphological differences between a developing flower and floral bud of Rafflesia is extremely pronounced, particularly in the extent of petal formation. As cell expansion in plants is highly dependent on cell wall structure and composition, genes encoding cell wall proteins and enzymes that are involved in such modifications would be differentially expressed during petal development [38]. Thus, this supports our observation of higher expression of cell wall associated genes in perigone tissue of the developing flower. Two such genes are extensin (XLOC_012915; 6.87-fold increase) and cellulose synthase (XLOC_007616; 4.12-fold increase). In Arabidopsis, expression of extensin has been detected in roots and inflorescences, and at regions of abscission and senescence [56]. Cellulose synthase is an enzyme involved in the synthesis of cellulose, which is the main component of plant cell walls [57].
Classes of transcription factors found in the developing flower transcriptome were described in a preceding section. Regulation of gene expression by transcription factors is instrumental during changes in plant development. XLOC_006977, which codes for a MYB transcription factor showed a greater than 5-fold increase in expression in the developing flower compared to the bud. In Antirrhinum majuspetals, there is evidence for control of different aspects of petal epidermal cell shape and subsequently overall petal presentation by a MYB-related transcription factor [58]. Homologs of this gene have also been identified in Arabidopsis and Petunia hybrid [59]. Changes in the shape of petal epidermal cells are believed to affect the perceived intensity of the colour signal that is important to attract pollinators. SEPALLATA (SEP) MADS box transcription factor was also identified in this study (XLOC_005948) and it showed a greater than 4-fold increase in gene expression in the flower compared to the bud. SEP appears to act in conjunction with another two MADS box transcription factors, namely APETALA3 (AP3) and PISTILLATA (PI), to regulate petal-specific differentiation [60].
Conclusions
The biology of Rafflesia flower development is fascinating yet elusive. This is largely due to challenges in collecting ample and suitable material in the wild, lack of analytical methodologies developed for this plant species and extremely limited molecular resources. This study is an effort to apply a transcriptomic approach to profile genes expressed in R. cantleyi, particularly those that are abundantly expressed in the developing flower. A total of 18,053 transcripts were assembled from RNA sequencing of perigone tissues, with close to 80% assigned putative functions. This data set forms the first in-depth sequence resource in Rafflesia from which we have highlighted important genes that could provide clues to elucidating Rafflesia flower development. In future, characterisation of transcriptomes of sequential developmental stages from bud to mature flower will provide valuable information to shed light on its species-specific gene functions and unique floral biology.
Supporting Information
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability
All reads produced in this study are available from the NCBI Sequence Read Archive (SRA) (accession number SRP075857). This Transcriptome Shotgun Assembly project has been deposited at DDBJ/EMBL/GenBank under the accession GFBU00000000. The version described in this paper is the first version, GFBU01000000.
Funding Statement
The work was supported financially by Research University Grants from Universiti Kebangsaan Malaysia (Grant No. UKM--‐AP--‐KPB--‐18--‐2010, LAUREATE--‐2013---‐001 and GUP--‐2016--‐008). The funder had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. Mote Research Limited provided support in the form of salaries for PHD, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Nais J. Rafflesia of the world. Malaysia: Natural History Publications; 2001. [Google Scholar]
- 2.Kuijt J. The biology of parasitic flowering plants. Berkeley: University of California Press; 1969. [Google Scholar]
- 3.Molina J, Hazzouri KM, Nickrent D, Geisler M, Meyer RS, Pentony MM, et al. Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae). Mol Biol Evol. 2014;31(4): 793–803. 10.1093/molbev/msu051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xi Z, Bradley RK, Wurdack KJ, Wong K, Sugumaran M, Bomblies K, et al. Horizontal transfer of expressed genes in a parasitic flowering plant. BMC Genomics. 2012;13: 227 10.1186/1471-2164-13-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meijer W. Rafflesiaceae. Flora Malesiana. 1997;1(13): 1–42. [Google Scholar]
- 6.López-Gomez R, Gomez-Lim MA. A method of extracting intact RNA from fruits rich in polysaccharides using ripe mango mesocarp. HortScience. 1992;27: 440–442. [Google Scholar]
- 7.Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28(8): 1086–1092. 10.1093/bioinformatics/bts094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol. 2011;29(7): 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18): 3674–3676. 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- 10.Conesa A, Götz S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int J Plant Genomic. 2008;2008: 619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li X, Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucl Acids Res. 2011;39(2): 316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo AY, Chen X, Gao G, Zhang H, Zhu QH, Liu XC, et al. PlantTFDB: a comprehensive plant transcription factor database. Nucl Acids Res. 2008;36: D966–D969. 10.1093/nar/gkm841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9): 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protocols. 2012;7(3): 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anastasiou E, Kenz S, Gerstung M, MacLean D, Timmer J, Fleck C, et al. Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev Cell. 2007;13(6): 843–856. 10.1016/j.devcel.2007.10.001 [DOI] [PubMed] [Google Scholar]
- 16.Sauret-Gueto S, Schiessl K, Bangham A, Sablowski R, Coen E. JAGGED controls Arabidopsis petal growth and shape by interacting with a divergent polarity field. PLoS Biol. 2013;11(4): e1001550 10.1371/journal.pbio.1001550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinneny JR, Yadegari R, Fischer RL, Yanofsky MF, Weigel D. The role of JAGGED in shaping lateral organs. Development. 2004;131(5): 1101–1110. 10.1242/dev.00949 [DOI] [PubMed] [Google Scholar]
- 18.Riechmann JL, Meyerowitz EM. Determination of floral organ identity by Arabidopsis MADS domain homeotic proteins AP1, AP3, PI, and AG is independent of their DNA-binding specificity. Mol Biol Cell. 1997;8(7): 1243–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zahn LM, Ma X, Altman NS, Zhang Q, Wall PK, Tian D, et al. Comparative transcriptomics among floral organs of the basal eudicot Eschscholzia californica as reference for floral evolutionary developmental studies. Genome Biol. 2010;11(10): R101 10.1186/gb-2010-11-10-r101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Running MP, Meyerowitz EM. Mutations in the PERIANTHIA gene of Arabidopsis specifically alter floral organ number and initiation pattern. Development. 1996;122(4): 1261–1269. [DOI] [PubMed] [Google Scholar]
- 21.Szecsi J, Joly C, Bordji K, Varaud E, Cock JM, Dumas C, et al. BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J. 2006;25(16): 3912–3920. 10.1038/sj.emboj.7601270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu XM, Moller SG. AtSufE is an essential activator of plastidic and mitochondrial desulfurases in Arabidopsis. EMBO J. 2006;25(4): 900–909. 10.1038/sj.emboj.7600968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye H, Pilon M, Pilon-Smits EA. CpNifS-dependent iron-sulfur cluster biogenesis in chloroplasts. New Phytol. 2006;171(2): 285–292. 10.1111/j.1469-8137.2006.01751.x [DOI] [PubMed] [Google Scholar]
- 24.M NM, Ollagnier-de-Choudens S, Sanakis Y, Abdel-Ghany SE, Rousset C, Ye H, et al. Characterization of Arabidopsis thaliana SufE2 and SufE3: functions in chloroplast iron-sulfur cluster assembly and NAD synthesis. J Biol Chem. 2007;282(25): 18254–18264. 10.1074/jbc.M701428200 [DOI] [PubMed] [Google Scholar]
- 25.Gross W, Lenze D, Nowitzki U, Weiske J, Schnarrenberger C. Characterization, cloning, and evolutionary history of the chloroplast and cytosolic class I aldolases of the red alga Galdieria sulphuraria. Gene. 1999;230(1): 7–14. [DOI] [PubMed] [Google Scholar]
- 26.Flechner A, Gross W, Martin WF, Schnarrenberger C. Chloroplast class I and class II aldolases are bifunctional for fructose-1,6-biphosphate and sedoheptulose-1,7-biphosphate cleavage in the Calvin cycle. FEBS Lett. 1999;447(2–3): 200–202. [DOI] [PubMed] [Google Scholar]
- 27.Lu W, Tang X, Huo Y, Xu R, Qi S, Huang J, et al. Identification and characterization of fructose 1,6-bisphosphate aldolase genes in Arabidopsis reveal a gene family with diverse responses to abiotic stresses. Gene. 2012;503(1): 65–74. 10.1016/j.gene.2012.04.042 [DOI] [PubMed] [Google Scholar]
- 28.Dure L 3rd, Greenway SC, Galau GA. Developmental biochemistry of cottonseed embryogenesis and germination: changing messenger ribonucleic acid populations as shown by in vitro and in vivo protein synthesis. Biochemistry. 1981;20(14): 4162–4168. [DOI] [PubMed] [Google Scholar]
- 29.Hundertmark M, Hincha DK. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics. 2008;9: 118 10.1186/1471-2164-9-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker J, dennSteele C, Dure L III. Sequence and characterization of 6 Lea proteins and their genes from cotton. Plant Mol Biol. 1988;11(3): 277–291. 10.1007/BF00027385 [DOI] [PubMed] [Google Scholar]
- 31.Skriver K, Mundy J. Gene expression in response to abscisic acid and osmotic stress. Plant Cell. 1990;2(6): 503–512. 10.1105/tpc.2.6.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ried JL, Walker-Simmons MK. Group 3 Late Embryogenesis Abundant proteins in desiccation-tolerant seedlings of wheat (Triticum aestivum L.). Plant Physiol. 1993;102(1): 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bostock RM, Quatrano RS. Regulation of Em gene expression in rice: Interaction between osmotic stress and abscisic acid. Plant Physiol. 1992;98(4): 1356–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bies N, Aspart L, Carles C, Gallois P, Delseny M. Accumulation and degradation of Em proteins in Arabidopsis thaliana: evidence for post-transcriptional controls. J Exp Bot. 1998;49(329): 1925–1933. [Google Scholar]
- 35.Hill JP, Lord EM. Floral development in Arabidopsis thaliana: a comparison of the wild type and the homeotic pistillata mutant. Can J Bot. 1989;67(10): 2922–2936. [Google Scholar]
- 36.Irish VF. Petal and stamen development. Curr Top Dev Biol. 1999;41: 133–161. [DOI] [PubMed] [Google Scholar]
- 37.Bey M, Stuber K, Fellenberg K, Schwarz-Sommer Z, Sommer H, Saedler H, et al. Characterization of antirrhinum petal development and identification of target genes of the class B MADS box gene DEFICIENS. Plant Cell. 2004;16(12): 3197–3215. 10.1105/tpc.104.026724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zik M, Irish VF. Global identification of target genes regulated by APETALA3 and PISTILLATA floral homeotic gene action. Plant Cell. 2003;15(1): 207–222. 10.1105/tpc.006353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dafny-Yelin M, Guterman I, Menda N, Ovadis M, Shalit M, Pichersky E, et al. Flower proteome: changes in protein spectrum during the advanced stages of rose petal development. Planta. 2005;222(1): 37–46. 10.1007/s00425-005-1512-x [DOI] [PubMed] [Google Scholar]
- 40.Melo-Oliveira R, Oliveira IC, Coruzzi GM. Arabidopsis mutant analysis and gene regulation define a nonredundant role for glutamate dehydrogenase in nitrogen assimilation. Proc Natl Acad Sci U S A. 1996;93(10): 4718–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terce-Laforgue T, Mack G, Hirel B. New insights towards the function of glutamate dehydrogenase revealed during source-sink transition of tobacco (Nicotiana tabacum) plants grown under different nitrogen regimes. Physiol Plant. 2004;120(2): 220–228. 10.1111/j.0031-9317.2004.0241.x [DOI] [PubMed] [Google Scholar]
- 42.Skopelitis DS, Paranychianakis NV, Paschalidis KA, Pliakonis ED, Delis ID, Yakoumakis DI, et al. Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis in tobacco and grapevine. Plant Cell. 2006;18(10): 2767–2781. 10.1105/tpc.105.038323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Restivo FM. Molecular cloning of glutamate dehydrogenase genes of Nicotiana plumbaginifolia: structure analysis and regulation of their expression by physiological and stress conditions. Plant Sci. 2004;166(4): 971–982. [Google Scholar]
- 44.Dubois F, Tercé-Laforgue T, Gonzalez-Moro M-B, Estavillo J-M, Sangwan R, Gallais A, et al. Glutamate dehydrogenase in plants: is there a new story for an old enzyme? Plant Physiol Biochem. 2003;41(6–7): 565–576. [Google Scholar]
- 45.Sawada K, Hasegawa M, Tokuda L, Kameyama J, Kodama O, Kohchi T, et al. Enhanced resistance to blast fungus and bacterial blight in transgenic rice constitutively expressing OsSBP, a rice homologue of mammalian selenium-binding proteins. Biosci Biotechnol Biochem. 2004;68(4): 873–880. 10.1271/bbb.68.873 [DOI] [PubMed] [Google Scholar]
- 46.Dutilleul C, Jourdain A, Bourguignon J, Hugouvieux V. The Arabidopsis putative selenium-binding protein family: expression study and characterization of SBP1 as a potential new player in cadmium detoxification processes. Plant Physiol. 2008;147(1): 239–251. 10.1104/pp.107.114033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hugouvieux V, Dutilleul C, Jourdain A, Reynaud F, Lopez V, Bourguignon J. Arabidopsis putative selenium-binding protein1 expression is tightly linked to cellular sulfur demand and can reduce sensitivity to stresses requiring glutathione for tolerance. Plant Physiol. 2009;151(2): 768–781. 10.1104/pp.109.144808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pearce G, Moura DS, Stratmann J, Ryan CA Jr.: RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc Natl Acad Sci U S A. 2001;98(22): 12843–12847. 10.1073/pnas.201416998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Covey PA, Subbaiah CC, Parsons RL, Pearce G, Lay FT, Anderson MA, et al. A pollen-specific RALF from tomato that regulates pollen tube elongation. Plant Physiol. 2010;153(2): 703–715. 10.1104/pp.110.155457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu J, Kurten EL, Monshausen G, Hummel GM, Gilroy S, Baldwin IT. NaRALF, a peptide signal essential for the regulation of root hair tip apoplastic pH in Nicotiana attenuata, is required for root hair development and plant growth in native soils. Plant J. 2007;52(5): 877–890. 10.1111/j.1365-313X.2007.03289.x [DOI] [PubMed] [Google Scholar]
- 51.Combier JP, Kuster H, Journet EP, Hohnjec N, Gamas P, Niebel A. Evidence for the involvement in nodulation of the two small putative regulatory peptide-encoding genes MtRALFL1 and MtDVL1. Mol Plant Microbe Interact. 2008;21(8): 1118–1127. 10.1094/MPMI-21-8-1118 [DOI] [PubMed] [Google Scholar]
- 52.Germain H, Chevalier E, Caron S, Matton DP. Characterization of five RALF-like genes from Solanum chacoense provides support for a developmental role in plants. Planta. 2005;220(3): 447–454. 10.1007/s00425-004-1352-0 [DOI] [PubMed] [Google Scholar]
- 53.Shiu SH, Bleecker AB. Plant receptor-like kinase gene family: diversity, function, and signaling. Sci STKE. 2001;2001(113): re22 10.1126/stke.2001.113.re22 [DOI] [PubMed] [Google Scholar]
- 54.Escobar-Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC, et al. The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science. 2007;317(5838): 656–660. 10.1126/science.1143562 [DOI] [PubMed] [Google Scholar]
- 55.Huck N, Moore JM, Federer M, Grossniklaus U. The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development. 2003;130(10): 2149–2159. [DOI] [PubMed] [Google Scholar]
- 56.Merkouropoulos G, Shirsat A. The unusual Arabidopsis extensin gene atExt1 is expressed throughout plant development and is induced by a variety of biotic and abiotic stresses. Planta. 2003;217(3): 356–366. 10.1007/s00425-003-1002-y [DOI] [PubMed] [Google Scholar]
- 57.Kumar M, Turner S. Plant cellulose synthesis: CESA proteins crossing the kingdoms. Phytochemistry. 2015;112: 91–99. 10.1016/j.phytochem.2014.07.009 [DOI] [PubMed] [Google Scholar]
- 58.Noda K, Glover BJ, Linstead P, Martin C. Flower colour intensity depends on specialized cell shape controlled by a Myb-related transcription factor. Nature. 1994;369(6482): 661–664. 10.1038/369661a0 [DOI] [PubMed] [Google Scholar]
- 59.Baumann K, Perez-Rodriguez M, Bradley D, Venail J, Bailey P, Jin H, et al. Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development. 2007;134(9): 1691–1701. 10.1242/dev.02836 [DOI] [PubMed] [Google Scholar]
- 60.Mara CD, Huang T, Irish VF. The Arabidopsis floral homeotic proteins APETALA3 and PISTILLATA negatively regulate the BANQUO genes implicated in light signaling. Plant Cell. 2010;22: 690–702. 10.1105/tpc.109.065946 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All reads produced in this study are available from the NCBI Sequence Read Archive (SRA) (accession number SRP075857). This Transcriptome Shotgun Assembly project has been deposited at DDBJ/EMBL/GenBank under the accession GFBU00000000. The version described in this paper is the first version, GFBU01000000.