Abstract
Introduction
Human papillomavirus (HPV) 52 is a carcinogenic, high-risk genotype frequently detected in cervical cancer cases from East Asia, including Korea.
Materials and Methods
Sequences of HPV52 detected in 91 cervical samples collected from women attending Seoul St. Mary’s Hospital were analyzed. HPV52 genomic sequences were obtained by polymerase chain reaction (PCR)-based sequencing and analyzed using Seq-Scape software, and phylogenetic trees were constructed using MEGA6 software.
Results
Of the 91 cervical samples, 40 were normal, 22 were low-grade lesions, 21 were high-grade lesions and 7 were squamous cell carcinomas. Four HPV52 variant lineages (A, B, C and D) were identified. Lineage B was the most frequently detected lineage, followed by lineage C. By analyzing the two most frequently detected lineages (B and C), we found that distinct variations existed in each lineage. We also found that a lineage B-specific mutation K93R (A379G) was associated with an increased risk of cervical neoplasia.
Conclusions
To our knowledge, we are the first to reveal the predominance of the HPV52 lineages, B and C, in Korea. We also found these lineages harbored distinct genetic alterations that may affect oncogenicity. Our findings increase our understanding on the heterogeneity of HPV52 variants, and may be useful for the development of new diagnostic assays and therapeutic vaccines.
Introduction
Human papillomavirus (HPV) is the major causative agent of cervical cancer, a leading cause of death among women worldwide [1]. The virus genome is divided into three functional regions: an upstream regulatory region, an early region, and a late region. The upstream regulatory region is a non-coding region, referred as the long control region (LCR), which regulates transcriptional and replication activities. In comparison, the early and late regions are coding regions. Early regions (E1, E2, E4, E5, E6 and E7) encode for non-structural proteins while late regions (L1 and L2) encode for structural proteins [2]. HPV is markedly heterogeneous with more than 200 genotypes which are classified into types, lineages, and sub-lineages based on the L1 sequence. The L1 sequences among different types differ by at least 10%, and those of lineages differ by >1% [3, 4]. The persistence of HPV contributes to the progression of cervical infection to cervical cancer. In particular, oncogenicity varies according to the HPV genotype, as well as the lineage of some genotypes [4–6]. Eight most common high-risk HPV genotypes (HPV16, HPV18, HPV31, HPV33, HPV35, HPV45, HPV52 and HPV58) are responsible for 91% of cervical cancers [7, 8]. Of these, HPV52 is a high-risk genotype and one of the nine HPV types (HPV6, HPV11, HPV16, HPV18, HPV31, HPV33, HPV45, HPV52 and HPV58) targeted by the recent Food and Drug Administration (FDA) approved HPV 9-valent vaccine [9].
Given that HPV52 is recognized as a high-risk genotype commonly found in cervical cancers from East Asia [10–12], we attempted to characterize HPV52 variants circulating in Korea and to investigate their association with cervical cancer development.
Materials and Methods
Cervical samples
Altogether, 91 cervical cytology/tissue samples that had tested positive for HPV52 were used for this study. These samples had been collected as part of the routine clinical management at Seoul St. Mary’s Hospital (Seoul, Korea) and all were treatment-naive. This study was approved by the institutional review board of the Catholic University of Korea, College of Medicine and the participants provided written informed consent. Pathologic features of patients are summarized in Table 1. The quality of DNA extracted from cytology/tissue samples was assessed by amplifying a 932-bp fragment of the long-control region (LCR). The HPV genotype was ascertained by demonstrating a nucleotide sequence similarity of >90%, compared with the HPV52 prototype (GenBank accession no. X74481).
Table 1. Cervical pathologies of study subject.
| Cervical pathology | Number of subjects (percentage) (n = 91) | Lineages | Mean age (years) (±SD) | ||||
|---|---|---|---|---|---|---|---|
| A (n = 5) | B (n = 79) | C n = 6) | D (n = 1) | ||||
| Normal | 40 (44.0) | 3 | 35 | 2 | 0 | 40.2 (±9.6) | |
| Low-grade lesions | 22 (24.2) | 2 | 19 | 0 | 1 | 39.6 (±10.7) | |
| ASCUS | 2 | ||||||
| LGSIL | 20 | ||||||
| High-grade lesions | 21 (23.1) | 0 | 17 | 4 | 0 | 46.7 (±11.9) | |
| HGSIL | 14 | ||||||
| CIN3 | 4 | ||||||
| CIS | 3 | ||||||
| SCC | 7 (7.7) | 0 | 7 | 0 | 0 | 57.6 (±18.0) | |
| Unknown | 1 (1.1) | 0 | 1 | 0 | 0 | ||
ASCUS: atypical squamous cells of undetermined significance; CIN3: cervical intraepithelial neoplasia 3; CIS: carcinoma in situ; LGSIL: low-grade squamous intraepithelial lesions; HGSIL: high-grade squamous intraepithelial lesions; SCC: squamous cell carcinoma.
Nucleotide sequencing
E6, E7, L1 and LCR sequences were amplified with long- or short-fragment polymerase chain reaction (PCR). Long-fragment PCR was performed on good-quality samples with primers 5′-ATG TCC ATT GAG TCA GGT CC-3′ and 5′-TGC ATT TTC ATC CTC GTC C-3′. When the first-round PCR product was not strong enough for sequencing, a second-round PCR was performed using inner primers 5′-GGT CCT GAC ATT CCA TTA CC-3′ and 5′-CCT CTA CTT CAA ACC AGC CT-3′ when necessary ′ (S1 Table). Each PCR was conducted in a 50-μL reaction mixture containing 1 unit of Phusion Hot Start II High-Fidelity DNA Polymerase (Thermo Fisher Scientific, Waltham, USA), 200 μM of dNTPs and 0.25 μM of each primer. An aliquot of 5 μL of extracted DNA was added as template. The thermal cycling began with a 30-sec initial denaturation and enzyme activation at 98°C, followed by 35 cycles of 10-sec denaturation at 98°C, 30-sec annealing at 62°C and 100-sec extension at 72°C, and ended with an 10-min final extension at 72°C. When long-fragment PCR was not successful, short-fragment PCR was performed with primer pairs E6/E7 (5′-TGC ACT ACA CGA CCG GTT A-3′ and 5′-CAT CCT CGT CCT CTG AAA TG-3′), L1A (5′-ATG TCC ATT GAG TCA GGT CC-3′ and 5′-GCA CAG GGT CAC CTA AGG TA-3′), L1B (5′-AGG ATG GGG ACA TGG TAG AT-3′ and 5′-CAC AGA CAA TTA CCC AAC AGA C-3′) and LCR (5′-GTC TGC ATC TTT GGA GGA CA-3′ and 5′-TGC GTT AGC TAC ACT GTG TTC-3′), respectively. When necessary, a second-round PCR, using inner primers E6/E7 (5′-TTA CCG TAC CCA CAA CCA CT-3′ and 5′-CCT CTA CTT CAA ACC AGC CT-3′), L1A (5′-GGT CCT GAC ATT CCA TTA CC-3′ and 5′-GGG CAC ATC ACT TTT ACT AGC-3′), L1B (5′-ACA GGA TTT GGT TGC ATG G-3′ and 5′-TTC TTT GTG GAG GTA CGT GG-3′) and LCR (5′-TTT GTT ACA GGC AGG GCT AC-3′ and 5′-CGT TTT CGG TTA CAC CCT A-3′), was performed (S2 Table). Each PCR was conducted in a 30-μL reaction mixture containing 0.75 unit of HotStarTaq Plus DNA Polymerase (QIAGEN, Hilden, Germany), 200 μM of dNTPs and 0.25 μM of each primer. An aliquot of 3 μL of extracted DNA was added as template. The thermal cycling began with a 5-min initial denaturation and enzyme activation at 95°C, followed by 35 cycles of 1-min denaturation at 94°C, 1-min annealing at 58°C and 40-sec extension at 72°C, and ended with an 8-min final extension at 72°C. PCR products were sequenced from both directions and analyzed using Seq-Scape software (version 2.5, Applied Biosystems, Foster City, CA, USA). Repeated sequencing was performed as a confirmation when mutations occurred only once.
Phylogenetic tree construction
A maximum-likelihood tree was constructed using MEGA6 (Molecular Evolutionary Genetic Analysis software program, version 6.0; http://www.megasoftware.net) [13]. The tree was comprised of concatenated E6-E7-L1-LCR sequences of unique HPV52 strains collected in this study and from a published reference strain of each lineage (A1: X74481, A2: HQ537739, B1: HQ537740, B2: HQ537743, C1: HQ537744, C2: HQ537746, D: HQ537748). Bootstrap values of key nodes were generated by 1000 resamplings. To root the tree, HPV67 prototype sequences (NCBI accession no. NC_004710) were set as the outgroup.
Statistical analysis
Statistical analysis was performed using a commercially available statistical software package (SPSS statistical software version 18.0 [SPSS Inc, Chicago, IL, USA]). Fisher’s exact test and logistic regression analysis were used to analyze categorical data. The level of significance was set at P < 0.05.
Results
Characteristics of cervical samples
Of the 91 cervical cytology samples, 40 (44.0%) showed normal pathologic findings (normal squamous cell epithelium; Table 1). We separated pre-malignant lesions into ‘low-grade lesions’ and ‘high-grade lesions’. ‘Low-grade lesions’ (n = 22) included ASCUS (atypical squamous cells of undetermined significance) and LGSIL (low-grade squamous intraepithelial lesions), while ‘high-grade lesions’ (n = 21) included HGSIL (high-grade squamous intraepithelial lesions), CIN3 (cervical intraepithelial neoplasia 3) and CIS (carcinoma in situ). For malignant lesions, seven (7.7%) squamous cell carcinomas (SCCs) were included.
Lineage identification
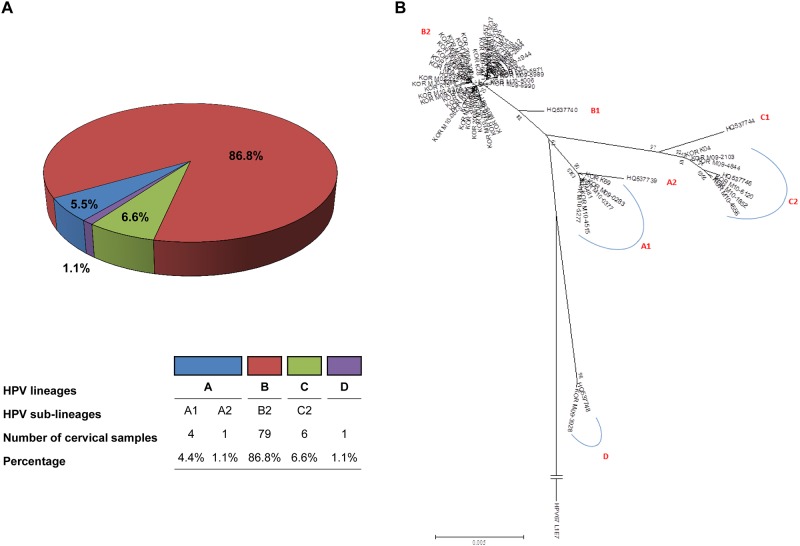
Altogether, four HPV52 variant lineages were identified based on the phylogenetic tree topology (Fig 1). Lineages A, B and C were closely related, while lineage D was relatively distant. Lineage B was most frequently detected (86.8%, 79 of 91 samples; Table 1). The majority of high-grade lesions (80.9%, 17 of 21 samples) belonged to lineage B, and the remaining high-grade lesions belonged to lineage C (19.1%). Lineage B harbored all of the seven SCCs and their association was significant (P = 0.02).
Fig 1. HPV52 variant lineage distribution of study samples.
(A) Lineages A (sublineages: A1 and A2), B (sublineage: B2), C (sublineage: C2) and D were detected. (B) A phylogenetic tree was constructed from 57 HPV52 variants using concatenated L1, LCR, E6 and E7. A maximum-likelihood tree was constructed using the program, MEGA6. Bootstrap values of key nodes generated by 1,000 resamplings are shown. The length of the scale bar represents 0.005 substitutions per nucleotide position. To root the tree, HPV67 prototype sequences (NCBI accession no. NC_004710) were set as outgroup. The GenBank accession no. of study samples are KY077824-KY077901.
HPV52 sequence variations
In this study, 40.6% (3226 nucleotides) of the HPV52 genome (7942bps, X74481) was sequenced (S3–S6 Tables). Nine E6 variants with 11 nucleotide positions showing sequence polymorphisms were identified, encompassing three nonsynonymous mutations (S3 Table). Five variants with 12 nucleotide positions showing sequence polymorphisms with seven nonsynonymous mutations were identified in E7 (S4 Table). L1 harbored 26 variants and 54 nucleotide sequence polymorphisms with 11 nonsynonymous mutations (S5 Table). The noncoding LCR was the most heterogeneous, encompassing 38 variants showing 87 nucleotide sequence variations (S6 Table).
An analysis of sequences for the E6, E7, L1 and LCR genes suggested that HPV52 harbors lineage-specific variations. Lineage B, the most frequently detected lineage, harbored a number of lineage-specific variations (Table 2). Of the 91 samples, K93R (A379G), the most frequently detected nonsynonymous mutation (85.71%) in E6, was only found in lineage B. In addition, a novel mutation, 7935_7936 insT in LCR was significantly associated with lineage B (P < 0.0001). Lineage C also harbored lineage-specific variations (Table 3), of which an E6 nonsynonymous mutation (L83V, concurrent mutations of C348G and G350T) and five E7 nonsynonymous mutations, S52D (concurrent mutations of A706G and G707A), Y55D (T727G), H61Y (C733T), D64N (G742A) and L99R (T848G), showed significant associations with the lineage (P < 0.0001).
Table 2. List of nucleotide variations associated with HPV52 lineage B.
| Non-B lineages (n = 12) | B lineage (n = 79) | P value* | Adjust odds ratio† (95% CI) | P value† | ||
|---|---|---|---|---|---|---|
| E6 nucleotide change | ||||||
| G350T | 7 | 79 | <0.0001 | 77.31 (12.39 –Infinity) | <0.0001 | |
| A379G | 0 | 78 | <0.0001 | 186.39 (48.04 –Infinity) | <0.0001 | |
| E7 nucleotide change | ||||||
| C751T | 0 | 79 | <0.0001 | 92.87 (35.35 –Infinity) | <0.0001 | |
| A801G | 7 | 79 | <0.0001 | 77.31 (12.39 –Infinity) | <0.0001 | |
| L1 nucleotide change | ||||||
| A5771G | 0 | 76 | <0.0001 | 350.56 (59.44–Infinity) | <0.0001 | |
| T5972C | 0 | 78 | <0.0001 | 186.39 (48.04– Infinity) | <0.0001 | |
| G6110A | 0 | 79 | <0.0001 | 92.87 (35.35–Infinity) | <0.0001 | |
| G6218A | 7 | 79 | <0.0001 | 77.31 (12.39–Infinity) | <0.0001 | |
| T6710G | 0 | 79 | <0.0001 | 92.87 (35.35–Infinity) | <0.0001 | |
| T6764C | 0 | 79 | <0.0001 | 92.87 (35.35–Infinity) | <0.0001 | |
| A6794G | 0 | 78 | <0.0001 | 186.25 (47.99–Infinity) | <0.0001 | |
| C6824T | 0 | 79 | <0.0001 | 92.87 (35.35–Infinity) | <0.0001 | |
| C6917A | 7 | 78 | <0.0001 | 84.04 (7.25–974.61) | 0.0004 | |
| G7052A | 2 | 78 | <0.0001 | 166.80 (21.96–Infinity) | <0.0001 | |
| LCR nucleotide change | ||||||
| G7168C | 6 | 71 | 0.0025 | 8.89 (2.31–34.20) | 0.0015 | |
| C7207A | 7 | 73 | 0.0051 | 9.32 (2.17–40.10) | 0.0027 | |
| G7371T | 4 | 77 | <0.0001 | 171.31 (14.34–Infinity) | <0.0001 | |
| G7622A | 7 | 79 | <0.0001 | 77.31 (12.39–Infinity) | <0.0001 | |
| T7624G | 7 | 79 | <0.0001 | 77.31 (12.39–Infinity) | <0.0001 | |
| A7657C | 0 | 79 | <0.0001 | 92.87 (35.35–Infinity) | <0.0001 | |
| T7659C | 6 | 79 | <0.0001 | 105.84 (17.45–Infinity) | <0.0001 | |
| G7712C | 7 | 79 | <0.0001 | 77.31 (12.39–Infinity) | <0.0001 | |
| G7861A | 7 | 79 | <0.0001 | 77.31 (12.39–Infinity) | <0.0001 | |
| A7865G | 1 | 77 | <0.0001 | 225.24 (41.70–Infinity) | <0.0001 | |
| 7935_7936 insT | 7 | 79 | <0.0001 | 77.31 (12.39–Infinity) | <0.0001 | |
| A7938G | 1 | 48 | <0.0001 | 18.05 (2.19–148.62) | 0.0071 | |
| T13C | 0 | 79 | <0.0001 | 92.87 (35.35–Infinity) | <0.0001 |
Statistical analyses were performed using *Fisher's exact test, †multivariable logistic regression (age-adjusted).
Table 3. List of nucleotide variations associated with HPV52 lineage C.
| Non-C lineages (n = 85) | C lineage (n = 6) | P value* | Adjust odds ratio† (95% CI) | P value† | ||
|---|---|---|---|---|---|---|
| E6 nucleotide change | ||||||
| A530G | 0 | 6 | <0.0001 | 41.78 (14.64–Infinity) | <0.0001 | |
| C348G and G350T | 0 | 3 | <0.0001 | 52.83 (7.31–Infinity) | 0.0011 | |
| E7 nucleotide change | ||||||
| T573A | 0 | 6 | <0.0001 | 41.78 (14.64–Infinity) | <0.0001 | |
| C662T | 0 | 6 | <0.0001 | 41.78 (14.64–Infinity) | <0.0001 | |
| A706G and G707A | 0 | 6 | <0.0001 | 41.78 (14.64–Infinity) | <0.0001 | |
| T727G | 0 | 6 | <0.0001 | 41.78 (14.64–Infinity) | <0.0001 | |
| C733T | 0 | 6 | <0.0001 | 41.78 (14.64–Infinity) | <0.0001 | |
| G742A | 0 | 6 | <0.0001 | 41.78 (14.64–Infinity) | <0.0001 | |
| T848G | 0 | 6 | <0.0001 | 41.78 (14.64–Infinity) | <0.0001 | |
| L1 nucleotide change | ||||||
| T5578C | 0 | 3 | <0.0001 | 140.86 (16.81–Infinity) | 0.0001 | |
| G5720A | 0 | 3 | <0.0001 | 52.83 (7.31–Infinity) | 0.0011 | |
| A5909G | 0 | 5 | <0.0001 | 155.81 (26.17–Infinity) | <0.0001 | |
| G6083A | 0 | 6 | <0.0001 | 41.78 (14.64–Infinity) | <0.0001 | |
| C6443T | 0 | 6 | <0.0001 | 41.78 (14.64–Infinity) | <0.0001 | |
| G6698A | 0 | 6 | <0.0001 | 41.78 (14.64–Infinity) | <0.0001 | |
| G7112A | 0 | 6 | <0.0001 | 41.78 (14.64–Infinity) | <0.0001 |
Statistical analyses were performed using *Fisher's exact test, †multivariable logistic regression (age-adjusted).
Discussion
HPVs are circular double-stranded DNA viruses that consist of heterogeneous variants with different pathogenicities [4, 14]. HPV52 is one of the most frequently detected carcinogenic high-risk genotypes in East Asia [15–17] and is one of the genotypes targeted by the recent, USA FDA-approved HPV 9-valent vaccine [9]. However, only a few studies on the pathogenicity of HPV52 are available [18]. In this regard, we attempted to achieve three aims. Firstly, we aimed at identifying HPV52 variants and lineages based on E6, E7, L1 and LCR sequences. Secondly, we attempted to delineate lineage-specific variations. Lastly, we sought to examine the risk association of various lineages, variants and mutations. As a result, we identified four HPV52 variant lineages (A, B, C and D), with lineage B (86.8%) being the most frequently detected, followed by lineages C (6.6%), A (5.5%), and D (1.1%). By further analyzing the two most frequently detected lineages, we found distinct sequence variations in each lineage. Of note, one of lineage B-specific mutations was found to associate with a higher risk for cervical cancer.
Our findings of the two most frequently detected lineages in HPV52, B and C, also confirm previous observations [19, 20]. These two lineages are associated with high-grade lesions [19, 21], and we have shown that they harbor a number of lineage-specific variations (Tables 2 and 3). Lineage B-specific mutations included the most frequently detected nonsynonymous mutation K93R (A379G) in E6, while lineage C-specific mutations included an E6 nonsynonymous mutation (L83V, concurrent mutations of C348G and G350T) and five E7 nonsynonymous mutations, S52D (concurrent mutations of A706G and G707A), Y55D (T727G), H61Y (C733T), D64N (G742A) and L99R (T848G)).
Our findings suggest that lineage-specific mutations may contribute to the carcinogenicity of each HPV52 lineage. E6 is an oncogene that interacts with a well-known tumor suppressor, TP53, increasing the risk for the accumulation of genetic changes [22] and inhibiting cellular responses such as cell cycle arrest, induction of apoptosis and DNA damage repair [23, 24]. K93R (A379G) is a nonsynonymous mutation located in the E6 oncogene, which may have a specific role in carcinogenesis: it is not only the most frequently detected variation [25], but is also independently associated with high-grade lesions [26]. An alteration at nucleotide position 350 in E6 was found for both lineages B and C. However, lineage C harbored an additional alteration at nucleotide position 348, yielding a nonsynonymous mutation, L83V. L83V have been reported not only in HPV52, but also in HPV16 and HPV33 [25, 27, 28]. In addition, an association of HPV16 L83V with high-grade lesions has been shown [29], suggesting that it may contribute to the pathogenicity of lineage C [19]. It is noticeable that E7 nonsynonymous mutations (S52D, 55D, H61Y, D64N and L99R) were harbored by lineage C, but not by lineage B. E7 interacts with the retinoblastoma (Rb) protein, causing uncontrolled cell division and inactivating its function as a tumor suppressor [30, 31].
Conclusions
HPV52 is the second to fifth most frequently detected high-risk HPV genotype in Korea [15, 32–34]. Our data demonstrated for the first time that HPV52 lineages (B and C) circulating in Korea harbor distinct genetic alterations that may affect pathogenicity. We also observed that most of the cervical samples (ranging from normal cervix to SCC) were infected with HPV52 lineages B or C that carried putative high-risk mutations. Our findings may provide a useful basis to understand the heterogeneity of HPV52 variants in Korea, and to assist the development of diagnostic assays and vaccines.
Supporting Information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
The work was supported by Grants of National R&D Program for Cancer Control (0720510), Ministry of Health and Welfare, Republic of Korea and Statistical consultations were supported by a Korean Health Technology R&D Project grant from the Ministry of Health & Welfare, Republic of Korea. (HI14C1731).
Abbreviations
- ASCUS
atypical squamous cells of undetermined significance
- CIN3
cervical intraepithelial neoplasia 3
- CIS
carcinoma in situ
- HGSIL
high-grade squamous intraepithelial lesions
- HPV
Human papillomavirus
- LGSIL
low-grade squamous intraepithelial lesions
- SCC
squamous cell carcinoma
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The work was supported by Grants of National R&D Program for Cancer Control (0720510), Ministry of Health and Welfare, Republic of Korea and Statistical consultations were supported by a Korean Health Technology R&D Project grant from the Ministry of Health & Welfare, Republic of Korea (HI14C1731).
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–50. 10.1038/nrc798 [DOI] [PubMed] [Google Scholar]
- 3.Bernard HU, Burk RD, Chen Z, van Doorslaer K, zur Hausen H, de Villiers EM. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401:70–9. 10.1016/j.virol.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. 10.1016/j.virol.2004.03.033 [DOI] [PubMed] [Google Scholar]
- 5.Bernard HU. The clinical importance of the nomenclature, evolution and taxonomy of human papillomaviruses. J Clin Virol. 2005;32 Suppl 1:S1–6. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z, Schiffman M, Herrero R, Desalle R, Anastos K, Segondy M, et al. Evolution and taxonomic classification of human papillomavirus 16 (HPV16)-related variant genomes: HPV31, HPV33, HPV35, HPV52, HPV58 and HPV67. PLoS One. 2011;6:e20183 10.1371/journal.pone.0020183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Sanjose S, Quint WG, Alemany L, Geraets DT, Klaustermeier JE, Lloveras B, et al. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol. 2010;11:1048–56. 10.1016/S1470-2045(10)70230-8 [DOI] [PubMed] [Google Scholar]
- 8.Muñoz N, Bosch FX, de Sanjosé S, Herrero R, Castellsagué X, Shah KV, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–27. 10.1056/NEJMoa021641 [DOI] [PubMed] [Google Scholar]
- 9.Serrano B, Alemany L, Tous S, Bruni L, Clifford GM, Weiss T, et al. Potential impact of a nine-valent vaccine in human papillomavirus related cervical disease. Infect Agent Cancer. 2012;7:38 10.1186/1750-9378-7-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sichero L, Ferreira S, Trottier H, Duarte-Franco E, Ferenczy A, Franco EL, et al. High grade cervical lesions are caused preferentially by non-European variants of HPVs 16 and 18. Int J Cancer. 2007;120:1763–8. 10.1002/ijc.22481 [DOI] [PubMed] [Google Scholar]
- 11.Chan PK, Ho WC, Chan MC, Wong MC, Yeung AC, Chor JS, et al. Meta-analysis on prevalence and attribution of human papillomavirus types 52 and 58 in cervical neoplasia worldwide. PLoS One. 2014;9:e107573 10.1371/journal.pone.0107573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang C, Park JS, Grce M, Hibbitts S, Palefsky JM, Konno R, et al. Geographical distribution and risk association of human papillomavirus genotype 52-variant lineages. J Infect Dis. 2014;210:1600–4. 10.1093/infdis/jiu310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giannoudis A, Herrington CS. Human papillomavirus variants and squamous neoplasia of the cervix. J Pathol. 2001;193:295–302. [DOI] [PubMed] [Google Scholar]
- 15.Bosch FX, Burchell AN, Schiffman M, Giuliano AR, de Sanjose S, Bruni L, et al. Epidemiology and natural history of human papillomavirus infections and type-specific implications in cervical neoplasia. Vaccine. 2008;26 Suppl 10:K1–16. [DOI] [PubMed] [Google Scholar]
- 16.Wheeler CM, Hunt WC, Joste NE, Key CR, Quint WG, Castle PE. Human papillomavirus genotype distributions: implications for vaccination and cancer screening in the United States. J Natl Cancer Inst. 2009;101:475–87. 10.1093/jnci/djn510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castle PE, Schiffman M, Wheeler CM, Wentzensen N, Gravitt PE. Human papillomavirus genotypes in cervical intraepithelial neoplasia grade 3. Cancer Epidemiol Biomarkers Prev. 2010;19:1675–81. 10.1158/1055-9965.EPI-10-0251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Z, Lu Z, Liu J, Wang G, Zhou W, Yang L, et al. Genomic polymorphism of human papillomavirus type 52 in women from Northeast China. Int J Mol Sci. 2012;13:14962–72. 10.3390/ijms131114962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang YJ, Chen HC, Lee BH, You SL, Lin CY, Pan MH, et al. Unique variants of human papillomavirus genotypes 52 and 58 and risk of cervical neoplasia. Int J Cancer. 2011;129:965–73. 10.1002/ijc.25724 [DOI] [PubMed] [Google Scholar]
- 20.Ishizaki A, Matsushita K, Hoang HT, Agdamag DM, Nguyen CH, Tran VT, et al. E6 and E7 variants of human papillomavirus-16 and -52 in Japan, the Philippines, and Vietnam. J Med Virol. 2013;85:1069–76. 10.1002/jmv.23566 [DOI] [PubMed] [Google Scholar]
- 21.Schiffman M, Rodriguez AC, Chen Z, Wacholder S, Herrero R, Hildesheim A, et al. A population-based prospective study of carcinogenic human papillomavirus variant lineages, viral persistence, and cervical neoplasia. Cancer Res. 2010;70:3159–69. 10.1158/0008-5472.CAN-09-4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Münger K, Baldwin A, Edwards KM, Hayakawa H, Nguyen CL, Owens M, et al. Mechanisms of human papillomavirus-induced oncogenesis. J Virol. 2004;78:11451–60. 10.1128/JVI.78.21.11451-11460.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheffner M, Whitaker NJ. Human papillomavirus-induced carcinogenesis and the ubiquitin-proteasome system. Semin Cancer Biol. 2003;13:59–67. [DOI] [PubMed] [Google Scholar]
- 24.Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2:594–604. 10.1038/nrc864 [DOI] [PubMed] [Google Scholar]
- 25.Xin CY, Matsumoto K, Yoshikawa H, Yasugi T, Onda T, Nakagawa S, et al. Analysis of E6 variants of human papillomavirus type 33, 52 and 58 in Japanese women with cervical intraepithelial neoplasia/cervical cancer in relation to their oncogenic potential. Cancer Lett. 2001;170:19–24. [DOI] [PubMed] [Google Scholar]
- 26.Formentin A, Archambault J, Koushik A, Richardson H, Brassard P, Franco EL, et al. Human papillomavirus type 52 polymorphism and high-grade lesions of the uterine cervix. Int J Cancer. 2013;132:1821–30. 10.1002/ijc.27874 [DOI] [PubMed] [Google Scholar]
- 27.Aho J, Hankins C, Tremblay C, Lang F, Forest P, Pourreaux K, et al. Molecular analysis of human papillomavirus type 52 isolates detected in the genital tract of human immunodeficiency virus-seropositive and -seronegative women. J Infect Dis. 2003;188:1517–27. 10.1086/379198 [DOI] [PubMed] [Google Scholar]
- 28.Gagnon S, Hankins C, Tremblay C, Forest P, Pourreaux K, Coutlée F. Viral polymorphism in human papillomavirus types 33 and 35 and persistent and transient infection in the genital tract of women. J Infect Dis. 2004;190:1575–85. 10.1086/424854 [DOI] [PubMed] [Google Scholar]
- 29.Grodzki M, Besson G, Clavel C, Arslan A, Franceschi S, Birembaut P, et al. Increased risk for cervical disease progression of French women infected with the human papillomavirus type 16 E6-350G variant. Cancer Epidemiol Biomarkers Prev. 2006;15:820–2. 10.1158/1055-9965.EPI-05-0864 [DOI] [PubMed] [Google Scholar]
- 30.Dyson N, Howley PM, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–7. [DOI] [PubMed] [Google Scholar]
- 31.Jones DL, Münger K. Interactions of the human papillomavirus E7 protein with cell cycle regulators. Semin Cancer Biol. 1996;7:327–37. 10.1006/scbi.1996.0042 [DOI] [PubMed] [Google Scholar]
- 32.Lee GH, Kang HJ, Kim SY, Park CM. The prevalence of human papilloma virus infections according to Pap smear results in Jeju island. Korean J Obstet Gynecol. 2011;54:689–95. [Google Scholar]
- 33.Choi IH, Jin SY, Lee DW, Kim DW, Jeen YM. Cytomorphologic features according to HPV DNA type in histologically proven cases of the uterine cervix. Korean J Pathol. 2011;45:612–20. [Google Scholar]
- 34.Kim TE, Kim HW, Lee KE. Distribution of human papillomavirus 52 and 58 genotypes, and their expression of p16 and p53 in cervical neoplasia. Korean J Pathol. 2014;48:24–9. 10.4132/KoreanJPathol.2014.48.1.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.