Abstract
Two thirds of Svalbard archipelago islands in the High Arctic are permanently covered with glacial ice and snow. Polar bacterial communities in the southern part of Svalbard were characterized using an amplicon sequencing approach. A total of 52,928 pyrosequencing reads were analyzed in order to reveal bacterial community structures in stream and lake surface water samples from the Fuglebekken and Revvatnet basins of southern Svalbard. Depending on the samples examined, bacterial communities at a higher taxonomic level mainly consisted either of Bacteroidetes, Betaproteobacteria, and Microgenomates (OP11) or Planctomycetes, Betaproteobacteria, and Bacteroidetes members, whereas a population of Microgenomates was prominent in 2 samples. At the lower taxonomic level, bacterial communities mostly comprised Microgenomates, Comamonadaceae, Flavobacteriaceae, Legionellales, SM2F11, Parcubacteria (OD1), and TM7 members at different proportions in each sample. The abundance of OTUs shared in common among samples was greater than 70%, with the exception of samples in which the proliferation of Planctomycetaceae, Phycisphaeraceae, and Candidatus Methylacidiphilum spp. lowered their relative abundance. A multi-variable analysis indicated that As, Pb, and Sb were the main environmental factors influencing bacterial profiles. We concluded that the bacterial communities in the polar aquatic ecosystems examined mainly consisted of freshwater and marine microorganisms involved in detritus mineralization, with a high proportion of zooplankton-associated taxa also being identified.
Keywords: cryophilic microorganisms, psychrophiles, polar freshwater environments, stream and lake water, 16S rRNA gene pyrosequencing
Several extreme environments are populated by a broad diversity of microorganisms that live at low temperatures, even below the pure water freezing point (6). These cold environments constitute an unexhausted source of vast metabolic potential that may be linked to the impact of climate change and used to exploit cold-active enzymes for biotechnological applications (5, 14).
Ambient cold environments are widely distributed on earth, including the alpine zones of the highest mountain ranges of the world, the deep ocean, the Arctic Circle, tundra, and Antarctica (44). Cold-adapted soil ecosystems must cope with wide temperature fluctuations and often radiation, whereas deep ocean microbes live at low, albeit constant temperatures (13). The prevailing low temperature and/or low liquid water availability in these ecosystems are considered to be major limiting factors for growth (44, 53). However, psychrophiles overcome these severe conditions through complex adaptive mechanisms. Increased membrane fluidity at lower temperatures (59), the accumulation of compatible osmolytes and secretion of anti-freezing proteins (16, 26), the induction of cold-shock proteins (32), and conformational flexibility of psychrophilic enzymes (56) are among the main adaptive strategies adopted by the cryophilic microbiota.
Polar zones, including Svalbard, are strongly affected by prolonged temperature increases in the globe, which are incommensurate in these regions and have led to the accelerating retreat of glaciers (34, 63). Nutrient fluxes from various sources, i.e. aeolian and guano inputs (60, 61), glacial flour particle release (39), and the transport of entrapped organic and inorganic compounds through cryoconite holes (61), influence in a complex matter biogeochemical cycles as well as the size and structure of microbial communities in the Arctic cryosphere (50). Interactions between ice-covered land and marine ecosystems (25), and freshwater from retreating glaciers (21, 62) also contribute to the Arctic microbial food web. On the other hand, cold ecosystems may act as ecological screens for immigrant microbiota, with the latter dealing with severe environmental conditions, such as successive freeze/thaw events, high surface radiation, and low nutrient and liquid water availability (25).
Members of the phyla Bacteroidetes, Actinobacteria, and Firmicutes together with the Alpha-, Beta-, and Gamma-subdivisions of Proteobacteria are the predominant taxa identified in cryophilic environments (27, 40, 62). The fjords and archipelago of Svalbard have recently attracted attention for assessing the impact of anthropogenic activities and global warming on the microbial ecology of the European part of the Arctic (20, 39, 57, 61). Microbial communities and glacial run-off to ice lakes have been extensively examined; however, information on the structures of microbial communities in the southern part of the Spitsbergen/Svalbard island complex is limited.
In the present study, the main objective was to characterize bacterial communities in the Fuglebekken and Revvatnet basins, two neighboring catchments located in geologically, hydrologically, and phytosociologically diverse regions of Svalbard, through pyrosequencing, and to link these bacterial profiles with various abiotic environmental factors.
Materials and Methods
Sampling procedure and site description
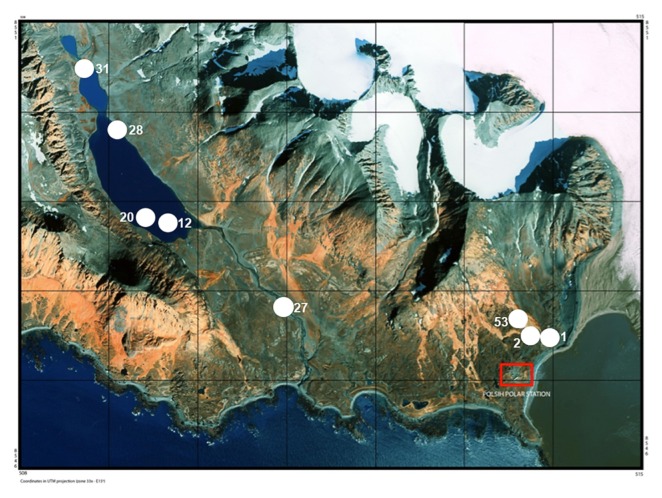
Surface water was collected from eight sites located in the Fuglebekken and Revvatnet basins between the 9th and 13th of August, 2010. Freshwater samples were obtained from the Fuglebekken stream (S1 [FS]), the Fuglebekken Lake (S2 [FL] and S53 [FL]), the Revvatnet lake (S12 [RL], S20 [RL], and S31 [RL]), the Revvatnet stream (S28 [RS]), and the Revelva river (S27 [RR]) (Fig. 1). Field research work did not include endangered or protected species and sampling permits were provided by The Governor of Svalbard. The Fuglebekken and Revvatnet basins, which drain to the Hornsund fjord, are situated in the southern part of the island of Spitsbergen and their substratum belongs to the Hecla Hoek Precambrian formations (28). Geographical coordinates are presented in Fig. 1, while a detailed description of the sampling sites is provided in Table S1.
Fig. 1.
Sampling site map (Kolondra L., Norway. Svalbard, Spitsbergen, Orthophotomap 1:10,000, NPI-Tromsø & University of Silesia). Sample S1 (FS): stream water (77 00.351, 15 33.203); Sample S2 (FL): lake (77 00.390, 15 32.928); Sample S53 (FL): lake (77 00.455, 15 32.928); Sample S20 (RL): lake (77 01.106, 15 22.401); Sample S27 (RR): river (77 01.630, 15 26.265); Sample S28 (RS): stream (77 01.662, 15 20.919), Sample S31 (RL): lake (77 01.989, 15 20.919). The Polish Polar Station is indicated in red.
All water samples (3×0.5 L each, 1.5 L in total) were collected from a depth of 20 to 50 cm into sterile polyethylene bottles and maintained at low temperatures (4°C) in the absence of preservatives, as described in detail by Larose et al. (35). In the streams, sampling sites were chosen in sections of fast, turbulent, flow, with care to avoid contamination from the disturbed bed sediment. Sample temperatures ranged between 0.8 and 1.6°C.
Chemical analysis
The pH and electrical conductivity (EC) of freshwater samples were assessed using an Elmetron CX-401 apparatus supported by an ESAgP-301W electrode and CD-2-conductivity meter. Standards for pH and EC calibrations were purchased from Metrohm. Solid particles were retained by filtration (0.45 μm, Millipore) where necessary.
A range of organic compounds were analyzed in the Department of Analytical Chemistry (Faculty of Chemistry) of Gdansk University of Technology: total anionic, cationic, and non-ionic surfactants, total phenols, and formaldehyde were measured spectrophotometrically (SQ 118, PHARO 100, Merck) according to Spectroquant methods. Total organic carbon was measured using the CM 5300 furnace (UIC Coulometrics) supplemented with a coulometric detector (CM 5014 CO2 Coulometer). The TOC device was operated under a high mineralization temperature (950°C), at which oxygen was served as the carrier gas at a flow rate of 100 mL min−1.
Anions and cations were analyzed in the Polish Polar Station (St. Siedlecki, Hornsund) using ion chromatography (IC 761 Compact, Metrohm). Anions and cations were measured using Metrosep A Supp 5 and Metrosep C 4 (Metrohm) chromatography columns (150×4.0 mm each), with sodium carbonate (3.2 mM)-sodium hydrogen carbonate (1.0 mM) and HNO3 (1.7 mM)-dipicolinic acid (0.7 mM) serving as eluents at flow rates of 0.7 and 0.9 mL min−1, respectively. Injection volumes were 20 μL and 100 μL for anions and cations, respectively. All standards (1,000 mg L−1 each), eluents, and reagents for the spectrophotometric analyses were purchased from Merck (Darmstadt, Germany). 18.2 MΩ deionized water was obtained using the Millipore Gradient A10 water purification system (Bedford, USA).
An analysis of metals was performed using inductively coupled plasma mass spectrometry (ICP-MS, Elan DRC, PerkinElmer). The operating parameters of ICP-MS are listed below: sample uptake rate, 1 mL min−1; nebulizer gas flow rate, 0.98 L min−1; plasma gas flow, 15 L min−1; RF power, 1,300 W.
Genomic DNA extraction and 16S rRNA gene pyrosequencing
In order to isolate genomic DNA, 0.5-L water samples were successively filtered through sterile filters of decreasing porosities (3-μm and 0.2-μm glass fiber and membrane filters, respectively, Whatman). Total DNA was extracted as previously described in Katsaveli et al. (33) and quantified with a Qubit fluorometer (Invitrogen). In brief, 1.2 mL of 10% w/v sodium dodecyl sulfate (SDS) and 175 μL of 20 mg mL−1 proteinase K were added to the filter and incubated at 37°C under periodic stirring. Four milliliters of 5 M NaCl and 3.3 mL of preheated 2% w/v cetyltrimethylammonium bromide (CTAB) solution (2% w/v CTAB, 1.4 M NaCl, 100 mM Tris-HCl, and 20 mM EDTA, pH 8) were added. Samples were then incubated at 65°C for 2 h and an equal volume of chloroform/isoamyl alcohol (24:1 [v/v]) was added for phase separation. Samples were subsequently centrifuged at 6,000×g for 30 min and the aqueous phase was collected. DNA was precipitated by adding an equal volume of isopropanol and centrifuging at 10,000×g at 4°C for 20 min. DNA was washed twice with 2 mL 70% v/v ethanol, left to dry in a vacuum desiccator, and dissolved in 200 μL of sterile Milli-Q water. The concentrations of the isolated DNA samples were: S1_FS: 25 ng μL−1, A260/A280=1.75; S2_FL: 42 ng μL−1, A260/A280=1.79; S53_FL: 38 ng μL−1, A260/A280=1.86; S12_RL: 67 ng μL−1, A260/A280=1.72; S20_RL: 54 ng μL−1, A260/A280= 1.67; S31_RL: 86 ng μL−1, A260/A280=1.81; S28_RS: 33 ng μL−1, A260/A280=1.83; S27_RR, 28 ng μL−1, A260/A280=1.68.
Bacterial diversity in stream and lake surface water samples was assessed by tag-encoded FLX amplicon pyrosequencing in 50 μL PCR reaction containing 100 ng genomic DNA each. The universal primers 926F (5′-CTYAAAKGAATTGRCGG-3′) and 1392R (5′-ACGGGCGGTGTGTRC-3′) were used to amplify the ~470-bp region of the bacterial 16S rRNA gene (47). PCR was performed through an initial denaturation period at 94°C for 3 min, followed by 32 cycles consisting of DNA denaturation at 94°C for 30 s, primer annealing at 55°C for 40 s, and an elongation step at 72°C for 1 min, with a final termination reaction at 72°C for 5 min. Amplicons from different samples were merged in equal concentrations and cleaned up using Agencourt Ampure beads (Agencourt Bioscience Corporation). Samples were sequenced using Roche’s 454 FLX titanium instrument technologies based on the manufacturer’s guidelines.
Sequence and statistical analyses
Sequences were analyzed using the QIIME package (10). Briefly, all sequences from a single pyrosequencing run entered a custom QIIME pipeline after denoising. Sample IDs were allocated using a mapping file and barcodes were assigned to each sample. Sequences were excluded from the analysis if they were less than 200 bp in size, the quality score was less than 25, they contained ambiguous characters or an uncorrectable barcode, or did not include the primer sequence. 16S rRNA gene sequences were clustered using uclust (19) and assigned to operational taxonomic units (OTUs) with a similarity threshold of 97%. Representative sequences from each OTU were selected and aligned via Pynast (11). Taxonomy was assigned using the SILVA111 16S rRNA gene database.
Based on square root-transformed read abundance data, a Bray-Curtis similarity matrix was calculated (9). Overall similarities in microbial communities were displayed using a multidimensional scaling plot (MDS) and tests on the multivariate null hypothesis were performed using the non-parametric statistical test PERMANOVA (2). Significant relationships among environmental factors and bacterial profiles were examined using distance-based multivariable (DISTLM) and redundancy (dbRDA) analyses. Analyses were performed using the PERMANOVA+ plugin option through PRIMER6 (3).
Data accession
The gene sequences obtained have been deposited in EMBL under study accession number PRJEB13414.
Results and Discussion
The physicochemical analysis revealed that samples S2 (FL) and S53 (FL) showed the highest pH, EC, TOC, anion, and cation values as well as the highest Cu, Se, Sr, Sb, and U concentrations among the stream and lake surface freshwaters examined (Table 1). In a comparison with the other aqueous specimens tested, sample S12 (RL) contained the highest Mn, Mo, Ni, and Zn concentrations and some of the highest As, Ba, and Rb concentrations (Table 1).
Table 1.
Physicochemical characteristics of stream and lake surface water samples obtained from Fuglebekken and Revvatnet basins
| Characteristic | S1 (FS) | S2 (FL) | S12 (RL) | S20 (RL) | S27 (RR) | S28 (RS) | S31 (RL) | S53 (FL) |
|---|---|---|---|---|---|---|---|---|
| pH | 7.92±0.05 | 9.45±0.05 | 7.01±0.07 | 7.11±0.06 | 7.06±0.07 | 7.05±0.07 | 7.09±0.06 | 9.48±0.05 |
| EC (μS cm−1) | 127±7.5 | 144±7.8 | 49.1±0.98 | 55.2±0.97 | 57.5±0.98 | 30.7±0.99 | 49.3±0.99 | 146±8.1 |
| TOC (mg L−1) | 82.1±0.88 | 88.7±0.84 | 56.5±0.92 | 50.2±0.99 | 58.9±0.89 | 27.1±0.98 | 39.7±0.96 | 89.2±0.81 |
| Total cationic surfactants (mg L−1) | 0.12±0.065 | 0.10±0.065 | 0.05±0.005 | <0.05 | 0.07±0.005 | 0.09±0.05 | <0.05 | 0.07±0.05 |
| Total anionic surfactants (mg L−1) | 0.06±0.005 | 0.08±0.005 | 0.11±0.055 | 0.07±0.005 | 0.10±0.055 | 0.14±0.055 | 0.17±0.055 | 0.06±0.005 |
| Total non-ionic surfactants (mg L−1) | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 |
| Formaldehyde (mg L−1) | <0.02 | <0.02 | <0.02 | 0.04±0.007 | 0.05±0.006 | <0.02 | 0.05±0.006 | 0.05±0.006 |
| Total phenols (mg L−1) | <0.025 | <0.025 | <0.025 | <0.025 | <0.025 | <0.025 | <0.025 | <0.025 |
| Anions | ||||||||
| F− (mg L−1) | 0.01±0.0088 | 0.01±0.0089 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 0.01±0.0093 |
| Cl− (mg L−1) | 6.16±0.055 | 10.2±1.4 | 5.31±0.059 | 5.42±0.055 | 6.09±0.051 | 3.51±0.061 | 4.08±0.049 | 10.6±0.44 |
| NO2− (mg L−1) | <0.01 | 0.014±0.0075 | <0.01 | <0.01 | <0.01 | <0.01 | 0.010±0.0054 | 0.013±0.0062 |
| NO3− (mg L−1) | 1.05±0.087 | 6.01±0.066 | 0.19±0.0061 | 0.17±0.0072 | 0.35±0.0037 | 0.17±0.0065 | 0.25±0.0045 | 6.53±0.071 |
| HCO3− (mg L−1) | 62±7.9 | 62±7.8 | 12±9.7 | 11±9.9 | 12±9.6 | 7.3±3.6 | 22±9.1 | 62±8.1 |
| SO42− (mg L−1) | 6.92±0.33 | 6.02±0.35 | 3.40±0.51 | 3.69±0.55 | 3.98±0.48 | 1.18±0.76 | 4.38±0.41 | 6.30±0.31 |
| Cations | ||||||||
| Na+ (mg L−1) | 3.64±0.44 | 5.70±0.36 | 3.16±0.47 | 3.20±0.43 | 3.76±0.44 | 2.13±0.51 | 2.51±0.44 | 5.76±0.37 |
| K+ (mg L−1) | 0.59±0.031 | 0.62±0.027 | 0.48±0.039 | 0.38±0.044 | 0.56±0.031 | 0.17±0.071 | 0.82±0.022 | 0.80±0.021 |
| NH4+ (mg L−1) | <0.01 | <0.01 | 0.01±0.008 | <0.01 | 0.01±0.009 | <0.01 | <0.01 | <0.01 |
| Mg2+ (mg L−1) | 0.99±0.023 | 1.84±0.55 | 0.73±0.031 | 0.62±0.041 | 0.88±0.036 | 0.40±0.052 | 0.89±0.038 | 1.87±0.61 |
| Ca2+ (mg L−1) | 22.2±2.31 | 22.7±2.45 | 4.44±0.64 | 4.35±0.62 | 5.14±0.49 | 2.29±0.88 | 7.27±0.33 | 22.6±2.4 |
| Metals | ||||||||
| Ag (μg L−1) | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| As (μg L−1) | <0.001 | 0.08±0.0045 | 0.21±0.041 | 0.20±0.044 | 0.22±0.028 | 0.11±0.041 | 0.20±0.044 | 0.20±0.049 |
| Ba (μg L−1) | 2.52±0.42 | 2.91±0.45 | 5.20±0.33 | 5.01±0.37 | 4.90±0.29 | 1.11±0.59 | 8.81±0.21 | 3.91±0.38 |
| Cd (μg L−1) | 0.051±0.0044 | 0.051±0.0041 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| Co (μg L−1) | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| Cs (μg L−1) | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| Cu (μg L−1) | 0.40±0.051 | 0.52±0.044 | 0.30±0.041 | 0.21±0.041 | 0.50±0.022 | <0.001 | 0.41±0.0032 | 0.71±0.0025 |
| Mn (μg L−1) | <0.001 | <0.001 | 1.01±0.31 | 0.23±0.052 | 0.61±0.051 | 0.11±0.062 | 0.40±0.051 | <0.001 |
| Mo (μg L−1) | 0.11±0.053 | 0.12±0.049 | 0.15±0.051 | 0.11±0.055 | 0.14±0.051 | <0.001 | 0.22±0.049 | 0.13±0.053 |
| Ni (μg L−1) | <0.001 | <0.001 | 0.61±0.033 | <0.001 | 0.091±0.0033 | <0.001 | <0.001 | <0.001 |
| Pb (μg L−1) | 0.021±0.0039 | 0.031±0.0029 | 0.031±0.0028 | 0.050±0.0020 | 0.041±0.0019 | <0.001 | <0.001 | 0.030±0.0021 |
| Rb (μg L−1) | 0.16±0.049 | 0.18±0.044 | 0.37±0.031 | 0.36±0.029 | 0.26±0.039 | 0.32±0.029 | 0.44±0.021 | 0.19±0.042 |
| Sb (μg L−1) | 0.021±0.0048 | 0.042±0.0033 | 0.030±0.0040 | 0.031±0.0030 | 0.051±0.0039 | 0.021±0.0051 | 0.022±0.0053 | 0.060±0.0031 |
| Se (μg L−1) | 0.27±0.041 | 0.29±0.043 | 0.11±0.052 | <0.001 | 0.21±0.048 | 0.15±0.050 | 0.16±0.051 | 0.30±0.041 |
| Sr (μg L−1) | 48.3±2.1 | 50.3±1.9 | 12.3±3.1 | 21.0±2.3 | 22.5±2.7 | 10.1±3.9 | 12.1±3.3 | 60.8±1.9 |
| Sn (μg L−1) | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| Th (μg L−1) | 0.034±0.0051 | 0.029±0.0055 | <0.001 | 0.011±0.0069 | 0.012±0.0071 | <0.001 | 0.011±0.0068 | 0.018±0.0066 |
| Tl (μg L−1) | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| U (μg L−1) | 0.101±0.049 | 0.105±0.050 | <0.001 | <0.001 | 0.034±0.0077 | <0.001 | <0.001 | 0.108±0.053 |
| Zn (μg L−1) | 0.71±0.023 | 0.31±0.055 | 1.31±0.23 | <0.001 | 0.81±0.024 | <0.001 | <0.001 | <0.001 |
Values are presented as the mean±standard deviation (mean±STDEV).
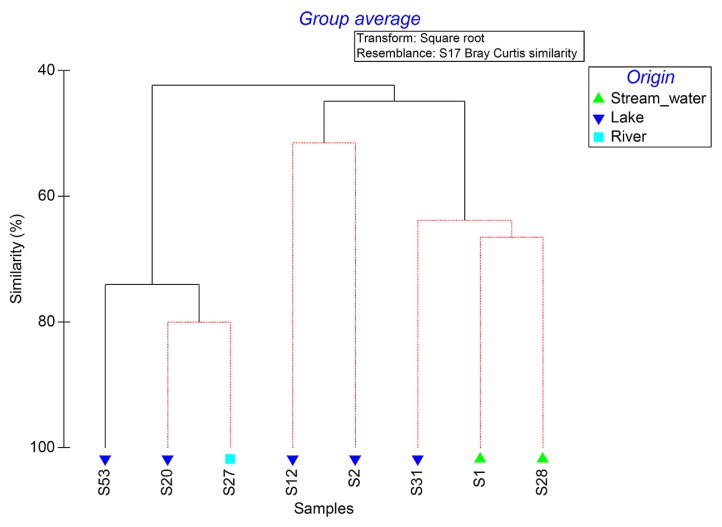
A total of 52,928 sequences (average length of 458 nucleotides) were obtained after the quality assessment. Samples S1 (FS) and S28 (RS) showed the highest bacterial diversity, while samples S2 (FL), S12 (RL), and S20 (RL) were the least diverse (Table 2). A clustering analysis of sequence data revealed that none of the samples examined were identical (Fig. 2). However, samples from Lakes Revvatnet and Fuglebekken together with sample S27 (RR), which was from a river fed with water from Lake Revvatnet, were more likely to be grouped together. The other group was comprised of the stream water samples S28 (RS) and S1 (FS) as well as sample S31 (RL), which was obtained nearby the delta of the Revelva river (Fig. 2).
Table 2.
Species richness and diversity among 16S rRNA gene pyrosequencing libraries
| Sample | Number of reads1 | Species diversity indices | |
|---|---|---|---|
|
| |||
| Simpson | Shannon | ||
| S1 (FS) | 10,567 | 86.90±0.99 | 6.34±0.03 |
| S2 (FL) | 1,912 | 39.50±1.15 | 4.13±0.06 |
| S12 (RL) | 2,783 | 42.20±1.02 | 4.71±0.04 |
| S20 (RL) | 15,759 | 44.10±1.18 | 4.89±0.05 |
| S27 (RR) | 7,620 | 51.70±1.25 | 5.22±0.04 |
| S28 (RS) | 5,417 | 87.07±1.00 | 6.38±0.02 |
| S31 (RL) | 4,788 | 76.00±0.89 | 5.96±0.04 |
| S53 (FL) | 4,082 | 48.30±1.24 | 5.03±0.06 |
Operational taxonomic units (OTUs) were defined at a 97% sequence identity threshold.
Fig. 2.
Clustering analysis of bacterial community structures of stream and lake surface water samples obtained from Fuglebekken and Revvatnet basins. Clusters drawn in red/black branches indicate significant/non-significant relationships.
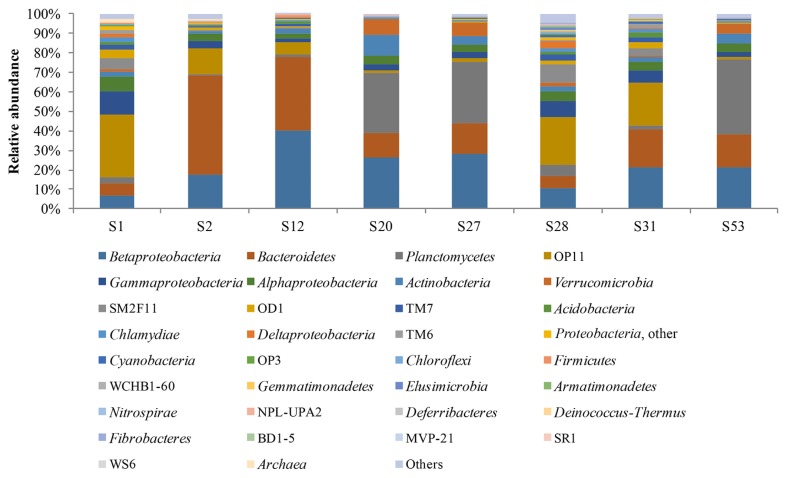
Samples S2 (FL), S12 (RL), and S31 (RL) were dominated by members of the phylum Bacteroidetes, the class Betaproteobacteria, and the Microgenomates (OP11) division, followed by the taxa of Alphaproteobacteria and Gammaproteobacteria (Fig. 3). However, the predominant taxa in these samples, i.e. Bacteroidetes, Betaproteobacteria, and Microgenomates, showed proportional differences (Fig. 3). In samples S20 (RL), S27 (RR), and S53 (FL), Planctomycetes, Betaproteobacteria, and Bacteroidetes were the major microbiota, followed by Actinobacteria, Alphaproteobacteria, Gammaproteobacteria, and Verrucomicrobia (Fig. 3). The dominance of Planctomycetes has not been previously identified in Arctic environments using universal primers; however, it was recently detected as a minor microbial component of the Arctic benthos (54, 55, 62). In a study performed in the Eastern Alps, Planctomycetes was identified as the predominant group in the microbiome of the arctic-alpine lichen Solorina crocea (23). The predominance of Microgenomates members was revealed in samples S1 (FS) and S28 (RS), followed by the lower abundances of the Alpha-, Beta-, and Gamma-subdivisions of Proteobacteria, Bacteroidetes, Planctomycetes, and candidate phylum SM2F11 (Fig. 3). The Microgenomates (OP11) division has rarely been reported in cold environments, e.g. in the polygonal tundra soils of Siberia (36), and mainly as a minor component of microbial diversity. Since Microgenomates was also among the main taxa identified in samples S2 (FL), S12 (RL), and S31 (RL), the ecological role of this microbial group appears to be significant in the biogeochemical cycles occurring in Arctic Circle habitats. This is the first study to show a dominant Microgenomates community in polar ecosystems. The Microgenomates and Parcubacteria communities were the main microbiota in Pavin Crater Lake (France), in which their possible involvement in sulfur and iron cycling was highlighted (8). Moreover, candidate division SM2F11 was also represented in an important proportion (up to 9% for sample S28 [RS]), which is among the highest reported in the literature. However, the function of this bacterial group remains unknown.
Fig. 3.
Bacterial community structures and relative abundances (based on the number of pyrosequencing reads) of major taxa identified in stream and lake surface water samples obtained from Fuglebekken and Revvatnet basins (Grouping is at the phylum level, except for Proteobacteria).
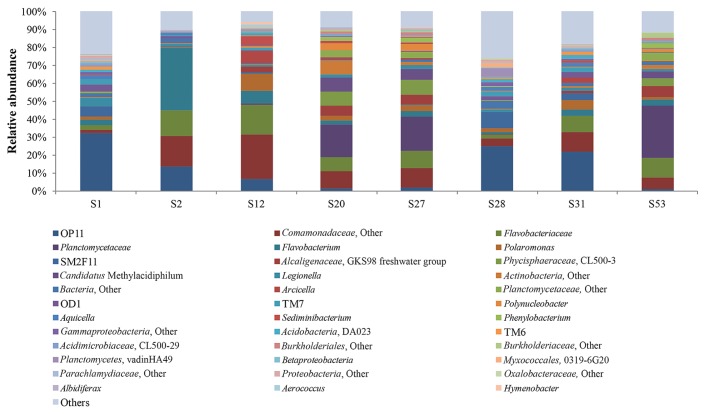
At the lower taxonomic levels, less than 20 taxa dominated in all the samples analyzed, with the sum of their abundances being in the range of 72–89% (Fig. 4 and Table 3). The most abundant taxa in samples S20 (RL), S27 (RR), and S53 (FL) were Planctomycetaceae, Comamonadaceae, Flavobacteriaceae, Phycisphaeraceae, Alcaligenaceae, and Candidatus Methylacidiphilum spp., while members of Microgenomates, Comamonadaceae, and Flavobacteriaceae, at various abundances, were the main taxa in the other samples examined (Fig. 4 and Table 3). The bacterial communities in the Fuglebekken and Revvatnet basins mainly comprised Microgenomates, Comamonadaceae, Flavobacteriaceae, Legionellales, SM2F11, Parcubacteria, and TM7 members as well as two unclassified bacterial and actinobacterial linkages, but at different proportions in each. The abundance of taxa that were shared in common among samples was greater than 70%, with the exception of samples S20 (RL), S27 (RR), and S53 (FL) in which the proliferation of Planctomycetaceae, Phycisphaeraceae, and Candidatus Methylacidiphilum members reduced the relative abundance of all OTUs shared in common (ranging in these samples between 33 and 38%) (Table 3). Skidmore et al. (52) showed that microbial populations and community structures in polar freshwater ecosystems are influenced by their aqueous geochemistries. Comamonadaceae and Flavobacteriaceae spp. were previously reported to be abundant in oligotrophic freshwater environments, including the Canadian High Arctic glaciers (15). Flavobacterium-rich communities have also been identified in Antarctic freshwater habitats (38), suggesting their possible involvement in the bacterioplankton mineralization process (1, 38). Similar to Flavobacteria, Planctomycetaceae and Phycisphaeraceae spp. are common inhabitants of detrital aggregates (22), which are linked to algal blooms and the degradation of algal sulfated polysaccharides (43). Planctomycetes has also been proposed to be indirectly involved in chemolithotrophic iron oxidation because of its prominent presence in the iron-hydroxide deposits of volcanic hydrothermal vents (54).
Fig. 4.
Relative abundance, based on the number of pyrosequencing reads, of major bacterial taxa in stream and lake surface water samples obtained from Fuglebekken and Revvatnet basins.
Table 3.
Major bacterial taxa identified in Fuglebekken and Revvatnet basins
| Taxon | Sample (relative abundance in %) | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| S1 (FS) | S2 (FL) | S12 (RL) | S20 (RL) | S27 (RR) | S28 (RS) | S31 (RL) | S53 (FL) | |
| Microgenomates (OP11)§ | 32.2 | 13.4 | 6.6 | 1.1 | 1.8 | 24.6 | 21.7 | 0.9 |
| Comamonadaceae, Other§ | 1.7 | 17.1 | 24.8 | 9.7 | 10.7 | 4.7 | 10.9 | 6.6 |
| Flavobacteriaceae§ | 2.6 | 14.2 | 16.6 | 7.8 | 9.8 | 1.9 | 9.2 | 10.8 |
| Planctomycetaceae | 0.2 | 0 | 0.6 | 18.3 | 18.9 | 0.4 | 0.1 | 29 |
| Flavobacterium§ | 3.1 | 34.8 | 7.2 | 2.2 | 3 | 0.9 | 3.6 | 3.6 |
| Polaromonas§ | 1.5 | 0.4 | 9.2 | 2.7 | 3.8 | 2.3 | 5.2 | 1.2 |
| Candidate division SM2F11§ | 5.9 | 0.7 | 0.9 | 0.1 | 0.1 | 9.2 | 3.8 | 0.2 |
| Alcaligenaceae, GKS98 freshwater group | 0 | 0 | 3 | 5.4 | 5.2 | 0 | 1.1 | 5.9 |
| Phycisphaeraceae, CL500-3 | 0 | 0 | 0.3 | 8 | 8.4 | 0 | 0 | 4.6 |
| Candidatus Methylacidiphilum | 0 | 0 | 0 | 7.7 | 6.1 | 0 | 0 | 4 |
| Legionella§ | 4.8 | 1.3 | 0.7 | 2 | 2 | 1.5 | 2.1 | 1.2 |
| Actinobacteria, Other§ | 0.1 | 0.4 | 0.5 | 7.6 | 1.9 | 0.4 | 0.6 | 2 |
| Bacteria, Other§ | 2.4 | 2.4 | 0.3 | 0.6 | 1.1 | 4.3 | 1.6 | 1.7 |
| Arcicella | 0.1 | 0 | 6.9 | 1 | 1.2 | 0 | 3.2 | 0.4 |
| Planctomycetaceae, Other | 0.7 | 0 | 0.1 | 4 | 3.3 | 0.3 | 0.1 | 4.9 |
| Parcubacteria (OD1)§ | 4 | 1.3 | 0.3 | 0.1 | 0.3 | 2.3 | 3 | 0.3 |
| Candidate division TM7§ | 2.8 | 0.4 | 1.6 | 0.2 | 0.1 | 2.6 | 2.8 | 0.2 |
| Polynucleobacter | 0 | 0 | 1 | 3.8 | 3.8 | 0 | 0.2 | 1.9 |
| Aquicella§ | 2.6 | 1 | 0.2 | 0.2 | 0.3 | 2.5 | 2.2 | 0.1 |
| Sediminibacterium | 0 | 0 | 5.4 | 1.1 | 0.9 | 0 | 1.4 | 0.1 |
| Phenylobacterium | 0.1 | 0 | 0.3 | 2.2 | 2.5 | 0.1 | 0.1 | 2.4 |
| Gammaproteobacteria, Other§ | 1.5 | 0.3 | 0.3 | 0.5 | 0.3 | 2.4 | 1 | 0.3 |
| Acidobacteria, DA023§ | 1.2 | 0.5 | 1.5 | 0 | 0.1 | 1.6 | 2.1 | 0.1 |
| Candidate division TM6§ | 1.9 | 0.3 | 0.4 | 0.1 | 0.3 | 0.8 | 1.6 | 0.7 |
| Acidimicrobiaceae, CL500-29§ | 0.9 | 0.1 | 0.8 | 1 | 0.8 | 0.9 | 0.6 | 0.6 |
| Burkholderiales, Other | 0 | 0 | 0.2 | 1.3 | 1.4 | 0.1 | 0.1 | 1.5 |
| Burkholderiaceae, Other | 0 | 0 | 0.1 | 1.1 | 1.1 | 0 | 0 | 2.7 |
| Planctomycetes, vadinHA49 | 0.5 | 0 | 0 | 0 | 0 | 4.2 | 0.4 | 0 |
| Betaproteobacteria, Other§ | 1.2 | 0.1 | 0.4 | 0.1 | 0.2 | 0.7 | 1 | 0.1 |
| Myxococcales, 0319-6G20 | 0.8 | 0 | 0 | 0 | 0 | 2.3 | 0.4 | 0 |
| Parachlamydiaceae, Other§ | 1.2 | 0.4 | 0.2 | 0 | 0.2 | 0.8 | 0.6 | 0 |
| Proteobacteria, Other§ | 1.1 | 0.4 | 0.3 | 0.1 | 0.2 | 0.9 | 0.2 | 0 |
| Oxalobacteraceae, Other§ | 0.7 | 0.1 | 0.4 | 0.1 | 0.1 | 1.1 | 0.7 | 0.3 |
| Albidiferax | 0 | 0.1 | 0.1 | 1.1 | 0.7 | 0 | 0.1 | 0.1 |
| Aerococcus | 0 | 0 | 1.5 | 0.2 | 0 | 0 | 0 | 0 |
| Hymenobacter | 0 | 0 | 1.1 | 0 | 0.1 | 0 | 0 | 0 |
|
| ||||||||
| Others | 24 | 10 | 6 | 9 | 9 | 26 | 18 | 12 |
|
| ||||||||
| Number of taxa with an abundance >1%* | 17 | 8 | 13 | 20 | 18 | 15 | 18 | 16 |
marked with bold-type case.
indicates taxa that were detected in all samples examined with an abundance >1% in at least one sample (20 phylogenetic linkages). Five more linkages with an abundance <1% (Chitinophagaceae, Pedobacter, and Sandarakinorhabdus spp. within the order Sphingobacteriales, a WCHB1-60 taxon, and a Coxiella sp. within the order Legionellales) were also detected in all samples of the present study.
Apart from the freshwater and marine microorganisms that were predominant in the samples examined, zooplankton-associated taxa were also identified. Legionellales members, i.e. Legionella, Coxiella, and Aquicella spp., comprised an important part of the bacterial communities in the Fuglebekken and Revvatnet basins. Legionellales spp. are potential intracellular parasites of free-living protozoa (12, 48, 51) and have been reported previously in polar environments, exhibiting host specificity (12). Chlamydiales, including Neochlamydia (29), Candidatus Metachlamydia (18), Candidatus Protochlamydia (41), and Candidatus Rhabdochlamydia spp. (17), and Rickettsiales, such as Rickettsia, Holospora, Caedibacter, Candidatus Captivus, and Candidatus Odyssella spp. (4, 7, 49), are obligate intracellular bacteria represented by nine and eleven OTUs, respectively, in all samples analyzed. The proportion of these taxa varied widely across sites and ranged between 1.4% (sample S12 [RL]) and 12.2% (sample S1 [FS]). Thus, endosymbiosis appears to be an important strategy in cryophilic microbiota of the Arctic and a mechanism to be adapted in polar temperatures (12).
The taxa identified also provide evidence for the biogeochemical pathways possibly occurring in the ecosystems of the Fuglebekken and Revvatnet regions. In the nitrogencycling process, the absence of the nitrite-oxidizing Nitrobacter spp. was notable. Nitrifiers were not detected in sample S27 (RR) or S53 (FL), whereas Nitrosomonadaceae and Nitrospirales spp. were the only ammonia and nitrite-oxidizing taxa identified in other samples, showing relatively low abundances (0.1–0.5% each of these functional groups). No nitrite oxidizers were identified in sample S2 (FL). Nitrospirae rather than proteobacterial nitrite oxidizers appears to be involved in the complete oxidation of ammonia in the Arctic cryosphere, and this is supported by the experimental findings of previous studies (30, 46, 55). Despite Candidatus Brocadia being a minor component of the microbial population in a single sample and no other known anammox bacteria being identified, the predominance of Planctomycetes in some of the samples does not exclude the presence of novel microbiota capable of removing nitrogen through anammox in the oxygen-deficient zones of polar environments.
Furthermore, sulfur and iron oxidation may have occurred in the stream and lake surface water samples analyzed because sulfur/sulfide and/or iron oxidizers, i.e. Gallionellaceae, Acidimicrobiaceae, Acidiferrobacter, Beggiatoa, Sulfuritalea, and Thiobacillus spp., were identified in most samples (with the exception of sample S2 [FL]), but at low abundances (0.1–1%, depending on the site examined). Filamentous sulfur bacteria of the genus Beggiatoa appear to play an important ecological role in the Arctic archipelago of Svalbard (31). Moreover, microbial ferrous oxidation was recently found to contribute to macroscopic aggregate formation in aquatic ecosystems (37). Dissimilatory sulfate reduction appears to be restricted because only a single OTU related to an Albidiferax sp. was detected in most samples (apart from S28 [RS]).
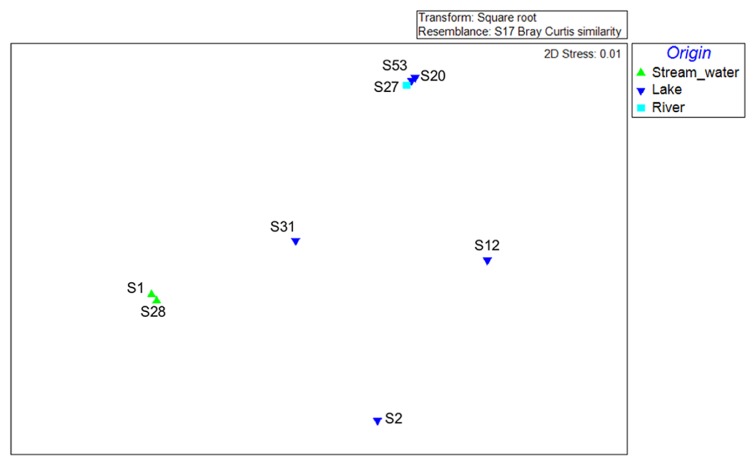
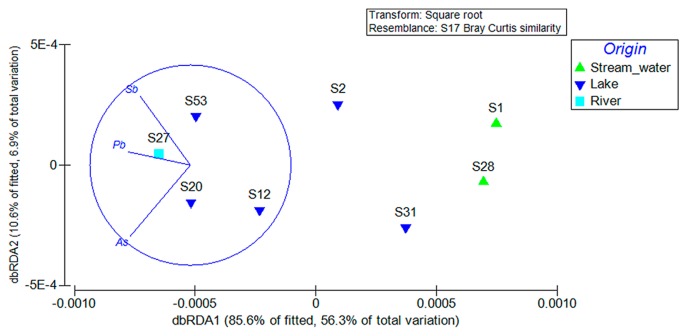
MDS was used to establish whether bacterial community structures changed as a function of the origin of the samples (lake, river, and stream water). MDS ordinations are typically interpreted based on the distance among ordinate points, with treatments that appear close together representing samples with similar bacterial community compositions. MDS ordinations based on Bray-Curtis similarities are illustrated in Fig. 5. Samples from the lake and the river fed with lake water were similar, while samples from stream water formed a separate distinct cluster (PERMDISP, F=35.529, P[perm]= 0.017). The stress values on the ordination plots were less than 0.01, indicating that the plots observed are realistic representations of the data obtained. A multivariable analysis (DISTLM) was performed in order to examine the influence of various environmental factors on the bacterial community composition (Table 1). Among the 39 factors examined, only As (p<0.05), Pb (p<0.05), and Sb (p<0.05) had a significant influence. The primary axis of dbRDA (Fig. 6) describes 56.3% of the total variation, but 85.6% of the variation within the linear model created by DistLM to analyze environmental variables (Fig. 6). Based on the dbRDA plot, increases in the concentration of As affected sampling sites S20 (RL) and S12 (RL), while Sb and Pb affected sampling sites S27 (RR) and S53 (FL). Despite S12 (RL) being relatively distinct in its bacterial composition with lake and river samples S20 (RL), S27 (RR), and S53 (FL) (Fig. 2–4), they formed a tight cluster as a consequence of their similar As, Sb, and Pb concentrations (Fig. 6).
Fig. 5.
Multidimensional scaling (MDS) plot for a graphical illustration of changes in the bacterial community structure. Green triangles represent stream water samples, Inverted blue triangles represent lake samples, Cyan squares represent rivers samples. Similarities in ribotype profiles were calculated by the Brey-Curtis algorithm.
Fig. 6.
dbRDA within a DistLM analysis of environmental factors. The fitted variation was predicted by the linear model adopted in the DistLM analysis, whereas the total variation referred to the original measurements.
Increases in As, Sb, and Pb concentrations appeared to negatively affect the Microgenomates population. Microgenomates (OP11) is not commonly found in heavy metal-contaminated environments. In samples S20 (RL), S27 (RR), and S53 (FL), Planctomycetes was more abundant than Microgenomates. Planctomycetes is an important constituent of As- and Pb-rich habitats (45), and is among the most active bacterial taxa in As-contaminated ecosystems (24). Moreover, an important Polaromonas and Flavobacterium-like population was detected in sample S12 (RL), which was recently found among the major microbiota in As-rich habitats (42, 58).
Conclusion
Wide bacterial diversity of an aquatic origin was detected, which appears to be involved in detritus mineralization processes. Moreover, a dominant Microgenomates population was identified in some of the polar freshwater ecosystems examined. This is the first study on a dominant Microgenomates community in polar ecosystems, and the ecological role of this microbial group in the biogeochemical cycles occurring in Arctic Circle habitats warrants further investigation. Based on the abiotic factors examined, only As, Pb, and Sb appear to have had significant effects on the structures of microbial communities in arctic samples. A high proportion of zooplankton-associated taxa was also identified, indicating that endosymbiosis is an adaptive mechanism by several microorganisms in the Arctic cryosphere.
Supplementary Information
Acknowledgements
This work was partially supported by the EU PEOPLE-2012-IAPP 324349 program, and by intramural funds of the University of Patras to George Tsiamis. The authors would like to thank the staff of the Polish Polar Station at Hornsund for the opportunity to carry out sampling and for their assistance with this work. The authors would also like to thank Katarzyna Cichała-Kamrowska for preparing the database. This project was also partially funded by the Polish National Science Centre (DEC-2013/09/N/ST10/04191) and the 4th International Polar Year (Special Project 1173/IPY/2007 Higher Education).
References
- 1.Abell G.C.J., Bowman J.P. Ecological and biogeographic relationships of class Flavobacteria in the Southern Ocean. FEMS Microbiol Ecol. 2005;51:265–277. doi: 10.1016/j.femsec.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Anderson M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 3.Anderson M.J., Gorley R.N., Clarke R.K. PERMANOVA+ for PRIMER: guide to software and statistical methods. PRIMER-E; Plymouth, UK: 2008. [Google Scholar]
- 4.Baker B.J., Hugenholtz P., Dawson S.C., Banfield J.F. Extremely acidophilic protists from acid mine drainage host Rickettsiales-Lineage endosymbionts that have intervening sequences in their 16S rRNA genes. Appl Environ Microbiol. 2003;69:5512–5518. doi: 10.1128/AEM.69.9.5512-5518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balcazar W., Rondón J., Rengifo M., et al. Bioprospecting glacial ice for plant growth promoting bacteria. Microbiol Res. 2015;177:1–7. doi: 10.1016/j.micres.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Bej A.K., Aislabie J., Atlas R.M. Polar Microbiology: The Ecology, Biodiversity, and Bioremediation Potential of Microorganisms in Extremely Cold Environments. Boca Raton, FL: CRC Press; 2009. [Google Scholar]
- 7.Birtles R.J., Rowbotham T.J., Michel R., et al. “Candidatus Odyssella thessalonicensis” gen. nov., sp. nov., an obligate intracellular parasite of Acanthamoeba species. Int J Syst Evol Microbiol. 2000;50:63–72. doi: 10.1099/00207713-50-1-63. [DOI] [PubMed] [Google Scholar]
- 8.Borrel G., Lehours A-C., Bardot C., Bailly X., Fonty G. Members of candidate divisions OP11, OD1 and SR1 are widespread along the water column of the meromictic Lake Pavin (France) Arch Microbiol. 2010;192:559–567. doi: 10.1007/s00203-010-0578-4. [DOI] [PubMed] [Google Scholar]
- 9.Bray J.R., Curtis J.T. An ordination of the upland forest communities of Southern Wisconsin. Ecol Monogr. 1957;27:325. [Google Scholar]
- 10.Caporaso J.G., Kuczynski J., Stombaugh J., et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caporaso J.G., Bittinger K., Bushman F.D., DeSantis T.Z., Andersen G.L., Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carvalho F.R.S., Nastasi F.R., Gamba R.C., Foronda A.S., Pellizari V.H. Occurrence and diversity of Legionellaceae in polar lakes of the antarctic peninsula. Curr Microbiol. 2008;57:294–300. doi: 10.1007/s00284-008-9192-y. [DOI] [PubMed] [Google Scholar]
- 13.Casanueva A., Tuffin M., Cary C., Cowan D.A. Molecular adaptations to psychrophily: the impact of “omic” technologies. Trends Microbiol. 2010;18:374–381. doi: 10.1016/j.tim.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Cavicchioli R., Charlton T., Ertan H., Omar S.M., Siddiqui K.S., Williams T.J. Biotechnological uses of enzymes from psychrophiles. Microb Biotechnol. 2011;4:449–460. doi: 10.1111/j.1751-7915.2011.00258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng S.M., Foght J.M. Cultivation-independent and -dependent characterization of bacteria resident beneath John Evans Glacier. FEMS Microbiol Ecol. 2007;59:318–330. doi: 10.1111/j.1574-6941.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 16.Collins R.E., Deming J.W. An inter-order horizontal gene transfer event enables the catabolism of compatible solutes by Colwellia psychrerythraea 34H. Extremophiles. 2013;17:601–610. doi: 10.1007/s00792-013-0543-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corsaro D., Feroldi V., Saucedo G., Ribas F., Loret J-F., Greub G. Novel Chlamydiales strains isolated from a water treatment plant. Environ Microbiol. 2009;11:188–200. doi: 10.1111/j.1462-2920.2008.01752.x. [DOI] [PubMed] [Google Scholar]
- 18.Corsaro D., Michel R., Walochnik J., Müller K-D., Greub G. Saccamoeba lacustris, sp. nov. (Amoebozoa: Lobosea: Hartmannellidae), a new lobose amoeba, parasitized by the novel chlamydia “Candidatus Metachlamydia lacustris” (Chlamydiae: Parachlamydiaceae) Eur J Protistol. 2010;46:86–95. doi: 10.1016/j.ejop.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 20.Edwards K.J., Wheat C.G., Sylvan J.B. Under the sea: microbial life in volcanic oceanic crust. Nat Rev Microbiol. 2011;9:703–712. doi: 10.1038/nrmicro2647. [DOI] [PubMed] [Google Scholar]
- 21.Fellman J.B., Spencer R.G.M., Hernes P.J., Edwards R.T., D’Amore D.V., Hood E. The impact of glacier runoff on the biodegradability and biochemical composition of terrigenous dissolved organic matter in near-shore marine ecosystems. Mar Chem. 2010;121:112–122. [Google Scholar]
- 22.Glöckner F.O., Kube M., Bauer M., et al. Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc Natl Acad Sci USA. 2003;100:8298–8303. doi: 10.1073/pnas.1431443100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grube M., Köberl M., Lackner S., Berg C., Berg G. Host–parasite interaction and microbiome response: effects of fungal infections on the bacterial community of the Alpine lichen Solorina crocea. FEMS Microbiol Ecol. 2012;82:472–481. doi: 10.1111/j.1574-6941.2012.01425.x. [DOI] [PubMed] [Google Scholar]
- 24.Halter D., Cordi A., Gribaldo A., et al. Taxonomic and functional prokaryote diversity in mildly arsenic-contaminated sediments. Res Microbiol. 2011;162:878–887. doi: 10.1016/j.resmic.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 25.Harding T., Jungblut A.D., Lovejoy C., Vincent W.F. Microbes in High Arctic snow and implications for the cold biosphere. Appl Environ Microbiol. 2011;77:3234–3243. doi: 10.1128/AEM.02611-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hashim N.H.F., Bharudin I., Nguong D.L.S., et al. Characterization of Afp1, an antifreeze protein from the psychrophilic yeast Glaciozyma antarctica PI12. Extremophiles. 2013;17:63–73. doi: 10.1007/s00792-012-0494-4. [DOI] [PubMed] [Google Scholar]
- 27.Hell K., Edwards A., Zarsky J., et al. The dynamic bacterial communities of a melting High Arctic glacier snowpack. ISME J. 2013;7:1814–1826. doi: 10.1038/ismej.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hjelle A. Geology of Svalbard. Norsk Polarinstitutt; Oslo: 1993. [Google Scholar]
- 29.Horn M., Wagner M., Müller K-D., et al. Neochlamydia hartmannellae gen. nov., sp. nov. (Parachlamydiaceae), an endoparasite of the amoeba Hartmannella vermiformis. Microbiology. 2000;146:1231–1239. doi: 10.1099/00221287-146-5-1231. [DOI] [PubMed] [Google Scholar]
- 30.Huang J.P., Swain A.K., Thacker R.W., Ravindra R., Andersen D.T., Bej A.K. Bacterial diversity of the rock-water interface in an East Antarctic freshwater ecosystem, Lake Tawani(P) Aquat Biosyst. 2013;9:4. doi: 10.1186/2046-9063-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jørgensen B.B., Dunker R., Grünke S., Røy H. Filamentous sulfur bacteria, Beggiatoa spp., in arctic marine sediments (Svalbard, 79°N) FEMS Microbiol Ecol. 2010;73:500–513. doi: 10.1111/j.1574-6941.2010.00918.x. [DOI] [PubMed] [Google Scholar]
- 32.Jung Y.H., Yi J-Y., Jung H.J., et al. Overexpression of cold shock protein A of Psychromonas arctica KOPRI 22215 confers cold-resistance. Protein J. 2010;29:136–142. doi: 10.1007/s10930-010-9233-9. [DOI] [PubMed] [Google Scholar]
- 33.Katsaveli K., Vayenas D., Tsiamis G., Bourtzis K. Bacterial diversity in Cr(VI) and Cr(III)-contaminated industrial wastewaters. Extremophiles. 2012;16:285–296. doi: 10.1007/s00792-012-0429-0. [DOI] [PubMed] [Google Scholar]
- 34.Kohler J., James T.D., Murray T., et al. Acceleration in thinning rate on western Svalbard glaciers. Geophys Res Lett. 2007;34:L18502. [Google Scholar]
- 35.Larose C., Berger S., Ferrari C., et al. Microbial sequences retrieved from environmental samples from seasonal Arctic snow and meltwater from Svalbard, Norway. Extremophiles. 2010;14:205–212. doi: 10.1007/s00792-009-0299-2. [DOI] [PubMed] [Google Scholar]
- 36.Liebner S., Harder J., Wagner D. Bacterial diversity and community structure in polygonal tundra soils from Samoylov Island, Lena Delta, Siberia. Int Microbiol. 2008;11:195–202. [PubMed] [Google Scholar]
- 37.Lu S., Chourey K., Reiche M., et al. Insights into the structure and metabolic function of microbes that shape pelagic iron-rich aggregates (“Iron Snow”) Appl Environ Microbiol. 2013;79:4272–4281. doi: 10.1128/AEM.00467-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michaud L., Caruso C., Mangano S., Interdonato F., Bruni V., Giudice A.L. Predominance of Flavobacterium, Pseudomonas, and Polaromonas within the prokaryotic community of freshwater shallow lakes in the northern Victoria Land, East Antarctica. FEMS Microbiol Ecol. 2012;82:391–404. doi: 10.1111/j.1574-6941.2012.01394.x. [DOI] [PubMed] [Google Scholar]
- 39.Mindl B., Anesio A.M., Meirer K., et al. Factors influencing bacterial dynamics along a transect from supraglacial runoff to proglacial lakes of a high Arctic glacier. FEMS Microbiol Ecol. 2007;59:307–317. doi: 10.1111/j.1574-6941.2006.00262.x. [DOI] [PubMed] [Google Scholar]
- 40.Møller A.K., Barkay T., Al-Soud W.A., Sørensen S.J., Skov H., Kroer N. Diversity and characterization of mercury-resistant bacteria in snow, freshwater and sea-ice brine from the High Arctic. FEMS Microbiol Ecol. 2011;75:390–401. doi: 10.1111/j.1574-6941.2010.01016.x. [DOI] [PubMed] [Google Scholar]
- 41.Nakamura S., Matsuo J., Hayashi Y., et al. Endosymbiotic bacterium Protochlamydia can survive in acanthamoebae following encystation. Environ Microbiol Rep. 2010;2:611–618. doi: 10.1111/j.1758-2229.2010.00182.x. [DOI] [PubMed] [Google Scholar]
- 42.Paul D., Kazy S.K., Gupta A.K., Pal T., Sar P. Diversity, metabolic properties and arsenic mobilization potential of indigenous bacteria in arsenic contaminated groundwater of West Bengal, India. PLoS ONE. 2015;10:e0118735. doi: 10.1371/journal.pone.0118735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pizzetti I., Fuchs B.M., Gerdts G., Wichels A., Wiltshire K.H., Amann R. Temporal variability of coastal Planctomycetes clades at Kabeltonne station, North Sea. Appl Environ Microbiol. 2011;77:5009–5017. doi: 10.1128/AEM.02931-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quesada A., Vincent W.F. Cyanobacteria in the cryosphere: snow, ice and extreme cold. In: Whitton B.A., editor. Ecology of Cyanobacteria II. Springer; Dordrecht, The Netherlands: 2012. pp. 387–399. [Google Scholar]
- 45.Rastogi G., Stetler L.D., Peyton B.M., Sani R.K. Molecular analysis of prokaryotic diversity in the deep subsurface of the former Homestake gold mine, South Dakota, USA. J Microbiol. 2009;47:371–384. doi: 10.1007/s12275-008-0249-1. [DOI] [PubMed] [Google Scholar]
- 46.Reigstad L.J., Jorgensen S.L., Lauritzen S-E, Schleper C., Urich T. Sulfur-oxidizing chemolithotrophic proteobacteria dominate the microbiota in high arctic thermal springs on Svalbard. Astrobiology. 2011;11:665–678. doi: 10.1089/ast.2010.0551. [DOI] [PubMed] [Google Scholar]
- 47.Rinke C., Schwientek P., Sczyrba A., et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 48.Santos P., Pinhal I., Rainey F.A., et al. Gamma-Proteobacteria Aquicella lusitana gen. nov., sp. nov., and Aquicella siphonis sp. nov. infect protozoa and require activated charcoal for growth in laboratory media. Appl Environ Microbiol. 2003;69:6533–6540. doi: 10.1128/AEM.69.11.6533-6540.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmitz-Esser S., Linka N., Collingro A., et al. ATP/ADP translocases: a common feature of obligate intracellular amoebal symbionts related to Chlamydiae and Rickettsiae. J Bacteriol. 2004;186:683–691. doi: 10.1128/JB.186.3.683-691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schostag M., Stibal M., Jacobsen C.S., et al. Distinct summer and winter bacterial communities in the active layer of Svalbard permafrost revealed by DNA- and RNA-based analyses. Terr Microbiol. 2015;6:399. doi: 10.3389/fmicb.2015.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scola B.L., Raoult D. Survival of Coxiella burnetii within free-living amoeba Acanthamoeba castellanii. Clin Microbiol Infect. 2001;7:75–79. doi: 10.1046/j.1469-0691.2001.00193.x. [DOI] [PubMed] [Google Scholar]
- 52.Skidmore M., Anderson S.P., Sharp M., Foght J., Lanoil B.D. Comparison of microbial community compositions of two sub-glacial environments reveals a possible role for microbes in chemical weathering processes. Appl Environ Microbiol. 2005;71:6986–6997. doi: 10.1128/AEM.71.11.6986-6997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skidmore M.L., Foght J.M., Sharp M.J. Microbial life beneath a high Arctic glacier. Appl Environ Microbiol. 2000;66:3214–3220. doi: 10.1128/aem.66.8.3214-3220.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Storesund J.E., Øvreås L. Diversity of Planctomycetes in iron-hydroxide deposits from the Arctic Mid Ocean Ridge (AMOR) and description of Bythopirellula goksoyri gen. nov., sp. nov., a novel Planctomycete from deep sea iron-hydroxide deposits. Antonie Van Leeuwenhoek. 2013;104:569–584. doi: 10.1007/s10482-013-0019-x. [DOI] [PubMed] [Google Scholar]
- 55.Tait K., Laverock B., Shaw J., Somerfield P.J., Widdicombe S. Minor impact of ocean acidification to the composition of the active microbial community in an Arctic sediment. Environ Microbiol Rep. 2013;5:851–860. doi: 10.1111/1758-2229.12087. [DOI] [PubMed] [Google Scholar]
- 56.Tang M.A.K., Motoshima H., Watanabe K. Fluorescence studies on the stability, flexibility and substrate-induced conformational changes of acetate kinases from psychrophilic and mesophilic bacteria. Protein J. 2012;31:337–344. doi: 10.1007/s10930-012-9408-7. [DOI] [PubMed] [Google Scholar]
- 57.Tian F., Yu Y., Chen B., Li H., Yao Y-F., Guo X-K. Bacterial, archaeal and eukaryotic diversity in Arctic sediment as revealed by 16S rRNA and 18S rRNA gene clone libraries analysis. Polar Biol. 2008;32:93–103. [Google Scholar]
- 58.Wang Y., Li P., Jiang D., Li B., Dai X., Jiang Z., Wang Y. Vertical distribution of bacterial communities in high arsenic sediments of Hetao Plain, Inner Mongolia. Ecotoxicology. 2014;23:1890–1899. doi: 10.1007/s10646-014-1322-7. [DOI] [PubMed] [Google Scholar]
- 59.White P.L., Wynn-Williams D.D., Russell N.J. Diversity of thermal responses of lipid composition in the membranes of the dominant culturable members of an Antarctic fellfield soil bacterial community. Antarct Sci. 2000;12:386–393. [Google Scholar]
- 60.Xiang W., Chai H., Tian D., Peng C. Marginal effects of silvicultural treatments on soil nutrients following harvest in a Chinese fir plantation. Soil Sci Plant Nutr. 2009;55:523–531. [Google Scholar]
- 61.Zarsky J.D., Stibal M., Hodson A., et al. Large cryoconite aggregates on a Svalbard glacier support a diverse microbial community including ammonia-oxidizing archaea. Environ Res Lett. 2013;8:035044. [Google Scholar]
- 62.Zeng Y-X., Yan M., Yu Y., et al. Diversity of bacteria in surface ice of Austre Lovénbreen glacier, Svalbard. Arch Microbiol. 2013;195:313–322. doi: 10.1007/s00203-013-0880-z. [DOI] [PubMed] [Google Scholar]
- 63.Zwally H.J., Giovinetto M.B. Overview and assessment of antarctic ice-sheet mass balance estimates: 1992–2009. Surv Geophys. 2011;32:351–376. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.