Abstract
Background
Nowadays, despite great progress in cancer research, the detailed mechanisms of colorectal cancer (CRC) are still poorly understood. Circular RNAs (circRNAs), a new star of the non-coding RNA network, have been identified as critical regulators in various cancers, including CRC.
Methods and results
In this study, by using unsupervised hierarchical clustering analysis, a novel dysregulated circRNA, hsa_circ_0000069, was found. The expression of hsa_circ_0000069 was measured in 30 paired CRC tissues and adjacent noncancerous tissues using quantitative polymerase chain reaction. A high expression of hsa_circ_0000069 was observed in CRC tissues and correlated with patients’ age and tumor, node, metastasis (TNM) stage (P<0.05). Furthermore, by using specifically designed siRNAs in CRC cells, a functional analysis was performed which revealed that hsa_circ_0000069 knockdown could notably inhibit cell proliferation, migration, and invasion, and induce G0/G1 phase arrest of cell cycle in vitro.
Conclusion
This study’s findings are the first to demonstrate that hsa_circ_0000069, an important regulator in cancer progression, could be a promising target in the diagnosis and therapy in colorectal cancer.
Keywords: circular RNA, colorectal cancer, regulation, hsa_circ_0000069
Introduction
Colorectal cancer (CRC) is the third most common cancer and cause of cancer-related deaths worldwide.1 Despite the tremendous advances in traditional surgery, radiochemotherapy, and targeted treatment, the exact mechanisms of CRC are still unclear. There is a pressing need to better understand the regulation network in CRC progression and identify novel biomarkers and therapy targets to improve tumor diagnosis and prognosis efficiently.
Circular RNAs (circRNAs) are new members of the non-coding RNAs network. Unlike regular linear RNAs, circRNAs have covalently linked ends.2 They are characterized as stable, abundant and conserved, and usually exhibit tissue/developmental-stage specific expression.3 Emerging evidence has revealed that circRNAs can serve as microRNA sponges, RNA-binding proteins or transcriptional regulators to participate in the initiation and development of multiple diseases, such as Alzheimer’s disease, Parkinson’s disease, atherosclerosis, and cancers.4,5 Several circRNAs have already been found to be important factors in cancers, such as Cdr1as in hepatocellular carcinoma,6 hsa_circ_002059 in gastric cancer,7 and cir-ITCH in esophageal squamous cancer.8 In this study, based on the unsupervised hierarchical clustering analysis, 892 dysregulated circRNAs were found including 412 upregulated circR-NAs and 480 downregulated circRNAs (fold >2, P<0.05) (Figure 1). Quantitative real-time polymerase chain reaction (qRT-PCR) was then used to detect the expression levels of several circRNAs in CRC tissues and found a significantly upregulated circRNA, hsa_circ_0000069, which had never been reported before. Further functional analysis revealed that hsa_circ_0000069 could affect CRC cell proliferation, migration, invasion, and cell cycle in vitro. This study’s results suggest that hsa_circ_0000069 could be a novel potential oncogene of CRC.
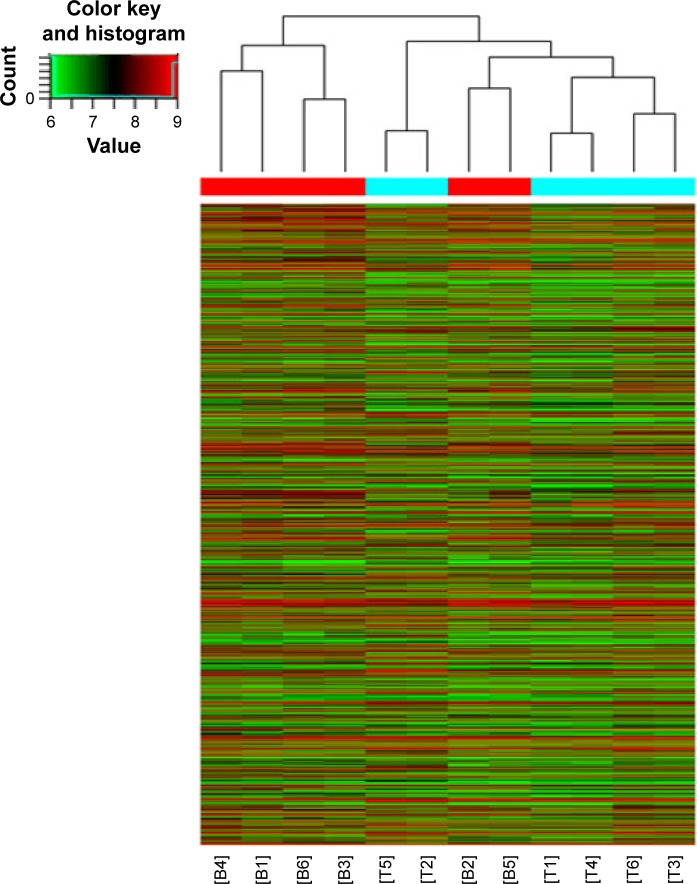
Figure 1.
Differentially expressed circRNA between 6 cases of CRC tissues and 6 cases of normal colorectal tissues were analyzed using hierarchical clustering.
Notes: The result from unsupervised hierarchical clustering analysis shows distinguishable circRNAs expression profiling among samples (T for CRC and B for normal adjacent tissues). Each column represents the expression profile of a tissue sample, and each row corresponds to a circRNAs. “Red” indicates higher expression level, and “green” indicates lower expression level.
Abbreviations: circRNA, circular RNA; CRC, colorectal cancer.
Materials and methods
Tissue samples and clinical data
A total of 30 paired CRC tissues and adjacent noncancerous tissues were obtained from patients who underwent routine surgical resection at Huai’an First People’s Hospital, Nanjing Medical University (Huai’an, People’s Republic of China) in 2015. The patients had not received chemoradiotherapy before surgery and were identified by pathological diagnosis. All tissue samples were quickly frozen in liquid nitrogen and stored at −80°C until further experiments. Written informed consent was obtained from all patients and this study was approved by the ethics committee of Huai’an First People’s Hospital, Nanjing Medical University (Huai’an, People’s Republic of China).
Cell lines
Four human CRC cell lines (HT-29, LoVo, HCT-116 and SW480) and one normal intestinal epithelial cell line (HCO) were purchased from Shanghai Institutes for Biological Science (Shanghai, People’s Republic of China). All cell lines were maintained in Roswell Park Memorial Institute (RPMI) 1640 medium (Hyclone, Logan, Utah, USA) with 10% FBS and kept in a 37°C incubator containing 5% CO2. The cells, after 2–3 stable generations, were used for subsequent analysis.
RNA extraction and qPCR analysis
To extract total RNA from CRC tissues and cultured cell lines, TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) was used by the manufacturer’s instructions. One μg total RNA was reverse transcribed using random primers with HiScript® II 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, People’s Republic of China), in a final volume of 10 μL. Then, an AceQ® qPCR SYBR® Green Master Mix (Vazyme, Nanjing, People’s Republic of China) was used to measure the expression levels of hsa_circ_0000069 in CRC tissues and cell lines according to the manufacturer’s protocol. The amplification was performed on an ABI PRISM 7500 Real-Time PCR System (Thermo Fisher Scientific) with 2 μL synthetic cDNA. Glyceraldehyde-3-phosphate dehydrogenase was used to normalize the expression of cir-cRNA or microRNAs. All of the experiments were performed in triplicate and data were analyzed using the 2−ΔΔCT method. The sequences for primers were listed in Table 1.
Table 1.
The sequences for primers and siRNAs
| Primers used for qRT-PCR | |
| GAPDH F | GGGAGCCAAAAGGGTCAT |
| GAPDH R | GAGTCCTTCCACGATACCAA |
| hsa_circ_0000069 F | CTACTTCAGGCACAGGTCTTC |
| hsa_circ_0000069 R | CTGACTCACTGGATGAGGACT |
| siRNAs oligonucleotides | |
| si-circ_0000069-1# | CTACTTCAGGCACAGGTCT |
| si-circ_0000069-2# | CTTCAGGCACAGGTCTTCC |
| si-circ_0000069-3# | AGGCACAGGTCTTCCCAAA |
Abbreviations: GAPDH, glyceraldehyde 3-phosphate dehydrogenase; qRT-PCR, quantitative real-time polymerase chain reaction.
Cell transfection
To efficiently knockdown the expression of hsa_circ_0000069, three different siRNAs were designed by Geneseed (Guangzhou, People’s Republic of China). Cells were planted on six well plated at a density of 5×105/well for 24 h. After reaching to 70%–80% fusion, cells were transfected with siRNAs or negative control with Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific) according to the instructions. After incubation for 48 h, cells were harvested to detect the efficiency of RNA knockdown by qRT-PCR. The sequences for siRNAs are listed in Table 1.
Cell proliferation assays
To investigate the effect of hsa_circ_0000069 knockdown on CRC cell proliferation, a Cell Counting Kit-8 (Dojindo, Japan) was used by the manufacturer’s protocol. In brief, 5×103/well cells were seeded into 96-well plates and maintained in a 37°C incubator. The cell viability was quantified at 0, 24, 48, 72, and 96 h with a microplate reader (Bio-Rad Laboratories Inc., Hercules, CA, USA) at 450 nm. For colony formation assay, transfected cells were placed in 6-well plates and maintained for 14 days, replacing the medium every 3 days. Colonies were then fixed with methanol, stained with crystal violet, and counted under the microscope.
Flow cytometric analysis
For cell cycle assay, transfected cells were collected and fixed with −20°C precooling 70% ethanol overnight at 4°C. Cells were then stained with propidium iodide (PI) using Cell Cycle Detection Kit (KeyGen, Nanjing, People’s Republic of China) by the manufacturer’s instructions. The PI-stained samples were run on a FACSCalibur (Becton, Dickinson and Company, FL, NJ, USA), and the percentage of cells in each phase was analyzed by FACScan (Becton, Dickinson and Company). For cell apoptosis assay, transfected cells were stained with Annexin V-FITC and PI using the Annexin V-FITC Apoptosis Detection Kit (KeyGen) according to the protocol and analyzed with CellQuest software (BD Bioscience, San Jose,CA, USA).
Cell migration and invasion assays
For migration assay, 1×105 cells after transfection were harvested and placed in the upper chamber (8 μm pore size, BD), and 600 μL proper medium was added into the lower chamber. For Transwell assay, 1×105 cells were placed in the upper chamber coated with Matrigel (BD Biosciences), and 600 μL proper medium was added into the lower chamber. After 24-h incubation, cells maintained on the upper membrane were removed with a cotton swab and cells migrated or invaded were fixed with methanol, stained with crystal violet, and counted under a microscope.
Statistical analysis
Statistical analyses were performed using SPSS 19.0 software (IBM Corporation, Armonk, NY, USA). The Student’s t-test, Wilcoxon test, or χ2 test were used to investigate the difference between groups, as appropriate. A two-tailed P-value <0.05 was considered to be statistically significant.
Results
Hsa_circ_0000069 expression is upregulated in CRC and associated with clinicopathological features
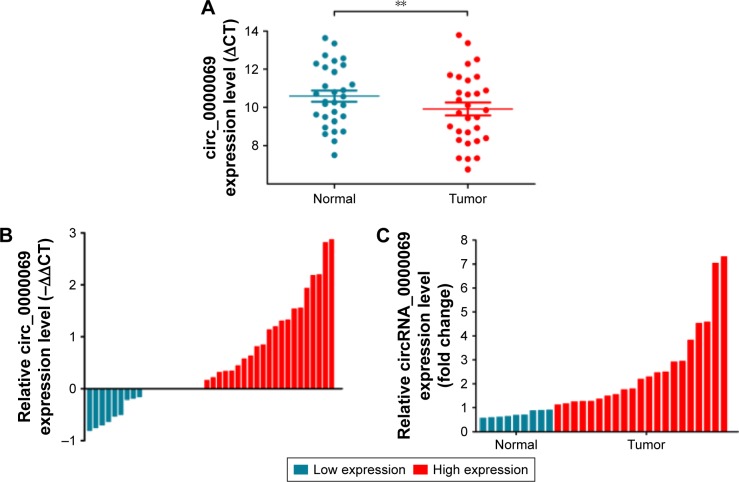
By analyzing the differentially expressed circRNAs in 6 paired CRC tissues and surrounding normal tissues, 412 upregulated circRNAs and 480 downregulated circRNAs were found (fold >2, P<0.05) (Figure 1). After the verification by qRT-PCR, hsa_circ_0000069 was found markedly upregulated in both CRC tissues and cell lines, which was consistent with the data of the unsupervised hierarchical clustering analysis. To be specific, in 70% CRC tissues (21 of 30, fold ≥1.0), the expression of hsa_circ_0000069 was significantly higher than adjacent normal tissues (P<0.01) (Figure 2A–C). Furthermore, the correlation between hsa_circ_0000069 expression and patients’ clinicopathological features were analyzed, which indicated that high expression of hsa_circ_0000069 was associated with patients’ age (P=0.036) and TNM stage (P=0.040) (Table 2). However, its expression was not correlated with gender (P=0.719), tumor size (P=0.211), and differentiation (P=0.185) of CRC patients.
Figure 2.
Relative hsa_circ_0000069 expression in CRC tissues and cell lines.
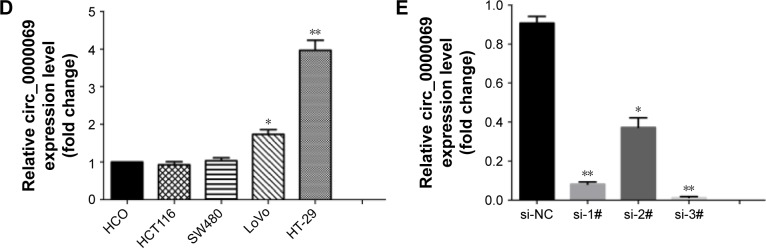
Notes: (A) Relative hsa_circ_0000069 expression in CRC tissues (n=30) compared with corresponding non-tumor tissues (n=30). Hsa_circ_0000069 expression was examined by qRT-PCR and normalized to GAPDH expression. Results were presented as ΔCT in tumor tissues relative to normal tissues. (B) Relative hsa_circ_0000069 expression in CRC tissues (n=30) compared with corresponding non-tumor tissues (n=30). Hsa_circ_0000069 expression was classified into two groups. Positive −ΔΔCT meant high hsa_circ_0000069 expression. Negative −ΔΔCT meant low hsa_circ_0000069 expression. (C) Relative hsa_circ_0000069 expression in CRC tissues (n=30) compared with corresponding non-tumor tissues (n=30). Hsa_circ_0000069 expression was classified into two groups. Fold change >1 meant high hsa_circ_0000069 expression. Fold change <1 meant low hsa_circ_0000069 expression. (D) Relative hsa_circ_0000069 expression levels of CRC cell lines (HCT116, SW480, LoVo, and HT-29) compared with that in the normal intestinal epithelial cell line (HCO). (E) The hsa_circ_0000069 expression level was determined by qRT-PCR when HT-29 cells transfected with si- circ_0000069 (si-1#, si-2# and si-3#). *P<0.05 and **P<0.01. Error bars show the uncertainty in 3 independent measurements. ΔCT is the difference between the CT value of hsa_circ_0000069 and the CT value of GAPDH. ΔΔCT is the difference between the ΔCT of CRC tissues and the ΔCT of adjacent normal tissues.
Abbreviations: CRC, colorectal cancer; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; qRT-PCR, quantitative real-time polymerase chain reaction.
Table 2.
Correlation between hsa_circ_0000069 expression and clinicopathological characteristics in colorectal cancer
| Clinical parameter | Hsa_circ_0000069
|
Chi-squared test P-value |
|
|---|---|---|---|
| High number of cases | Low number of cases | ||
| Gender | 0.719 | ||
| Male | 15 | 7 | |
| Female | 6 | 2 | |
| Age (years) | 0.036* | ||
| ≤50 | 6 | 4 | |
| 50–70 | 5 | 5 | |
| >70 | 10 | 0 | |
| Tumor size | 0.211 | ||
| ≤3 cm | 5 | 1 | |
| 3–5 cm | 9 | 7 | |
| >5 cm | 7 | 1 | |
| Differentiation | 0.185 | ||
| Poor | 6 | 0 | |
| Moderate | 14 | 8 | |
| Well | 1 | 1 | |
| Tumor stage | 0.040* | ||
| I–II | 11 | 1 | |
| III–IV | 10 | 8 | |
Note:
Statistically significant P-value<0.05.
Knockdown of hsa_circ_0000069 inhibits CRC cell proliferation and induce G0/G1 phase arrest in vitro
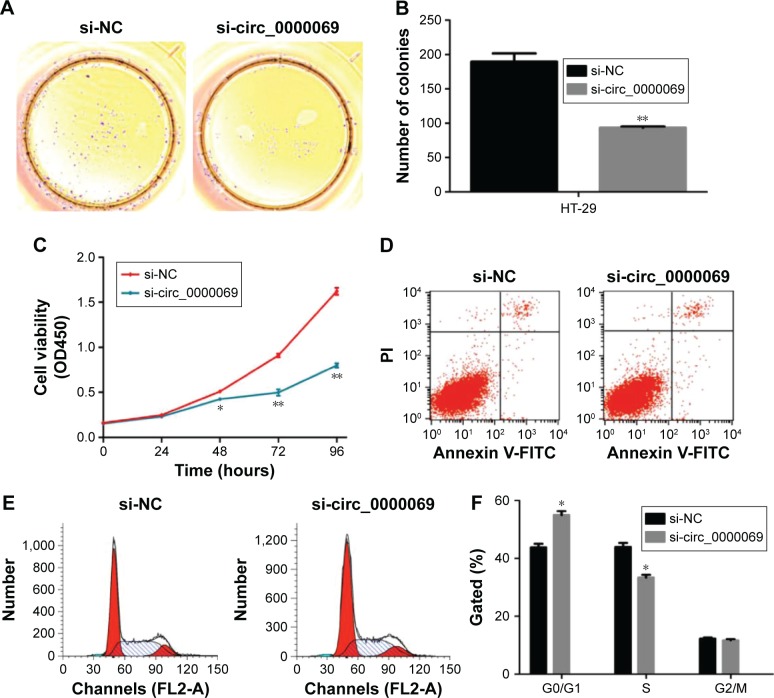
To further explore the functional role of hsa_circ_0000069 in CRC cells, its expression levels in four CRC cell lines (HT-29, LoVo, HCT-116 and SW480) and one normal intestinal epithelial cell line (HCO) were measured. The results showed that hsa_circ_0000069 was obviously elevated in HT-29 (P<0.01) and LoVo (P<0.05) (Figure 2D). Specifically designed siR-NAs were used to knockdown hsa_circ_0000069 expression in HT-29 cells (Figure 2E). Verification by qRT-PCR revealed that the levels of hsa_circ_0000069 were significantly reduced in HT-29 cells transfected with si-circ_0000069-3#, which were used in further analyses. Cell Counting Kit-8 assays showed that knockdown of hsa_circ_0000069 could obviously inhibit cell proliferation in HT-29 cells (Figure 3C). Similarly, colony formation assays indicated that clonogenic survival was decreased following inhibition of hsa_circ_0000069 in HT-29 cells (Figure 3A and B). A flow cytometry analysis was performed to investigate the effect of hsa_circ_0000069 knockdown on cell cycle and apoptosis. The results revealed that, compared with si-NC group, the cell cycle progression of HT-29/si-circ_0000069 was stalled at the G0/G1 phase (P<0.05) (Figure 3E and F). However, hsa_circ_0000069 knockdown had no obvious effect on cell apoptosis in HT-29 cells (Figure 3D). Further experiments showed that the knockdown of hsa_circ_0000069 had no significant effect on cell proliferation, apoptosis, and cell cycle of other cell lines (HCT-116, SW480, LoVo).
Figure 3.
Effects of hsa_circ_0000069 knockdown on CRC cell proliferation, apoptosis, and cell cycle in vitro.
Notes: (A, B) Colony-forming assays were conducted to determine the proliferation of si-circ_0000069-transfected HT-29 cells. (C) CCK8 assays were used to determine the cell viability for si-circ_0000069-transfected HT-29 cells. (D) Flow cytometry assays were performed to analyse the cell apoptosis when HT-29 cells transfected with si-circ_0000069 48 hours later. (E, F) Flow cytometry assays were performed to analysis the cell cycle when HT-29 cells transfected with si-circ_0000069 48 hours later. Values represented the mean ± standard deviation from three independent experiments. *P<0.05, **P<0.01.
Abbreviation: CRC, colorectal cancer; si-NC, cells transfected with negative control siRNAs.
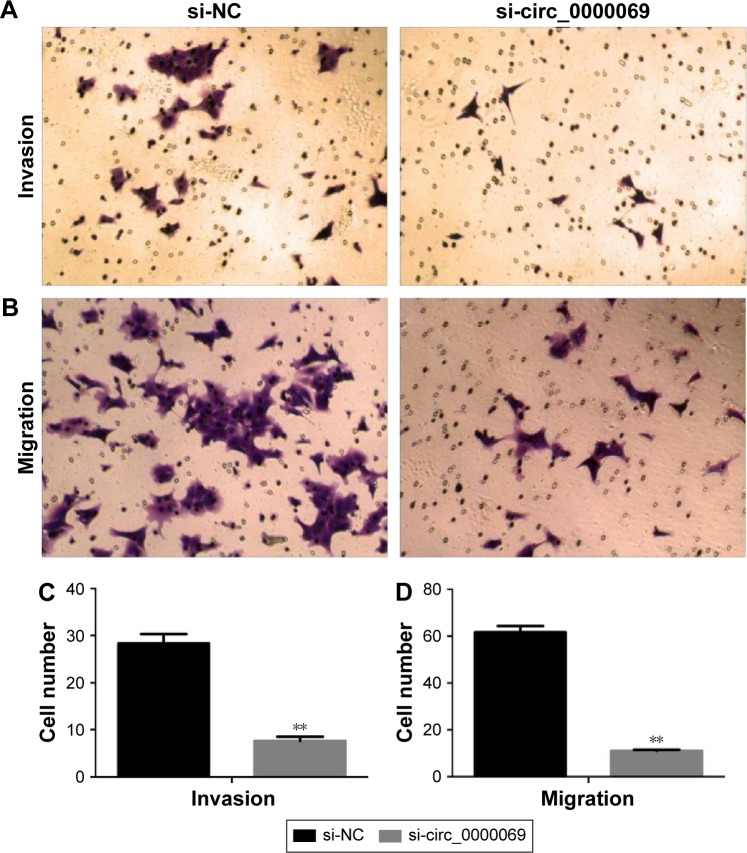
Knockdown of hsa_circ_0000069 inhibited cell migration and invasion in vitro
Invasion and migration assays were performed to detect whether hsa_circ_0000069 had a direct functional role on CRC progression. The results revealed that knockdown of hsa_circ_0000069 expression could prominently reduce cell invasion and migration in CRC cells, compared with negative control groups (Figure 4). In addition, invasion and migration of other cell lines (HCT-116, SW480, LoVo) were not significantly affected after hsa_circ_0000069 knockdown.
Figure 4.
Effects of hsa_circ_0000069 knockdown on CRC cell invasion and migration in vitro.
Notes: Transwell assays were used to determine the invasion (A, C) and migration (B, D) ability of si-circ_0000069-transfected HT-29 cells. The results showed that inhibition of hsa_circ_0000069 could significantly impair HT-29 cell invasion and migration ability when compared with control cells. **P<0.01. Magnification 1:100.
Abbreviations: CRC, colorectal cancer; si-NC, cells transfected with negative control siRNAs.
Discussion
Recently, a subset of highly conserved and stable endogenous non-coding RNAs have emerged as new and crucial layers of gene regulators.9 CircRNAs have the remarkable characteristics of covalently closed loop structures without 5′–3′ polarities, which provide them with the potential to serve as specific markers in various diseases, including cancers.2 Several researchers have found that circRNAs could be novel and stable biomarkers for cancer diagnosis, such as hsa_circRNA_104912 in laryngeal squamous cell cancer (LSCC),10 hsa_circ_0001649 in hepatocellular carcinoma,11 and hsa_circ_001988 in CRC.12 Furthermore, it has been demonstrated that there are novel regulatory mechanisms of circRNA in the initiation and progression of cancers. The majority of previous studies suggest that circRNAs could act as microRNA sponges to exert their functions. For instance, a classic circRNA, Cdr1as (ciRS-7), was proved to be a “super sponge” of miR-7 that competitively inhibits its activity.13 Another circRNA cir-ITCH was also found to have an inhibitory effect on esophageal squamous cancer cells. As a sponge of miR-7, miR-17, and miR-214, cir-ITCH could increase the level of ITCH and thereby inhibit the Wnt/β-catenin pathway.8 Xie et al14 indicated that hsa_ circ_001569 was identified as a sponge of miR-145 and could upregulate miR-145 functional targets E2F5, BAG4, and FMNL2 expression to effect tumor progression in CRC.
In this study, through unsupervised hierarchical clustering analysis, the differentially expressed circRNAs in 6 paired CRC tissues and matched normal tissues were analyzed. When comparing 2 groups of profile differences (such as disease versus control), the “fold change” (ie, the ratio of the group averages) between the groups for each circRNA was computed. The statistical significance of the difference may be conveniently estimated by Student’s t-test. CircRNAs having fold changes ≥2 and P≤0.05 are selected as the significantly differentially expressed. Using qRT-PCR to measure the expression of several selected circRNAs, this study found that hsa_circ_0000069 was a significantly upregulated circRNA with effective regulating functions. Loss of function assays revealed that hsa_circ_0000069 could promote CRC cell proliferation, invasion, and migration. Knockdown of hsa_circ_0000069 could notably induce G0/G1 phase arrest of cell cycle in CRC cells in vitro.
In summary, this study’s results demonstrated that hsa_circ_0000069 is markedly elevated in CRC tissues and promotes cell proliferation, invasion, and migration in CRC, which makes it a promising therapeutic target for further treatment. However, the exact regulatory mechanism of hsa_circ_0000069 in CRC progression needs further investigation.
Acknowledgments
This study was supported by the grants from Project Fund of Health Bureau of Jiangsu Province (No H201358) and Project Fund of Sci & Tech Bureau of Huai’an City (Nos HAS2015001, HAS2012003, and HAP201206)
Footnotes
Author contributions
JG and JL designed this study, detected the cell biological function test, and conducted the qRT-PCR assays. CZ performed the statistical analysis and drafted the manuscript. WF provided the tissue samples and clinical data. JS and LW participated in the data analysis. JH and MH conceived the study, participated in its design and coordination, and helped to draft the manuscript. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Chen LL, Yang L. Regulation of circrna biogenesis. RNA Biol. 2015;12(4):381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang F, Nazarali AJ, Ji S. Circular rnas as potential biomarkers for cancer diagnosis and therapy. Am J Cancer Res. 2016;6(6):1167–1176. [PMC free article] [PubMed] [Google Scholar]
- 4.Li J, Yang J, Zhou P, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5(2):472–480. [PMC free article] [PubMed] [Google Scholar]
- 5.Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One. 2016;11(7):e0158347. doi: 10.1371/journal.pone.0158347. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 8.Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β -catenin pathway. Oncotarget. 2015;6(8):6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 10.Xuan L, Qu L, Zhou H, et al. Circular RNA: A novel biomarker for progressive laryngeal cancer. Am J Transl Res. 2016;8(2):932–939. [PMC free article] [PubMed] [Google Scholar]
- 11.Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16(1):161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Zhang Y, Huang L, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8(12):16020–16025. [PMC free article] [PubMed] [Google Scholar]
- 13.Peng L, Yuan XQ, Li GC. The emerging landscape of circular RNA ciRS-7 in cancer (review) Oncol Rep. 2015;33(6):2669–2674. doi: 10.3892/or.2015.3904. [DOI] [PubMed] [Google Scholar]
- 14.Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]