Abstract
Eimeria tenella is an obligate intracellular parasite that actively invades cecal epithelial cells of chickens. The basis of cell invasion is not completely understood, but some key molecules of host cell invasion have been discovered. This paper investigated the characteristics of calcium-dependent protein kinase 4 (EtCDPK4), a critical molecule in E. tenella invasion of host cells. A full-length EtCDPK4 cDNA was identified from E. tenella using rapid amplification of cDNA ends. EtCDPK4 had an open reading frame of 1803 bp encoding a protein of 600 amino acids. Quantitative real-time PCR and western blotting were used to explore differences in EtCDPK4 transcription and translation in four developmental stages of E. tenella. EtCDPK4 was expressed at higher levels in sporozoites, but translation was higher in second-generation merozoites. In vitro invasion inhibition assays explored whether EtCDPK4 was involved in invasion of DF-1 cells by E. tenella sporozoites. Polyclonal antibodies against recombinant EtCDPK4 (rEtCDPK4) inhibited parasite invasion, decreasing it by approximately 52%. Indirect immunofluorescence assays explored EtCDPK4 distribution during parasite development after E. tenella sporozoite invasion of DF-1 cells in vitro. The results showed that EtCDPK4 might be important in sporozoite invasion and development. To analyze EtCDPK4 functional domains according to the structural characteristics of EtCDPK4 and study the kinase activity of rEtCDPK4, an in vitro phosphorylation system was established. We verified that rEtCDPK4 was a protein kinase that was completely dependent on Ca2+ for enzyme activity. Specific inhibitors of rEtCDPK4 activity were screened by kinase activity in vitro. Some specific inhibitors were applied to assays of DF-1 cell invasion by E. tenella sporozoites to confirm that the inhibitors functioned in vitro. W-7, H-7, H-89, and myristoylated peptide inhibited DF-1 invasion by E. tenella sporozoites. The experimental results showed that EtCDPK4 may be involved in E. tenella invasion of chicken cecal epithelial cells.
Introduction
The protozoan phylum Apicomplexa comprises thousands of obligate intracellular parasites, many of which cause significant human and animal health problems. For example, Toxoplasma gondii infects approximately one-third of the global human population and causes severe disease in immunocompromised patients and pregnant women [1]. Other examples are Plasmodium falciparum, the causative agent of malaria, which causes more than 1 million deaths per year [2] and Eimeria species, the protozoan parasites that cause the severe intestinal disease coccidiosis [3, 4].
Avian coccidiosis is a major disease of poultry caused by parasitic Eimeria species including Eimeria tenella, Eimeria necatrix, Eimeria acervulina, Eimeria maxima, Eimeria brunetti, Eimeria mitis and Eimeria praecox. Coccidiosis causes severe economic losses in the poultry industry every year [5]. Eimeria have complex life cycles and need to invade the intestinal epithelium of chickens to develop and propagate. The invasion of host gut epithelial cells by Eimeria species is a complex, multistep process that begins with the apical attachment of the parasite to the host cell. This is followed by rapid internalization to form an intracellular, parasitophorous vacuole (PV) that encloses the newly invaded parasite, enabling its survival within the host [6]. To perpetuate the infection, Eimeria need to egress from infected cells and then reinvade uninfected cells. In response to these events, parasites have developed regulatory mechanisms for self-proliferation and invasion. During these processes, specialized secretory organelles known as micronemes, rhoptries and dense granules deliver cargo proteins in a coordinated fashion. Secreted proteins are thought to be central to invasion and the establishment of infection [7, 8]. However, secretion by these organelles is controlled by intracellular calcium as a second messenger, which is important in signal transduction cascades, including for protein secretion, gliding motility, invasion of and egress from host cells, proliferation and differentiation [9].
In Apicomplexan parasites, calcium-dependent protein kinases (CDPKs) are main receptors of Ca2+ signals [10, 11]. CDPKs have been identified throughout the plant kingdom and in some protozoans, but not in animals or fungi [12]. CDPKs have two key domains, a Ser/Thr kinase domain and an EF-hand-type calcium-binding domain. They also contain an N-terminal variable domain, an auto-inhibitory junction region and a C-terminus [13]. The N-terminal domain shows the highest sequence divergence among CDPKs and often contains myristoylation or palmitoylation sites that are believed to be associated with subcellular targeting [14]. The C-terminal domain is also variable and differs in lengths and amino acid compositions among CDPKs. The N- and C-terminal variable domains are suggested to determine the specific function of individual CDPKs [15].
Increasing evidence suggests that CDPKs control important physiological events in Apicomplexan parasite life cycles. For example, conditional suppression of T. gondii CDPK1 (TgCDPK1) weakens microneme protein secretion, parasite gliding motility, host cell invasion and egress ability [16, 17]. PbCDPK4 from Plasmodium berghei, an ortholog of TgCDPK1, regulates cell cycle progression in the male gametocyte [18]. Genetic disruption of TgCDPK3 demonstrates that it has a regulatory function in parasite physiology in addition to the ionophore-induced egress process [19, 20, 21]. Apicomplexa parasites contain multiple CDPK genes; Plasmodium species possess seven [22], Billker et al. found that T. gondii contains 12 [23]. To our knowledge, only three E. tenella CDPK members (EtCDPK1, EtCDPK2 and EtCDPK3) have been studied [24, 25], and the physiological functions of most EtCDPKs remain unclear. Studies suggest that CDPKs regulate biological functions in Apicomplexa parasites. However, CDPKs regulatory mechanisms and targets remain unclear in Apicomplexa parasites. Nonetheless, this family of CDPKs has received attention as potential drug targets in Apicomplexan parasites.
Because the CDPKs have essential functions in Apicomplexan parasites and are absent in mammalian and avian hosts, CDPKs are promising targets for research on drugs against Eimeria species and related Apicomplexans parasites [26]. Some selective inhibitors against their kinase activity have been generated [27, 28].
We studied new members of the E. tenella CDPK family. We carried out a comprehensive analysis including the cloning, sequencing, protein expression and characterization of a novel EtCDPK4 gene and protein. We provided novel insights into E. tenella invasion and development from a detailed study of the expression of EtCDPK4. This study aimed to provide information for further research and discovery of other members of the CDPK family of E. tenella.
Materials and Methods
Ethics Statement
This research with chickens was approved by the Shanghai Administration Committee of Laboratory Animals (GB14925-2010) and performed in accordance with the Chinese Academy of Agricultural Sciences Institutional Animal Care and Use Committee guidelines.
E. tenella Propagation and Purification
The Shanghai strain of E. tenella was isolated from a sample collected from a chicken farm in Shanghai, China in 1985 and was maintained in our laboratory (Resource Number: CAAS21111601, Shanghai Veterinary Research Institute innovation team of protozoosis preservation, Chinese Academy of Agricultural Sciences). 60 healthy AA chickens were fed with coccidian-free water and feed. E. tenella was propagated as previously described [29] by passage through 2-week-old coccidian-free chickens. Unsporulated oocysts were obtained from the cecal contents of chickens at 8 days post-infection (p.i.). Some unsporulated oocysts was purified and stored in liquid nitrogen. The rest were incubated in 2.5% potassium dichromate to induce sporulation. After sporulation, oocysts were collected and purified. Sporozoites were prepared from cleaned, sporulated oocysts by excystation in vitro. Second-generation merozoites were collected from ceca at 120 h p.i. from chickens inoculated with 8.0 × 104 sporulated oocysts per bird. Isolation was carried out as previously described [30]. Isolated sporozoites and second-generation merozoites were stored in liquid nitrogen.
The chicken embryo fibroblast cell line DF-1, derived from East Lansing Line (ELL-0) chicken embryos, was used for infection, inhibition assays and immunofluorescence experiments [31].
Molecular cloning of the EtCDPK4 full-length cDNA by RACE
Total RNA was extracted from E. tenella sporozoites using TRIzol reagent (TaKaRa, Tokyo, Japan) according to the manufacturer’s protocol. RNA quality was analyzed by 1% agarose gel electrophoresis and visualization with Ethidium Bromide staining. Total RNA concentration was quantified by UV spectrophotometry (Eppendorf, Hamburg, Germany). Rapid amplification of cDNA ends (RACE) was carried out with GeneRacer kits (Invitrogen, Carlsbad, CA, USA) to obtain the full-length 5' -and 3' -termini. RACE primers were designed based on gene sequences of the CDPK family (ETH_00010685, http://www.genedb.org/Homepage/Etenella). Approximately 10 μg of total RNA was used to synthesize 5' -and 3' -RACE-Ready cDNA. Sequences were amplified by Touchdown PCR with either 5'- RACE or 3'- RACE gene-specific primers (Table 1) and GeneRacer 5'- or 3'- primers. Nested PCRs were performed with nested gene-specific primers and 5'- or 3'- RACENEST primers (Table 1). Amplification products were subjected to electrophoresis using 1% agarose gels. Single bands were extracted, purified, cloned into the pGEM-T Easy vector (Promega, Madison, WI, USA), and propagated in Escherichia coli TOP10 (TIANGEN, Beijing, China) competent cells. Clones were sequenced by Shanghai Sunny Biotechnology Co., Ltd. Sequences of 5'-RACE and 3'-RACE were compared with the original EtCDPK4 open reading frame (ORF) sequence using DNAstar software (Promega) to determine overlap. Full-length cDNA sequences were submitted to GenBank (Accession No. KU925778).
Table 1. Primer sequences used in this study.
| Primer ID | Primer Sequences |
|---|---|
| 5'-RACE Primer | 5'-TCGAATATCTTCCGCAGGGCCGACTCA-3' |
| 5'-RACENEST Primer | 5'-CGACTCATCCACTGGGGGAAGCTGCAA-3' |
| 3'-RACE Primer | 5'-CGAAGCTGTCCTGGGCGGCGAAGACTA-3' |
| 3'-RACENEST Primer | 5'-GGCGATGGGCAGATTGACTGGGACGAA-3' |
| EtCDPK4UP | 5'-CGGGATCCGAGCAGGTGATGGGTGGGCGGGAGGT-3' |
| EtCDPK4LOW | 5'-GCGTCGACAATTCGTCCCAGTCAATCTGCCCAT-3' |
| EtCDPK4-RT(UP) | 5'-GACCATCAGATTCACAAG-3' |
| EtCDPK4-RT(LOW) | 5'-CTCAAAGACATCCACATC-3' |
| 18Sr RNA sense | 5'-TGTAGTGGAGTCTTGGTGATTC-3' |
| 18Sr RNA antisense | 5'-CCTGCTGCCTTCCTTAGATG-3' |
Bioinformatics analysis of EtCDPK4
The sequence of full-length EtCDPK4 cDNA was analyzed. Signal peptide sequences, transmembrane and hydrophobic regions, genetically mobile domains and conservative structure predictions were identified using SignalP (http://www.cbs.dtu.dk/), TMPRED (http://www.ch.embnet.org/), Hydrophobic (http://web.expasy.org/), SMART (http://smart.embl-heidelberg.de/) and CDD (http://www.ncbi.nlm.nih.gov/Structure/) computational tools. Secondary structure, antigen index, flexible region and surface probability of EtCDPK4 were analyzed using DNAstar.
Expression and Purification of Recombinant CDPK4 Proteins
Total RNA was extracted from sporozoites and the first-strand cDNA templates were generated by M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA, USA) using oligo (dT) as primer. A 1660-bp (from 303 bp to 1962 bp) fragment of EtCDPK4 encoding 550 amino acids with a calculated molecular weight of 62.6 kDa was amplified using primers EtCDPK4UP and EtCDPK4LOW according to the ORF sequence. Sequence-specific primers were designed to contain sites for Bam HI in the forward primer (EtCDPK4UP) and Sal I in the reverse primer (EtCDPK4LOW) (Table 1). PCR amplification was performed as follows program: 94°C for 5 min, 38 cycles 94°C for 90 s, 56°C for 45 s and 72°C 90 s, followed by 10 min at 72°C. After sequencing, fragments were digested with Bam HI and Sal I, purified from agarose gels by TIANgel Midi Purification Kits (TIANGEN) and ligated into the corresponding sites of the expression vector pCold I and sequenced. The pCold I-EtCDPK4 plasmid was transformed into E. coli (BL21) to produce a recombinant protein (rEtCDPK4) with a 6×-His tag at the N- terminus. rEtCDPK4 protein was induced by 1.0 mM IPTG (Sigma, St. Louis, MO, USA) at 16°C. Induced bacterial cells were incubated for 24 h and harvested by centrifugation. Cell pellets were lysed by sonication and the insoluble portion of the pellet was suspended in 10 mM imidazole binding buffer and purified by His Bind Resin (Merck, Darmstadt, Germany). Yield of the affinity-purified protein was estimated using a Biophotometer (Eppendorf, Hamburg, Germany). Purified rEtCDPK4 protein was visualized by 12% SDS-PAGE. Then purified protein was stored in aliquots at– 20°C.
Production of Anti-rEtCDPK4 Serum and Identification of rEtCDPK4
Two 2-month-old rabbits were immunized with 0.2 mg rEtCDPK4 emulsified in Freund Complete Adjuvant (Sigma) by intraperitoneal injection. Rabbits were boosted three times with Freund’s incomplete adjuvant at 2-week intervals. Eight days after final immunization, polyclonal antibody serum was separated from two rabbits blood.
rEtCDPK4 was resolved by 12% SDS-PAGE and transferred to PVDF membranes (Millipore, Bellerica, MA, USA). Western blots were performed according to standard procedures by using rabbit anti-merozoite protein sera (1:500) previously obtained in our lab or anti-His-tag monoclonal antibody (1:2000). Native rabbit IgG (1:1000) was the negative control. IRDye 800CW goat anti-rabbit IgG (LI-COR, Lincoln, NE, USA) and IRDye 680RD donkey anti-mouse IgG (1:25,000) (LI-COR, Lincoln, NE, USA) were used as the secondary antibody. Indirect Enzyme Linked Immunosorbent Assay (ELISA) was used to determine rabbit anti-rEtCDPK4 serum titers.
EtCDPK4 Transcription and Translation Analysis in E. tenella Life Stages
Total RNAs isolated from four life stages of E. tenella (unsporulated oocysts, sporulated oocysts, sporozoites and second-generation merozoites) were treated with DNase I (Invitrogen) according to the protocol. Quality and quantity of total RNAs were assessed as described above. The first cDNAs were generated by SuperScript II reverse transcriptase (Invitrogen) using random primers. Quantitative real-time PCR (qRT-PCR) was performed on an Eppendorf Mastercycler ep Realplex (Eppendorf, Hamburg, Germany) using the SYBR1 green I dye method. Negative (no template) controls were included. A fragment encoding the E. tenella 18S ribosomal RNA was used as a control. Reactions were carried out in triplicate and experiments were performed six times. Primers for real-time PCR are in Table 1. Primers for EtCDPK4 (EtCDPK4-RT[UP] and EtCDPK4-RT[LOW]) and 18S rRNA were designed by the Beacon Designer program (Corbett Robotics, USA). Relative mRNA expression level was determined as the ratio of EtCDPK4 to 18S rRNA.
To investigate expression of EtCDPK4 in developmental stages, lysates from E. tenella unsporulated oocysts, sporulated oocysts, sporozoites and second-merozoites were prepared using cell lysis buffer for western Blot and IP (Beyotime, Haimen, China). Protein concentrations were determined using BCA Protein Assay kits (Beyotime) and separated on 12% SDS-PAGE. Western blots were performed according to standard procedures [32]. Anti-rEtCDPK4 antibodies were used at 1:100 and mouse monoclonal anti-α-tubulin antibodies (Sigma) at 1:1000 were controls. Secondary antibodies were used as above. IRDyes were detected using an ODYSSEY Infrared Imaging System (LI-COR).
Assays for rEtCDPK4 protein kinase activity
To calculate rEtCDPK4 activity units in catalytic reactions, we defined one unit rEtCDPK4 activity as a nanomole of phosphate group transferred to a substrate per minute per milliliter. In vitro phosphorylation reactions were performed using Non-radioactive PepTag assays (Promega). Assays (25 μL) were carried out in 20 mM HEPES-KOH, pH 7.4, 1.3 mM CaCl2, 1 mM DTT, 10 mM MgCl2, 1 mM ATP, 5 μg sonicated phosphatidylserine, 1 mM phenylmethylsulfonyl fluoride, 5 ng leupeptin, 5 ng aprotinin, 2 mM mercaptoethanol, 0.05% Triton X-100, 47.5 μM PepTag®C1-Peptide, 2 μL peptide protective solution, with an aliquot of purified rEtCDPK4 (10 μg, initial concentration about 1.0 mg/mL). Phosphorylation reactions were performed for 30 min at 30°C and stopped by heating at 95°C for 15 min. Phosphorylated C1 peptides were separated from non-phosphorylated peptides by electrophoresis on 0.8% agarose gels at 140 V for 30 min. To estimate the amount of phosphorylated peptide, bands were excised from gels under UV light, melted at 95°C and mixed (325 μL) with gel solubilization solution (75 μL, Promega) and glacial acetic acid (100 μL). Optical absorbance values of solutions were measured with a NanoDrop2000/2000C spectrophotometer at 570 nm. rEtCDPK4 enzyme activity was measured using Beer’s Law as described above.
Effect of Ca2+ Concentrations on rEtCDPK4 Activity
Based on rEtCDPK4 kinase activity assays, the initial reaction system was improved. The kinase activity assay had two portions: qualitative and quantitative detection. Assays (30 μL) were carried out in 20 mM HEPES-KOH, pH 7.4, 1 mM DTT, 10 mM MgCl2, 1 mM ATP, 5 μg sonicated phosphatidylserine, 1 mM phenylmethylsulfonyl fluoride, 5 ng leupeptin, 5 ng aprotinin, 2 mM mercaptoethanol, 0.05% Triton X-100 (v/v), 47.5 μM PepTagC1-Peptide, 2 μL peptide protective solution, and an aliquot of purified rEtCDPK4 (10 μg, initial concentration about 1 mg/mL), and five concentrations of CaCl2: 0 μM, 10 μM, 50 μM, 100 μM, or 1000 μM. Negative controls and rEtCDPK4 sample controls were used. Phosphorylation reactions were for times and temperatures as above. Phosphorylated-C1-peptides were separated and amounts of phosphorylated peptide estimated as above. rEtCDPK4 kinase activity was measured by spectrophotometer and experiments were performed twice.
Screening and Analysis of rEtCDPK4 Specific Inhibitors
By bioinformatics analysis of EtCDPK4 functional domains, we chose seven inhibitors: W-7 (Sigma), H-7 (Sigma), H-89 (Beyotime), staurosporine (Beyotime), D-sphingosine (Sigma), Ro-31-8220 (Sigma) and myristoylated peptide (Promega). These inhibitors belonged to three categories: ATP competitive inhibitor, hypothetical-substrate inhibitor and Ca2+-binding-domain inhibitor. Staurosporine, H-7, H-89, W-7 and Ro-31-8220 belong to the category of ATP competitive inhibitor; Myristoylated peptide belongs to the hypothetical-substrate inhibitor; D-sphingosine belongs to the Ca2+-binding-domain inhibitor. Inhibitory concentrations were as recommended by the supplier instructions. The inhibitors’ final concentrations were adjusted to 100 μM in the reaction system. Assays (30 μL) were carried out in 20 mM HEPES-KOH, pH 7.4, 1.3 mM CaCl2, 1 mM DTT, 10 mM MgCl2, 1 mM ATP, 5 μg sonicated phosphatidylserine, 1 mM phenylmethylsulfonyl fluoride, 5 ng leupeptin, 5 ng aprotinin, 2 mM mercaptoethanol, 0.05% Triton X-100, 47.5 μM PepTag C1-Peptide, 2 μL peptide protective solution, and purified rEtCDPK4 as above and specific inhibitor. Negative controls and rEtCDPK4 samples without inhibitors were included. Phosphorylation reactions, separation of phosphorylated-C1-peptides and estimation of amounts were as above. Experiments were performed three times. Data differences among groups were analyzed by one-way analysis of variance (ANOVA) Duncan test.
Inhibition of DF-1 Invasion by Anti-rEtCDPK4 Polyclonal Antibody or rEtCDPK4 Specific Inhibitors
The chicken embryo fibroblast cell line DF-1 was used for inhibition assays [33]. Antibodies were purified with Protein A+G Agarose (Beyotime). E. tenella sporozoites were labeled using carboxyfluorescein diacetate, succinimidyl ester (CFDA SE, Beyotime) according to the manufacturer’s instructions. Labeled sporozoites (1.0 × 108) were resuspended in 1 mL CM (Gibco, Grand Island, NY, USA), purified anti-rEtCDPK4 polyclonal antibody, IgG from native rabbit serum (negative control), or an equivalent volume of PBS (normal control) was added to labeled sporozoites to final concentrations of 50, 100, 200, 300 or 400 μg/mL respectively. Sporozoites were incubated at 37°C for 2 h, washed twice in sterile phosphate buffered saline, then infected 2.0 × 105 DF-1 cells in 24-well plates (Corning, NY, USA). After cultured 16 h at 41°C, cells were collected and analyzed by flow cytometry (Beckman Coulter, USA). Uninfected DF-1 cells were the control. Infected cells, uninfected cells, and free sporozoites were gated using RXP software (Beckman Coulter, USA) to count infected (labeled sporozoites) and uninfected (fluorescence-free) cells. All of invasion-inhibition assays were performed in triplicate. Percentages of cells infected in the presence or absence of anti-rEtCDPK4 polyclonal antibody were used to calculate inhibition rate, as previously described: inhibition = 100% × (1− [% (infected cellsAntibody treatment) /% (infected cellsnegative control)]) [34].
E. tenella sporozoites were labeled as above. Labeled sporozoites (4.0 × 108) were resuspended in 1 mL of DMEM and incubated with specific inhibitors of rEtCDPK4 (100 μM) or dimethyl sulfoxide (DMSO; negative control) (100 μM) for 2 h at 37°C. All assays were performed in triplicate. Uninfected DF-1 cells were controls. Infected cells, uninfected cells, and free sporozoites were gated as above to count infected (labeled sporozoites) and uninfected (fluorescence-free) cells. Percentages of infected cells in the presence or absence of rEtCDPK4 specific inhibitors were used to calculate inhibition rates as described.
Indirect Immunofluorescence Assays of EtCDPK4 Expression During First Schizogony
DF-1 cells were infected with sporozoites at 41°C, 5% CO2 at one sporozoite per cell in DMEM (Gibco) supplemented with 5% FBS, 100 U/mL penicillin/streptomycin, 2 mM L-glutamine. Purified freshly excysted sporozoites were incubated in PBS or complete medium for 2 h at 41°C and washed before transferring to a glass slide and air-dried as previously described [35, 36]. Sporozoites incubated in CM for 2 h at 41°C were used to infect cells. After infection 12, 24, 36, 48, 60 and 72 h, cells were collected and washed before transferred to glass slides and air-dried. Slides were fixed in 2% paraformaldehyde in PBS for 30 min and permeabilized using 1% Triton X-100 in PBS for 30 min. Slides were blocked with PBS containing 2% (w/v) bovine serum albumin at room temperature for 2.5 h. A 1:500 dilution of anti-rEtCDPK4 polyclonal antibody was added and incubated for 2 h at 37°C. Then a 1:500 dilution of a goat anti-rabbit IgG fluorescein isothiocyanate (FITC)-conjugated antibody (Sigma-Aldrich, St. Louis, State of Missouri, USA) was added and incubated for 2.5 h at 37°C. Nuclei were stained with 15 μg/mL 4, 6-diamidino-2-phenylindole (DAPI) (Beyotime) at room temperature for 10 min. After each step, slides were washed six times with PBS containing 0.5% (v/v) Tween-20. Finally, slides were mounted using 100 μL Fluoromount Aqueous Mounting Medium (Sigma-Aldrich). Before observation under a fluorescence microscope (Olympus, Tokyo, Japan), 50 μL 1, 4—diazabicyclo [2. 2. 2] octane (DABCO; Sigma) was added.
Statistical Analysis
Statistical analysis was performed using Microsoft Office Excel for Windows version 2013 (Redmond, Washington, USA) and GraphPad Prism 5.0 (GraphPad, La Jolla, CA, USA). All data, including EtCDPK4 Transcription and Translation Analysis in E. tenella Life Stages, Inhibition of DF-1 Invasion by Anti-rEtCDPK4 Polyclonal Antibody or rEtCDPK4 Specific Inhibitors, Effect of Ca2+ Concentrations on rEtCDPK4 Activity, Screening and Analysis of rEtCDPK4 Specific Inhibitors, were analyzed. Differences among groups were tested by one-way analysis of variance (ANOVA) Duncan test. The data are presented as mean ± standard deviation (SD). P < 0.05 was considered significant and P < 0.01 highly significant.
Results
Characteristics of EtCDPK4 Sequence by Bioinformatics
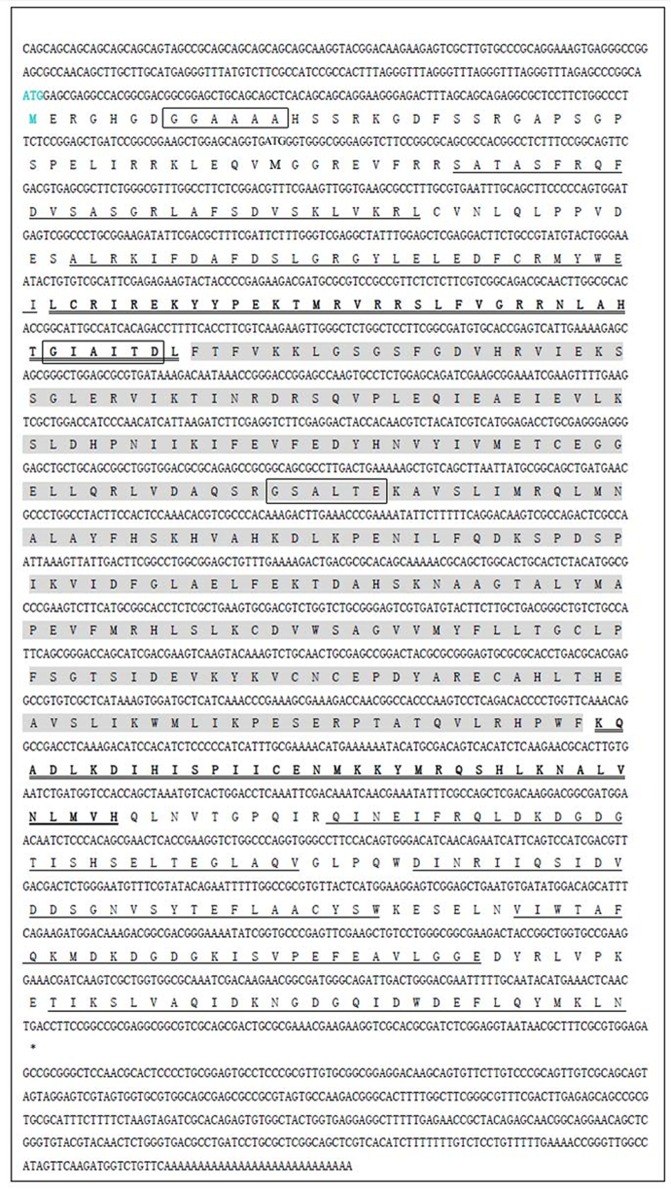
The full-length EtCDPK4 cDNA was 2499 bp with a single ORF of 1803 bp (positions 186 bp– 1988 bp) encoding a polypeptide of 600 amino acid residues with a calculated molecular mass of approximately 68.3 kDa. Fig 1 shows the complete nucleotide sequence of EtCDPK4 and the deduced amino acids. The cDNA contained a 5'- untranslated region (UTR) of 185 bp and a 3'- UTR of 511 bp. The 3'- UTR contained a characteristic poly A tail (AAAAAA) but without a classic polyadenylation signal (AATAAA).
Fig 1. Complete sequence of EtCDPK4 cDNA and deduced amino acids.
Single underlined, six calcium-binding EF-hand motifs; Double underlined, junction domain; Shaded, serine/threonine kinase domain; Rectangular box, N-myristoylation site.
TMPRED analysis of the transmembrane regions of the EtCDPK4 amino acid sequence found a transmembrane region in the Val302—Ile324 position. SignalP analysis revealed that the protein most likely does not contain a signal peptide. Structural module and conservative structure predictions indicated that the protein contained distinct domains characteristic of a calcium-dependent protein kinase, including an amino-terminal kinase domain with a typical serine/threonine-kinase active site, a carboxy-terminal calmodulin-like domain with four EF-hand motifs and an amino-terminal calmodulin-like domain with two EF-hand motifs for calcium-binding (Fig 2). Also predicted were three N-myristoylation sites and two N-glycosylation sites. The gene was designated EtCDPK4 (GenBank Accession No. KU925778). DNAstar analysis of the secondary structure, antigen index, flexible regions and surface probability of EtCDPK4 showed a polypeptide rich in α-helix, β-sheet, β-turn, random coil and flexible regions that might contribute to polypeptide chain folding to tertiary structures. The polypeptide antigen-index score was relatively high at 1.7. Therefore, the EtCDPK4 polypeptide had antigenic sites and was predicted to be immunogenic. Surface probability analysis of the EtCDPK4 polypeptide had high scores of about 6.0, suggesting that the function of the EtCDPK4 was involved in the E. tenella membrane surface.
Fig 2. Genetically mobile domain and conservative structure analysis of the EtCDPK4.
S_TKc, Serine/Threonine protein kinases, catalytic domain; EFh, calcium binding motif.
Expression and Purification of Recombinant CDPK4
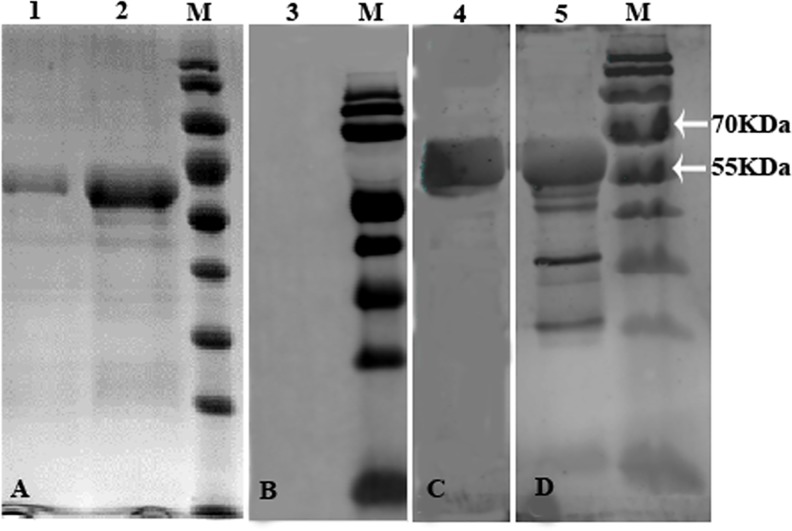
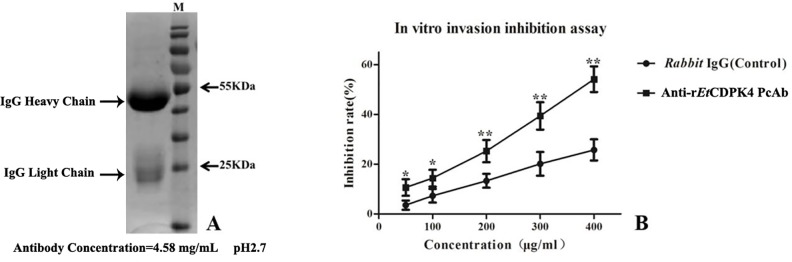
The 1660-bp fragment (location: 118 bp—1777 bp) of the EtCDPK4 ORF was amplified by RT-PCR according to the structural and functional domains, because it is difficult to amplify the full-length of ORF of EtCDPK4. The protein encoded by this fragment contained all distinct domains, including serine/threonine-kinase active site, EF-hand motifs for calcium-binding. Then the fragment was ligated with expression vector pColdⅠto create an expression plasmid pCold-EtCDPK4. After sequencing, pCold-EtCDPK4 was transformed into E. coli BL21 to induce expression of recombinant protein. Electrophoresis results showed that rEtCDPK4 was a soluble protein. Lysates of E. coli included a band on SDS-PAGE gels with a molecular weight (MW) close to the theoretical 62.6 kDa MW of EtCDPK4. rEtCDPK4 was purified using column affinity chromatography and identified by SDS-PAGE (Fig 3A).
Fig 3. Rabbit anti-serum against E. tenella second merozoites or anti-His monoclonal as the primary antibody.
Reactive bands were visualized using IRDye 800CW goat anti-rabbit IgG and IRDye 680RD donkey anti-mouse IgG. M, protein standard molecular weight (15.0 kDa to 170.0 kDa). A, Lane 1, imidazole elution buffer 150 mM; elution of rEtCDPK4 proenzyme. Lane 2, imidazole elution buffer 250 mM; elution of rEtCDPK4 proenzyme; B, Lane 3, native rabbit serum IgG. C, Lane 4, anti-His tag monoclonal antibody. D, Lane 5, rabbit anti-merozoites whole serum.
Production of Anti-rEtCDPK4 and Identification of rEtCDPK4
Serum was generated using rabbits. Western blots showed that rEtCDPK4 was recognized by rabbit anti-merozoites serum or anti-His tag monoclonal antibody (Fig 3B, 3C and 3D). The rabbit anti-rEtCDPK4 serum titer was 1:12, 800 by indirect ELISA.
EtCDPK4 Transcripts and Protein in E. tenella Developmental Stages
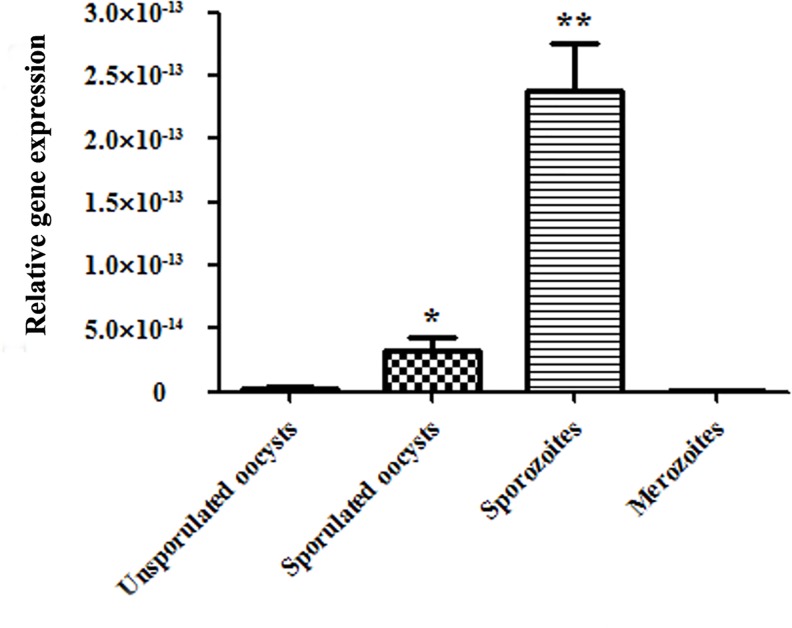
To determine the mRNA level of EtCDPK4 in unsporulated oocysts, sporulated oocysts, sporozoites, and second-generation merozoites of E. tenella, total RNA was subjected to qPCR analysis. Among the four development stages, EtCDPK4 mRNA was highest in sporozoites; transcripts were nearly undetectable in unsporulated oocysts and merozoites (Fig 4).
Fig 4. EtCDPK4 transcription at different E. tenella life stages.
Bars not sharing the same symbol were significantly different (P < 0.05) and the error bars indicate standard deviations.
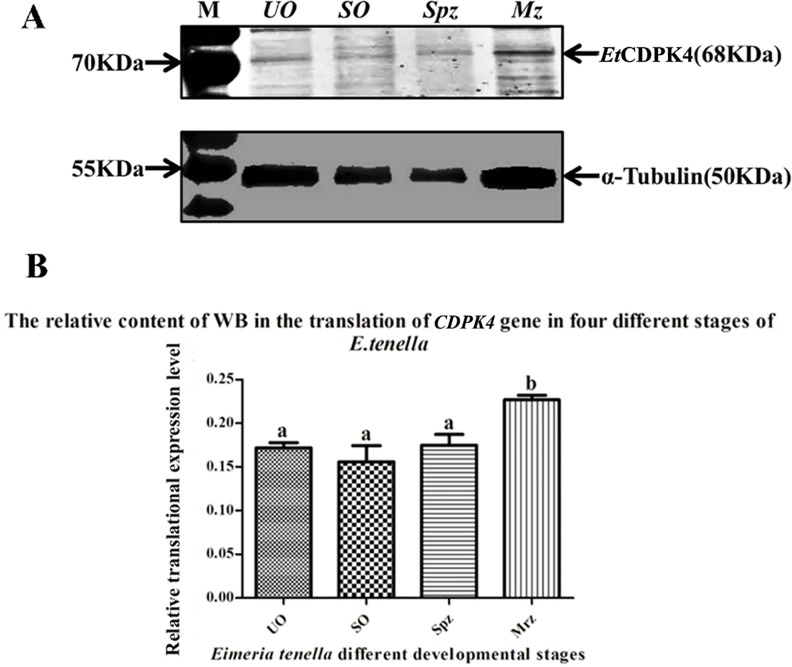
The presence of EtCDPK4 protein in the four developmental stages was determined by immunoblotting using antibodies from rabbits immunized with rEtCDPK4. Anti-α-tubulin monoclonal antibodies were used as internal reference controls. Anti-rEtCDPK4 labeled the same 68.3-kDa band in second-generation merozoites, unsporulated oocysts and sporozoites with weak reactivity in sporulated oocysts (Fig 5A). Thus, the EtCDPK4 had a high expression level in stage of the second-generation merozoites, while the EtCDPK4 in other three developmental stages of the sporozoites, unsporulated oocysts and sporulated oocysts were approximately at the same level (Fig 5B).
Fig 5. Analysis of western blots of EtCDPK4 at different of E. tenella stages probed with anti-rEtCDPK4.
A, qualitative analysis of EtCDPK4 at different stages of E. tenella by western blot; α-tubulin, internal reference protein. B, relative quantitative analysis of EtCDPK4 expression difference at different stages of E. tenella. Lanes: UO, unsporulated oocysts; SO, sporulated oocysts; Spz, sporozoites; Mz or Mrz, second-generation merozoites. Bars with different letters were significantly different (P < 0.05) and the error bars indicate standard deviations. WB, western blot. M, protein weight standard (from top to bottom: 170 kDa, 130 kDa, 100 kDa, 70 kDa, 55 kDa, 40 kDa, 35 kDa, 25 kDa, 15 kDa).
Protein Kinase Activity of rEtCDPK4
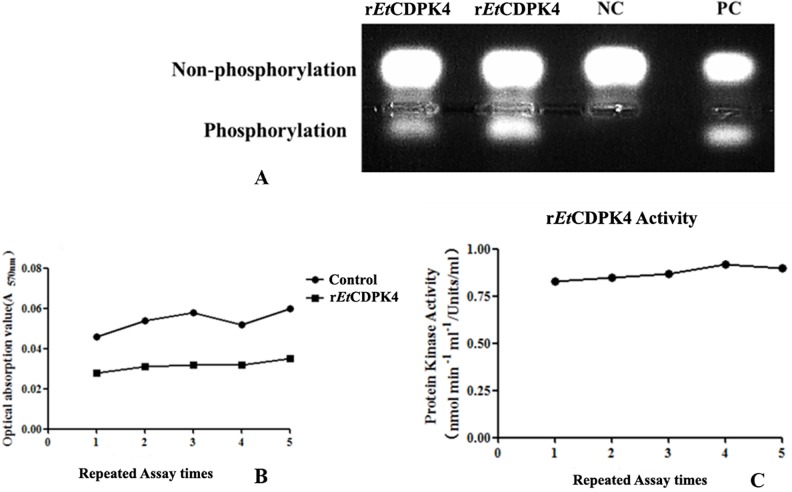
rEtCDPK4 was purified by column affinity chromatography (Fig 3A) and protein kinase activity was measured based on nonradioactive detection of in vitro phosphorylation of a PKC-specific fluorescent synthetic peptide substrate. Phosphopeptides were detected under UV light and quantified spectrophotometrically (Fig 6A, 6B and 6C). rEtCDPK4 had protein kinase activity with a reaction rate of about 0.87 nmol•min-1 •mL-1 protein or 0.87 units/mL.
Fig 6. rEtCDPK4 protein kinase activity assays.
A, Qualitative detection of rEtCDPK4 kinase activity. NC, negative control (no rEtCDPK4); PC, positive control (PKC). B, Quantitative detection of five times the optical absorption value (A570nm value). C, Quantitative detection of rEtCDPK4 kinase activity. Phosphorylation, the gel band that the PepTag®C1-Peptide was phosphorylated by rEtCDPK4. Non-phosphorylation, the gel band that the remainder PepTag®C1-Peptide was not phosphorylated by rEtCDPK4.
Effect of Ca2+Concentration on rEtCDPK4 Activity
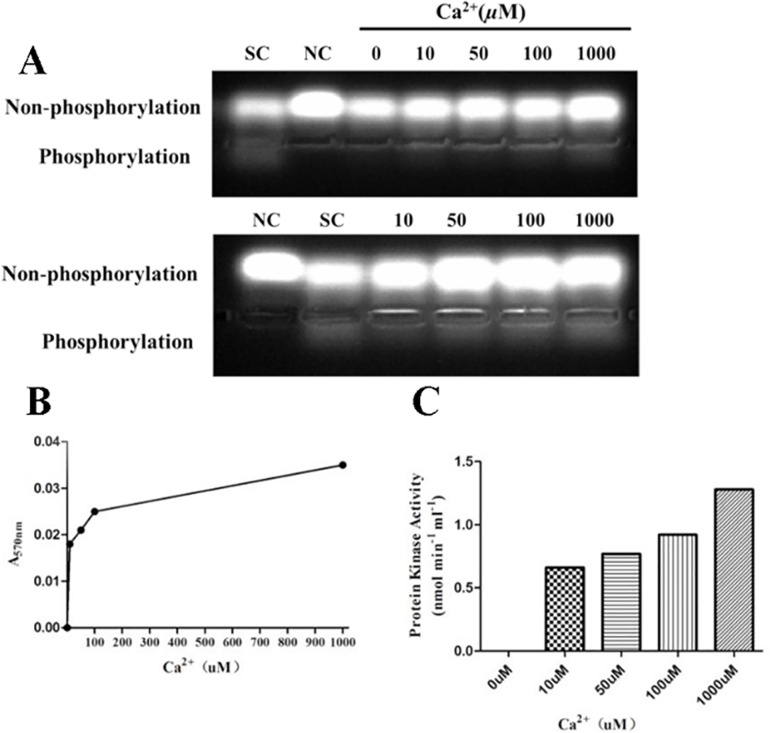
Enzyme activity was strongly dependent on Ca2+ concentrations with a calculated Ka of 10–1000 μM based on incubating rEtCDPK4 with various Ca2+ concentrations for in vitro phosphorylation assays (Fig 7A, 7B and 7C). With no Ca2+, rEtCDPK4 was inactivated and no substrate was phosphorylated. With increasing Ca2+ concentrations, the activity of rEtCDPK4 was enhanced in spite of the band relatively weak.
Fig 7. Kinase activity dependence on Ca2+ concentration by qualitative assays.
SC, rEtCDPK4 sample control; NC, negative control. Five Ca2+ concentrations were: 0, 10, 50, 100, and 1000 μM. A, Qualitative detection assay; B, A570nm value of phosphorylated short peptides at indicated calcium concentrations; C, rEtCDPK4 enzyme activity under indicated Ca2+ concentrations. Phosphorylation, the gel band that the PepTag®C1-Peptide was phosphorylated by rEtCDPK4. Non-phosphorylation, the gel band that the remainder PepTag®C1-Peptide was not phosphorylated by rEtCDPK4.
Analysis of rEtCDPK4 Specific Inhibitors
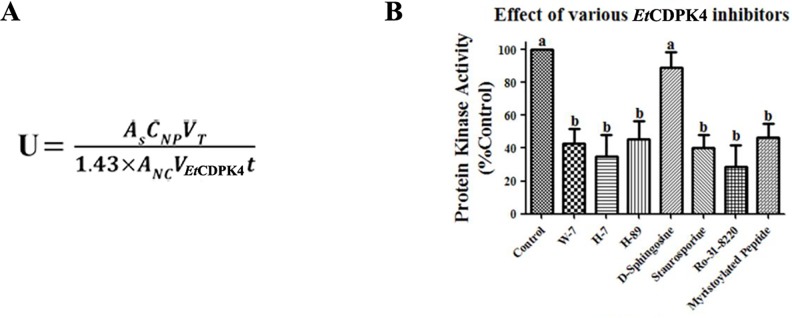
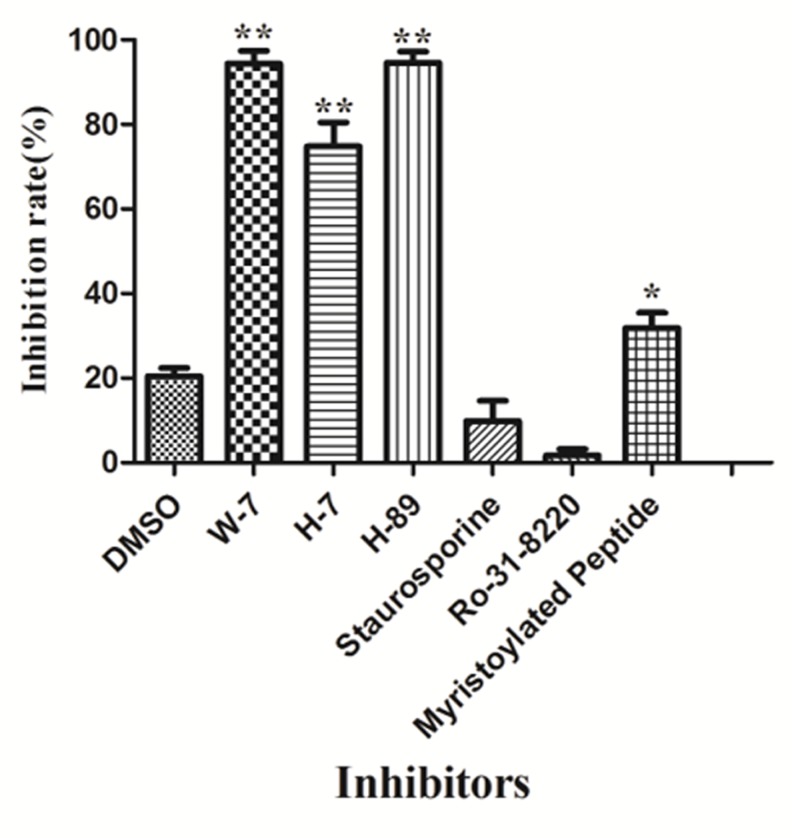
Seven inhibitors were applied to rEtCDPK4 activity assays with quantitative detection (Fig 8B). The activity of rEtCDPK4 in the reaction system with different inhibitors was detected by measuring the absorbance value of the cutting gel band at the 570 nm. The results showed that the absorbance values of phosphorylation reaction products treated by W-7, H-7, H-89, staurosporine, Ro-31-8220 and myristoylated peptide were lower than the sample control, but the absorbance value of phosphorylation reaction products treated by D-sphingosine was very close to the sample control. The calculation formula of kinase activity was showed in Fig 8A. Calculations for rEtCDPK4 activity showed that W-7, H-7, H-89, staurosporine, Ro-31-8220 and myristoylated peptide significantly decreased enzyme activity, while D-sphingosine had no effect.
Fig 8. Quantitative analysis for seven inhibitors in rEtCDPK4 activity assays.
A, formula for calculating rEtCDPK4 activity, rEtCDPK4 enzyme activity units were nmol×min-1×mL-1. AS, optical absorption for sample at 570 nm wavelength; CNP, standard concentration of PepTag®-C1; VT, total volume of samples in cuvette (500 μL); 1.43, correction coefficient for measuring absorbance (250/175); ANC, 570 nm optical absorption value of non-phosphorylated peptides in the negative control; VEtCDPK4, total volume of rEtCDPK4 in the reaction system (10 μL); t, time of phosphorylation reaction (30 min). B, Bars with different letters were significantly different (P < 0.05) and the error bars indicate standard deviations.
EtCDPK4 Localization During In Vitro Infection by Immunofluorescence
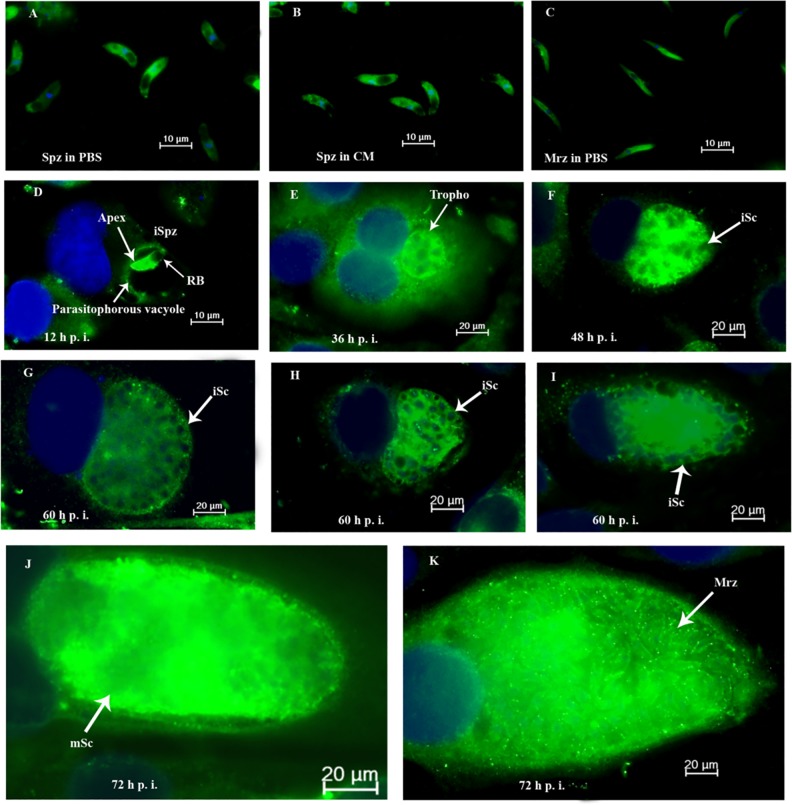
Anti-rEtCDPK4 was used to localize EtCDPK4 in sporozoites and during first schizogony. EtCDPK4 exhibited a homogenous distribution pattern throughout the cytoplasm of sporozoites except for the refractive body and second-generation merozoites incubated in PBS for 2 h (Fig 9A and 9C). In contrast, when sporozoites were incubated in culture medium, EtCDPK4 expression did not change significantly (Fig 9B). After sporozoites invaded host cells, EtCDPK4 was mainly localized to the cytoplasm of parasites, except for the refractive body. Green fluorescence intensity was enhanced at this phase. Foci of intense EtCDPK4 staining were closely associated with the parasitophorous vacuole membrane (Fig 9D, 9H and 9J). Observations at 36 h p.i. showed that EtCDPK4 protein increased and was distributed in trophozoites (Fig 9E). Labeled EtCDPK4 was eventually uniformly dispersed in immature and mature schizonts and decreased in immature schizonts (Fig 9F, 9G, 9H, 9I and 9J). After formation of first-generation merozoites from mature schizonts in DF-1 cells, labeling increased (Fig 9K).
Fig 9. EtCDPK4 localization in DF-1 cell infection by immunofluorescence.
Parasites were immune-stained with anti-rEtCDPK4 and visualized with FITC (green) and counterstained with DAPI (blue). A, Sporozoites (Spz) were incubated in PBS for 2 h or B, complete medium (CM) at 41°C. C, second-generation merozoites (Mrz) incubated in PBS for 2 h. Infected DF-1 cells were collected at indicated time points p.i. D, 12 h p.i. intracellular sporozoites (iSpz); E, 36 h p.i. intracellular trophozoite (Tropho); F, 48 h p.i. immature schizont (iSc); G, H, I, 60 h p.i. immature schizont (iSc); J, 72 h p.i. mature schizont (mSc); K, 72 h p.i., first-generation merozoites (Mrz) released from mature schizont.
Anti-rEtCDPK4 and rEtCDPK4 Specific Inhibitors Inhibited DF-1 Cell Invasion
To evaluate EtCDPK4 effects on invasion of DF-1 cells by E. tenella sporozoites, invasion inhibition assays were performed. Protein function was blocked by pre-incubation of sporozoites with purified antibody against rEtCDPK4 (Fig 10A) before DF-1 cell infection. AbEtCDPK4 inhibited invasion to 52% at an antibody concentration of 400 μg/mL, compared to infection with non-treated sporozoites. Inhibition was dose dependent. By comparison, native rabbit-sera IgG did not have a significant effect on invasion by E. tenella sporozoites (Fig 10B).
Fig 10. Inhibition of sporozoite invasion in vitro.
A, Purification of rabbit anti-rEtCDPK4 serum by SDS-PAGE. B, Inhibition by anti-rEtCDPK4 serum and rabbit IgG. All assays were performed in triplicate. Anti-rEtCDPK4 PcAb, rabbit antiserum generated against rEtCDPK4 protein; rabbit IgG, IgG from native rabbit serum. The symbol “**” representability P < 0.01 and the “*” representability P < 0.05 for comparison of treatment with antibody against rEtCDPK4 and native rabbit serum at the same IgG concentration and the error bars indicate standard deviations. M, protein weight standard (from top to bottom: 170 kDa, 130 kDa, 100 kDa, 70 kDa, 55 kDa, 40 kDa, 35 kDa, 25 kDa, 15 kDa).
Flow cytometry used to determine the effect of specific inhibitors on labeled sporozoites is in Fig 11. W-7, H-7, H-89 and myristoylated peptide significantly decreased invasion, while staurosporine and Ro-31-8220 had no effect.
Fig 11. Inhibition of DF-1 invasion by E. tenella sporozoites using specific inhibitors of rEtCDPK4.
The symbol “**” representability P < 0.01, very significant difference; “*” representability P < 0.05, significant difference. And the error bars indicate standard deviations.
Discussion
In this work, the calcium-dependent protein kinase 4 of E. tenella was cloned and identified. The full-length cDNA of EtCDPK4 was 2499 bp, with a 1803 bp ORF encoding a protein of 600 amino acids. The 5'-UTR was 185 bp. The 3'-UTR of 511 nucleotides ended with a poly (A) tail. Sequence analysis indicated that the protein contained domains characteristic of CDPKs: an N-terminal protein kinase domain, a C-terminal calmodulin-like domain with four calcium-binding EF-hand motifs, an N-terminal calmodulin-like domain of two calcium-binding EF-hand motifs, a junction domain and a very short N-terminal variable region. Searching the E. tenella genome database, the deduced amino acid sequence had 100% identity with a gene (ETH_00010685) encoding a putative CDPK. These results suggested that EtCDPK4 was a member of the E. tenella CDPK family.
CDPKs, encoded by multigene families, are widespread in plants and some Apicomplexan parasites. However, only four genes encoding for putative CDPKs of E. tenella are in the E. tenella genome database [24], including EtCDPK4. According to the number of calcium-binding EF-hand motifs, Apicomplexan parasite CDPKs are classified into four major categories [23]. The first category contains proteins with canonical CDPK structures containing four C-terminal EF-hand motifs. The second category contains proteins with three C-terminal EF hands. The other two groups of CDPKs have one or more N-terminal EF hands followed by a Ser/Thr kinase domain and three or four C-terminal EF-hand motifs. Motif scan results classified EtCDPK4 in the other group of canonical Apicomplexan parasite CDPKs. The EF-hand motif structure of Cryptosporidium parvum CDPK6 was also similar to EtCDPK4 [37]. The arrangement of the N-terminal EF domains is unusual and their role in the regulation of kinase activity has not been examined. Bioinformatic analysis on EtCDPK4 showed that it has three N-myristoylation sites. Several canonical CDPKs in parasites contain consensus motifs for N-myristoylation or palmitoylation, a feature also seen in plant CDPKs, many of which show membrane localization [38, 39]. This result suggests that association with membranes may be important in localization, as has been shown for PfCDPK1, which is modified by both palmitate and myristate [40]. The role of such modifications in regulation of activity has not been explored in parasites.
The mRNA and protein levels of EtCDPK4 were examined in four developmental stages of E. tenella. Results from qPCR showed that mRNA for EtCDPK4 was highest in the sporozoites and the lowest in the unsporulated oocysts and second-generation merozoites. These results showed that the EtCDPK4 gene was transcribed predominantly at a distinct phase of the E. tenella life cycle. However, western blots showed that protein levels were highest for EtCDPK4 in second-generation merozoites and weakest in sporulated oocysts. This difference may be the result of underlying molecular mechanisms and signaling pathways in the four stages of E. tenella and should be investigated in more detail in future studies [41].
Previous studies of Apicomplexans parasites found that each CDPK gene is expressed predominantly at a distinct phase of the parasite life cycle. For example, another CDPK isoform of E. tenella, EtCDPK1, is expressed in sporulated oocysts, sporozoites and merozoites, but not in unsporulated oocysts, as determined by western blots [25]. EtCDPK3 has the highest expression in sporozoites by qPCR or western blots compared with other stages [24]. In P. falciparum, PfCDPK1 is mainly expressed in the asexual blood stages of the parasite, particularly in late-stage schizonts [40]. In contrast, PfCDPK3 is expressed specifically in the sexual erythrocytic stage [42]. PfCDPK4 is detected only in P. falciparum gametocytes [32]. In the Apicomplexan parasite T. gondii, TgCDPK1 and TgCDPK3 are produced in the tachyzoite stage, but TgCDPK2 protein is not detectable in this stage [43, 44].
In our study, mRNA for EtCDPK4 was highest in sporozoites, but EtCDPK4 protein was higher in second-generation merozoites than in the other three stages. A high number of transcripts does not necessarily indicate corresponding amounts of translated protein, which is related to gene functions in the various the parasite stages [37]. Nonetheless, we propose a biological significance for CDPK4 in the E. tenella life cycle. Sporulated oocysts in the environment require material for metabolism from the storage of oocysts [45]. Without an energy supply, sporulated oocysts maintain a low metabolic rate until they can infect fresh host cells [46]. However, during this period, sporozoites are present in sporulated oocysts, and the energy required by sporozoites is provided by oocysts, so metabolism is moderate [47, 48]. To adapt to this mechanism, physiological activity of sporozoites is regulated at the gene level [49]. Protein translation consumes more energy in sporozoites, therefore mRNA for EtCDPK4 was significantly higher in the sporozoites stage. When the merozoite stage invades the host cell and obtains nutrients, merozoites are in schizogony. In this period, the merozoites have higher metabolism and translation is activated [50], so merozoites can invade cells for schizogony and gametogony functions. At this time, EtCDPK4 has physiological and biochemical functions, so parasites generate a large amount of EtCDPK4 protein. EtCDPK4 protein expression was increased sufficiently to be detected by western blot. Therefore, we propose that although EtCDPK4 transcripts were moderately expressed in merozoites, EtCDPK4 protein could have a high expression in the same stage.
Protein phosphorylation on Ser-/Thr- residues is a key post-translational modification required for signal transduction in eukaryotes [51]. CDPKs have a Ser/Thr kinase domain that phosphorylates downstream substrates. CDPK activity can be detected by either radioactive or non-radioactive methods [52, 53]. In this experiment, the rEtCDPK4 activity was detected by non-radioactive methods. rEtCDPK4 catalyzed the phosphorylation of the C1-peptide PKC substrate in vitro and rEtCDPK4 kinase activity could be maintained in a stable range. The kinase activity of rEtCDPK4 depended on Ca2+ concentration. In the absence of calcium, rEtCDPK4 did not have activity. This result was in agreement with results of CDPK kinase activity assays in Digitaria sanguinalis mesophyll cells [54]. Therefore, we propose that EtCDPK4 activity is completely dependent on calcium in the cytoplasm of E. tenella.
From the pharmacological perspective, EtCDPK4 has unique structural features that could be the target of specific inhibitors in parasites because these targets do not exist in the host. Crystal structures of TgCDPK1 and CpCDPK1 show an enlarged ATP-binding pocket due to glycine at the “gatekeeper” position adjacent to an adenine recognition site [55, 56]. Therefore, the activity of CDPK4 could be blocked by inhibitors. In this study, using a functional module of EtCDPK4, we selected seven specific inhibitors: W-7, H-7, H-89, staurosporine, Ro-31-8220, myristoylated peptide and D-sphingosine. The effect of Ca2+-concentrations on rEtCDPK4 kinase activity and specific inhibitors was determined. The organic compounds W-7, H-7, H-89, staurosporine, Ro-31-8220 and myristoylated peptide significantly decreased enzyme activity, while D-sphingosine had no effect on rEtCDPK4 kinase activity. Previous studies reported that activity of D. sanguinalis CDPKs and the rice CDPK OsCDPK14 can be effectively inhibited by W-7 [54, 57] and H-7, staurosporine, Ro-31-8220, and myristoylated peptide inhibit CDPK kinase activity in D. sanguinalis mesophyll cells [54]. H-89 blocks CDPK kinase activity of French beans in enzyme activity assays in vitro [58]. D-sphingosine, a well-known physiological inhibitor of PKC is a broad-spectrum PKC inhibitor. However, the EtCDPK4 structure was much more complex than PKC and therefore, the inhibitory effect of D-sphingosine on rEtCDPK4 was likely blocked by calcium ions in the reaction system. This might be why D-sphingosine did not inhibit rEtCDPK4 activity. This result was similar to findings from previous studies [59] that D-sphingosine does not affect the activity of protein kinase C in porcine theca cells. D-sphingosine did not inhibit enzyme activity but stimulates phospholipase D activity in 7721 human hepatocarcinoma cells [60].
Inhibition of DF-1cell invasion by E. tenella sporozoites using specific inhibitors of rEtCDPK4 showed that W-7, H-7, H-89 and myristoylated peptide significantly decreased sporozoite invasion activity, while staurosporine and Ro-31-8220 had no effect. This result might be because the compounds inhibited the activity of rEtCDPK4 in vitro but were removed by a regulation mechanism [61] in E. tenella sporozoites and could not block the kinase activity of EtCDPK4 in vivo. This would explain the lack of effect on invasion.
We determined experimental conditions for in vitro screening of four specific inhibitors rEtCDPK4 activity. W-7, H-7, and H-89 are broad-spectrum kinase inhibitors and that affected DF-1 invasion by E. tenella sporozoites. Myristoylated peptide derived from the pseudosubstrate sequence of β-PKC is a selective inhibitor of PKC subtypes in human fibroblasts [62]. In our study, inhibition by myristoylated peptide was modest but significant for DF-1 invasion by E. tenella sporozoites. These results showed that the inhibitors effectively blocked the activity of EtCDPK4 kinase and inhibited invasion by E. tenella sporozoite of DF-1 cells.
Expression of EtCDPK4 was detected by indirect immunofluorescence assays. Expression of EtCDPK4 was high in mature schizonts and forming first-generation merozoites. Therefore, suitable organic compounds that block the EtCDPK4 activity may inhibit development of E. tenella in host cecum epithelial cells. We conclude that EtCDPK4 is likely to be an excellent molecular target candidate for anti-coccidiosis drug or vaccine research.
Using antibodies raised against rEtCDPK4, we showed by indirect immunofluorescence assays that EtCDPK4 kinase was located on the surface and in the cytoplasm of E. tenella sporozoites and merozoites. CDPKs were found in several subcellular localizations including membranes and cytoplasm and some isoforms were in more than one compartment [40]. Indirect immunofluorescence assays also showed that EtCDPK1 [25] and EtCDPK3 [24] were near the apical end and on the surface of E. tenella sporozoites. PfCDPK1 is present in the membrane and organelle fractions of blood-stage parasites and the membrane fraction of ring-stage infected erythrocytes [63, 64]. EtCDPK4 has a transmembrane region and three N-myristoylation sites (positions: 8 to 13, 152 to 157, and 253 to 258). Myristoylation sites are vital for membrane targeting and signal transduction in plants and P. falciparum response to environmental stress or the host cell environment [40, 65]. This might be one reason that EtCDPK4 was found on sporozoite and merozoite membranes. EtCDPK4 localization in intracellular parasites showed mobilization to the membranes of refractile bodies in the anterior of sporozoites. When E. tenella developed in DF-1 cells, specific staining was more intense than in trophozoites, mature schizonts or first-generation merozoites, but decreased in immature schizonts by fluorescence intensity. Later during sporozoite development in DF-1 cells, EtCDPK4 was found in PV membranes. The PV is a crucial structure that protects the parasite against the host environment [7]. Invasion and the formation of PVs is mediated by different parasite organelles, including micronemes and rhoptries [66]. PV composition evolves during infection according to metabolite exchange between the parasite and host cell [67]. When merozoites escape from mature schizonts to invade new host cells, they must pass through the PV membrane. Thus, we postulated that EtCDPK4 was important in merozoite maturity and release. Western blots showed that EtCDPK4 protein increased significantly in merozoite stages. This result was similar to localization of EtCDPK1 at the apical end of sporozoites after addition to Mardin-Darby bovine kidney cells. EtCDPK1 protein was higher in mature schizonts than in immature schizonts. EtCDPK1 appeared to be specifically involved in sporozoite invasion of host cells and in release of merozoites from mature E. tenella schizonts [25].
In P. falciparum, PfCDPK1 has a key role in schizont development, microneme protein secretion, invasion of host erythrocytes and regulating mRNA expression to assure timely and stage-specific protein expression [68, 69, 70]. TgCDPK7 is crucial for T. gondii differentiation, growth and proper maintenance of centrosomes [71]. Knock out of PbCDPK3 leads to a pronounced defect in ookinete transmission to the mosquito midgut epithelium and terminates oocysts production [72, 73]. PfCDPK5 is essential for regulating parasite egress from erythrocytes [74], whereas PbCDPK6 is critical for controlling the sporozoite switch from a migratory to an invasion phenotype [75]. Finally, Sharma et al. demonstrated that PfCDPK7 binds to PI (4, 5) P2 and controls P. falciparum development in host erythrocytes [76]. These results suggest that EtCDPK4 is related to the invasion and survival of E. tenella intracellular stages.
Previous in vitro invasion inhibition assays showed reduced sporozoite invasion in the presence of monoclonal or specific polyclonal antibodies [34, 36, 77]. In our study, in vitro invasion inhibition assays using specific antibody against rEtCDPK4 showed partial blockage of the invasion of sporozoites into cells. Inhibition of sporozoites was modest at 52%. Therefore, EtCDPK4 might be a key factor in host cell invasion by E. tenella sporozoites. While further studies are needed to determine the exact function of EtCDPK4, because this kinase family is absent from the parasite’s hosts, it represents a target that might be exploited for chemotherapy against Apicomplexans parasites.
In conclusion, we cloned, expressed and characterized a CDPK4 from E. tenella, adding substantially to the current understanding of its function in the E. tenella invasion process. Given the importance of EtCDPK4 in invasion, host cell adhesion and E. tenella development in cecum epithelial cells, the results of this study have implications for both novel chemotherapeutic and immunotherapeutic approaches to interfering with EtCDPK4 function in E. tenella.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 31572266, No. 31272557) and Central Public Welfare Research Institutions and Basic Scientific Research Business Expenses (No. 2016JB10). We would like to thank Prof. Chan Ding (CAAS, Beijing, China) for generously providing the DF-1 cell line used in this study.
Data Availability
All relevant data are within the manuscript and supporting information files.
Funding Statement
This work was supported by the National Natural Science Foundation of China (Grant No. 31572266, No. 31272557) and Central Public Welfare Research Institutions and Basic Scientific Research Business Expenses (No. 2016JB10).
References
- 1.Elmore SA, Jones JL, Conrad PA, Patton S, Lindsay DS, Dubey JP. Toxoplasma gondii: epidemiology, feline clinical aspects, and prevention. Trends Parasitol. 2010; 26: 190–196. 10.1016/j.pt.2010.01.009 [DOI] [PubMed] [Google Scholar]
- 2.Sachs J, Malaney P. The economic and social burden of malaria. Nature. 2002; 415: 680–685. 10.1038/415680a [DOI] [PubMed] [Google Scholar]
- 3.Alcala-Canto Y, Ramos-Martinez E, Tapia-Perez G, Gutierrez L, Sumano H. Pharmacodynamic evaluation of a reference and a generic toltrazuril preparation in broilers experimentally infected with Eimeria tenella or E. acervulina. Br Poult Sci. 2014; 55: 44–53. 10.1080/00071668.2013.872770 [DOI] [PubMed] [Google Scholar]
- 4.Matsubayashi M, Hatta T, Miyoshi T, Anisuzzaman, Alim MA, Yamaji K, et al. Synchronous development of Eimeria tenella in chicken caeca and utility of laser microdissection for purification of single stage schizont RNA. Parasitology. 2012; 139: 1553–1561. 10.1017/S0031182012001072 [DOI] [PubMed] [Google Scholar]
- 5.Shirley MW, Ivens A, Gruber A, Madeira AM, Wan KL, Dear PH, et al. The Eimeria genome projects: a sequence of events. Trends Parasitol. 2004; 20: 199–201. 10.1016/j.pt.2004.02.005 [DOI] [PubMed] [Google Scholar]
- 6.Tabarés E, Ferguson D, Clark J, Soon PE, Wan KL, Tomley F. Eimeria tenella sporozoites and merozoites differentially express glycosylphosphatidylinositol-anchored variant surface proteins*1. Mol Biochem Parasitol. 2004; 135:123–132. [DOI] [PubMed] [Google Scholar]
- 7.Daszak P. Zoite migration during Eimeria tenella infection: parasite adaptation to host defences. Parasitol Today. 1999; 15: 67–72. [DOI] [PubMed] [Google Scholar]
- 8.Sasai K, Fetterer RH, Lillehoj H, Matusra S, Constantinoiu CC, Matsubayashi M, et al. Characterization of monoclonal antibodies that recognize the Eimeria tenella microneme protein MIC2. J Parasitol. 2008; 94: 1432–1434. 10.1645/GE-1558.1 [DOI] [PubMed] [Google Scholar]
- 9.Moreno SN, Docampo R. Calcium regulation in protozoan parasites. Curr Opin Microbiol. 2003; 6: 359–364. [DOI] [PubMed] [Google Scholar]
- 10.Sanders D, Pelloux J, Brownlee C, Harper JF. Calcium at the crossroads of signaling. Plant Cell. 2002; 14: S401–S417. 10.1105/tpc.002899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kudla J, Oliver B, Kenji H. Calcium signals: the lead currency of plant information processing. Plant Cell. 2010; 22: 541–563. 10.1105/tpc.109.072686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hettenhausen C, Sun G, He Y, Zhuang H, Sun T, Qi J, et al. Genome-wide identification of calcium-dependent protein kinases in soybean and analyses of their transcriptional responses to insect herbivory and drought stress. Sci Rep. 2016; 6: 18973 10.1038/srep18973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harper JF, Sussman MR, Schaller GE, Putnam-Evans C, Charbonneau H, Harmon AC. A calcium-dependent protein kinase with regulatory domain similar to calmodium. Science. 1991; 252: 951–954. [DOI] [PubMed] [Google Scholar]
- 14.Cheng SH, Willmann MR, Chen HC, Sheen J. Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 2002; 129: 469–485. 10.1104/pp.005645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harper JF, Huang JF, Lloyd SJ. Genetic identification of an autoinhibitor in CDPK, a protein kinase with a calmodulin-like domain. Biochemistry. 1994; 33:7267–7277. [DOI] [PubMed] [Google Scholar]
- 16.Lourido S, Shuman J, Zhang C, Shokat KM, Hui R, Sibley LD. Calcium-dependent protein kinase 1 is an essential regulator of exocytosis in Toxoplasma. Nature. 2010; 465: 359–362. 10.1038/nature09022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lourido S, Tang K, Sibley LD. Distinct signalling pathways control Toxoplasma egress and host-cell invasion. EMBO J. 2012; 31: 4524–4534. 10.1038/emboj.2012.299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Billker O, Dechamps S, Tewari R, Wenig G, Franke-Fayard B. Calcium and a calcium-dependent protein kinase regulate gamete formation and mosquito transmission in a malaria parasite. Cell. 2004; 117: 503–514. [DOI] [PubMed] [Google Scholar]
- 19.Mccoy JM, Whitehead L, van Dooren GG, Tonkin CJ. Correction: TgCDPK3 Regulates Calcium-Dependent Egress of Toxoplasma gondii from Host Cells. PLoS Pathog. 2013; 8: 155–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erin G, Moritz T, Emma E, Butz H, Garbuz T, Oswald BP, et al. A Forward Genetic Screen Reveals that Calcium-dependent Protein Kinase 3 Regulates Egress in Toxoplasma. PLoS Pathog. 2012; 8: e1003049–e1003049 10.1371/journal.ppat.1003049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moritz T, Sanders JL, Gaji RY, Lafavers KA, Child MA. The calcium-dependent protein kinase 3 of Toxoplasma influences basal calcium levels and functions beyond egress as revealed by quantitative phosphoproteome analysis. PLoS Pathog. 2014; 10: e1004197–e1004197. 10.1371/journal.ppat.1004197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagamune K, Sibley LD. Comparative genomic and phylogenetic analyses of calcium ATPases and calcium-regulated proteins in the Apicomplexa. Mol Biol Evol. 2006; 23: 1613–1627. 10.1093/molbev/msl026 [DOI] [PubMed] [Google Scholar]
- 23.Billker O, Lourido S, Sibley LD. Calcium-dependent signaling and kinases in Apicomplexan parasites. Cell Host Microbe. 2009; 5: 612–622. 10.1016/j.chom.2009.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han HY, Zhu SH, Jiang LL, Li Y, Dong H, Zhao QP, et al. Molecular characterization and analysis of a novel calcium-dependent protein kinase from Eimeria tenella. Parasitology. 2013; 140: 746–755. 10.1017/S0031182012002107 [DOI] [PubMed] [Google Scholar]
- 25.Dunn PPJ, Bumstead JM, Tomley FM. Sequence, expression and localization of calmodulin-domain protein kinases in Eimeria tenella and Eimeria maxima. Parasitology. 1996; 113: 439–448. [DOI] [PubMed] [Google Scholar]
- 26.Harper JF, Harmon A. Plants, symbiosis and parasites: a calcium signalling connection. Nat Rev Mol Cell Biol. 2005; 6: 555–566. 10.1038/nrm1679 [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z, Ojo KK, Vidadala R, Huang W, Geiger JA, Scheele S, et al. Potent and selective inhibitors of CDPK1 from T. gondii and C. parvum based on a 5-aminopyrazole-4-carboxamide scaffold. ACS Med Chem Lett. 2014; 5:40–44. 10.1021/ml400315s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hui R, Bakkouri ME, Sibley LD. Designing selective inhibitors for calcium-dependent protein kinases in apicomplexans. Trends Pharmacol Sci. 2015; 36: 452–460. 10.1016/j.tips.2015.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomley F. Techniques for isolation and characterization of apical organelles from Eimeria tenella sporozoites. Methods. 1997; 13: 171–176. 10.1006/meth.1997.0509 [DOI] [PubMed] [Google Scholar]
- 30.Xie MQ, Gilbert JM, Fuller AL, McDougald LR. A new method for purification of Eimeria tenella merozoites. Parasitol Res. 1990; 76: 566–569. [DOI] [PubMed] [Google Scholar]
- 31.Jiang L, Lin J, Han H, Zhao Q, Dong H, Zhu S, et al. Identification and partial characterization of a serine protease inhibitor (serpin) of Eimeria tenella. Parasitol Res. 2012; 110: 865–874. 10.1007/s00436-011-2568-0 [DOI] [PubMed] [Google Scholar]
- 32.Ranjan R, Ahmed A, Gourinath S, Sharma P. Dissection of mechanisms involved in the regulation of Plasmodium falciparum calcium-dependent protein kinase 4. J Biol Chem. 2009; 284: 15267–15276. 10.1074/jbc.M900656200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang LL, Lin JJ, Han HY, Dong H, Zhao QP, Zhu SH, et al. Establishment and application of DF-1 cell culture system for the sporozoites of Eimeria tenella. Vet Sci China. 2011; 41: 551–556. [Google Scholar]
- 34.Jahn D, Matros A, Bakulina AY, Tiedemann J, Schubert U, Giersberg M, et al. Model structure of the immunodominant surface antigen of Eimeria tenella identified as a target for sporozoite-neutralizing monoclonal antibody. Parasitol Res. 2009; 105: 655–668. 10.1007/s00436-009-1437-6 [DOI] [PubMed] [Google Scholar]
- 35.Labbé M, Péroval M, Bourdieu C, Girard-Misguich F, Péry P. Eimeria tenella enolase and pyruvate kinase: A likely role in glycolysis and in others functions. Int J Parasitol. 2006; 36: 1443–1452. 10.1016/j.ijpara.2006.08.011 [DOI] [PubMed] [Google Scholar]
- 36.Péroval M, Péry P, Labbé M. The heat shock protein 90 of Eimeria tenella is essential for invasion of host cell and schizont growth. Int J Parasitoly. 2006; 36: 1205–1215. [DOI] [PubMed] [Google Scholar]
- 37.Etzold M, Lendner M, Daugschies A, Dyachenko V. CDPKs of Cryptosporidium parvum—stage-specific expression in vitro. Parasitol Res. 2014; 113: 2525–2533. 10.1007/s00436-014-3902-0 [DOI] [PubMed] [Google Scholar]
- 38.Dammann C, Ichida A, Hong B, Romanowsky SM, Hrabak EM. Subcellular targeting of nine calcium-dependent protein kinase isoforms from Arabidopsis. Plant Physiol. 2003; 132: 1840–1848. 10.1104/pp.103.020008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hrabak EM, Dickmann LJ, Satterlee JS, Sussman MR. Characterization of eight new members of the calmodulin-like domain protein kinase gene family from Arabidopsis thaliana. Plant Mol Biol. 1996; 31: 405–412. [DOI] [PubMed] [Google Scholar]
- 40.Möskes C, Burghaus PA, Wernli B, Sauder U, Dürrenberger M, Kappes B. Export of Plasmodium falciparum calcium-dependent protein kinase 1 to the parasitophorous vacuole is dependent on three N-terminal membrane anchor motifs. Mol Microbiol. 2004; 54: 676–691. 10.1111/j.1365-2958.2004.04313.x [DOI] [PubMed] [Google Scholar]
- 41.De Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009; 5: 1512–1526. 10.1039/b908315d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li JL, Baker DA, Cox LS. Sexual stage-specific expression of a third calcium-dependent protein kinase from Plasmodium falciparum. Biochim Biophys Acta. 2000; 1491: 341–349. [DOI] [PubMed] [Google Scholar]
- 43.Kieschnick H, Wakefield T, Narducci CA, Beckers C. Toxoplasma gondii attachment to host cells is regulated by a calmodulin-like domain protein kinase. J Biol Chem. 2001; 276: 12369–12377. 10.1074/jbc.M011045200 [DOI] [PubMed] [Google Scholar]
- 44.Donald RG, Zhong T, Wiersma H, Nare B, Yao D, Lee A, et al. Anticoccidial kinase inhibitors: identification of protein kinase targets secondary to cGMP-dependent protein kinase. Mol Biochem Parasitol. 149: 86–98. 10.1016/j.molbiopara.2006.05.003 [DOI] [PubMed] [Google Scholar]
- 45.Wilson PAG, Fairbairnz D. Biochemistry of Sporulation in Oocysts of Eimeria acervulina*. J Eukaryot Microbiol. 2007; 8: 410–416. [Google Scholar]
- 46.Belli SI, Walker RA, Flowers SA. Global protein expression analysis in apicomplexan parasites: current status. Proteomics. 2005; 5: 918–924. 10.1002/pmic.200401161 [DOI] [PubMed] [Google Scholar]
- 47.Michalski WP, Edgar JA, Prowse SJ. Mannitol metabolism in Eimeria tenella. Int J Parasitol. 1992; 22: 1157–1163. [DOI] [PubMed] [Google Scholar]
- 48.Mielke D, Alabdul RG. The sporogony of Eimeria tenella. Angew Parasitol. 1991; 32: 39–41. [PubMed] [Google Scholar]
- 49.Katrib M, Ikin RJ, Brossier F, Robinson M, Slapetova I, Sharman PA, et al. Stage-specific expression of protease genes in the apicomplexan parasite, Eimeria tenella. BMC genomics. 2012; 13: 685 10.1186/1471-2164-13-685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fetterer RH, Miska KB, Lillehoj H, Barfield RC. Serine protease activity in developmental stages of Eimeria tenella. J Parasitol. 2007; 93: 333–340. 10.1645/GE-824R1.1 [DOI] [PubMed] [Google Scholar]
- 51.Nemoto K, Takemori N, Seki M, Shinozaki K, Sawasaki T. Members of the Plant CRK-superfamily are Capable of trans-/auto-Phosphorylation of Tyrosine Residues. J Biol Chem. 2015; 290: 16665–16677. 10.1074/jbc.M114.617274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keyloun KR, Reid MC, Choi R, Song Y, Fox AM, Hillesland HK, et al. The gatekeeper residue and beyond: homologous calcium-dependent protein kinases as drug development targets for veterinarian Apicomplexa parasites. Parasitology. 2014; 141: 1499–1509. 10.1017/S0031182014000857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leclercq J, Ranty B, Sanchez-Ballesta MT, Li Z, Jones B, Jauneau A, et al. Molecular and biochemical characterization of LeCRK1, a ripening-associated tomato CDPK-related kinase. J Exp Bot. 2005; 56: 25–35. 10.1093/jxb/eri003 [DOI] [PubMed] [Google Scholar]
- 54.Osuna L, Coursol S, Pierre JN, Vidal J. A Ca2+-dependent protein kinase with characteristics of protein kinase C in leaves and mesophyll cell protoplasts from Digitaria sanguinalis: possible involvement in the C4-phosphoenolpyruvate carboxylase phosphorylation cascade. Biochem Bioph Res Co. 2004; 314: 428–433. [DOI] [PubMed] [Google Scholar]
- 55.Ojo KK, Larson ET, Keyloun KR, Castaneda LJ, Derocher AE, Inampudi KK, et al. Toxoplasma gondii calcium-dependent protein kinase 1 is a target for selective kinase inhibitors. Nat Struct Mol Biol. 2010; 17: 602–607. 10.1038/nsmb.1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wernimont AK, Artz JD, Finerty P Jr, Lin YH, Amani M, Allali-Hassani A, et al. Structures of Apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium. Nat Struct Mol Biol. 2010; 17: 596–601. 10.1038/nsmb.1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang T, Wang Q, Chen X, Tian C, Wang X, Xing T, et al. Cloning and biochemical properties of CDPK gene OsCDPK14 from rice. J Plant Physiol. 2005; 162: 1149–1159. 10.1016/j.jplph.2004.12.010 [DOI] [PubMed] [Google Scholar]
- 58.Allwood EG, Davies DR, Gerrish C, Bolwell GP. Regulation of CDPKs, including identification of PAL kinase, in biotically stressed cells of French bean. Plant Mol Biol. 2002; 49: 533–544. [DOI] [PubMed] [Google Scholar]
- 59.Kaminski T. The response of phospholipase C/protein kinase C and adenylyl cyclase/protein kinase A pathways in porcine theca interna cells to opioid agonist FK 33–824. Domest Anim Endocrinol. 2004; 27: 379–396. 10.1016/j.domaniend.2004.05.001 [DOI] [PubMed] [Google Scholar]
- 60.Huang Y, Zhang X, Chen H. Regulation of phospholipase D activity in human hepatocacinoma cells by protein kinases and D-sphingosine. Acta Bioch Bioph Sin. 1998; 31: 572–576. [PubMed] [Google Scholar]
- 61.Salowe SP, Judyann W, Liberator PA, Donald RGK. The role of a parasite-specific allosteric site in the distinctive activation behavior of Eimeria tenella cGMP-dependent protein kinase. Biochemistry. 2002; 41: 4385–4391. [DOI] [PubMed] [Google Scholar]
- 62.Eichholtz T, de Bont DB, de Widt J, Liskamp RM, Ploegh HL. A myristoylated pseudosubstrate peptide, a novel protein kinase C inhibitor. J Biol Chem. 1993; 268: 1982–1986. [PubMed] [Google Scholar]
- 63.Yi Z, Franklin RM, Kappes B. Plasmodium falciparum calcium-dependent protein kinase phosphorylates proteins of the host erythrocytic membrane. Mol Biochem Parasitol. 1994; 66: 329–343. [DOI] [PubMed] [Google Scholar]
- 64.Zhao Y, Pokutta S, Maurer P, Lindt M, Franklin RM, Kappes B. Calcium-binding properties of a calcium-dependent protein kinase from Plasmodium falciparum and the significance of individual calcium-binding sites for kinase activation. Biochemistry. 1994; 33: 3714–3721. [DOI] [PubMed] [Google Scholar]
- 65.Podell S, Gribskov M. Predicting N-terminal myristoylation sites in plant proteins. BMC Genomics. 2004; 5: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carruthers VB, Sibley LD. Sequential protein secretion from three distinct organelles of Toxoplasma gondii accompanies invasion of human fibroblasts. Eur J Cell Biol. 1997; 73:114–23. [PubMed] [Google Scholar]
- 67.Entzeroth R, Mattig FR, Wernermeier R. Structure and function of the parasitophorous vacuole in Eimeria species. Int J Parasitol. 1998; 28: 1015–1018. [DOI] [PubMed] [Google Scholar]
- 68.Abhisheka B, Shailja S, More KR, Dhiraj H, Kuldeep N. Characterization of Plasmodium falciparum calcium-dependent protein kinase 1 (PfCDPK1) and its role in microneme secretion during erythrocyte invasion. J Biol Chem. 2013; 288: 1590–1602. 10.1074/jbc.M112.411934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sebastian S, Brochet M, Collins M, Schwach F, Jones ML, Goulding D, et al. A Plasmodium Calcium-Dependent Protein Kinase Controls Zygote Development and Transmission by Translationally Activating Repressed mRNAs. Cell Host Microbe. 2012; 12: 9–19. 10.1016/j.chom.2012.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Azevedo MF, Sanders PR, Efrosinia K, Nie CQ, Fu P, Bach LA, et al. Inhibition of Plasmodium falciparum CDPK1 by conditional expression of its J-domain demonstrates a key role in schizont development. Biochem J. 2013; 452: 433–441. 10.1042/BJ20130124 [DOI] [PubMed] [Google Scholar]
- 71.Juliette M, Laurence B, Chen C, Gubbels MJ, Lebrun M, Daher W. The Toxoplasma gondii calcium-dependent protein kinase 7 is involved in early steps of parasite division and is crucial for parasite survival. Cell Microbiol. 2014; 16: 95–114. 10.1111/cmi.12186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ishino T, Orito Y, Chinzei Y, Yuda M. A calcium-dependent protein kinase regulates Plasmodium ookinete access to the midgut epithelial cell. Mol Microbiol. 2006; 59: 1175–1184. 10.1111/j.1365-2958.2005.05014.x [DOI] [PubMed] [Google Scholar]
- 73.Siden-Kiamos I, Ecker A, Nybäck S, Louis C, Sinden RE, Billker O. Plasmodium berghei calcium-dependent protein kinase 3 is required for ookinete gliding motility and mosquito midgut invasion. Mol Microbiol. 2006; 60: 1355–1363 10.1111/j.1365-2958.2006.05189.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dvorin JD, Martyn DC, Patel SD, Grimley JS, Collins CR, Hopp CS, et al. A plant-like kinase in Plasmodium falciparum regulates parasite egress from erythrocytes. Science. 2010; 328: 910–912. 10.1126/science.1188191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Coppi A, Tewari R, Bishop JR, Bennett BL, Lawrence R, Esko JD, et al. Heparan sulfate proteoglycans provide a signal to Plasmodium sporozoites to stop migrating and productively invade host cells. Cell Host Microbe. 2007; 2: 316–327. 10.1016/j.chom.2007.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kumar P, Tripathi A, Ranjan R, Halbert J, Gilberger T, Doerig C, et al. Regulation of Plasmodium falciparum development by calcium-dependent protein kinase 7 (PfCDPK7). J Biol Chem. 2014; 289: 20386–20395. 10.1074/jbc.M114.561670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jiang L, Lin J, Han H, Dong H, Zhao Q, Zhu S, et al. Identification and Characterization of Eimeria tenella Apical Membrane Antigen-1 (AMA1). PLoS One. 2012; 7:362–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the manuscript and supporting information files.