Abstract
The intestinal microbiome modulates host susceptibility to enteric pathogens, but the specific protective factors and mechanisms of individual bacterial species are not fully characterized. We show that secreted antigen A (SagA) from Enterococcus faecium is sufficient to protect Caenorhabditis elegans against Salmonella pathogenesis by promoting pathogen tolerance. The NlpC/p60 peptidoglycan hydrolase activity of SagA is required and generated muramyl-peptide fragments that are sufficient to protect C. elegans against Salmonella pathogenesis in a tol-1-dependent manner. SagA can also be expressed and secreted in other bacteria and improve the protective activity of probiotics against Salmonella pathogenesis in C. elegans and mice. Our study highlights how protective intestinal bacteria can modify microbial-associated molecular patterns to enhance pathogen tolerance.
Dysbiosis of the gut microbiota is associated with metabolic disorders, inflammatory bowel disease and increased pathogen susceptibility (1). Nonetheless, individual bacterial species and factors involved in host protection have been difficult to characterize (2). Enterococci are lactic acid bacteria associated with the intestinal microbiome of diverse species ranging from humans to flies and can attenuate host susceptibility to enteric pathogens, including Salmonella (3, 4). Non-pathogenic strains of E. faecium have been used as probiotics, but their protection mechanisms are unclear (5). Since E. faecium can colonize the C. elegans intestine without causing apparent disease (6), we employed C. elegans as a model organism (7) to elucidate the protective mechanism(s) underlying E. faecium probiotic activity. To investigate whether E. faecium can attenuate enteric bacterial pathogenesis in C. elegans, we developed a treatment-infection assay with Salmonella enterica serovar Typhimurium, (fig. S1A), which causes persistent intestinal infection and death in C. elegans (8–10). In our assay, E. faecium-treated animals appeared less fragile and more motile than control Escherichia coli OP50-treated animals after S. Typhimurium infection (fig. S1B). C. elegans survival was increased in animals fed E. faecium prior to infection as compared to animals fed E. coli OP50 or Bacillus subtilis 168 (Fig. 1A, fig. S1C). Multiple strains of E. faecium, including a pathogenic strain, were able to inhibit S. Typhimurium pathogenesis (fig. S1D–E). E. faecium-treated animals were also more resistant to the intrinsic pathogenesis of E. coli OP50 (fig. S1F) as well as pathogenesis caused by Enterococcus faecalis V583 (11) (fig. S1G). These results suggest that the mechanism of protection is conserved amongst E. faecium strains and is active against diverse enteric pathogens.
Figure 1. E. faecium induces host tolerance to S. Typhimurium.
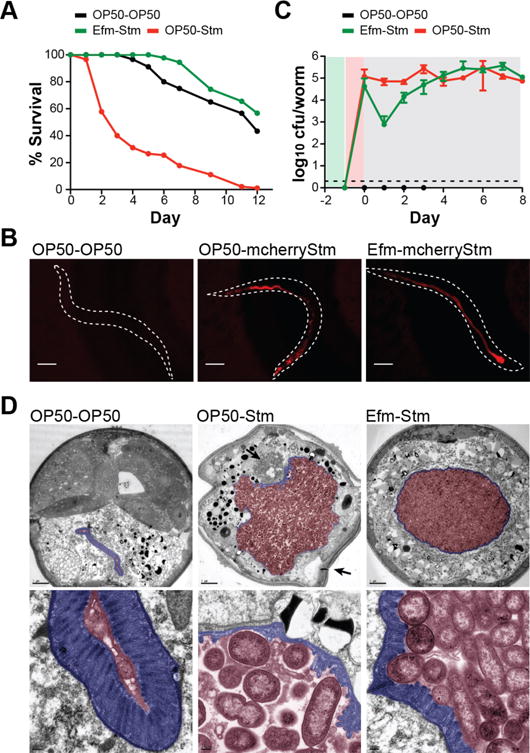
(A) Survival curve showing E. faecium (Efm, Com15)-mediated inhibition of S. Typhimurium (Stm, 14028) pathogenesis (p<10−10). The legend indicates treatment-infection. Control worms were fed E. coli OP50 for both the treatment and infection stages of the assay. For C. elegans survival curves in all figures, significance was calculated by log-rank test with Bonferroni correction for multiple comparisons. Data points represent mean survival from 90 worms from a representative experiment independently replicated at least twice. (B) Fluorescence images of C. elegans infected with Stm-expressing plasmid-encoded mcherry (mcherryStm) at 3 dpi. The dotted lines indicate an outline of the worm body. Scale bar = 100 μm. (C) Stm CFUs measured in C. elegans throughout the infection assay. Data points represent average CFUs from 5 worms ± standard deviation of two independent experiments. The dotted line indicates detection limit. The background shading represents stage of the treatment-infection assay. Green indicates treatment, red indicates infection, and grey indicates E. coli (OP50) feeding. (D) Electron microscopy of transverse sections of C. elegans (top), and magnification of intestinal region (bottom) at 4 dpi. The intestinal microvilli are highlighted blue; the intestinal lumen is highlighted red. In the top middle panel, the top arrow indicates bacteria that have breached the epithelial barrier, and the bottom arrow indicates loss of overall turgidity. Scale bar (top row) = 5 μm. Scale bar (bottom row) = 200 nm.
We next analyzed the effect of E. faecium on S. Typhimurium colonization and persistence. Fluorescence imaging of mCherry-S. Typhimurium 3 days post-infection (dpi) showed comparable S. Typhimurium colonization with or without E. faecium treatment (Fig. 1B, fig. S1H). Viable S. Typhimurium (CFUs) recovered from lysed worms revealed a ~2 log decrease in S. Typhimurium colonization 1 dpi in E. faecium-treated S. Typhimurium-infected animals (Fig. 1C). However, by 3 dpi, S. Typhimurium titer was similar in OP50- and E. faecium-treated S. Typhimurium-infected animals (Fig. 1C). To determine if this transient decrease in S. Typhimurium colonization represented niche competition early in our assay, we monitored E. faecium CFUs throughout the infection assay (fig. S1I). While E. faecium initially colonized worms to ~105 CFUs/worm, E. faecium numbers decreased to ~10–102 CFUs/worm 1 dpi, demonstrating that the transient decrease in S. Typhimurium colonization was not concomitant with an increase in E. faecium load. Electron microscopy of worm transverse sections 4 dpi revealed substantial degradation of the intestinal microvilli in OP50-treated S. Typhimurium-infected animals as compared to uninfected or E. faecium-treated animals (Fig. 1D). In OP50-treated S. Typhimurium-infected animals, bacteria had escaped the intestinal lumen and caused extensive tissue damage (Fig. 1D, middle panel). In contrast, E. faecium-treated S. Typhimurium-infected animals contained a similar bacterial load to the intestinal lumen and showed no apparent tissue damage (Fig. 1D, right panel), suggesting improved epithelial barrier integrity. These results demonstrate that E. faecium does not prevent S. Typhimurium colonization or replication, but may enhance host tolerance to pathogens.
We next explored whether specific factors produced by E. faecium were sufficient for protection against S. Typhimurium pathogenesis. E. faecium culture supernatant was as effective as live bacterial cultures in inhibiting S. Typhimurium pathogenesis (Fig. 2A). Activity of the supernatant was sensitive to proteinase-K treatment, trichloro-acetic acid precipitation, and 10-kDa size exclusion (fig. S2A–C), leading us to analyze the protein composition of E. faecium culture supernatant by mass spectrometry (fig. S2D–E, table S1). This revealed a number of secreted proteins and an enrichment of peptidoglycan remodeling factors (Fig. 2B). We focused on secreted antigen A (SagA), the most abundant protein identified in the supernatant (Fig. 2B), which encodes a putative secreted NlpC/p60 peptidoglycan hydrolase that is essential for E. faecium viability (12). Imaging of animals treated with E. faecium-expressing mCherry under the sagA promoter (psagA:mcherry) showed that E. faecium expresses SagA in vivo (Fig. 2C). Treatment of animals with recombinant SagA-His6 purified from either E. coli BL21-RIL(DE3) or E. faecium Com15 was sufficient to inhibit S. Typhimurium pathogenesis (Fig. 2, D–E, fig. S3, table S2). All sequenced E. faecium strains encode a sagA ortholog in their genomes whereas sequenced E. faecalis strains do not. We inserted sagA-his6 into the E. faecalis OG1RF chromosome to generate E. faecalis-sagA (fig. S4, fig. S5). Treatment of C. elegans with E. faecalis-sagA attenuated S. Typhimurium pathogenesis comparably to E. faecium, while treatment with wild-type E. faecalis was not protective (fig. S6A, Fig. 2F). S. Typhimurium load was similar across all infected conditions, demonstrating that E. faecalis-sagA does not inhibit S. Typhimurium colonization in vivo but rather affects pathogen tolerance (fig. S6B). SagA expression also counteracted the intrinsic pathogenesis of E. faecalis OG1RF (6) (fig. S6C). These results demonstrate that SagA is sufficient to enhance host tolerance against distinct bacterial pathogens.
Figure 2. SagA is sufficient for inducing pathogen tolerance in a tol-1-dependent manner.
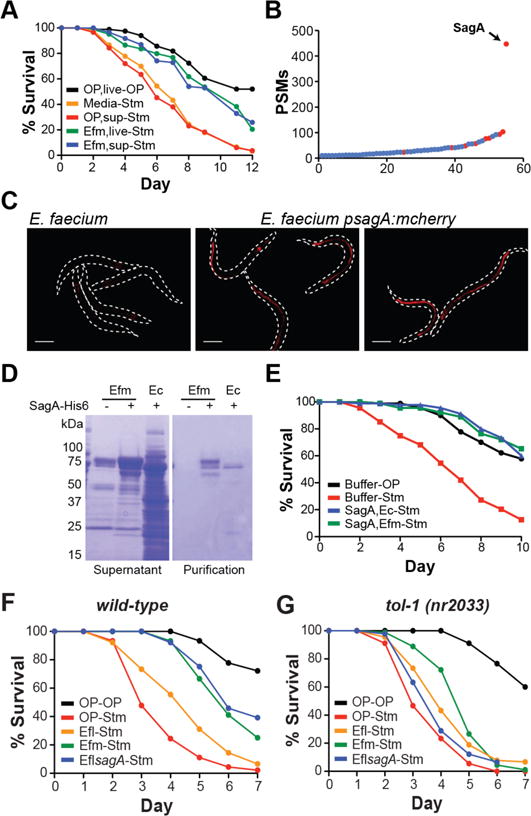
(A) Survival curve showing that both E. faecium culture supernatant (Efm, sup) (p<10−6) and live E. faecium culture (Efm, live) (p<10−7) inhibit S. Typhimurium (Stm)-induced death. OP50 culture supernatant (OP, sup) is not protective (p=1). (B) Summary of proteins identified in Efm culture supernatant by mass spectrometry with at least 10 peptide spectrum matches (PSMs). Proteins involved in peptidoglycan remodeling are in red (See Supplementary Table 1). The x-axis represents arbitrary protein number. (C) Fluorescence images of C. elegans treated for 1 day with wild-type Efm or Efm-expressing mcherry under the sagA promoter (psagA:mcherry). The dotted lines indicate an outline of the worm body. Scale bar = 200 μm. (D) Coomassie stained SDS-PAGE of culture supernatants and SagA-His6 purifications from E. faecium Com15 (Efm) and E. coli BL21-RIL(DE3) (Ec). (E) Survival curve showing that SagA-His6 purified from either E. coli BL21-RIL(DE3) (SagA, Ec) (p<10−10) or E. faecium Com15 (SagA, Efm) (p<10−10) inhibits Stm pathogenesis. (F) Survival curve from a continuous infection assay (see fig. S6A) showing that E. faecalis (Efl, OG1RF)-sagA inhibits Stm pathogenesis (p<10−10) similarly to Efm (Com15) (p=1) compared to E. faecalis (Efl, OG1RF) and OP50. (G) Survival curve from a continuous infection assay showing that Efl-sagA (p=0.053) does not inhibit Stm pathogenesis in tol-1(nr2033) C. elegans.
The protective activity of E. faecium against multiple enteric pathogens suggested that SagA may engage host pathways to limit pathogenesis. A survey of C. elegans immunity-associated mutants indicated no major role for the p38 MAPK/Pmk-1 pathway (13, 14), the TGF-β-like/Dbl-1 pathway (15), the insulin-like receptor/Daf-2 pathway (16), or the Npr-1 mediated pathogen avoidance pathway (17, 18) (fig. S7). C. elegans encodes one homologue of Toll-like receptor, tol-1 (19). C. elegans lacking the tol-1 TIR signaling domain [tol-1(nr2033)] exhibit defective pathogen avoidance to S. marsescens (20) and increased susceptibility to S. Typhimurium infection (21). We assessed SagA-mediated protection in tol-1(nr2033) animals and found that neither E. faecium nor E. faecalis-sagA were protective against S. Typhimurium infection in this mutant background, suggesting SagA enhances pathogen tolerance through tol-1 signaling (Fig. 2G).
To evaluate the mechanism of SagA protection (22), we generated an active site mutant as well as a C-terminal domain truncation of the NlpC/p60 hydrolase domain (Fig. 3A, fig. S8A). Neither mutant was able to inhibit S. Typhimurium pathogenesis, indicating that the NlpC/p60 hydrolase activity is required (Fig. 3B). SagA did not affect S. Typhimurium colonization of C. elegans or directly attenuate S. Typhimurium growth or virulence mechanisms (fig S8B–E). In culture, recombinant SagA had no effect E. coli growth rate (fig. S9A), but induction of SagA expression caused a decrease in culture optical density (OD) (Fig. 3C, S9B–C), indicating cell lysis. In contrast, expression of the active site mutant or cytoplasmically-localized SagA did not induce E. coli cell lysis (Fig. 3C, fig. S9B–C). These data suggest that while exogenous addition of SagA is not bacteriolytic, SagA is a functional hydrolase that can cleave peptidoglycan when targeted to the periplasm.
Figure 3. Enzymatic activity of SagA is required for enhancing pathogen tolerance.
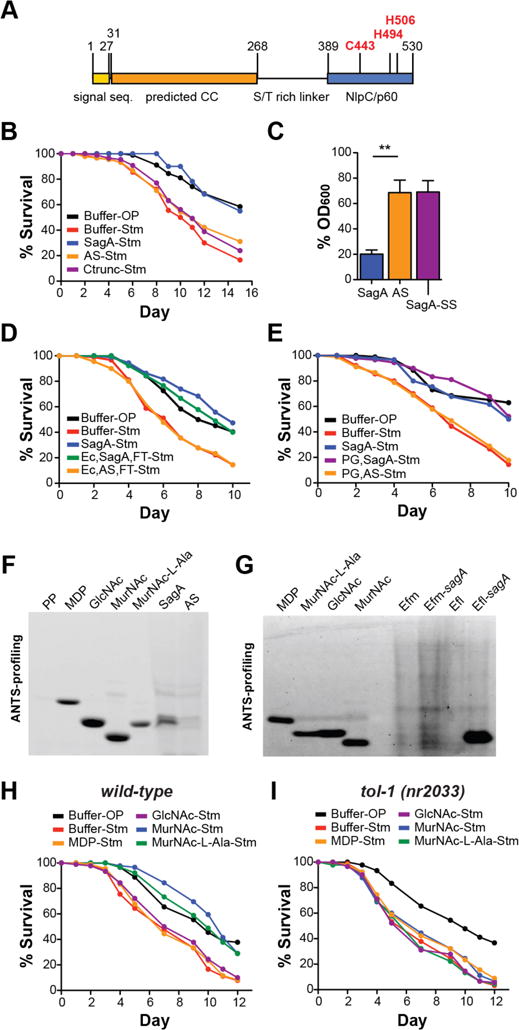
(A) Schematic of SagA domain organization: the signal sequence is yellow, a predicted coiled-coil (CC) domain is orange, and the NlpC/p60 type hydrolase domain is blue. Active site residues are in red type. (B) Survival curve showing that SagA inhibits S. Typhimurium (Stm) pathogenesis (p<10−10) while an active site mutant (AS) and C-terminal truncation mutant (Ctrunc) do not (p=0.42 and 0.98 respectively). (C) OD600 of E. coli BL21-RIL(DE3) expressing SagA, the active site mutant, or cytoplasmically-localized SagA (SagA-SS) 1 hour post-induction. Bars represent mean ± s.e.m. from three independent experiments. Significance was calculated by unpaired t test. For **, p < 0.01. (D) Survival curve showing that 5-kDa MWCO column filtered E. coli culture supernatants expressing SagA-His6 (Ec, sagA-FT) inhibit Stm pathogenesis (p<10−4), while filtered E. coli culture supernatants expressing the active site mutant (Ec, AS-FT) do not (p=1). (E) Survival curve showing that purified E. coli peptidoglycan treated with SagA (PG, SagA) can inhibit Stm pathogenesis (p<10−10), while E. coli peptidoglycan treated with the active site mutant (PG, AS) cannot (p=1). (F) ANTS visualization of E. coli culture supernatants expressing SagA-His6 or the active site mutant. A sugar-less pentapeptide (PP) shows UV signal specificity. (G) ANTS visualization of peptidoglycan fragments in Efm, Efm-sagA, Efl, and Efl-sagA culture supernatants. (H) Survival curve showing that treatment with MurNAc (p<10−5) or MurNAc-L-Ala (p<10−10) can inhibit Stm pathogenesis, while MDP (p=1) and GlcNAc (p=1) are not protective. (I) Survival curve showing that MurNAc (p=1) and MurNAc-L-Ala (p=0.61) do not inhibit pathogenesis in tol-1(nr2033) C. elegans.
We hypothesized that SagA generates peptidoglycan fragments responsible for enhancing pathogen tolerance. Consistent with this hypothesis, we found that the flow-thru from 5 kDa-MWCO column-filtered culture supernatants of E. coli expressing SagA, but not the active site mutant, protected C. elegans from S. Typhimurium pathogenesis (Fig. 3D, fig. S10A), suggesting that lower molecular weight products of SagA enzymatic activity are sufficient for protection. To test if SagA-generated E. coli peptidoglycan fragments can protect C. elegans from S. Typhimurium, we digested purified E. coli peptidoglycan with lysozyme and either SagA or the active site mutant, then filtered the digests to exclude protein. C. elegans treated with the SagA peptidoglycan digests survived similarly to SagA-treated animals, whereas active site mutant digests failed to attenuate pathogenesis (Fig. 3E). These results suggest that SagA-generated peptidoglycan fragments, and not SagA itself, are responsible for enhancing pathogen tolerance.
To identify the peptidoglycan fragment(s) generated by SagA, we analyzed filtered bacterial culture supernatants by ANTS labeling and gel-based profiling (23, 24). From E. coli expressing SagA, we detected a SagA-specific product that migrated similarly to the synthetic peptidoglycan fragments MurNAc-L-Ala and GlcNAc, but not to MurNAc-L-Ala-D-Glu (MDP) or MurNAc (Fig. 3F). ANTS analysis of E. faecium, E. faecalis, and E. faecalis-sagA peptidoglycan extracts revealed that SagA expression alters the muropeptide profile (fig. S11). From E. faecalis-sagA culture supernatant, we detected an ANTS-labeled product that co-migrates with MurNAc (Fig. 3G), suggesting that heterologous SagA expression induces muropeptide shedding in both E. coli and E. faecalis. In contrast, 10 kDa-MWCO filtered E. faecium culture supernatant did not yield detectable levels of MurNAc-L-Ala or MurNAc (Fig. 3G) and was not protective when administered to C. elegans (fig. S2C). E. faecium that expresses SagA endogenously is likely resistant to SagA-induced peptidoglycan shedding. As SagA is abundantly secreted by E. faecium (fig. S4, tables S1, S4) and is protective after purification (Fig. 2D–E), soluble SagA may hydrolyze extracellular peptidoglycan fragments derived from digested bacteria in vivo. Indeed, incubation of purified E. coli peptidoglycan with lysozyme and recombinant SagA, but not the active site mutant, yielded a peptidoglycan cleavage product with similar mobility to MurNAc-L-Ala (fig. S10B). These data suggest that heterologous expression of SagA in bacteria can remodel bacterial peptidoglycan (fig. S11), induce shedding of small muropeptide fragments (Fig. 3F–G), and cleave extracellular peptidoglycan when secreted (fig. S10B). We next assessed the protective activity of SagA-generated peptidoglycan fragments, MurNAc and MurNAc-L-Ala, as well as GlcNAc and MDP. Treatment of C. elegans with either MurNAc or MurNAc-L-Ala was sufficient to inhibit S. Typhimurium pathogenesis, while MDP or GlcNAc were not (Fig. 3H). MurNAc and MurNAc-L-Ala were not protective in tol-1(nr2033) animals (Fig. 3I), suggesting that tol-1 is required for mediating host protection in response to these peptidoglycan fragments. These data are consistent with the activity of muropeptides in mammals (25, 26), but show MurNAc-L-Ala and MurNAc are the minimal peptidoglycan components that enhance pathogen tolerance in C. elegans.
We next evaluated SagA-mediated protection against Salmonella pathogenesis in mice. Germ-free mice were mono-colonized with E. faecium, E. faecalis, or E. faecalis-sagA 7 days prior to infection with S. Typhimurium. Enterococcus and Salmonella load were measured in the feces, and mouse survival was tracked. All Enterococcus strains were similarly recovered from the feces after gavage, indicating efficient intestinal colonization (fig. S12). Consistent with our results in C. elegans, S. Typhimurium CFUs in the feces were similar across all conditions throughout infection (Fig. 4A), suggesting that E. faecium does not inhibit Salmonella colonization. Remarkably, mice gavaged with E. faecium or E. faecalis-sagA prior to infection exhibited reduced weight loss and prolonged survival, with a median survival of 9 days, as compared to E. faecalis-treated mice (Fig. 4B–C). Although Enterococci are used as probiotics in livestock, their pathogenic potential makes them problematic for use in humans (27). We thus introduced sagA into a non-pathogenic probiotic, Lactobacillus plantarum (28), and confirmed its expression and secretion (fig. S13). sagA-expressing L. plantarum significantly prevented weight loss and improved survival in an antibiotic-induced S. Typhimurium infection model compared to L. plantarum (Fig. 4D–F, fig. S14). These results indicate that SagA is sufficient to attenuate Salmonella pathogenesis in mammals and is protective even when expressed by other probiotic bacteria.
Figure 4. E. faecium and SagA enhance pathogen tolerance in mice.

(A–C) Germ-free (GF) C57BL/6 mice were orally gavaged with 108 CFU E. faecalis (Efl), Efl-expressing sagA (EflsagA) or E. faecium (Efm) 7 days before oral infection with 102 CFU S. Typhimurium (Stm). (A) Stm CFU in feces, (B) weight loss, and (C) survival are shown. Pooled data from 4 independent experiments, n=10–14 mice/group. (D–F) Mice were given AMNV-antibiotic cocktail for 14 days and colonized with 108 CFU L. plantarum (Lpl) harboring an empty plasmid vector (Lpl-vector) or a sagA plasmid (Lpl-sagA) or 108 CFU Efm prior to oral infection with 106 Stm. (D) Stm CFU in feces, (E), Weight loss and (F) survival are shown. Pooled data from 2 independent experiments, n=2–5 mice/group. (A, B, D and E) mean±SEM, 2-way ANOVA, p-value shown comparing sagA-expressing Efl or Lpl to WT or vector controls, respectively (A, B, D and E). n.s.=not significant. (C and F) Log-rank analysis, p-value shown comparing Efm, sagA-expressing Efl or Lpl to WT or vector controls, respectively (C and F). **p≤0.01, ***p≤0.001 for all analyses. Comparisons with no (*) had p>0.05 and were not considered significant.
Here we demonstrate that C. elegans is an effective model to explore the protective mechanisms of intestinal bacteria and find that SagA from E. faecium is sufficient to protect C. elegans and mice from enteric pathogens. Our results suggest that the NlpC/p60 hydrolase activity of SagA generates unique peptidoglycan fragments that may activate host immune pathways to enhance epithelial barrier integrity and confine pathogens to the intestinal lumen, ultimately promoting tolerance to infection (fig. S15). Our analysis of E. faecium and engineered SagA-expressing bacterial strains in mice suggests SagA also improves intestinal epithelial barrier integrity to limit bacterial pathogenesis in mammals (29). The protective activity of E. faecium and SagA in mice requires the TLR signaling adaptor MyD88, the peptidoglycan pattern recognition receptor NOD2, and the C-type lectin RegIIIγ (29). These results together suggest that E. faecium and SagA may function through evolutionarily conserved pathways to enhance epithelial barrier integrity and protect animals from enteric pathogens. Finally, this study suggests that bacterial NlpC/p60-type peptidoglycan hydrolases (30–33) can enhance host tolerance to pathogens and that these enzymes could be used to improve the activity of existing probiotics.
Supplementary Material
Acknowledgments
We thank M. Tesic for LC/MS-MS analyses, S.T. Chen for cloning pET21a-SagA-SS, A. Rogoz and T. Rendon for assistance with germ-free mouse care and the Bargmann lab for reagents and helpful discussions. We also thank M.S. Gilmore, B.E. Murray, J.T. Singer, A.J. Bäumler and B. Sartor for reagents. The data presented in this manuscript are tabulated in the main paper and in the supplementary materials. All C. elegans strains were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). K.J.R. received support from the David Rockefeller Graduate Program and a Helmsley Graduate Fellowship. V.A.P. thanks Center for Basic and Translational Research on Disorders of the Digestive System Pilot Award through the generosity of the Leona M. and Harry B. Helmsley Charitable Trust. Y-C.W. is a Cancer Research Institute Irvington Fellow supported by the Cancer Research Institute. This work was supported by the NIH-NIGMS R01GM103593 and Robertson Therapeutic Development Fund to H.C.H and D. M. H.C.H also thanks Lerner Trust for support. K.J.R, V.A.P., D.M., and H.C.H. are inventors on patent PCT/US2016/028836 submitted by The Rockefeller University that covers modified microorganisms expressing SagA as anti-infective agents, probiotics, and food components.
Footnotes
The authors declare no competing financial interests.
References
- 1.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013 Nov;13:790. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lebreton F, Willems RJL, Gilmore MS. In: Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Boston: 2014. [PubMed] [Google Scholar]
- 4.Staley C, Dunny GM, Sadowsky MJ. Environmental and animal-associated enterococci. Adv Appl Microbiol. 2014;87:147. doi: 10.1016/B978-0-12-800261-2.00004-9. [DOI] [PubMed] [Google Scholar]
- 5.Franz CM, Huch M, Abriouel H, Holzapfel W, Galvez A. Enterococci as probiotics and their implications in food safety. Int J Food Microbiol. 2011 Dec 2;151:125. doi: 10.1016/j.ijfoodmicro.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 6.Garsin DA, et al. A simple model host for identifying Gram-positive virulence factors. Proc Natl Acad Sci U S A. 2001 Sep 11;98:10892. doi: 10.1073/pnas.191378698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irazoqui JE, Urbach JM, Ausubel FM. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat Rev Immunol. 2010 Jan;10:47. doi: 10.1038/nri2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nat Rev Microbiol. 2008 Jan;6:53. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- 9.Aballay A, Yorgey P, Ausubel FM. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Current biology : CB. 2000 Nov 30;10:1539. doi: 10.1016/s0960-9822(00)00830-7. [DOI] [PubMed] [Google Scholar]
- 10.Labrousse A, Chauvet S, Couillault C, Kurz CL, Ewbank JJ. Caenorhabditis elegans is a model host for Salmonella typhimurium. Curr Biol. 2000 Nov 30;10:1543. doi: 10.1016/s0960-9822(00)00833-2. [DOI] [PubMed] [Google Scholar]
- 11.Garsin DA, et al. A simple model host for identifying Gram-positive virulence factors. Proc Natl Acad Sci U S A. 2001 Sep 11;98:10892. doi: 10.1073/pnas.191378698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teng F, Kawalec M, Weinstock GM, Hryniewicz W, Murray BE. An Enterococcus faecium secreted antigen, SagA, exhibits broad-spectrum binding to extracellular matrix proteins and appears essential for E. faecium growth. Infect Immun. 2003 Sep;71:5033. doi: 10.1128/IAI.71.9.5033-5041.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim DH, et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002 Jul 26;297:623. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- 14.Shivers RP, Kooistra T, Chu SW, Pagano DJ, Kim DH. Tissue-specific activities of an immune signaling module regulate physiological responses to pathogenic and nutritional bacteria in C. elegans. Cell Host Microbe. 2009 Oct 22;6:321. doi: 10.1016/j.chom.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mallo GV, et al. Inducible antibacterial defense system in C. elegans. Curr Biol. 2002 Jul 23;12:1209. doi: 10.1016/s0960-9822(02)00928-4. [DOI] [PubMed] [Google Scholar]
- 16.Garsin DA, et al. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003 Jun 20;300:1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- 17.Macosko EZ, et al. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature. 2009 Apr 30;458:1171. doi: 10.1038/nature07886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Styer KL, et al. Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science. 2008 Oct 17;322:460. doi: 10.1126/science.1163673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pujol N, et al. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol. 2001 Jun 5;11:809. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- 20.Pradel E, et al. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2007 Feb 13;104:2295. doi: 10.1073/pnas.0610281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tenor JL, Aballay A. A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity. EMBO reports. 2008 Jan;9:103. doi: 10.1038/sj.embor.7401104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Firczuk M, Bochtler M. Folds and activities of peptidoglycan amidases. FEMS Microbiol Rev. 2007 Nov;31:676. doi: 10.1111/j.1574-6976.2007.00084.x. [DOI] [PubMed] [Google Scholar]
- 23.Li SY, Holtje JV, Young KD. Comparison of high-performance liquid chromatography and fluorophore-assisted carbohydrate electrophoresis methods for analyzing peptidoglycan composition of Escherichia coli. Anal Biochem. 2004 Mar 1;326:1. doi: 10.1016/j.ab.2003.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young KD. A simple gel electrophoretic method for analyzing the muropeptide composition of bacterial peptidoglycan. J Bacteriol. 1996 Jul;178:3962. doi: 10.1128/jb.178.13.3962-3966.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Philpott DJ, Sorbara MT, Robertson SJ, Croitoru K, Girardin SE. NOD proteins: regulators of inflammation in health and disease. Nat Rev Immunol. 2014 Jan;14:9. doi: 10.1038/nri3565. [DOI] [PubMed] [Google Scholar]
- 26.Royet J, Dziarski R. Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nat Rev Microbiol. 2007 Apr;5:264. doi: 10.1038/nrmicro1620. [DOI] [PubMed] [Google Scholar]
- 27.Lund B, Edlund C. Probiotic Enterococcus faecium strain is a possible recipient of the vanA gene cluster. Clin Infect Dis. 2001 May 1;32:1384. doi: 10.1086/319994. [DOI] [PubMed] [Google Scholar]
- 28.Dicks LM, Botes M. Probiotic lactic acid bacteria in the gastro-intestinal tract: health benefits, safety and mode of action. Benef Microbes. 2010 Mar;1:11. doi: 10.3920/BM2009.0012. [DOI] [PubMed] [Google Scholar]
- 29.L AK, Pedicord VA, Rangan KJ, Craig JW, Loschko J, Rogoz A, Hang HC, Mucida D. Exploiting a host-commensal interaction to promote intestinal barrier function limits enteric infections. Science Immunology. 2016 doi: 10.1126/sciimmunol.aai7732. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vollmer W, Joris B, Charlier P, Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008 Mar;32:259. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 31.Yan F, et al. Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism. J Clin Invest. 2011 Jun;121:2242. doi: 10.1172/JCI44031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan F, et al. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology. 2007 Feb;132:562. doi: 10.1053/j.gastro.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan F, et al. A Lactobacillus rhamnosus GG-derived soluble protein, p40, stimulates ligand release from intestinal epithelial cells to transactivate epidermal growth factor receptor. J Biol Chem. 2013 Oct 18;288:30742. doi: 10.1074/jbc.M113.492397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stiernagle T. Maintenance of C. elegans. WormBook. 2006:1. doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winter SE, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010 Sep 23;467:426. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 37.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991 May 24;252:1162. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 38.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 39.Nallapareddy SR, Singh KV, Murray BE. Construction of improved temperature-sensitive and mobilizable vectors and their use for constructing mutations in the adhesin-encoding acm gene of poorly transformable clinical Enterococcus faecium strains. Appl Environ Microbiol. 2006 Jan;72:334. doi: 10.1128/AEM.72.1.334-345.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singer JT, et al. Broad-host-range plasmids for red fluorescent protein labeling of gram-negative bacteria for use in the zebrafish model system. Applied and environmental microbiology. 2010 Jun;76:3467. doi: 10.1128/AEM.01679-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Podbielski A, Spellerberg B, Woischnik M, Pohl B, Lutticken R. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS) Gene. 1996 Oct 24;177:137. doi: 10.1016/0378-1119(96)84178-3. [DOI] [PubMed] [Google Scholar]
- 42.Dunny GM, Lee LN, LeBlanc DJ. Improved electroporation and cloning vector system for gram-positive bacteria. Appl Environ Microbiol. 1991 Apr;57:1194. doi: 10.1128/aem.57.4.1194-1201.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raffatellu M, et al. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009 May 8;5:476. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang JS, et al. OASIS: online application for the survival analysis of lifespan assays performed in aging research. PLoS One. 2011;6:e23525. doi: 10.1371/journal.pone.0023525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Powell JR, Ausubel FM. Models of Caenorhabditis elegans infection by bacterial and fungal pathogens. Methods Mol Biol. 2008;415:403. doi: 10.1007/978-1-59745-570-1_24. [DOI] [PubMed] [Google Scholar]
- 46.Garsin DA, et al. A simple model host for identifying Gram-positive virulence factors. Proc Natl Acad Sci U S A. 2001 Sep 11;98:10892. doi: 10.1073/pnas.191378698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaham S. Methods in cell biology. WormBook. 2006 Jan 12;:1551. 2006. [Google Scholar]
- 48.Lundquist EA, Reddien PW, Hartwieg E, Horvitz HR, Bargmann CI. Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development. 2001 Nov;128:4475. doi: 10.1242/dev.128.22.4475. [DOI] [PubMed] [Google Scholar]
- 49.Rangan KJ, Yang YY, Charron G, Hang HC. Rapid visualization and large-scale profiling of bacterial lipoproteins with chemical reporters. J Am Chem Soc. 2010 Aug 11;132:10628. doi: 10.1021/ja101387b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuhner D, Stahl M, Demircioglu DD, Bertsche U. From cells to muropeptide structures in 24 h: peptidoglycan mapping by UPLC-MS. Scientific reports. 2014;4:7494. doi: 10.1038/srep07494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young KD. A simple gel electrophoretic method for analyzing the muropeptide composition of bacterial peptidoglycan. J Bacteriol. 1996 Jul;178:3962. doi: 10.1128/jb.178.13.3962-3966.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hesek D, et al. Synthetic peptidoglycan substrates for penicillin-binding protein 5 of Gram-negative bacteria. J Org Chem. 2004 Feb 6;69:778. doi: 10.1021/jo035397e. [DOI] [PubMed] [Google Scholar]
- 53.Mugunthan G, Sriram D, Yogeeswari P, Ravindranathan Kartha KP. Synthesis and biological evaluation of sugar-derived chiral nitroimidazoles as potential antimycobacterial agents. Carbohydr Res. 2011 Sep 27;346:1760. doi: 10.1016/j.carres.2011.05.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


