Abstract
SWI/SNF chromatin-remodeling complexes are key regulators of the epigenetic modifications that determine whether stem cells maintain pluripotency or commit toward specific lineages through development and during postnatal life. Dynamic combinatorial assembly of multiple variants of SWI/SNF subunits is emerging as the major determinant of the functional versatility of SWI/SNF. Here, we summarize the current knowledge on the structural and functional properties of the alternative SWI/SNF complexes that direct stem cell fate toward skeletal muscle lineage and control distinct stages of skeletal myogenesis. In particular, we will refer to recent evidence pointing to the essential role of two SWI/SNF components not expressed in embryonic stem cells—the catalytic subunit BRM and the structural component BAF60C—whose induction in muscle progenitors coincides with the expansion of their transcriptional repertoire.
Keywords: Epigenetics, Stem cells, Skeletal muscle, SWI/SNF, BRM, BAF60C
Introduction
Chromatin is the central component of the nuclear landscape that controls the expression of genes, which regulate stem cell pluripotency and ability to differentiate into specialized cell types that compose tissues and organs of multicellular organisms. As such, chromatin modifications are regarded as major epigenetic underpinning that orchestrates transcription in stem cells during developmental transitions.
Chromatin results from the 3D arrangement of DNA and histones in the nuclei of eukaryotic cells. The basic unit of chromatin is the nucleosome, which consists of a segment of duplex DNA wrapped around a histone octamer comprised of two of each of the conventional histone proteins: H2A, H2B, H3, and H4 [1]. The histone proteins help compacting and strengthening the DNA, and are subjected to conformational changes upon a variety of post-translational modifications to ultimately alter nucleosome structure and position, thereby regulating gene expression and DNA replication. Chromatin remodeling is an active process, which represses or enables access of the transcription machinery to genes. Chromatin-remodeling complexes (CRCs) provide specialized enzymatic activities that are able to disrupt the chromatin structure using ATP hydrolysis that destabilizes nucleosomes and regulates DNA accessibility to transcription factors and other proteins [2].
There are four major families of ATP-dependent CRCs: SWI/SNF (switch/sucrose non-fermentable), ISWI (imitation switch), CHD (chromodomain helicase DNA-binding), and INO80 (inositol). The SWI/SNF complex was first identified in 1984 from genetic screens in yeast [3]. Following this discovery, homologs of the SWI/SNF subunits in higher organisms were identified and purified leading to the biochemical isolation of multi-protein complexes. The Drosophila SWI2/SNF2 homolog Brahma (BRM) was identified as one of the trithorax group proteins, which antagonize the function of polycomb group (PcG) proteins in repressing gene expression [4, 5]. In 1996, Wang et al. purified mammalian SWI/SNF complexes and identified approximately ten subunits [6]. SWI/SNF complexes typically contain one enzymatic subunit—either the ATPase Brahma (Brm) or Brg1—and a collection of Brg1/Brm-associated factors (BAFs) [7]. Many of the subunits are conserved from yeast to human, with an increased number of components and the appearance of multiple variants for each subunit. The evolutionarily expansion of SWI/SNF subunits indicates that dynamic combinatory assembly can accommodate the demand of transcriptional control of a more complex genome. Currently, there are at least 14 documented subunits of the mammalian SWI/SNF complex, also known as mBAF complex (BRG1/BRM-associated factors), encoded by 25 genes, whose products can be combinatorially assembled in multiple ways to promote hundreds combinations of distinct SWI/SNF complexes [8, 9]. The catalytic core of the SWI/SNF complex consists of two mutually exclusive ATPase subunits, Brg1 and Brm, which are alternatively incorporated into large complexes that also include non-enzymatic, structural subunits. While the presence of one of these catalytic subunits endows the SWI/SNF complex with the ATPase activity necessary to remodel the chromatin, the presence of few “core” structural components—Baf47, Baf155, and Baf170—appear required for minimal SWI/SNF enzymatic activity in vitro [10]. Still, the precise composition of SWI/SNF complexes present in different cell types has not conclusively been determined.
Combinatorial SWI/SNF assembly controls stem cell pluripotency and lineage specification
ATP-dependent CRCs have been shown to be involved in most biological processes and studies over the last decade unraveled their fundamental role in regulating crucial aspects of embryonic stem cell (ESC) pluripotency and differentiation [11, 12]. In particular, the contribution of SWI/SNF-mediated chromatin remodeling in directing the alternative ability of ESC to either maintain pluripotency or to undergo lineage commitment and differentiation has been intensively investigated.
Seminal studies have revealed a peculiar feature that defines the chromatin structure in ESCs, consisting of bivalent domains at the regulatory regions of developmental genes that poise them for activation upon specific developmental cues [13–15]. Bivalent domains are typically defined by large regions of tri-methylation of histone 3 lysine 27 (H3K27me3) harboring smaller regions of tri-methylation of histone 3 lysine 4 (H3K4me3). Promoters under these conditions are kept transcriptionally silenced, yet prone to activation. ATP-dependent CRCs contribute to generate this unique chromatin structure, thereby participating both to transcriptional repression of genes involved in early development and to the chromatin reorganization that allows specific patterns of gene expression upon differentiation [16, 17]. Accordingly, changes in SWI/SNF subunit composition appear to determine tissue or cell-type specific gene expression, thereby regulating lineage commitment during embryonic development. As such, the incorporation or exchange of specific subunits generates cell context-dependent sub-complexes that are competent to trigger different transcriptional programs, following specific differentiation stimuli. For instance, the specific assembly of chromatin-remodeling complex that has been detected in ESC (esBAF) includes the core components BRG1, BAF155, BAF250a, BAF60a/b, and BAF53a [18]. EsBAFs are devoid of two alternative variants—the ATPase and Brm—and the structural component—Baf60c—and play a crucial role to maintain pluripotency in ESCs, as indicated by its detection on the regulatory elements of genes downstream of the master pluripotency factors Oct4, Sox2, and Nanog [8, 19]. Surprisingly, experiments of genetic inactivation of SWI/SNF subunits indicate that SWI/SNF-mediated chromatin remodeling in pluripotent ESCs is predominantly directed at establishing repressive chromatin toward preventing differentiation [16]. An additional “repressive” function of esBAF has been revealed by a recent study reporting on the suppression of pervasive transcription from open chromatin regions in ESCs [20]. This study revealed that esBAF generates an open nucleosome-depleted region (NDR) at many of its binding sites throughout the genome, to permit binding of transcription factors. Within this context, esBAF operates to suppress transcription of ncRNAs, via maintenance of defined nucleosome-free and nucleosome-bound segments within small open chromatin domains (~200 bp NDRs plus a few flanking nucleosomes) throughout the genome. Given that esBAF complexes interact directly with key regulators of pluripotency in ESC [18], the function of BAF complexes in pluripotency might be to coordinate access of pluripotency factors and pervasive transcription, by generating specific patterns of chromatin conformation that support the maintenance of pluripotency.
In response to developmental cues and generation of the three germ layers, different progenitor cells from ectoderm, endoderm, and mesoderm show distinct SWI/SNF complexes with a specific combination of components that imparts cell-type specific patterns of chromatin remodeling, in concert with tissue-specific transcriptional activators [16]. The heterogeneity in composition and dynamic exchange of subunits conceivably endows the SWI/SNF complex to adopt the conformation for an optimal response to developmental cues instructing stem cells to commit toward specific lineages.
A striking example is provided by the incorporation of BAF60c subunit that coincides with the competence to activate mesoderm-derived lineages, such as skeletal and cardiac muscle. During cardiac development, BAF60c marks the region of the embryo with cardiogenic potential [21]. Complexes containing BAF60C promote the formation of heart tissue from pre-cardiac mesoderm by interacting with cardiogenic transcriptional activators, in response to coordinate changes of Nodal and BMP signaling [21–23]. In this circumstance, the expression of homologous subunits could not compensate for the loss of the correct subunit, highlighting the importance of the specificity of each subunit in instructing cell fate decisions. On the other hand, the absence of BAF60c in ESCs restricts their competence to activate skeletal myogenesis [23, 24]. The role of BAF60c in the activation of the myogenic program is described in detail in the following sections.
Likewise, the expression of BAF250a and BAF250b—two other mutually exclusive subunits of the BAF complex—appears to influence the formation of SWI/SNF complexes with specific functions. Indeed, BAF250a-null ES cells fail to differentiate into mesodermal lineages, including cardiomyocytes, adipocytes, and, to a lesser extent, skeletal muscles [25], suggesting that BAF250a is a key component of the SWI/SNF complex that controls gene expression during ESC cell lineage commitment and differentiation.
Moreover, during neural differentiation, esBAF undergoes a dramatic change in composition, accompanied by an extensive switch of BAF components, including the repression of BAF53a in post-mitotic neurons and the induction of BAF53b, BAF45b, and BAF45c variants, which are required for activity-dependent dendrite growth [26, 27]. Importantly, this essential transition is mediated by miR-9* and miR-124 which are selectively expressed in post-mitotic neurons and target BAF53a. These miRs are repressed in proliferating neural progenitors by the repressor-element-1 silencing transcription factor (REST, also known as NRSF). This pivotal study shed light on the crosstalk among these different epigenetic proteins to form the critical regulatory network that regulates neuronal development, by translating environmental cues into the control of the neurogenic process [26, 28].
Finally, studies with mouse ESCs showed that members of SWI/SNF complexes, Brg1, BAF57, BAF155, and BAF47, are required to repress Nanog expression during lineage formation, with Baf155 expression being necessary for heterochromatin formation and chromatin compaction during mESC differentiation [29].
Thus, in stem cells, the coordinated activation and repression of pluripotency and cell-type specific genes, respectively, relies on the assembly of specific SWI/SNF complexes. It is likely that signal-dependent regulation of SWI/SNF activity plays a fundamental role in this process. This concept has been well illustrated by studies from Crabtree lab showing that Brg1 “instructs” STAT3 binding across the genome of pluripotent stem cells, by establishing the chromatin accessibility and binding to target genes in response to the activation of leukemia inhibitory factor (LIF) signaling, thereby promoting pluripotency. At the same time, Brg1 also cooperate with PcG to repress developmental genes in ESCs [30].
SWI/SNF and embryonic stem cell commitment to the myogenic lineage
Exchange of SWI/SNF subunits plays a pivotal role in establishing muscle during embryonic development [19]. Although cardiac and skeletal muscles arise from different embryonic compartments, they share a common mechanism for the activation of their respective muscle-specific loci. This relies on the function of the SWI/SNF-associated BAF60c variant, encoded by SMARCD3 gene that, unlike the other BAF60 variants (Baf60a and Baf60b), is preferentially expressed in embryonic heart and somites [23]. Remarkably, Baf60c knockout mice show embryos with impaired cardiac and skeletal myogenesis [23]. Consistently, Baf60c has been further identified as the factor required to “prime” the chromatin of cardiac and skeletal muscle regulatory region to be accessed by tissue-specific transcription factors and actively transcribed [21, 24].
The skeletal muscle differentiation program is activated by the members of the basic helix-loop-helix (bHLH) myogenic regulatory factors (MRFs) [31]. MyoD and Myf5 act as the master transcriptional activators of the myogenic program in Pax3/Pax7-expressing muscle progenitors from paraxial mesoderm to promote differentiation into myotubes [31, 32]. MRFs activate transcription by heterodimerization with ubiquitously expressed bHLH proteins—termed E-proteins (E12/E47) [33]—that facilitates binding to consensus E-box sequence (CANNTG) found in the regulatory region of many muscle-specific genes [31]. By contrast, commitment to cardiac lineage relies on the activity of a transcriptional network composed by GATA4, HAND2, MEF2C, MESP1, NKX2.5, and TBX5 [34].
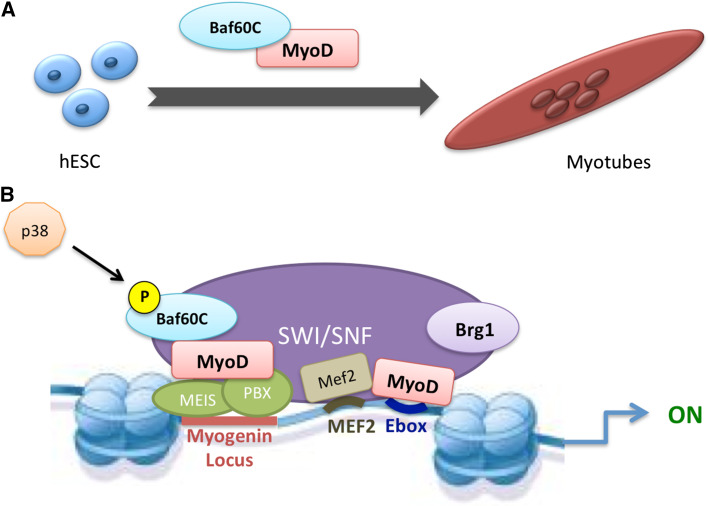
The functional relevance of SWI/SNF in the activation of the myogenic program has been anticipated by former seminal studies, showing the requirement of SWI/SNF complexes in MyoD-mediated activation of muscle-specific genes during fibroblast conversion into muscle cells [35, 36]. Further studies led to the discovery that Baf60c is the subunit that converts myogenic extracellular cues, such as those transduced by the p38 kinases alpha and beta, into changes in chromatin conformation, by promoting the recruitment of the Brg1-containing containing complex on muscle-specific loci [37, 38]. In proliferating myoblasts, BAF60C associates with MyoD to form “pioneer” complexes that pre-assemble on the chromatin at regulatory elements of muscle genes, such as myogenin. Upon differentiation, BAF60c phosphorylation by the p38 kinase leads to the incorporation of pre-assembled BAF60c/MyoD sub-complex into a Brg1-based SWI/SNF complex enabling MyoD to activate muscle-specific genes [37, 38]. This two-step model (Fig. 1) highlights the importance of BAF60c in priming the chromatin of differentiation genes for subsequent activation via a prior interaction with MyoD on E-box sequences [31]. This evidence paved the way to further investigate BAF60C role in muscle lineage determination during embryonic development. Albini and colleagues identified BAF60C as a key epigenetic determinant of human ESC commitment to the myogenic lineage [24]. Indeed, ESCs are resistant to the myogenic conversion upon the ectopic expression of physiological amounts of MyoD that are otherwise capable of converting a variety of somatic cells into muscle [39]. ESC resistance to myogenic conversion is conferred by the absence of BAF60C, which unlike the other BAF60 variants, is not expressed in ESCs [18, 24]. Forced expression of BAF60C enables MyoD to directly activate skeletal myogenesis in hESCs, by instructing MyoD positioning and allowing chromatin remodeling at target genes [24] (Fig. 2). Importantly, ESC epigenetically reprogrammed with BAF60C and MyoD that were competent to generate 3D contractile myospheres [19, 24].
Fig. 1.
Schematic representation of muscle gene activation in hESC. a hESC is resistant to MyoD-mediated conversion and the ectopic expression of both MyoD and Baf60C to be converted into myotubes [24]. b Molecular mechanism of the Baf60C-mediated MyoD activation of myogenin (and possibly other muscle genes) expression. MyoD–BAF60C complex is pre-assembled at E-box sequences, and together with additional pioneer factors (e.g., PBX/Meis) [36] favors the recruitment of SWI/SNF chromatin-remodeling complex. Signal-dependent phosphorylation of Baf60C (e.g., by cytokine-activated p38 kinases alpha/beta) promotes MyoD–BAF60C incorporation into the SWI/SNF complex that remodels chromatin and disrupts nucleosome to allow the activation of gene expression [37, 38]
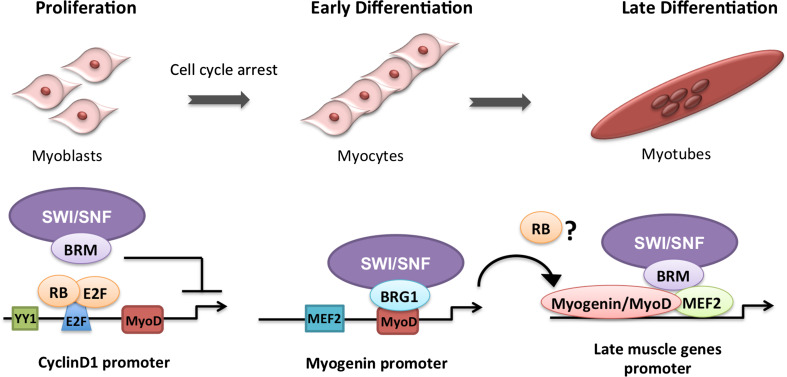
Fig. 2.
Schematic representation of skeletal muscle differentiation steps mediated by Brm and Brg1-SWI/SNF complexes. During proliferation of myoblasts Brm is required for Cyclin D1 repression possibly in cooperation with Rb-E2F factors. At the onset of differentiation, Brg1 is required for the activation of Myogenin, which in turn cooperates with other muscle bHLH proteins (i.e., MyoD) and MEF2 to activate late muscle genes. Brm is also involved in the activation of late muscle genes. A potential interaction between Brm, myogenin/MyoD, MEF2 is proposed in this illustration. RB has been postulated to contribute to late myogenesis [74–76] and can potentially cooperate with SWI/SNF on the activation of muscle late genes [78, 79]
A similar role for BAF60c as a key SWI/SNF target of signaling has been found in the heart, where BAF60c is essential to commit mesodermal-derived multipotent progenitors into cardiomyocytes, by mediating interactions between lineage-specific transcription factors Gata4 and Tbx5 and SWI/SNF to promote the expression of cardiogenic genes, upon nodal and BMP inhibition by endodermal-derived Cerberus 1 [22, 40].
The exchange of BAF60 variants can also impact cell fate decision of mesoderm-derived cells in both embryo and adults [9]. Recent works showed that the selection of specific BAF60 variants determines the myogenic lineage during somitogenesis [41] and directs the fate and the biological activity of fibro-adipogenic progenitors (FAPs)—a population of mesenchimal cells reside in the interstitium between myofibers and contributes to the regenerative environment of skeletal muscles [42, 43]. In particular, the alternative usage of BAF60 variants determines whether FAPs adopt a pathogenic phenotype, which mediates the formation of fibrotic scars and fatty deposition in dystrophic muscles, or a pro-myogenic fate, which promotes compensatory regeneration. Interestingly, BAF60 variant selection and incorporation into SWI/SNF complex are regulated by the expression of muscle-specific micro-RNA—the myomiRs miR-1.2, miR-133, and miR-206 [41, 44]. In FAPs, from dystrophic muscles, BAF60C and myomiRs are repressed by HDAC activity, and therefore, the SWI/SNF complexes are largely based on BAF60A or B variants, which appear to promote their pro-fibrotic and adipogenic phenotype; however, upon exposure to HDAC inhibitors (HDACi), the de-repression of BAF60C and myomiRs activates a feed forward circuit, whereby selective targeting of BAF60A and B by myomiRs favors the formation of BAF60C-based SWI/SNF complex, which further amplifies this process by inducing the expression BAF60C and myomiRs [44]. These events account for the observed ability of HDACi to promote regeneration and prevents fibrosis and fat deposition in dystrophic muscles [45, 46], activating a pro-myogenic program that antagonizes the constitutive fibro-adipogenic lineage adopted by FAPs in dystrophic muscles [43].
SWI/SNF complex and control of gene expression in adult muscle (satellite) stem cells
Both embryonic and adult skeletal myogenesis rely on two sequential and interconnected events: the expansion of committed skeletal muscle progenitors (myoblasts) and their subsequent differentiation into multinucleated myotubes. These events are coordinated by molecular networks that are responsive to developmental cues (for embryonic myogenesis) or regenerative signals (for adult myogenesis), which regulate the activity of the MRFs [31]. Among MRFs, MyoD and its functional paralog Myf5 are expressed in proliferating myoblasts, prior to the activation of the differentiation program. For instance, during muscle regeneration, MyoD and Myf5 expression identifies the fraction of “activated” muscle stem (satellite) cells (MuSCs) that proliferate toward differentiating into new fibers [32, 47]. As MyoD and Myf5 are transcriptionally “latent” in proliferating myoblasts [31], a currently unsolved issue regards the mechanism by which these two transcriptional activators establish and maintain the myogenic lineage without activating transcription of muscle genes in proliferating myoblasts.
A number of studies revealed the potential contribution of SWI/SNF-mediated chromatin remodeling at different stages along the transition from MuSCs to terminally differentiated skeletal myofibers. The involvement of specific combinations of SWI/SNF subunits in the establishment of the myogenic lineage during myoblast proliferation has been suggested by the finding that MyoD and BAF60C associate to form a complex detected on the regulatory elements of MyoD-regulated genes, without activating transcription [38]. While this complex does not appear to have any chromatin remodeling activity, as it is devoid of the enzymatic SWI/SNF subunits, it likely provides the “platform” for signal-activated recruitment of Brg1-based SWI/SNF complex upon exposure to regeneration cues that elicit the p38 signaling [37]. As such, BAF60C–MyoD complex might “pre-set” the optimal nuclear landscape for the activation of the myogenic program in the fraction of committed muscle progenitors. This hypothesis fits with the preferential segregation of active p38 in the subsets of activated MuSCs [48, 49] and suggests that asymmetric partitioning of SWI/SNF components might regulate division symmetry in MuSC exposed to regeneration cues. An additional indirect evidence supporting the potential contribution of SWI/SNF heterogeneity to the regulation of MuSC division symmetry is the recent finding that cytokine-activated JAK-STAT3 signaling is preferentially activated in differentiation-committed MuSCs [50, 51]. Indeed, studies in ESCs have revealed a role of Brg1-based SWI/SNF in directing STAT3 genome-wide chromatin binding, by establishing the chromatin accessibility at STAT3 binding targets, thereby preparing for a proper response to regeneration signals that activate JAK-STAT pathway (i.e., IL6 signaling) [30]. Experimental deletion of Brg1 precludes STAT3-mediated activation of target genes, by promoting polycomb (PcG) binding and H3K27me3-mediated silencing [30]. Since STAT3 activates MyoD expression in MuSCs [51], it is possible that the mechanism by which Brg1/SWI/SNF-mediated chromatin remodeling at STAT3 binding sites can be extended to the early commitment of MuSCs to the myogenic lineage. On the other hand, Brg1 is required for maintaining Pax7 expression that supports proliferation and survival of the expanding population of MuSCs that has broken quiescence [52]. Interestingly, it appears that the SWI/SNF complex implicated in establishing the landscape conducive for MuSC quiescence relies on the activity of the other ATPase and Brm, which has the unique ability to interact with critical components of the notch signaling [53]—an essential pathway for keeping MuSC quiescence [54–56]. Consistently, recent evidence has revealed a specific role of Brm in the control of cell cycle in MuSCs, via CyclinD1 repression [57]—see paragraph below for details.
Overall, distinct SWI/SNF complexes appear to regulate the nuclear landscape conducive for cell cycle arrest and retention of stemness in quiescent MuSCs or for the commitment to the myogenic lineage in the fraction of MuSCs activated during regeneration [58]. This regulation occurs in concert with the activity of Polycomb Repressive Complex 2 (PRC2) [59], to coordinate gene expression, as in the case of differentiation-committed MuSCs, in which p38-mediated targeting of PgC to Pax7 locus mediates the repression of Pax7 transcription [60]. The concerted activities of SWI/SNF and PgC are complemented with that of other chromatin-modifying complexes activated in response to regeneration cues [61–64], thereby generating a network of functional interactions that shape the epigenome of asymmetrically dividing satellite stem cells [65, 66]. Impairment of these networks might be implicated in loss of symmetric division that has been associated to the functional exhaustion of MuSCs during chronic muscular diseases and aging [65, 67, 68].
Stage-specific functions of the two SWI/SNF ATPases Brg1 and Brm during skeletal myogenesis
As skeletal myogenesis proceeds through multiple stages, including myoblast proliferation, differentiation-associated irreversible cell cycle arrest and formation of terminally multinucleated myotubes, a longstanding question regards the potential existence of stage-specific SWI/SNF complexes with defined combinations of subunits. In particular, an individual role of the two alternative SWI/SNF ATPases Brg1 and Brm has been postulated, based on the different domains that they contain, on their non-redundant function observed in the previous studies and on the different phenotypes shown by Brg1 and Brm null mice [53, 69, 70].
Seminal studies from Imbalzano and Tapscott labs have revealed an essential role of Brg1-based SWI/SNF in mediating the ability of MyoD to remodel the chromatin at target genes [35, 71]. Further evidence indicated that Brg1-remodeling activity is required to activate the transcription of muscle-specific genes (myogenin) and myogenic microRNAs (myomiRs) miR-1 and miR-133 [72, 73] by allowing MyoD access on their regulatory regions. By contrast, the specific role of the Brm ATPase has not been addressed by parallel studies. Only recently, our group unveiled a previously unrecognized role for Brm during skeletal myogeneis, distinct from that played by Brg1. Indeed, C2C12 depleted of either Brm or Brg1 displayed distinct phenotypes. While Brg1 knockdown completely inhibited myoblast differentiation into multinucleated myotubes, Brm knockdown led to incomplete myogenesis, with the formation of sporadic and shorter myotubes [57]. Importantly, Brm depleted myoblasts failed to undergo cell cycle arrest, leading to the presence of an abundant population of proliferating cells even upon serum withdrawal, which typically triggers irreversible cell cycle arrest in wild-type myoblasts. Overall, these studies showed that Brg1 is required at the early stage of differentiation, by activating a key MyoD downstream target—myogenin—while Brm is required at two distinct stages of myogenesis: at the onset of differentiation by regulating cell cycle arrest through Cyclin D1 repression, and at the later stage by activating the expression of late muscle genes. Importantly, these two functions appear to be independent from each other [57]. In vivo analysis of Brm null mice shows impaired muscle regeneration after injury, with aberrant proliferation of muscle stem cells, satellite cells, and delayed formation of new myofibers. This phenotype coincides with the intrinsic defect of muscle stem cells to arrest the cell cycle and complete the differentiation program in culture, which was due to a deregulated expression of Brm-target genes. This evidence supports the conclusion that Brm plays an essential role in the activation of the myogenic program of muscle stem cells at both early and late stages of adult myogenesis [57]. It also emphasizes once again the functional versatility of SWI/SNF complexes conferred by the alternative incorporation of Brg1 or Brm (Fig. 2).
The function of Brm during skeletal muscle differentiation is somehow reminiscent of that one described for the retinoblastoma protein (pRb) in the previous studies [73–76]. Indeed, during skeletal myogenesis, pRb is a critical regulator of myogenesis, as it directs the irreversible withdrawal of myoblasts, presumably through the repression of E2F genes, [74] and controls the late phase of myogenesis by activating the transcriptional function of myocyte enhancer family 2 (Mef2) in cooperation with MyoD [75]. While its role in the establishment of the post-mitotic state has been documented [74], the role of pRb in promoting terminal differentiation remains puzzling [76, 77]. Similar to Brm null muscles, pRb −/− muscle cells show the normal expression of p21 and myogenin, but no expression of late markers of muscle differentiation and compromised formation of myotubes [75]. As such, such as Brm, pRb appears to be specifically required for the execution of the later steps in skeletal mitogenesis and its differentiation-promoting function might not be linked to its cell-cycle regulatory activity. An interesting hypothesis would be that pRb and Brm cooperate to coordinate the activity of bHLH and MEF2 muscle regulatory factors, possibly by assembling transcriptional co-factors. Moreover, the functional link between Brm and pRb could also be extended to the repression of cell-cycle-related genes [78–80]. Our and other studies have recently shown the importance of SWI/SNF complex in mediating CyclinD1 repression [57, 81]. Albini et al. showed that in differentiating myoblasts, Cyclin D1 promoter is occupied specifically by Brm [57]; in contrast, Joliot et al. reported on the enrichment in Brg1 at the same elements [81]. While future studies should better establish the relative enrichment of Brg1 and Brm at Cyclin D1 promoter, it should be noted that the Brm-mediated repression of Cyclin D1 is consistent with the reported anti-proliferative activity of Brm in myoblasts [57]. Interestingly, Brm binding to the chromatin at Cyclin D1 promoter correlated with an enriched in H3K27 tri-methylation (H3K27me3) in correspondence of putative YY1 and E2F binding sites [57]. It is, therefore, tempting to speculate that Brm recruitment on Cyclin D1 promoter could coincide with the formation of a repressive complex also containing pRb and PRC2 [82, 83]. This hypothesis is supported by recent evidence showing that SWI/SNF complexes can act in cooperation with PRC2 to repress certain target genes by facilitating PRC2 binding and activity [30].
Interestingly, in a very recent study, Ruijtenberg and Heuvel have uncovered in C. elegans a tissue-type specific regulatory mechanism for cell-cycle arrest that depends on multiple functional interactions between the SWI/SNF and the cell-cycle machinery. When disrupted, these interactions can cause tumorous over-proliferation of somatic cells [84]. These findings further reinforce the idea that SWI/SNF chromatin-remodeling complex regulates cell cycle and gene expression independently and possibly through distinct subunits that are capable of mediating interactions with different proteins.
Conclusion and perspectives
Collectively, the information reported in this review demonstrates the importance of a balanced and coordinated activity of the different SWI/SNF assemblies in regulating stem cell lineage-specific gene expression. Currently, the importance of CRCs subunit heterogeneity to drive lineage specification seems well established. However, we lack a better understanding of the complete mechanism and signals that controls the subunit selection during this process. Advance in new technologies, as genetic models, genome-wide binding studies and more accurate and sensitive biochemical analysis, may provide us the tools necessary to identify the precise composition of SWI/SNF assembly at a particular stage and the gene targets. Answering to those open questions will anticipate future directions directed toward the identification of the specific regulation of SWI/SNF dynamic exchange. Given the well-established role of SWI/SNF complexes during development, adult homeostasis and cancer progression under different signaling and cellular contexts [85], further elucidations of the mechanisms that regulate SWI/SNF composition, may provide a new foundation for the development of drugs targeting CRCs in human disorders, including muscular diseases.
Acknowledgments
PLP is an Associate Professor in the Sanford Children’s Health Research Center at the Sanford-Burnham Medical Research Institute (SBMRI) and acknowledges support from the NIH (R01AR056712, R01AR052779, and P30AR061303), from MDA, EPIGEN, FILAS and from the European Community’s Seventh Framework Program in the project FP7-Health—2009 ENDOSTEM 241440. SA was supported by CIRM training fellowship (TG2-01162). PCT is supported by NIH diversity supplement to 5 R01 AR052779. PLP dedicates this work to the memory of Dr Piccinelli, president of Fondazione Sovena, which has been supporting research in Puri’s lab.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest regarding the publication of this paper.
Contributor Information
Pier Lorenzo Puri, Phone: +39-06-50170-3266, Email: lpuri@sbdiscobvery.org.
Sonia Albini, Phone: 858-646-3000, Email: salbini@sbpdiscovery.org.
References
- 1.Kornberg R (1974) Chromatin structure: a repeating unit of histones and DNA. Science (New York) [DOI] [PubMed]
- 2.Cairns B. The logic of chromatin architecture and remodelling at promoters. Nature. 2009 doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 3.Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- 4.Tamkun JW, Deuring R, Scott MP, et al. Brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-E. [DOI] [PubMed] [Google Scholar]
- 5.Elfring L, Deuring R, McCallum C et al (1994) Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol Cell Biol [DOI] [PMC free article] [PubMed]
- 6.Wang W, Xue Y, Zhou S, et al. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 7.Ho L, Crabtree G. Chromatin remodelling during development. Nature. 2010 doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho L, Ronan JL, Wu J, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci USA. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puri P, Mercola M. BAF60 A, B, and Cs of muscle determination and renewal. Genes Dev. 2012;26:2673–2683. doi: 10.1101/gad.207415.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–253. doi: 10.1016/S1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 11.Lessard J, Crabtree G. Chromatin regulatory mechanisms in pluripotency. Annu Rev Cell Dev Biol. 2010;26:503–532. doi: 10.1146/annurev-cellbio-051809-102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singhal N, Graumann J, Wu G, et al. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell. 2010;141:943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- 13.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 14.Azuara V, Perry P, Sauer S et al (2016) Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8:532–8. doi:10.1038/ncb1403 [DOI] [PubMed]
- 15.Boyer L, Plath K, Zeitlinger J et al (2016) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441:349–53. doi:10.1038/nature04733 [DOI] [PubMed]
- 16.Saladi SV, de la Serna IL. ATP dependent chromatin remodeling enzymes in embryonic stem cells. Stem Cell Rev. 2010;6:62–73. doi: 10.1007/s12015-010-9120-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J, Chu J, Shen X, et al. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho L, Jothi R, Ronan JL, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci USA. 2009;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albini S, Puri P. SWI/SNF complexes, chromatin remodeling and skeletal myogenesis: it’s time to exchange! Exp Cell Res. 2010;316:30733080. doi: 10.1016/j.yexcr.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hainer SJ, Gu W, Carone BR, et al. Suppression of pervasive noncoding transcription in embryonic stem cells by esBAF. Genes Dev. 2015;29:362–378. doi: 10.1101/gad.253534.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai W, Albini S, Wei K, et al. Coordinate nodal and BMP inhibition directs Baf60c-dependent cardiomyocyte commitment. Genes Dev. 2013;27:2332–2344. doi: 10.1101/gad.225144.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lickert H, Takeuchi JK, Von Both I, et al. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 24.Albini S, Coutinho P, Malecova B, et al. Epigenetic reprogramming of human embryonic stem cells into skeletal muscle cells and generation of contractile myospheres. Cell Rep. 2013;3:661–670. doi: 10.1016/j.celrep.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao X, Tate P, Hu P, et al. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci USA. 2008;105:6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lessard J, Wu JI, Ranish JA, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu JI, Lessard J, Olave IA, et al. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 28.Yoo A, Staahl B, Chen L, Crabtree G. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009 doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaniel C, Ang Y-SS, Ratnakumar K, et al. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells. 2009;27:2979–2991. doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho L, Miller EL, Ronan JL, et al. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol. 2011;13:903–913. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puri Sartorelli. Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J Cell Physiol. 2000 doi: 10.1002/1097-4652(200011)185:2<155::AID-JCP1>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 32.Zammit PS, Golding JP, Nagata Y, et al. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J Cell Biol. 2004;166:347–357. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lassar AB, Davis RL, Wright WE, et al. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-E. [DOI] [PubMed] [Google Scholar]
- 34.Kathiriya IS, Nora EPP, Bruneau BG. Investigating the transcriptional control of cardiovascular development. Circ Res. 2015;116:700–714. doi: 10.1161/CIRCRESAHA.116.302832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serna I, Ohkawa Y, Berkes C, et al. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol Cell Biol. 2005;25:3997–4009. doi: 10.1128/MCB.25.10.3997-4009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berkes CA, Bergstrom DA, Penn BH, et al. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell. 2004;14:465–477. doi: 10.1016/S1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
- 37.Simone C, Forcales SV, Hill DA, et al. p38 pathway targets SWI–SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet. 2004;36:738–743. doi: 10.1038/ng1378. [DOI] [PubMed] [Google Scholar]
- 38.Forcales SV, Albini S, Giordani L, et al. Signal-dependent incorporation of MyoD–BAF60c into Brg1-based SWI/SNF chromatin-remodelling complex. EMBO J. 2012;31:301–316. doi: 10.1038/emboj.2011.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weintraub H, Tapscott SJ, Davis RL, et al. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci USA. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D’Aniello C, Fiorenzano A, Iaconis S, et al. The G-protein-coupled receptor APJ is expressed in the second heart field and regulates Cerberus-Baf60c axis in embryonic stem cell cardiomyogenesis. Cardiovasc Res. 2013;100:95–104. doi: 10.1093/cvr/cvt166. [DOI] [PubMed] [Google Scholar]
- 41.Goljanek-Whysall K, Mok GF, Fahad Alrefaei A, et al. myomiR-dependent switching of BAF60 variant incorporation into Brg1 chromatin remodeling complexes during embryo myogenesis. Development. 2014;141:3378–3387. doi: 10.1242/dev.108787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joe AW, Yi L, Natarajan A, et al. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uezumi A, Ito T, Morikawa D, et al. Fibrosis and adipogenesis originate from a common mesenchymal progenitor in skeletal muscle. J Cell Sci. 2011;124:3654–3664. doi: 10.1242/jcs.086629. [DOI] [PubMed] [Google Scholar]
- 44.Saccone V, Consalvi S, Giordani L, et al. HDAC-regulated myomiRs control BAF60 variant exchange and direct the functional phenotype of fibro-adipogenic progenitors in dystrophic muscles. Genes Dev. 2014;28:841–857. doi: 10.1101/gad.234468.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minetti GC, Colussi C, Adami R, et al. Functional and morphological recovery of dystrophic muscles in mice treated with deacetylase inhibitors. Nat Med. 2006;12:1147–1150. doi: 10.1038/nm1479. [DOI] [PubMed] [Google Scholar]
- 46.Consalvi S, Saccone V, Mozzetta C. Histone deacetylase inhibitors: a potential epigenetic treatment for Duchenne muscular dystrophy. Epigenomics. 2014;6:547–560. doi: 10.2217/epi.14.36. [DOI] [PubMed] [Google Scholar]
- 47.Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Troy A, Cadwallader AB, Fedorov Y, et al. Coordination of satellite cell activation and self-renewal by Par-complex-dependent asymmetric activation of p38α/β MAPK. Cell Stem Cell. 2012;11:541–553. doi: 10.1016/j.stem.2012.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones NC, Tyner KJ, Nibarger L, et al. The p38alpha/beta MAPK functions as a molecular switch to activate the quiescent satellite cell. J Cell Biol. 2005;169:105–116. doi: 10.1083/jcb.200408066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price FD, von Maltzahn J, Bentzinger CF, et al. Inhibition of JAK-STAT signaling stimulates adult satellite cell function. Nat Med. 2014;20:1174–1181. doi: 10.1038/nm.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tierney MT, Aydogdu T, Sala D, et al. STAT3 signaling controls satellite cell expansion and skeletal muscle repair. Nat Med. 2014;20:1182–1186. doi: 10.1038/nm.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Padilla-Benavides T, Nasipak BT, Imbalzano AN. Brg1 controls the expression of Pax7 to promote viability and proliferation of mouse primary myoblasts. J Cell Physiol. 2015;230:2990–2997. doi: 10.1002/jcp.25031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell. 2003;11:377–389. doi: 10.1016/S1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 54.Bjornson CRR, Cheung TH, Liu L, et al. Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem Cells. 2012;30:232–242. doi: 10.1002/stem.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mourikis P, Gopalakrishnan S, Sambasivan R, Tajbakhsh S. Cell-autonomous notch activity maintains the temporal specification potential of skeletal muscle stem cells. Development. 2012;139:4536–4548. doi: 10.1242/dev.084756. [DOI] [PubMed] [Google Scholar]
- 56.Mourikis P, Sambasivan R, Castel D, et al. A critical requirement for notch signaling in maintenance of the quiescent skeletal muscle stem cell state. Stem Cells. 2012;30:243–252. doi: 10.1002/stem.775. [DOI] [PubMed] [Google Scholar]
- 57.Albini S, Coutinho Toto P, Dall’Agnese A, et al. Brahma is required for cell cycle arrest and late muscle gene expression during skeletal myogenesis. EMBO Rep. 2015;16:1037–1050. doi: 10.15252/embr.201540159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu M, Wu R, Yang F, et al. Identification of FN1BP1 as a novel cell cycle regulator through modulating G1 checkpoint in human hepatocarcinoma Hep3B cells. PLoS One. 2013;8:e57574. doi: 10.1371/journal.pone.0057574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Juan AH, Derfoul A, Feng X, et al. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 2011;25:789–794. doi: 10.1101/gad.2027911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Palacios D, Mozzetta C, Consalvi S, et al. TNF/p38α/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell. 2010;7:455–469. doi: 10.1016/j.stem.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Serra C, Palacios D, Mozzetta C, et al. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol Cell. 2007;28:200–213. doi: 10.1016/j.molcel.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rampalli S, Li L, Mak E, et al. p38 MAPK signaling regulates recruitment of Ash2L-containing methyltransferase complexes to specific genes during differentiation. Nat Struct Mol Biol. 2007;14:1150–1156. doi: 10.1038/nsmb1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McKinnell IW, Ishibashi J, Le Grand F, et al. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat Cell Biol. 2008;10:77–84. doi: 10.1038/ncb1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kawabe Y-I, Wang YX, McKinnell IW, et al. Carm1 regulates Pax7 transcriptional activity through MLL1/2 recruitment during asymmetric satellite stem cell divisions. Cell Stem Cell. 2012;11:333–345. doi: 10.1016/j.stem.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu L, Cheung TH, Charville GW, et al. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 2013;4:189–204. doi: 10.1016/j.celrep.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dilworth F, Blais A. Epigenetic regulation of satellite cell activation during muscle regeneration. Stem Cell Res Therapy. 2011;2:18. doi: 10.1186/scrt59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sousa-Victor P, Gutarra S, García-Prat L, et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature. 2014;506:316–321. doi: 10.1038/nature13013. [DOI] [PubMed] [Google Scholar]
- 68.Sacco A, Puri PL. Regulation of muscle satellite cell function in tissue homeostasis and aging. Cell Stem Cell. 2015;16:585–587. doi: 10.1016/j.stem.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bultman S, Gebuhr T, Yee D, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000 doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 70.Reyes J, Barra J, Muchardt C, et al. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α) EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gerber A, Klesert T, Bergstrom D, Tapscott S Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev 11:436–50 [DOI] [PubMed]
- 72.Mallappa C, Nasipak B, Etheridge L, et al. Myogenic microRNA expression requires ATP-dependent chromatin remodeling enzyme function. Mol Cell Biol. 2010 doi: 10.1128/MCB.00214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ohkawa Y, Yoshimura S, Higashi C, et al. Myogenin and the SWI/SNF ATPase Brg1 maintain myogenic gene expression at different stages of skeletal myogenesis. J Biol Chem. 2007 doi: 10.1074/jbc.M608898200. [DOI] [PubMed] [Google Scholar]
- 74.Schneider JW, Gu W, Zhu L, et al. Reversal of terminal differentiation mediated by p107 in Rb−/− muscle cells. Science. 1994;264:1467–1471. doi: 10.1126/science.8197461. [DOI] [PubMed] [Google Scholar]
- 75.Novitch BG, Spicer DB, Kim PS et al (1999) pRb is required for MEF2-dependent gene expression as well as cell-cycle arrest during skeletal muscle differentiation. Curr Biol [DOI] [PubMed]
- 76.Sellers W, Novitch B, Miyake S et al Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev 12:95–106 [DOI] [PMC free article] [PubMed]
- 77.Puri PL, Iezzi S, Stiegler P, et al. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol Cell. 2001;8:885–897. doi: 10.1016/S1097-2765(01)00373-2. [DOI] [PubMed] [Google Scholar]
- 78.Strobeck M, Knudsen K, Fribourg A et al BRG-1 is required for RB-mediated cell cycle arrest. Proc Natl Acad Sci USA 97:7748–7753 [DOI] [PMC free article] [PubMed]
- 79.Zhang H, Gavin M, Dahiya A et al Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 101:79–89. doi:10.1016/S0092-8674(00)80625-X [DOI] [PubMed]
- 80.Zhang M, Chen M, Kim J-R, et al. SWI/SNF complexes containing brahma or brahma-related gene 1 play distinct roles in smooth muscle development. Mol Cell Biol. 2011;31:2618–2631. doi: 10.1128/MCB.01338-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Joliot V, Ait-Mohamed O, Battisti V, et al. The SWI/SNF subunit/tumor suppressor BAF47/INI1 is essential in cell cycle arrest upon skeletal muscle terminal differentiation. PLoS One. 2014;9:e108858. doi: 10.1371/journal.pone.0108858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blais A, van Oevelen C, Margueron R, et al. Retinoblastoma tumor suppressor protein–dependent methylation of histone H3 lysine 27 is associated with irreversible cell cycle exit. J Cell Biol. 2007 doi: 10.1083/jcb.200705051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Caretti G, Di Padova M, Micales B, et al. The polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ruijtenberg S, van den Heuvel S. G1/S inhibitors and the SWI/SNF complex control cell-cycle exit during muscle differentiation. Cell. 2015 doi: 10.1016/j.cell.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 85.Wu JI. Diverse functions of ATP-dependent chromatin remodeling complexes in development and cancer. Acta Biochim Biophys Sin (Shanghai) 2012;44:54–69. doi: 10.1093/abbs/gmr099. [DOI] [PubMed] [Google Scholar]