Abstract
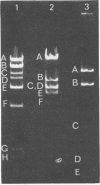
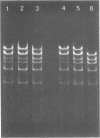
The structure of DNA from the temperate Bacillus subtilis phage phi105 was examined by using the restriction endonuclease EcoRI and by sedimentation analysis. The DNA contains six EcoRI cleavage sites. Although eight DNA fragments were identified in the EcoRI digests, the largest of these was shown to consist of the two fragments that carry the cohesive ends of the phage DNA. In neutral gradients, the majority of whole phi105 DNA sedimented as nicked circles and the remainder as oligomers. No unit-length linear structures were detected. The associated cohesive ends could be sealed by DNA ligase from Escherichia coli and could be cleaved by S1 nuclease. On the basis of these results and previously reported studies, it appears that, as isolated from phage particles, phi105 DNA is a circular molecule that is formed from the linear structure by the association of complementary single-stranded DNA.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armentrout R. W., Rutberg L. Mapping of prophage and mature deoxyribonucleic acid from temperate Bacillus bacteriophage phi 105 by marker rescue. J Virol. 1970 Dec;6(6):760–767. doi: 10.1128/jvi.6.6.760-767.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armentrout R. W., Skoog L., Rutberg L. Structure and biological activity of deoxyribonucleic acid from Bacillus bacteriophage phi 105: effects of Escherichia coli exonucleases. J Virol. 1971 Mar;7(3):359–371. doi: 10.1128/jvi.7.3.359-371.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BURGI E., HERSHEY A. D. Sedimentation rate as a measure of molecular weight of DNA. Biophys J. 1963 Jul;3:309–321. doi: 10.1016/s0006-3495(63)86823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdsell D. C., Hathaway G. M., Rutberg L. Characterization of Temperate Bacillus Bacteriophage phi105. J Virol. 1969 Sep;4(3):264–270. doi: 10.1128/jvi.4.3.264-270.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutlag D., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. 36. A proofreading function for the 3' leads to 5' exonuclease activity in deoxyribonucleic acid polymerases. J Biol Chem. 1972 Jan 10;247(1):241–248. [PubMed] [Google Scholar]
- Chow L. T., Boice L., Davidson N. Map of the partial sequence homology between DNA molecules of Bacillus subtilis bacteriophages SPO2 and phi105. J Mol Biol. 1972 Jul 28;68(3):391–400. doi: 10.1016/0022-2836(72)90093-9. [DOI] [PubMed] [Google Scholar]
- Chow L. T., Davidson N. Electron microscope study of the structures of the Bacillus subtilis prophages, SPO2 and phi105. J Mol Biol. 1973 Apr 5;75(2):257–264. doi: 10.1016/0022-2836(73)90019-3. [DOI] [PubMed] [Google Scholar]
- Couturier M. The integration and excision of the bacteriophage Mu-1. Cell. 1976 Feb;7(2):155–163. doi: 10.1016/0092-8674(76)90015-5. [DOI] [PubMed] [Google Scholar]
- Dean D. H., Arnaud M., Halvorson H. O. Genetic evidence that Bacillus bacteriophage phi 105 integrates by insertion. J Virol. 1976 Oct;20(1):339–341. doi: 10.1128/jvi.20.1.339-341.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donelson J. E., Wu R. Nucleotide sequence analysis of deoxyribonucleic acid. VII. Characterization of Escherichia coli exonuclease 3 activity for possible use in terminal nucleotide sequence analysis of duplex deoxyribonucleic acid. J Biol Chem. 1972 Jul 25;247(14):4661–4668. [PubMed] [Google Scholar]
- Garro A. J. Isolation and properties of Bacillus subtilis strains lysogenized by a clear plaque mutant of bacteriophage phi 105. J Virol. 1973 Jul;12(1):13–17. doi: 10.1128/jvi.12.1.13-17.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgpeth J., Goodman H. M., Boyer H. W. DNA nucleotide sequence restricted by the RI endonuclease. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3448–3452. doi: 10.1073/pnas.69.11.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling R. B., Goodman H. M., Boyer H. W. Analysis of endonuclease R-EcoRI fragments of DNA from lambdoid bacteriophages and other viruses by agarose-gel electrophoresis. J Virol. 1974 Nov;14(5):1235–1244. doi: 10.1128/jvi.14.5.1235-1244.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J., Kawamura F., Yanofsky S. Analysis of phi 29 and phi 15 genomes by bacterial restriction endonucleases, EcoR1 and Hpal. Virology. 1976 Mar;70(1):37–51. doi: 10.1016/0042-6822(76)90234-8. [DOI] [PubMed] [Google Scholar]
- Masamune Y., Fleischman R. A., Richardson C. C. Enzymatic removal and replacement of nucleotides at single strand breaks in deoxyribonucleic acid. J Biol Chem. 1971 Apr 25;246(8):2680–2691. [PubMed] [Google Scholar]
- Modrich P., Anraku Y., Lehman I. R. Deoxyribonucleic acid ligase. Isolation and physical characterization of the homogeneous enzyme from Escherichia coli. J Biol Chem. 1973 Nov 10;248(21):7495–7501. [PubMed] [Google Scholar]
- Moore S. K., James E. Purification and electrophoretic assay of T4-induced polynucleotide ligase for the in vitro construction of recombinant DNA molecules. Anal Biochem. 1976 Oct;75(2):545–554. doi: 10.1016/0003-2697(76)90109-3. [DOI] [PubMed] [Google Scholar]
- Polisky B., Greene P., Garfin D. E., McCarthy B. J., Goodman H. M., Boyer H. W. Specificity of substrate recognition by the EcoRI restriction endonuclease. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3310–3314. doi: 10.1073/pnas.72.9.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzaki T., Bukhari A. I. Events following prophage Mu induction. J Bacteriol. 1975 May;122(2):437–442. doi: 10.1128/jb.122.2.437-442.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A. J., Bellett J. D. A circular DNA-protein complex adenoviruses and its possible role in DNA replication. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):523–531. doi: 10.1101/sqb.1974.039.01.064. [DOI] [PubMed] [Google Scholar]
- Rutberg L., Armentrout R. W. Low-frequency rescue of a genetic marker in deoxyribonucleic acid from Bacillus bacteriophage phi 105 by superinfecting bacteriophage. J Virol. 1970 Dec;6(6):768–771. doi: 10.1128/jvi.6.6.768-771.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutberg L. Heat induction of prophage phi 105 in Bacillus subtilis: bacteriophage-induced bidirectional replication of the bacterial chromosome. J Virol. 1973 Jul;12(1):9–12. doi: 10.1128/jvi.12.1.9-12.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STUDIER F. W. SEDIMENTATION STUDIES OF THE SIZE AND SHAPE OF DNA. J Mol Biol. 1965 Feb;11:373–390. doi: 10.1016/s0022-2836(65)80064-x. [DOI] [PubMed] [Google Scholar]
- Sgaramella V. Enzymatic oligomerization of bacteriophage P22 DNA and of linear Simian virus 40 DNA. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3389–3393. doi: 10.1073/pnas.69.11.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A., Dean D. H., Halvorson H. O. Low-frequency specialized transduction with Bacillus subtilis bacteriophage phi 105. Virology. 1974 Dec;62(2):393–403. doi: 10.1016/0042-6822(74)90401-2. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Shenk T. E., Rhodes C., Rigby P. W., Berg P. Biochemical method for mapping mutational alterations in DNA with S1 nuclease: the location of deletions and temperature-sensitive mutations in simian virus 40. Proc Natl Acad Sci U S A. 1975 Mar;72(3):989–993. doi: 10.1073/pnas.72.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Thuring R. W., Sanders J. P., Borst P. A freeze-squeeze method for recovering long DNA from agarose gels. Anal Biochem. 1975 May 26;66(1):213–220. doi: 10.1016/0003-2697(75)90739-3. [DOI] [PubMed] [Google Scholar]
- Thurm P., Garro A. J. Isolation and characterization of prophage mutants of the defective Bacillus subtilis bacteriophage PBSX. J Virol. 1975 Jul;16(1):184–191. doi: 10.1128/jvi.16.1.184-191.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J., Radloff R., Watson R., Laipis P. The twisted circular form of polyoma viral DNA. Proc Natl Acad Sci U S A. 1965 May;53(5):1104–1111. doi: 10.1073/pnas.53.5.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt V. M. Purification and further properties of single-strand-specific nuclease from Aspergillus oryzae. Eur J Biochem. 1973 Feb 15;33(1):192–200. doi: 10.1111/j.1432-1033.1973.tb02669.x. [DOI] [PubMed] [Google Scholar]
- Wilson G. A., Williams M. T., Baney H. W., Young F. E. Characterization of temperate bacteriophages of Bacillus subtilis by the restriction endonuclease EcoRI: evidence for three different temperate bacteriophages. J Virol. 1974 Oct;14(4):1013–1016. doi: 10.1128/jvi.14.4.1013-1016.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]