Abstract
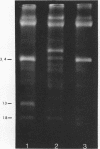
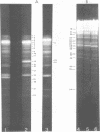
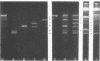
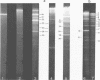
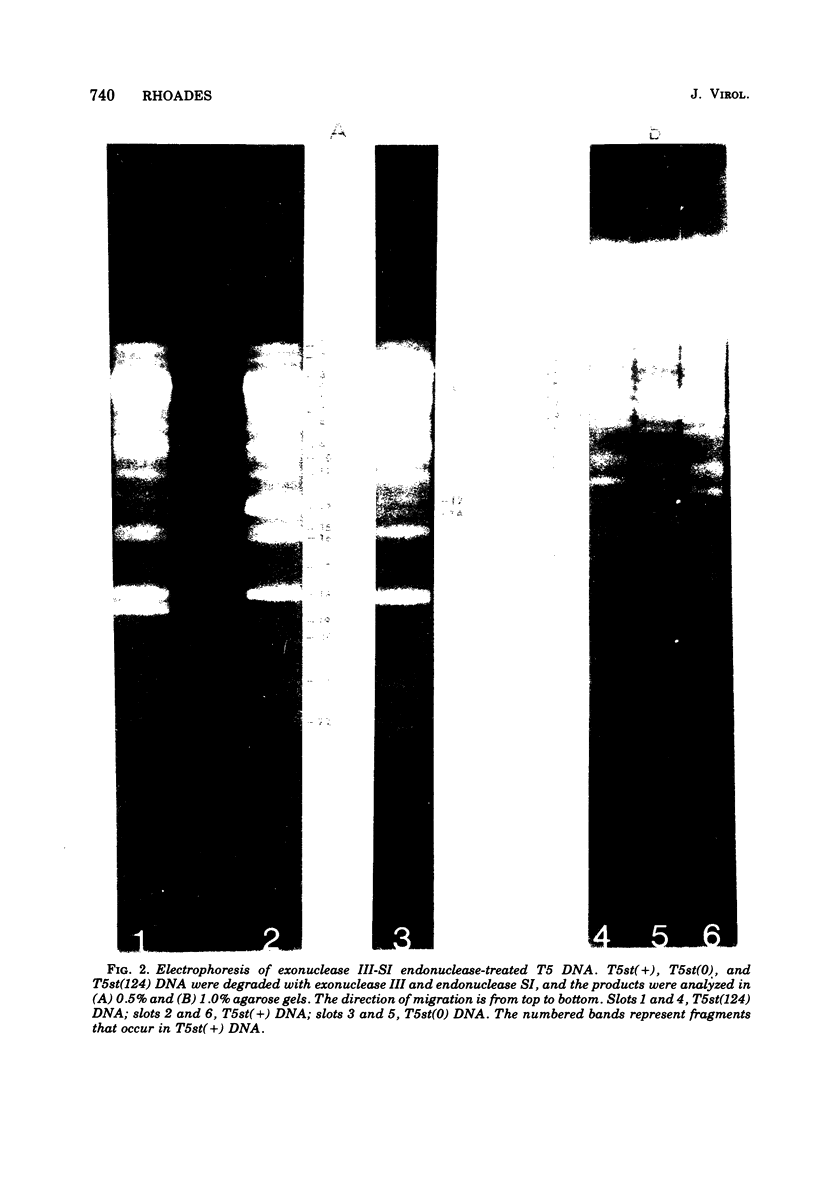
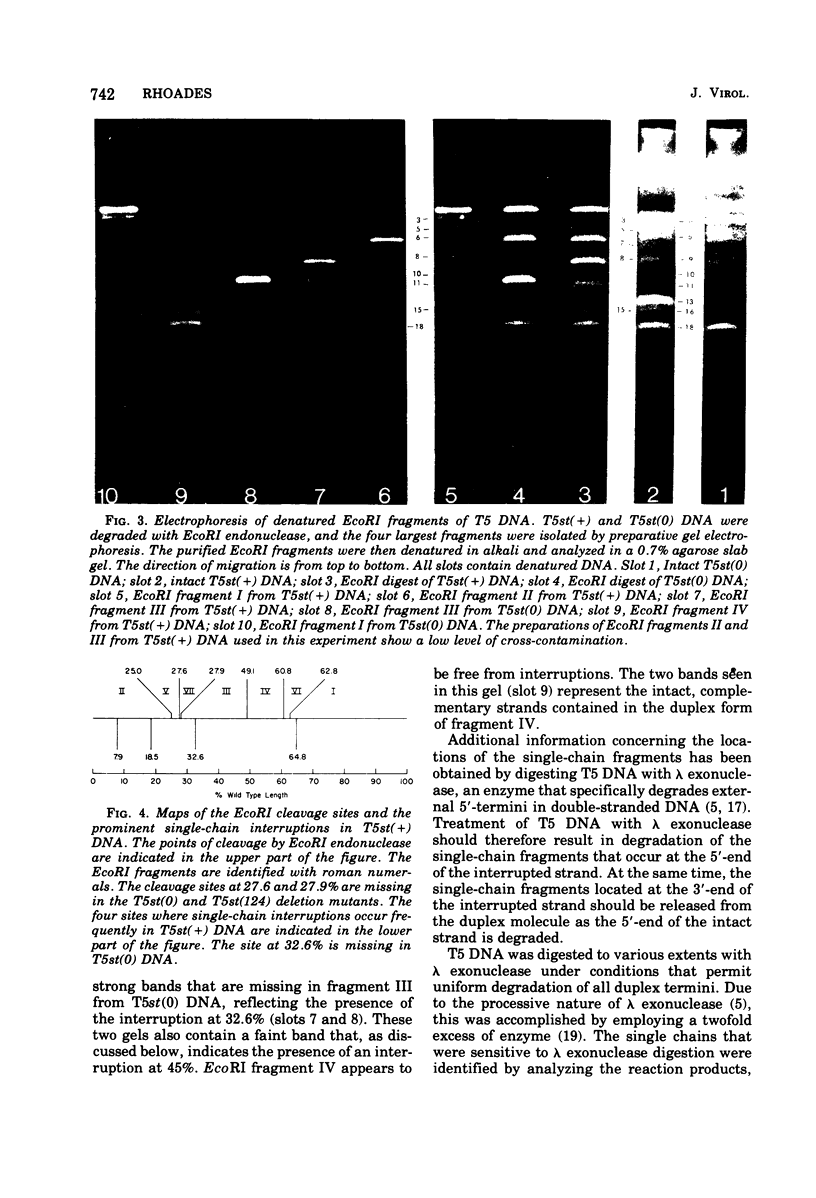
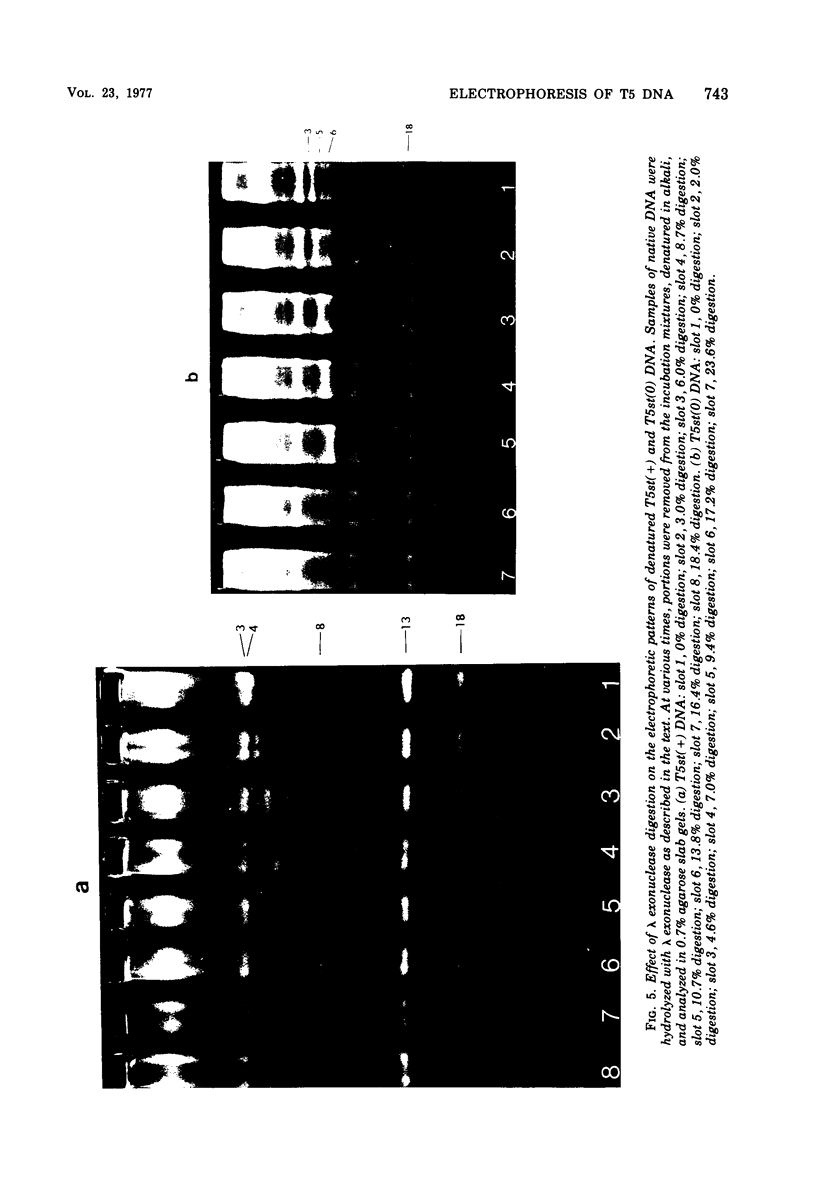
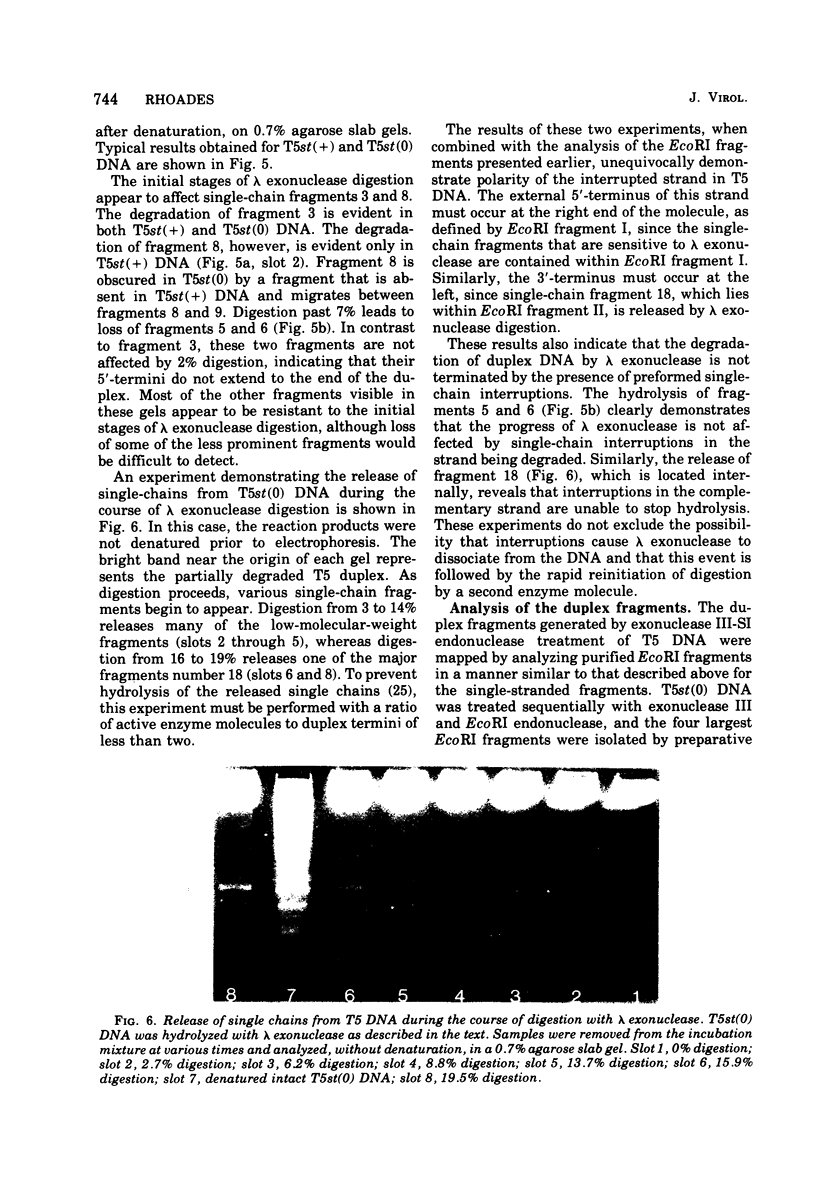
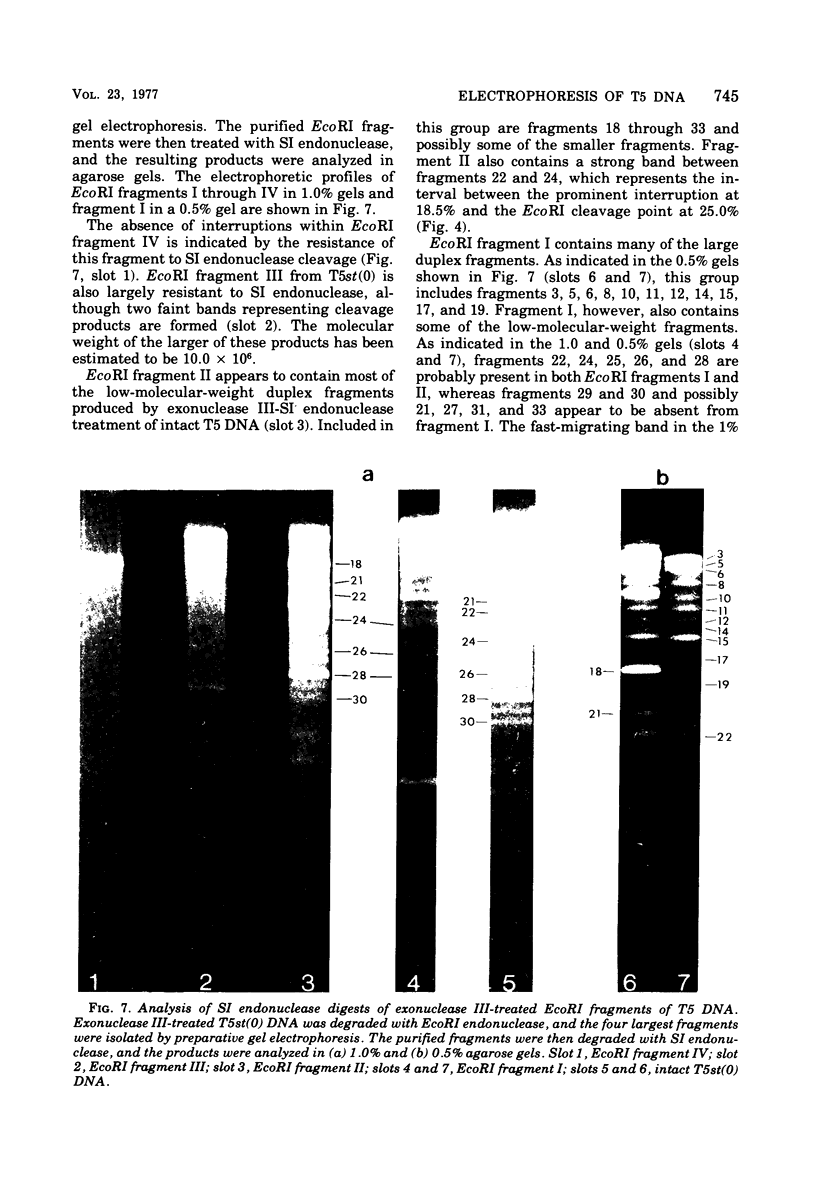
Upon denaturation, T5 DNA yields a large number of discrete, single-chain fragments that can be resolved by agarose gel electrophoresis. The positions of the more prominent of these fragments in the T5 duplex were determined by analyzing their sensitivity to digestion with λ exonuclease and their distribution among EcoRI fragments of T5 DNA. These experiments also provide firm evidence concerning the polarity of the strands in T5 DNA. An analogous study was carried out on the fragments produced by treating exonuclease III-degraded T5 DNA with the single-strand-specific SI endonuclease. This procedure yielded over 40 discrete duplex fragments that could be resolved with considerable precision by agarose gel electrophoresis. The positions of most of these fragments were determined by analyzing EcoRI fragments of T5st(+) and T5st(0) DNA. Over 20 sites where single-chain interruptions can occur in T5 DNA were identified, and the distribution of interruptions within the terminal repetition was shown to be identical at both ends of the molecule. A precise value for the size of the terminal repetition in T5 DNA was obtained by analyzing SI endonuclease digests of ligase-repaired, circular T5 DNA in agarose gels. The repeated segment represented 8.3% of the T5st(+) DNA. The results of this study also provide information concerning the properties of λ exonuclease. Hydrolysis by this enzyme was not terminated when single-chain interruptions were encountered either in the strand being degraded or in the complementary strand.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allet B., Jeppesen P. G., Katagiri K. J., Delius H. Mapping the DNA fragments produced by cleavage by lambda DNA with endonuclease RI. Nature. 1973 Jan 12;241(5385):120–123. doi: 10.1038/241120a0. [DOI] [PubMed] [Google Scholar]
- Bourguignon G. J., Sweeney T. K., Delius H. Multiple origins and circular structures in replicating T5 bacteriophage DNA. J Virol. 1976 Apr;18(1):245–259. doi: 10.1128/jvi.18.1.245-259.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujard H. Location of single-strand interruptions in the DNA of bacteriophage T5. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1167–1174. doi: 10.1073/pnas.62.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter D. M., Radding C. M. The role of exonuclease and beta protein of phage lambda in genetic recombination. II. Substrate specificity and the mode of action of lambda exonuclease. J Biol Chem. 1971 Apr 25;246(8):2502–2512. [PubMed] [Google Scholar]
- FATTIG W. D., LANNI F. MAPPING OF TEMPERATURE-SENSITIVE MUTANTS IN BACTERIOPHAGE T5. Genetics. 1965 Jan;51:157–166. doi: 10.1093/genetics/51.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W., Dressler D. DNA replication: the rolling circle model. Cold Spring Harb Symp Quant Biol. 1968;33:473–484. doi: 10.1101/sqb.1968.033.01.055. [DOI] [PubMed] [Google Scholar]
- Hayward G. S. Gel electrophoretic separation of the complementary strands of bacteriophage DNA. Virology. 1972 Jul;49(1):342–344. doi: 10.1016/s0042-6822(72)80042-4. [DOI] [PubMed] [Google Scholar]
- Hayward G. S., Smith M. G. The chromosome of bacteriophage T5. I. Analysis of the single-stranded DNA fragments by agarose gel electrophoresis. J Mol Biol. 1972 Feb 14;63(3):383–395. doi: 10.1016/0022-2836(72)90435-4. [DOI] [PubMed] [Google Scholar]
- Hayward G. S. Unique double-stranded fragments of bacteriophage T5 DNA resulting from preferential shear-induced breakage at nicks. Proc Natl Acad Sci U S A. 1974 May;71(5):2108–2112. doi: 10.1073/pnas.71.5.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward S. D., Smith M. G. The chromosome of bacteriophage T5. 3. Patterns of transcription from the single-stranded DNA fragments. J Mol Biol. 1973 Oct 25;80(2):345–359. doi: 10.1016/0022-2836(73)90177-0. [DOI] [PubMed] [Google Scholar]
- Hendrickson H. E., Bujard H. Structure and function of the genome of coliphage T5. 2. Regions of transcription of the chromosome. Eur J Biochem. 1973 Mar 15;33(3):529–534. doi: 10.1111/j.1432-1033.1973.tb02712.x. [DOI] [PubMed] [Google Scholar]
- Hendrickson H. E., McCorquodale D. J. Genetic and physiological studies of bacteriophage t5 I. An expanded genetic map of t5. J Virol. 1971 May;7(5):612–618. doi: 10.1128/jvi.7.5.612-618.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemin-Sablon A., Richardson C. C. Analysis of the interruptions in bacteriophage T5 DNA. J Mol Biol. 1970 Feb 14;47(3):477–493. doi: 10.1016/0022-2836(70)90316-5. [DOI] [PubMed] [Google Scholar]
- Knopf K., Bujard H. Structure and function of the genome of coliphage T5. Transcription in vitro of the "nicked" and "nick-free" T5+ DNA. Eur J Biochem. 1975 May 6;53(2):371–385. doi: 10.1111/j.1432-1033.1975.tb04077.x. [DOI] [PubMed] [Google Scholar]
- Lanni Y. T. First-step-transfer deoxyribonucleic acid of bacteriophage T5. Bacteriol Rev. 1968 Sep;32(3):227–242. doi: 10.1128/br.32.3.227-242.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masamune Y., Fleischman R. A., Richardson C. C. Enzymatic removal and replacement of nucleotides at single strand breaks in deoxyribonucleic acid. J Biol Chem. 1971 Apr 25;246(8):2680–2691. [PubMed] [Google Scholar]
- Rhoades M. Cleavage of T5 DNA by the Escherichia coli R-I restriction endonuclease. Virology. 1975 Mar;64(1):170–179. doi: 10.1016/0042-6822(75)90089-6. [DOI] [PubMed] [Google Scholar]
- Rhoades M., Rhoades E. A. Terminal repetition in the DNA of bacteriophage T5. J Mol Biol. 1972 Aug 21;69(2):187–200. doi: 10.1016/0022-2836(72)90224-0. [DOI] [PubMed] [Google Scholar]
- Rogers S. G., Rhoades M. Bacteriophage T5-induced endonucleases that introduce site-specific single-chain interruptions in duplex DNA. Proc Natl Acad Sci U S A. 1976 May;73(5):1576–1580. doi: 10.1073/pnas.73.5.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible P. P., Rhoades E. A., Rhoades M. Localization of single-chain interruptions in bacteriophage T5 DNA I. Electron microscopic studies. J Virol. 1977 Sep;23(3):725–736. doi: 10.1128/jvi.23.3.725-736.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible P. P., Rhoades M. Heteroduplex mapping of heat-resistant deletion mutants of bacteriophage t5. J Virol. 1975 May;15(5):1276–1280. doi: 10.1128/jvi.15.5.1276-1280.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Shenk T. E., Rhodes C., Rigby P. W., Berg P. Biochemical method for mapping mutational alterations in DNA with S1 nuclease: the location of deletions and temperature-sensitive mutations in simian virus 40. Proc Natl Acad Sci U S A. 1975 Mar;72(3):989–993. doi: 10.1073/pnas.72.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriprakash K. S., Lundh N., Huh MM-O, Radding C. M. The specificity of lambda exonuclease. Interactions with single-stranded DNA. J Biol Chem. 1975 Jul 25;250(14):5438–5445. [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Vogt V. M. Purification and further properties of single-strand-specific nuclease from Aspergillus oryzae. Eur J Biochem. 1973 Feb 15;33(1):192–200. doi: 10.1111/j.1432-1033.1973.tb02669.x. [DOI] [PubMed] [Google Scholar]