Summary
CC chemokine receptor 2 (CCR2) is one of 19 members of the chemokine receptor subfamily of human Class A G protein-coupled receptors (GPCRs). CCR2 is expressed on monocytes, immature dendritic cells and T cell subpopulations, and mediates their migration towards endogenous CC chemokine ligands such as CCL21. CCR2 and its ligands are implicated in numerous inflammatory and neurodegenerative diseases2 including atherosclerosis, multiple sclerosis, asthma, neuropathic pain, and diabetic nephropathy, as well as cancer3. These disease associations have motivated numerous preclinical studies and clinical trials4 (see ClinicalTrials.gov) in search of therapies that target the CCR2:chemokine axis. To aid drug discovery efforts5, we solved a structure of CCR2 in a ternary complex with an orthosteric (BMS-6816) and allosteric (CCR2-RA-[R]7) antagonist. BMS-681 inhibits chemokine binding by occupying the orthosteric pocket of the receptor in a previously unseen binding mode. CCR2-RA-[R] binds in a novel, highly druggable pocket that is the most intracellular allosteric site observed in Class A GPCRs to date; this site spatially overlaps the G protein-binding site in homologous receptors. CCR2-RA-[R] inhibits CCR2 non-competitively by blocking activation-associated conformational changes and formation of the G protein-binding interface. The conformational signature of the conserved microswitch residues observed in double-antagonist-bound CCR2 resembles the most inactive GPCR structures solved to date. Like other protein:protein interactions, receptor:chemokine complexes are considered challenging therapeutic targets for small molecules, and the present structure suggests diverse pocket epitopes that can be exploited to overcome drug design obstacles.
Main
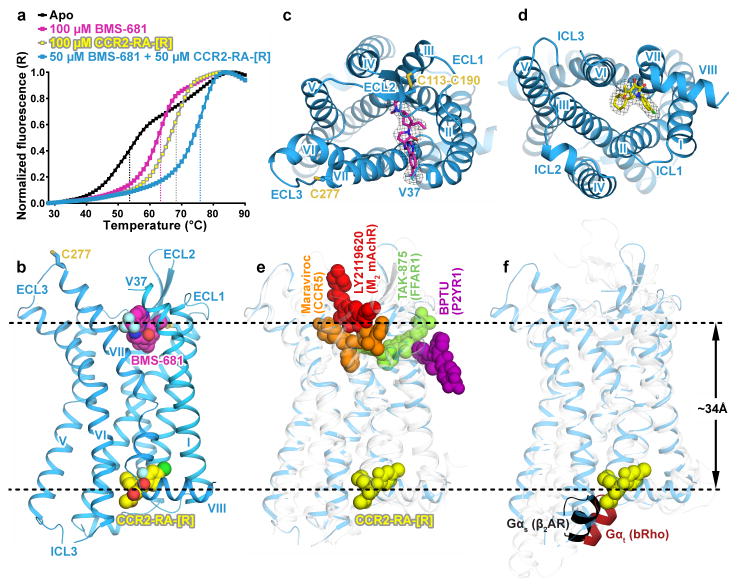
A ternary complex between an engineered construct of human CCR2 isoform b (further referred to as CCR2-T4L or simply CCR2), an orthosteric antagonist BMS-681 (compound 13d in6), and an allosteric antagonist CCR2-RA-[R]7 was crystallized using the lipidic cubic phase method8, and the structure was determined to 2.8 Å resolution (Extended Data Table 1 and Extended Data Fig. 1). Simultaneous addition of two compounds dramatically stabilized detergent-solubilized CCR2-T4L in comparison with twice the concentration of each compound individually (Fig. 1a), suggesting concurrent binding of CCR2-RA-[R] and BMS-681 to the receptor. The presence of both compounds was critical for crystallization.
Figure 1. Structure of the CCR2:BMS-681:CCR2-RA-[R] complex and comparison to other allosteric modulators of class A GPCRs.
a, Thermal denaturation curves demonstrate higher stability of CCR2-T4L in the presence of both BMS-681 and CCR2-RA-[R] compared to each compound individually. Data is representative of three independent experiments conducted on different days. b, Overall view of double-antagonist-bound CCR2. c, d, Structure viewed from the extracellular (c) and intracellular (d) side with simulated annealing omit maps of BMS-681 (c) and CCR2-RA-[R] (d) shown at 3σ. e, CCR2-RA-[R] compared to other allosteric ligands crystallized with Class A GPCRs (PDBIDs 4mbs, 4xnv, 4phu, and 4mqt). f, CCR2-RA-[R] compared with the C-terminal helix of Gαs bound to the β2 adrenergic receptor and transducin peptide bound to rhodopsin (PDBIDs 3sn6 and 4x1h).
In the structure, CCR2 adopts the canonical fold of class A GPCRs with seven transmembrane (TM) helices connected by three extracellular (EC) and three intracellular (IC) loops (Fig. 1b). Both compounds are visible (Fig. 1b–d); BMS-681 binds in the extracellular orthosteric pocket (Fig. 1b, c) while CCR2-RA-[R] is located more than 30Å away (Fig. 1b, d), in a site that is the most intracellular allosteric pocket observed in Class A GPCRs to date (Fig 1e). The binding site of CCR2-RA-[R] spatially overlaps with the G protein-binding site in homologous receptors (Fig 1f). As for other chemokine receptors9–12, CCR2 is expected to have two conserved disulfide bonds in its extracellular domains, with Cys32-Cys277 connecting the N-terminus (NT) to ECL3, and Cys113-Cys190 connecting TM3 to ECL2. Electron density is apparent for the ECL2-TM3 disulfide bond but not for the NT residues 1–36 or the NT-ECL3 disulfide bond (Fig. 1b, c). Because the NT-ECL3 disulfide bond has been shown important for CCR2 signaling13, its absence is unlikely to be an inherent feature of the receptor; instead, it might be caused by strain of the bond in the ligand-bound state of the receptor14, possibly exacerbated by solvent exposure and radiation damage of the crystals15.
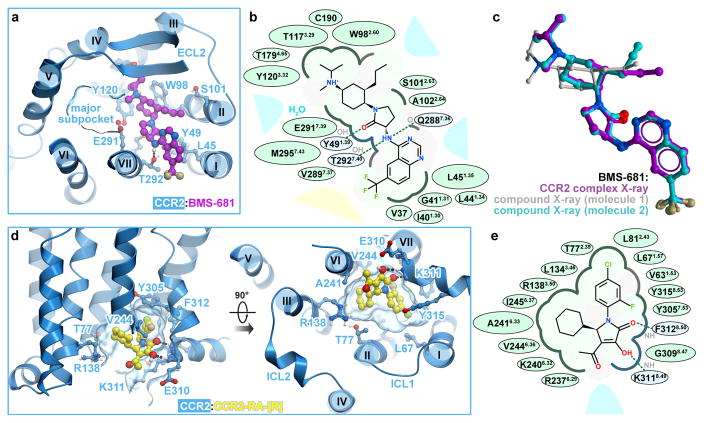
As with other chemokine receptors, the extracellular orthosteric pocket of CCR2 can be divided into a major and a minor subpocket, defined by helices III–VII and helices I–III, VII, respectively, and separated by residues Y1203.32 and E2917.39 (superscript indicates residue number as per Ballesteros-Weinstein nomenclature). BMS-681 binds predominantly in the minor subpocket (Fig. 2a,b) and buries 366.3 Å2 of surface area. The 6-trifluoromethyl quinazoline moiety protrudes between helices I and VII towards the lipid bilayer, while the tri-substituted cyclohexane packs against W982.60. The γ-lactam secondary exocyclic amine forms a hydrogen bond with the hydroxyl of T2927.40, which is critical for binding of chemically related compounds such as BMS-558 (compound 22 in 16) and the Teijin lead series17,18. This amine is also within hydrogen bonding distance from the backbone carbonyl of Q2887.36. The carbonyl oxygen of the γ-lactam forms a hydrogen bond with Y491.39, which itself is hydrogen-bonded to the side chain of T2927.40. The N1 nitrogen of the quinazoline is within 4 Å of the Q2887.36 side chain. The protonated tertiary amine on the cyclohexane ring is proximal to a structured water molecule in the binding site. Some CCR2 antagonists, particularly those containing a basic amine, are known to depend on the conserved E2917.39 in the receptor19; however, no direct interaction is observed between E2917.39 and BMS-681. The receptor-bound, bioactive conformation of BMS-681 is strikingly similar to the crystallographic conformation of free BMS-681 (Fig. 2c, Extended Data Table 2) suggesting the absence of internal strain in the bound state.
Figure 2. Ligand binding sites and receptor interactions.
a,b BMS-681 interactions with CCR2 viewed in 3D from the extracellular side (a) and in a 2D schematic depiction (b). c, Of the two conformers in the free BMS-681 structure, one is almost identical to the CCR2-complexed conformation. d,e CCR2-RA-[R] interactions viewed in 3D along the plane of the membrane and from the intracellular side (d), and in a 2D depiction (e). In (b, e), polar and non-polar residue contacts are shown as blue and green, respectively. Bulk solvent and lipid are represented by blue and yellow shading, respectively.
BMS-681 engages several residues that are critical for CCL2 binding and/or activation of CCR217,18 including Y491.39, W982.60, Y1203.32, and T2927.40. Thus, it seems to directly compete with chemokine binding to the orthosteric pocket. Additionally, by inserting between helices I and VII, BMS-681 may put strain onto residues C32-V37 connecting TM1 to ECL3, destabilize the conserved NT-ECL3 disulfide bond (absent in the structure), and prevent the N-terminus and TM1 from adopting a productive chemokine binding conformation observed in homologous receptor:chemokine structures11,12 (Extended Data Fig. 2).
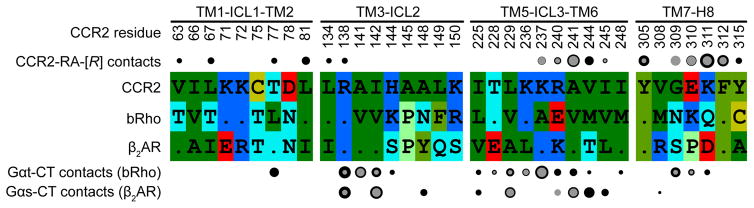
On the opposite side of the receptor, CCR2-RA-[R] is caged by the intracellular ends of helices I–III and VI–VIII and buries 297.8 Å2 of surface area. The inner hydrophobic part of the cage is made by V631.53, L671.57, L812.43, L1343.46, A2416.33, V2446.36, I2456.37, Y3057.53, and F3128.50, while the outer (cytosol-facing) polar part consists of T772.39, R1383.50, G3098.47, K3118.49, and Y3158.53 (Fig. 2d, e), as well as the backbones of engineered R2376.29 and K2406.32. The binding pocket of CCR2-RA-[R] is highly enclosed and possesses a balanced combination of hydrophobic and polar features, all of which favors pocket “druggability”5. Owing to the lack of a side-chain on G3098.47, the hydroxyl and pyrrolone carbonyl groups of CCR2-RA-[R] can hydrogen-bond to the exposed backbone amides of E3108.48, K3118.49, and F3128.50 (Fig. 2d, e). The acetyl group of the compound resides near the terminal amine of K3118.49. The critical roles of V2446.36, Y3057.53, K3118.49, and F3128.50 in CCR2-RA-[R] binding were established by an earlier mutagenesis study20. Because homologs of several residues in the CCR2-RA-[R] binding pocket directly couple to the G protein in bovine rhodopsin21 and the β2 adrenergic receptor (β2AR)22 structures (Extended Data Fig. 3), CCR2-RA-[R] appears to sterically interfere with G protein binding to CCR2.
The structure suggests an interesting symmetric mechanism for the concurrent antagonistic action of the two compounds. BMS-681 interferes with chemokine binding directly and with G protein coupling indirectly, by stabilizing an inactive, presumably G protein-incompatible6 conformation of the receptor. Conversely, CCR2-RA-[R] directly prevents G protein coupling and allosterically inhibits binding of the CCL2 chemokine23, which, like most GPCR agonists, requires an active, G protein-associated receptor for high affinity binding23. Bi-directional allosteric communication between the extra- and intracellular sides of the receptor is reminiscent of that previously observed in adenosine A2A receptor (AA2AR)24 and β2AR25 using allosteric inverse agonist antibodies/nanobodies that target the same epitope as CCR2-RA-[R]. Similar to these antibodies, CCR2-RA-[R] was previously shown to allosterically enhance, and to be allosterically enhanced by, binding of orthosteric antagonists23, i.e. to demonstrate positive binding cooperativity.
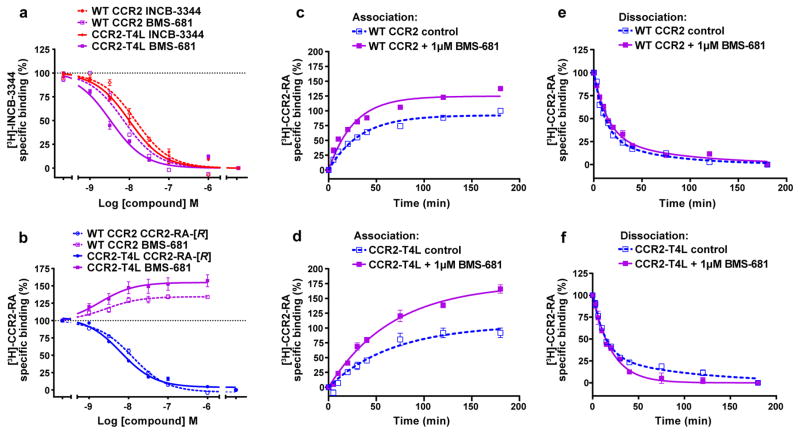
We further characterized this cooperativity by studying the binding of BMS-681 to WT CCR2 and the crystallization construct CCR2-T4L using previously characterized radioactive probes [3H]-INCB-334423 (orthosteric) and [3H]-CCR2-RA (allosteric). In equilibrium competition binding assays on WT CCR2, both INCB-3344 and CCR2-RA-[R] displaced their homologous radioligand with IC50 values of 17 and 13 nM, respectively (Extended Data Fig. 4a, b and Extended Data Table 3), comparable to previously reported values23. Compared to WT CCR2, the affinity of both antagonists towards CCR2-T4L was improved by approximately 2-fold, suggesting a slight engineering-related shift towards the inactive state. BMS-681 fully displaced [3H]-INCB-3344 with nanomolar affinities for both constructs, but did not displace [3H]-CCR2-RA. Instead, at 1 μM concentration it enhanced the binding of [3H]-CCR2-RA by >30% (Extended Data Fig. 4a, b and Extended Data Table 3).
In kinetic radioligand experiments, the presence of BMS-681 also increased total binding of [3H]-CCR2-RA to both WT CCR2 and CCR2-T4L, with the increase as high as 62% in the case of CCR2-T4L (Extended Data Fig. 4c–d, and Extended Data Table 4). 1μM BMS-681 decreased the dissociation rate constant of [3H]-CCR2-RA, while producing a slight increase (WT CCR2) or no change (CCR2-T4L) in the observed association rate constants. Moreover, in CCR2-T4L, the presence of BMS-681 changed the biphasic dissociation profile of [3H]-CCR2-RA to monophasic, suggesting stabilization of the receptor population in a homogenous conformational state (Extended Data Table 4). Along with the stability and equilibrium binding data, these results further corroborate the hypothesis that BMS-681 and CCR2-RA-[R] cooperatively stabilize a preferred inactive conformation of CCR2-T4L.
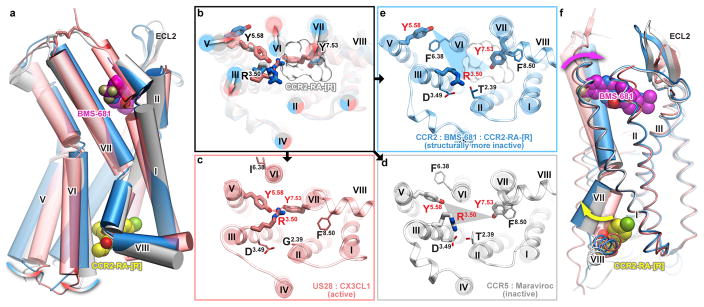
We next analyzed the structure of double-antagonist-bound CCR2-T4L to better understand this conformation. The plethora of existing class A GPCR structures suggests a conserved conformational signature of an active receptor state26. This signature involves separation between the intracellular end of helix VI and the rest of the TM bundle, an inward repositioning and rotation of helix VII, and concerted repacking of the highly conserved microswitches R3.50 (of the DR3.50Y motif), Y5.58, and Y7.53 (of the NPxxY7.53 motif) (Fig. 3a, b) to form an intracellular binding interface for G protein. Furthermore, rather than adopting either an “on” or “off” state, receptors can occupy an ensemble of intermediate conformations27. The active state signature is fully represented in US28, the only agonist-bound chemokine receptor crystallized to date12 (Fig. 3a–c). By contrast, the double-antagonist-bound CCR2 structure appears to occupy the opposite end of the activation spectrum as it shares the conformational microswitch signatures of the most inactive GPCR structures observed thus far (Fig. 3a–e).
Figure 3. The crystallographic conformation of double antagonist bound CCR2 has pronounced structural signatures of an inactive state.
Structures of active (US28, salmon), inactive (CCR5, grey), and apparently more inactive (CCR2, blue) chemokine receptors viewed along the plane of the membrane (a) and across the membrane from the intracellular side (b–e). b, Overlay of structures; arrows show the direction of activation-associated conformational changes; sticks show conserved Y5.58, R3.50 and Y7.53; the white mesh is CCR2-RA-[R]. c–e, Detailed, single-receptor depictions of (b). (f) Although located ~30 Å apart, the orthosteric (BMS-681, magenta) and the allosteric (CCR2-RA-[R], yellow) ligands cooperate in stabilizing an inactive conformation of CCR2 through helix VII.
As in the inactive CCR5:Maraviroc complex10, the intracellular ends of CCR2 helices III and VI are close together, and the conserved R3.50 interacts with D3.49 and T2.39, effectively disrupting the G protein binding pocket (Fig. 3b, d, e). Similarly, in both CCR2 and CCR5 structures, the intracellular end of helix VII is in the inactive outward-facing conformation with Y7.53 pointing towards helix II rather than the center of the bundle. However, in CCR5, Y5.58 is oriented towards the center of the bundle, whereas in the present CCR2 structure, it faces the lipid and is sterically blocked from approaching R3.50 and Y7.53 by F6.38 (Fig. 3d, e). The net result of these interactions is that the crystallographically observed conformation of CCR2 appears to be even more inactive than that of CCR5 and most similar to dark rhodopsin28 and Fab-bound β2AR29. Although receptor construct engineering appears to contribute to stabilization of this inactive state, the ligand binding and thermal denaturation data suggest that the concerted action of the two antagonists is also important. By directly interacting with the conserved activation microswitch residues, CCR2-RA-[R] is perfectly positioned to stabilize this inactive state: it sterically blocks Y7.53 from populating the active conformation and is propped against R3.50, restricting its orientation away from the G protein interface (Fig. 3b). Although located 30 Å away, BMS-681 appears to cooperate with CCR2-RA-[R] through their common interactions with helix VII, which moves outward on the intracellular side (opposite to its movement during activation) and inward on the extracellular side (relative to CCR5 and US28) (Fig. 3f).
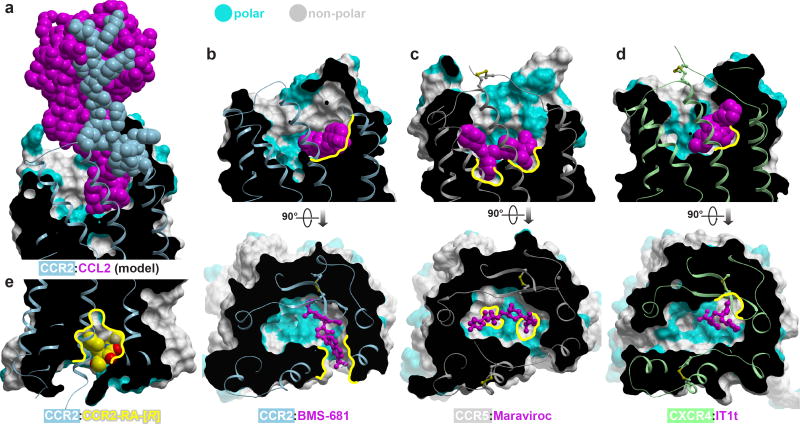
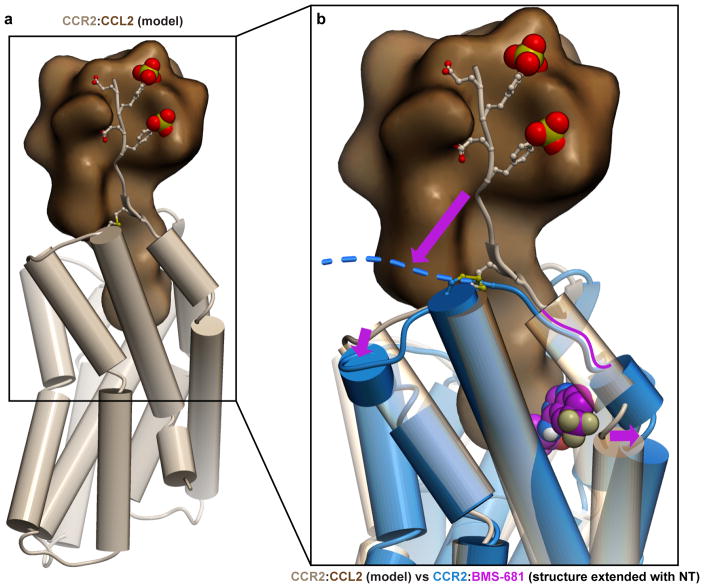
The CCR2 structure has general implications for the design of drugs targeting chemokine receptors as a family. As with most protein:protein interfaces, the orthosteric binding pockets of chemokine receptors are large, wide open, and highly polar. Chemokines explore numerous hotspots within these pockets and their binding is additionally reinforced by the interaction with the flexible N-termini of the receptors11,12 (Fig. 4a), collectively making for an extensive and versatile interaction that is conceptually difficult to inhibit with small molecules. The CCR2:BMS-681:CCR2-RA-[R] structure extends the repertoire of ideas that can be employed to overcome these obstacles. The binding mode of BMS-681 (Fig. 4b) contrasts with both the binding mode of Maraviroc to CCR5 (Fig. 4c) where the ligand spans the major and the minor subpocket of the receptor, and that of IT1t to CXCR4 (Fig. 4d) where the ligand is entirely accommodated in the minor subpocket. While occupying the minor subpocket of CCR2, BMS-681 protrudes between helices I and VII towards the lipid bilayer (Fig. 4b) in an interaction facilitated by the trifluoromethyl group that is often present in CCR2 antagonists30. This interaction enables hydrophobic anchoring of BMS-681 to the otherwise polar and open binding site of CCR2; by doing so, it parallels the role of other unique non-polar subpockets exploited by crystallized small molecule antagonists of CCR5 and CXCR4 (Fig. 4b–d). The novel subpocket explored by BMS-681 may have an additional advantage of disrupting the chemokine-compatible conformation of the receptor N-terminus (Extended Data Fig. 2).
Figure 4. Structural motifs exploited by small molecule antagonists of chemokine receptors.
Receptor surface meshes are colored by polarity (cyan–polar, grey–nonpolar). a, Modeled CCR2:CCL2 complex illustrates the extensive receptor:chemokine interface. b–d, Structures CCR2:BMS-681 (b), CCR5:Maraviroc (PDBID 4mbs, (c), and CXCR4:IT1t (PDBID 3odu, (d). Compounds utilize unique non-polar subpockets (yellow contours) within the open polar binding pockets of their target receptors. e, The allosteric pocket possesses a balanced combination of hydrophobic and polar features, making it a promising target for drug development.
CCR2-RA-[R] demonstrates a previously unseen binding mode within an allosteric pocket on the intracellular side of CCR2. Although relatively small, this pocket has a desirable balance of polarity and hydrophobicity (Fig. 2e and 4e). Homologous pockets may be present in other chemokine receptors, owing to a conserved G8.47; in fact, compound binding in homologous regions has been indirectly demonstrated for CCR1 and CCR5, and directly for CCR431, CXCR1, and CXCR232. In most other receptors that have been crystallized thus far, the non-glycine residue at position 8.47 appears to both reduce the pocket volume and block access to the backbone amides of helix 8; consequently, the homologous pockets in these receptors may not be druggable although negative allosteric modulation with antibodies and nanobodies targeting the same region has been reported24,25. By simultaneously competing with G protein and blocking activation-related conformational changes, compound binding in the allosteric pocket appears a powerful way to antagonize the receptor. Therefore, for those receptors in which the allosteric pocket is druggable, targeting it with small molecules may open new avenues for GPCR drug discovery.
Methods
Design and Expression of CCR2-T4L fusion constructs
The sequence of human CCR2 isoform b (Uniprot ID P41597-2) was engineered for crystallization by truncation of C-terminal residues 329–360 and by grafting T4 lysozyme (T4L) into the ICL3. In the process of construct optimization, the native CCR2 residues between L2265.62 and R2406.32 (L2265.62-KTLLRCRNEKKRH-R2406.32) were removed and replaced with corresponding residues from the crystallized structure of M2 muscarinic acetylcholine receptor (PDB ID 3UON, resulting amino-acid sequence S2265.62-RASKSRI-T4L-PPPSREK-K2406.32). The presence of T4L in ICL3 is expected to prevent receptor activation; however the similar affinities of BMS-681 and CCR2-RA-[R] for both WT CCR2 and CCR2-T4L (Extended Data Fig. 4a,b and Extended Data Table 3) suggest that the fusion construct is a good surrogate of WT CCR2 for understanding ligand recognition.
The CCR2-T4L coding sequence was cloned into a modified pFastBac1 vector (Invitrogen) with an HA signal sequence followed by a FLAG tag at the N-terminus and a PreScission protease site followed by a 10xHis tag and another FLAG tag at the C-terminus. The receptor was expressed in Spodoptera frugiperda (Sf9) cells. High-titer recombinant baculovirus (>109 viral particles per ml) was obtained using the Bac-to-Bac Baculovirus Expression System (Invitrogen) as previously described11. Sf9 cells at a cell density of 2–3 × 106 cells ml−1 were infected with P1 virus at an MOI (multiplicity of infection) of 5. Cells were harvested by centrifugation 48 h post-infection and stored at −80 °C until use.
Purification of CCR2-T4L
Insect cell membranes were prepared by thawing frozen cell pellets in a hypotonic buffer containing 10 mM HEPES (pH 7.5), 10 mM MgCl2, 20 mM KCl and EDTA-free complete protease inhibitor cocktail tablets (Roche). Extensive washing of the raw membranes was performed by repeated douncing and centrifugation in the same hypotonic buffer (2–3 times) and then in a high osmotic buffer containing 1.0 M NaCl, 10 mM HEPES (pH 7.5), 10 mM MgCl2, 20 mM KCl and EDTA-free complete protease inhibitor cocktail tablets (3–4 times), thereby separating soluble and membrane associated proteins from integral transmembrane proteins. 40 mM stock solutions of BMS-681 and CCR2-RA-[R] were made in isopropanol. Washed membranes were resuspended into a buffer containing 50 μM BMS-681, 2 mg/ml iodoacetamide, and EDTA-free complete protease inhibitor cocktail tablets, and incubated at 4 °C for 1 h before solubilization. The membranes were then solubilized in 50 mM HEPES (pH 7.5), 400 mM NaCl, 1% (w/v) n-dodecyl-β-D-maltopyranoside (DDM, Anatrace), 0.2% (w/v) cholesteryl hemisuccinate (CHS, Sigma) at 4 °C for 3 h. The supernatant was isolated by centrifugation at 50,000g for 30 min, and incubated in 20 mM HEPES (pH 7.5), 400 mM NaCl with TALON IMAC resin (Clontech) overnight at 4 °C. After binding, the resin was washed without addition of ligands with 10 column volumes of Wash I Buffer (50 mM HEPES (pH 7.5), 400 mM NaCl, 10% (v/v) glycerol, 0.1% (w/v) DDM, 0.02% (w/v) CHS, 10 mM imidazole), followed by 4 column volumes of Wash II Buffer (50 mM HEPES (pH 7.5), 400 mM NaCl, 10% (v/v) glycerol, 0.02% (w/v) DDM, 0.01% (w/v) CHS, 50mM imidazole). The protein was then eluted with 3–4 column volumes of Elution Buffer (50 mM HEPES (pH 7.5), 1 μM BMS-681, 400 mM NaCl, 10% (v/v) glycerol, 0.02% (w/v) DDM, 0.01% (w/v) CHS, and 250 mM imidazole). PD MiniTrap G-25 columns (GE Healthcare) were used to remove imidazole. The protein was then treated overnight with His-tagged PreScission protease to cleave the C terminal His-tag and Flag-tag. PreScission protease and the cleaved C-terminal fragment were removed by binding to TALON IMAC resin for 2 h at 4 °C. The protein was collected as the TALON IMAC column flow-through. The protein was supplemented with 75 μM each of BMS-681 and CCR2-RA-[R] before being concentrated to 30mg/ml with a 100 kDa molecular weight cut-off Amicon centrifuge concentrator (Millipore). The estimated final compound concentrations were ~ 1–2 mM for both compounds.
Protein stability assays
The thermostability of CCR2-T4L was analyzed by a differential scanning fluorimetry (DSF) assay adapted from previous publications33 using a RotorGene Q 6-plex RT-PCR machine (Qiagen). Briefly, 1–5 μg of protein was mixed with 3 μM 7-Diethylamino-3-(4′-Maleimidylphenyl)-4-Methylcoumarin (CPM) dye (2.5 mM stock in DMSO) in 25 mM HEPES pH 7.5, 400 mM NaCl, 0.02% DDM, 0.004% CHS, 10% glycerol, and indicated concentrations of compounds to a final volume of 20 μL; samples were incubated for 5 min at room temperature and then heated gradually from 28 °C to 90 °C at a rate of 0.8 °C/min, with CPM fluorescence (excitation 365 nm, emission 460 nm) recorded every 1 °C. The melting temperature (Tm) was determined from the first derivative of the denaturation curve, using the Rotor-Gene Q – Pure Detection software (version 2.0.3).
Crystallization
Purified CCR2 in complex with BMS-681 and CCR2-RA-[R] was reconstituted into lipidic cubic phase (LCP) by mixing with molten lipid using a mechanical syringe mixer8. The protein-LCP mixture contained 40% (w/w) receptor solution, 54% (w/w) monoolein and 6% (w/w) cholesterol. Crystallization trials were performed in 96-well glass sandwich plates (Hampton research) using a Mosquito LCP robot (TTP Labtech) by dispensing 45 nL of protein-laden LCP and 800 nL of precipitant solution per well. Plates were incubated and imaged at 20 °C. Initial crystal hits were found from a precipitant condition containing 100 mM MES, pH 6.5, 30% (v/v) PEG400, 100 mM Li2SO4. After optimization, diffraction quality crystals were obtained from 100 mM MES, pH 6.5, 30–32% (v/v) PEG400, 75–85 mM Li2SO4. Crystals usually grew to a maximum size of 60 μm × 10 μm × 10 μm in one week, and were harvested directly from the LCP matrix using MiTeGen micromounts and flash cooled in liquid nitrogen.
Data collection and structure determination
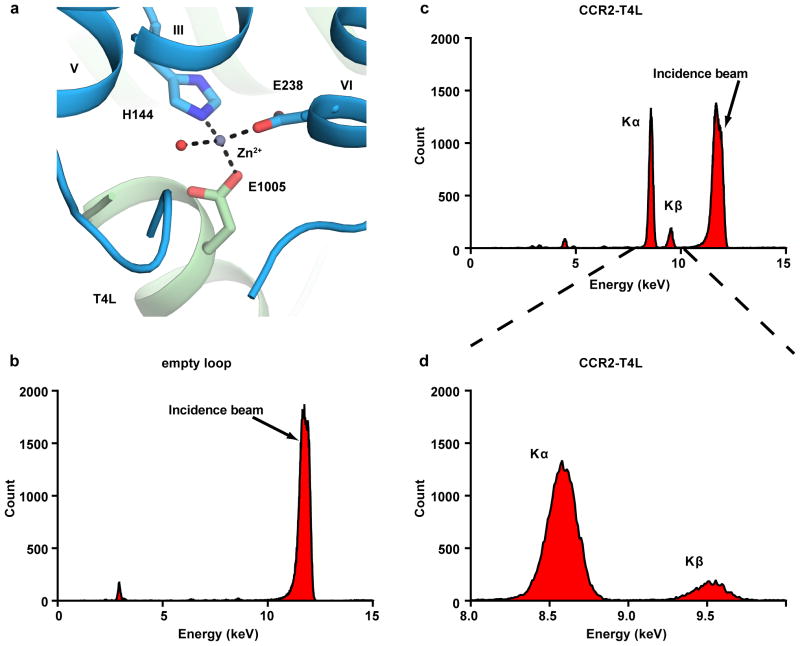
X-ray diffraction data were collected using a 10 μm collimated minibeam at a wavelength of 1.0332 Å with a Pilatus3 6M direct detector on the 23ID-D beamline (GM/CA CAT) of the Advanced Photon Source at the Argonne National Laboratory. Crystals were located and aligned by the rastering strategy34. Among the several hundred crystal samples screened, most crystals diffracted to 2.8–3.5 Å resolution when exposed to 0.3 s of unattenuated beam using 0.3° oscillations. A 93.1% complete data set at 2.80 Å resolution was obtained by merging data from 17 crystals, using XDS35 and Aimless36. As the data showed anisotropy, the UCLA Diffraction Anisotropy Server (http://services.mbi.ucla.edu/anisoscale/) was used to truncate the data to 3.0 Å along both a* and b* axes, and to 2.81 Å along the c* axis. Initial phase information was obtained by molecular replacement (MR) with the program Phaser37 using the receptor portion of the CCR5 structure (PDB ID: 4MBS) converted to polyalanines, and the T4L portion of the CXCR4 structure (PDB ID: 3ODU) as search models. The correct MR solution (TFZ=14.8) contained one CCR2-T4L molecule in the asymmetric unit. Refinement was performed with Phenix38 followed by manual examination and rebuilding of the refined coordinates in the program COOT39 using both |2Fo| - |Fc| and |Fo| - |Fc| maps, as well as omit maps. The final model includes 295 residues (37 to 225 and 241–319) of the 360 residues of CCR2 and residues 2 to 161 of T4L plus 16 residues of two 8-residue linkers. The remaining N- and C-terminal residues are disordered and were not built. Strong electron density for one metal ion was observed. The identity of the ion was determined to be Zn2+ by X-ray fluorescence scans (Extended Data Fig. 5). The zinc ion is coordinated by a water molecule as well as side chains of H1443.56, E238, and E1005. Data collection and refinement statistics is shown in Extended Data Table 1.
Crystallization and structure determination of BMS-681
BMS-681 was dissolved in a minimal amount of CH3CN and then 15% water was added. After standing overnight, the resulting crystals were harvested. Data were collected on a Bruker-AXS X8-Proteum Kappa goniometer and APEXII detector. Intensities were measured using Cu Kα radiation (λ = 1.5418 Å) with the crystal kept at a constant temperature using an Oxford cryo system during data collection. Indexing and processing of the measured intensity data were carried out with the SAINT-APEX2 (Bruker-AXS) program suite, structure solution with SHELXS-97 and structure refinement with SHELXL-97.
The derived atomic parameters (coordinates and temperature factors) were refined through full matrix least-squares. The function minimized in the refinements was Σw (|Fo| - |Fc|)2. R is defined as Σ ||Fo| - |Fc||/Σ |Fo| while Rw = [Σw (|Fo| - |Fc|)2/ Σw |Fo|2]1/2 where w is an appropriate weighting function based on errors in the observed intensities. Hydrogens were introduced in idealized positions with isotropic temperature factors, but no hydrogen parameters were varied. It should be noted that the refinement model illustrates significant disorder and partial occupancy factors of ‘guest’ solvent/water molecules within the crystalline lattice. The atomic positions of these disordered molecules were taken from the difference map analysis, which showed peaks of electron density of varying intensities at the refined positions representing the disordered solvent/water molecules. Data collection and refinement statistics is shown in Extended Data Table 2.
Cell culture and transfections
Chinese hamster ovary (CHO) cells (provided by Hans den Dulk, Leiden University, The Netherlands; originally obtained from and certified by ATCC) were cultured in Dulbecco’s Modified Eagle Medium/F-12 Nutrient Mixture (DMEM/F-12) supplemented with 10% (v/v) newborn calf serum, 50 IU/mL penicillin and 50μg/mL streptomycin; and maintained at 37°C and 5% CO2. Cells were subcultured twice a week at a ratio of 1:30 to 1:50 by trypsinization. Transient transfection of CHO cells with WT CCR2 and CCR2-T4L constructs was performed using a polyethylenimine (PEI) method, as described previously23. Briefly, CHO cells were grown on 15 cm ø to around 50% confluence and then transfected with a DNA/PEI mixture containing 10 μg plasmid DNA – previously diluted in 150 mM NaCl solution – mixed with PEI solution (1 mg/mL) at a 1:6 DNA:PEI mass ratio. Before adding 1 mL of the transfection mixture to each plate, the culture medium of the cells was refreshed and the mixture incubated for 20 min at room temperature. Following transfection, cells were incubated for 48 h at 37°C and 5% CO2 before membrane preparation. 24 h after transfection, sodium butyrate was added to each plate at a final concentration of 3 mM to increase receptor expression. CHO cells were tested for mycoplasma contamination before use, the outcome of which was negative.
Membrane preparation
Membranes from CHO cells transiently expressing the WT CCR2 or CCR2-T4L were prepared as described previously23. Briefly, cells were detached from 15-cm ø plates using 5 mL of phosphate-buffered saline and centrifuged for 5 min at 3000g. The membranes were separated from the cytosolic fractions by several centrifugation and homogenization steps. First, the pellets were resuspended and homogenized in ice-cold membrane buffer (50 mM Tris-HCl buffer, supplemented with 5 mM MgCl2, pH 7.4) using an Ultra Thurrax Homogenizer (IKA-Werke GmbH & Co. KG; Staufen, Germany). Homogenized membranes were then centrifuged in an Optima LE-80 K ultracentrifuge (Beckman Coulter, Inc., Fullerton, CA) at 31,000g for 20 min at 4°C. The final membrane pellet was resuspended also in ice-cold membrane buffer and aliquoted before storage. Membrane aliquots were stored at −80°C and protein concentrations were measured using a standard BCA protein determination assay (Pierce Chemical Company, Rockford, IL USA).
Radioligand binding assays
[3H]-INCB-3344 (specific activity 32 Ci mmol−1) and [3H]-CCR2-RA (specific activity 63 Ci mmol−1) were custom-labeled by Vitrax (Placentia, CA). JNJ-27141491 was synthesized as described previously40. INCB-3344 and CCR2-RA-[R] were synthesized in-house as described previously7,41.
All radioligand binding assays were performed at 25 °C in a 100 μL reaction volume containing assay buffer (50 mM Tris-HCl buffer (pH 7.4), 5 mM MgCl2, 0.1% CHAPS) and 30 μg of membrane protein from CHO cells transiently expressing WT CCR2 or CCR2-T4L. For competition binding assays with [3H]INCB-3344, a concentration of 5 nM [3H]INCB-3344 was used, and non-specific binding was determined with 10 μM of unlabeled INCB-3344. In the case of [3H]-CCR2-RA competition binding assays, a radioligand concentration of 3 nM was used and non-specific binding was determined with 10 μM of JNJ-27141491. In all cases, homologous or competition displacement assays were carried out using six increasing concentrations of competing ligands. Kinetic experiments were also performed at 25 °C using 7 nM [3H]-CCR2-RA and 30 μg of membrane protein in a 100 μL reaction volume. For association experiments, CHO-CCR2 or CHO-CCR2-T4L membranes were added to the reaction at 8 different time points, in the absence or presence of 1μM BMS-681. For dissociation experiments, membranes were first incubated with radioligand for 90 min, dissociation was then initiated by addition of 10 μM of CCR2-RA-[R] at 12 different time points, in the presence or absence of 1μM BMS-681. A higher number of time points were used in the dissociation assays, in order to characterize the biphasic profile of [3H]-CCR2-RA dissociation. In all cases, total radioligand binding did not exceed 10% of the total radioligand added to avoid ligand depletion. For all experiments, incubation was terminated by dilution with ice-cold wash buffer (50 mM Tris-HCl buffer (pH 7.4), 5 mM MgCl2, 0.05% CHAPS). Separation of bound from free radioligand was achieved by rapid filtration over a 96-well GF/B filter plate using a Perkin Elmer Filtermate-harvester (Perkin Elmer, Groningen, the Netherlands) and filter-bound radioactivity was determined in a Perkin Elmer 2450 Microbeta2 plate counter after addition of 25 μL Microscint scintillation cocktail per well (Perkin-Elmer, Groningen, the Netherlands).
Statistical Methods
All radioligand binding data were analysed using Prism 6.0 and 7.0 (GraphPad software, San Diego, CA, USA). The pIC50 values were obtained by non-linear regression analysis of competition displacement assays. Apparent association rate constants (kobs) and maximum binding (Bmax, used to calculate %B/Bcontrol) were determined by fitting the association data to a one-phase exponential association function. Dissociation rate constants were determined by fitting the dissociation data to a monophasic (koff) or biphasic (koff,fast and koff,slow) exponential decay model. All data shown represent means ± S.E.M of at least three independent experiments performed in duplicate. An unpaired, two-tailed Student’s t-test was used to compare differences in pIC50 as well as differences in kinetic parameters. Differences in binding enhancement (%Binding) in the absence (set at 100%) or presence of BMS-681 were analyzed using a one-way ANOVA with Dunnett’s post-hoc test. Significant differences are denoted as follows: *p < 0.05, **p < 0.01, ***p < 0.001 ****p < 0.0001.
Data deposition statement
The atomic coordinates and structure factors for CCR2:BMS-681:CCR2-RA-[R] complex have been deposited in the Protein Data Bank under accession number 5T1A. The structure of free BMS-681 is deposited in the Cambridge Crystallographic Data Centre (CCDC) under accession number 1479580.
Extended Data
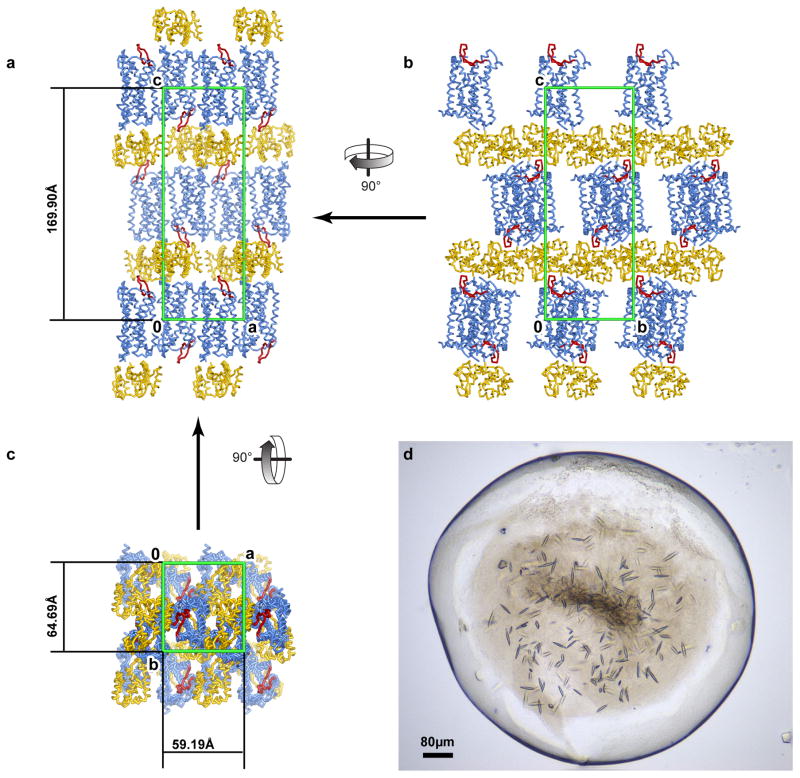
Extended Data Figure 1. CCR2-T4L crystals and crystal packing.
a–c, Crystal packing of CCR2-T4L. CCR2 is a blue ribbon with ECL2 colored red and T4L yellow. The unit cell is shown as a green box. CCR2-T4L molecules are arranged in a type I packing with hydrophilic stacking mediated by T4L and T4L-ECL2 interactions along axis c. a, Crystal packing in the ac plane. CCR2 makes abundant hydrophobic contacts with its neighbor via an interface mediated by antiparallel helix IV–helix VI interactions related by a screw axis along axis a. b, Crystal packing in the bc plane. Contacts between receptors and T4L involve ECL2 and the intracellular surface of CCR2 including helix VIII. Direct contacts between T4L are along axis b. One layer of CCR2-T4L molecules at the very top of the stacking column are omitted for clarity. c, Crystal packing in the ab plane. There are no direct interactions between T4L along axis a. d, Crystals of CCR2-T4L in the LCP bolus. Average crystals grew to 60 μm × 10 μm × 10 μm before harvesting.
Extended Data Figure 2. BMS-681 binding may disrupt a chemokine-recognizing conformation of the CCR2 N-terminus and helix I.
a, Model of CCR2:CCL2 built by homology from the structure of CXCR4:vMIP-II11 suggests that a productive chemokine-compatible conformation of the receptor requires re-orientation of the N-terminus from almost parallel to almost perpendicular to the membrane plane, and formation of an extra helical turn in helix I to bring it closer to helix VII and ECL3. b, Binding of BMS-681 may disrupt this chemokine-compatible conformation by inserting between helices I and VII.
Extended Data Figure 3. CCR2-RA-[R] directly binds to CCR2 residues that are homologous to those involved in G protein coupling in other GPCRs.
Partial alignment of intracellular regions of CCR2 and homologous regions in bovine Rho (bRho) and β2 adrenergic receptor (β2AR), alongside profile of contacts that CCR2-RA-[R], the Gαt C-terminal peptide21, and Gαs C-terminus22 make with the three respective receptors. Contacts are shown by circles above and below the alignment, with circle area indicative of contact strength. Backbone and side-chain contacts are gray and black, respectively. Assuming structural homology between the CCR2:G protein interface and at least one of the bRho:Gαt and β2AR:Gαs interfaces, several residue positions appear to be involved in binding both CCR2-RA-[R] and the G protein C-terminus.
Extended Data Figure 4. Equilibrium binding and binding kinetics of BMS-681 and CCR2-RA-[R] with WT CCR2 and CCR2-T4L.
a,b, Displacement of [3H]-INCB-3344 (5nM, a) and [3H]-CCR2-RA-[R] (3 nM, b) from WT CCR2 and CCR2-T4L in CHO cells by increasing concentrations of unlabeled INCB-3344, CCR2-RA-[R] and BMS-681. c, d, Association and e, f, dissociation of 7nM [3H]-CCR2-RA from CHO cell membranes transiently expressing WT CCR2 (c, e) or CCR2-T4L (d, f) at 25°C, in the absence or presence of 1 μM BMS-681. Figures represent normalized and combined data from three independent experiments performed in duplicate, with results presented as mean ± S.E.M percentage of specific [3H]-CCR2-RA binding.
Extended Data Figure 5. A Zn2+ binding site was identified by X-ray fluorescence emission (Xfe) analysis of the CCR2-T4L:BMS-681:CCR2-RA-[R] crystals.
a, View of the Zn2+ ion at an interface formed by CCR2 helices III and VI and the N-terminus of T4L. The Zn2+ ion is coordinated by side chains of H1443.56 (from WT receptor), E2386.30 (from the engineered part of the receptor), and E1005 (from T4L) as well as a structured water. b, The background fluorescence signal of an empty MiTeGen micromount is low, indicating the absence of metal ion. Excitation at 12 keV results in a peak at 11.7 keV (due to the incidence beam). c, Xfe signal from a wide fluorescence scan of the CCR2-T4L crystal. The fluorescence peaks at 8.60 keV and 9.53 keV correspond to X-ray emission lines Kα (8.64 keV) and Kβ (9.57 keV) and indicate the presence of Zn2+ bound to CCR2-T4L. d, A zoomed-in view of the Xfe signal from (c).
Extended Data Table 1.
Data collection and refinement statistics (molecular replacement)
| CCR2-T4L:BMS-681:CCR2-RA-[R]a | |
|---|---|
| Data collectionb wavelength (Å) | 1.03319 |
| Space group | P212121 |
| Unit cell parameters a,b,c (Å) | 59.19 64.69 169.90 |
| Number of reflections measured | 82,111 |
| Number of unique reflections | 15,550 |
| Resolution (Å) | 48-2.8 (2.95-2.8) |
| Rmerge (%) | 22.5(101) |
| Rpim (%) | 12.8(88.4) |
| Mean I/s(I) | 6.9(0.8) |
| Completeness (%) | 93.1(66.6) |
| Redundancy | 5.3(1.8) |
| Refinement | |
| Resolution (Å) | 25-2.81 (3.0, 3.0, 2.81) |
| Number of reflections (test set) | 14515 (746) |
| Rwork/Rfree | 0.227/0.272 (0.319/0.389) |
| Number of atoms | 3,580 |
| CCR2 | 2,215 |
| T4L | 1,243 |
| BMS-681 | 35 |
| CCR2-RA-[R] | 24 |
| Monoolein | 25 |
| Sulfate | 20 |
| Water | 17 |
| Zn | 1 |
| Mean overall B value (Å2) | 41.4 |
| Wilson B | 40.4 |
| Protein | 41.5 |
| Ligands | 41.3 |
| Water | 22.9 |
| Root mean square deviation | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.84 |
| Ramanchandran plot statisticsc (%) | |
| Favored regions | 97.3 |
| Allowed regions | 2.7 |
| Disallowed regions | 0 |
Diffraction data from 17 crystals were merged into a complete data set
Highest resolution shell statistics are shown in parentheses
As defined in MolProbity42
Extended Data Table 2.
Small Molecule (BMS-681) X-ray data collection and refinement.
| Empirical formula | C26 H36 F3N 5 O3.58 |
|---|---|
| Formula weight | 532.80 |
| Temperature | 173(2) K |
| Wavelength | 1.54178 Å |
| Crystal system | Tetragonal |
| Space group | P43212 |
| Unit cell dimensions | a = 20.4436(4)Å α= 90°. b = 20.4436(4)Å β= 90°. c = 28.9325(7)Å γ = 90°. |
| Volume | 12092.1(4) Å3 |
| Z | 16 |
| Density (calculated) | 1.171 Mg/m3 |
| Absorption coefficient | 0.768 mm−1 |
| F(000) | 4522 |
| Crystal size | 0.46 × 0.18 × 0.16 mm3 |
| Theta range for data collection | 2.65 to 58.78°. |
| Resolution range | 16.7 to 0.9 Å |
| Index ranges | −22 ≤ h ≤ 22, −21 ≤ k ≤ 22, −31 ≤ l ≤ 14 |
| Reflections collected | 108743 |
| Independent reflections | 8518 [R(int) = 0.1259] |
| Completeness to theta = 58.78° | 98.6 % |
| Absorption correction | None |
| Refinement method | Full-matrix least-squares on F2 |
| Data / restraints / parameters | 8518 / 22 / 713 |
| Goodness-of-fit on F2 | 1.058 |
| Final R indices [I>2sigma(I)] | R1 = 0.0770, wR2 = 0.2087 |
| R indices (all data) | R1 = 0.0860, wR2 = 0.2178 |
| Absolute structure parameter; Flack(x) | 0.1(2) |
| Absolute structure parameter; Hooft(y), P3true | 0.03(5), 1.000 |
| Largest diff. peak and hole | 0.543 and −0.405 e.Å−3 |
Extended Data Table 3.
Displacement of specific [3H]-INCB-3344 (5 nM) and [3H]-CCR2-RA (3 nM) binding from CCR2 constructs transiently expressed on CHO cells.
| [3H]-INCB-3344 displacement by INCB-3344 | [3H]-INCB-3344 displacement by BMS-681 | [3H]-CCR2-RA displacement by CCR2-RA-[R] | [3H]-CCR2-RA enhancement by BMS-681 | |
|---|---|---|---|---|
| Construct | pIC50 ± S.E.M (IC50, nM) | %Binding | ||
| WT CCR2 | 7.8 ± 0.0 (17) | 8.1 ± 0.0 (8) | 7.9 ± 0.0 (13) | 134 ± 3%a ** |
| CCR2-T4L | 8.1 ± 0.1* (8) | 8.6 ± 0.1** (3) | 8.2 ± 0.0** (6) | 157 ± 13%a **** |
Values represent mean ± S.E.M of at three independent experiments performed in duplicate.
Percentage of [3H]-CCR2-RA (3 nM) binding in presence of BMS-681 (1 μM). Values higher than 100% represent binding enhancement compared to the 100% control without BMS-681.
Differences in pIC50 values between constructs were analyzed using a Student’s t-test, with significant differences noted as follows: *p < 0.05, **p < 0.01.
Differences in %Binding in the absence (100%) and presence of BMS-681 were analyzed using a one-way ANOVA with Dunnett’s post-hoc test, with significant differences noted as follows: **p < 0.01, ****p < 0.0001.
Extended Data Table 4.
Observed association and dissociation rate constants of [3H]-CCR2-RA (7 nM) on membranes from CHO cells transiently expressing WT CCR2 and CCR2-T4L, in the absence or presence of 1 μM BMS-681.
| CHO-CCR2 | CHO-CCR2-T4L | |||
|---|---|---|---|---|
| Control | +1 μM BMS-681 | Control | +1 μM BMS-681 | |
| kobs(min−1) | 0.031 ± 0.002 | 0.038 ± 0.003* | 0.015 ± 0.003 | 0.015 ± 0.001 |
| %B/Bcontrola | 100 ± 0.0 | 135 ± 2.0**** | 100 ± 0.0 | 162 ± 8.4** |
| koff,fast (min−1) | 0.089 ± 0.015 | 0.069 ± 0.012* | 0.077 ± 0.013 | 0.049 ± 0.003b |
| koff,slow (min−1) | 0.016 ± 0.005 | 0.012 ± 0.004 | 0.010 ± 0.003 | |
| %fast | 70 ± 10 | 71 ± 11 | 69 ±8 | N/Ab |
Values represent mean ± S.E.M of three independent experiments performed in duplicate.
% B/Bcontrol represents the % of maximum binding in absence (Bcontrol) or presence (B) of BMS-681 (1 μM).
For CHO-CCR2-T4L only, dissociation kinetics of [3H]CCR2-RA (7 nM) in presence of BMS-681 (1 μM) fitted best with a monophasic exponential decay model, resulting in a single koff value, as shown in the table. Thus for CHO-CCR2-T4L, the statistical significance between koff measurements with and without BMS-681 could not be calculated.
Statistical significance was analyzed using a Student’s t-test, with significant differences versus control noted as follows: *p < 0.05, **p < 0.01, ****p<0.0001
Acknowledgments
The authors thank A. Ishchenko and H. Zhang for help with x-ray data collection, C. Wang and H.X. Wu for suggestions on construct design, F. Li for help with data processing, and M. Galella for assistance with BMS compound data and statistics. We thank C. Ogata, R. Sanishvili, N. Venugopalan, M. Becker and S. Corcoran at beamline 23ID at GM/CA CAT Advanced Photon Source. Funding for this research was provided by National Institutes of Health grants R01 GM071872, R01 GM117424, R01 AI118985, R21 AI121918, R21 AI122211, U01 GM094612, and U54 GM094618. GM/CA@APS has been funded in whole or in part with federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Footnotes
Competing interest declaration
R.A. has an equity interest in Molsoft, LLC. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. R.C., P.C., and A.T. are employees of Bristol-Myers Squibb Company. D.S. is an employee of Vertex Pharmaceuticals, Inc.
Author contributions
I.K. and T.M.H. designed the study and coordinated all experiments. Y.Z. designed and engineered protein constructs, performed crystallization experiments, collected the diffraction data, and determined the structure. L.Q., M.G., and C.Z. assisted with protein engineering and crystallization. G.W.H. assisted with structure determination and refinement. A.P.I. and L.H.H. designed and N.V.O.Z. and H.d.V. performed equilibrium and kinetics binding experiments. I.K. performed computational and bioinformatics analyses. R.C., P.C., and A.T. synthesized, characterized, and crystallized the BMS compound analogs. M.D. assisted with compound crystallization. D.S. assisted with the allosteric compound characterization. R.A. assisted with structure analysis. V.C. and R.C.S. assisted with crystallization. Y.Z., N.V.O.Z., A.P.I., L.H.H., I.K., and T.M.H. wrote the paper.
References
- 1.Scholten DJ, et al. Pharmacological modulation of chemokine receptor function. Br J Pharmacol. 2012;165:1617–1643. doi: 10.1111/j.1476-5381.2011.01551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Connor T, Borsig L, Heikenwalder M. CCL2-CCR2 Signaling in Disease Pathogenesis. Endocr Metab Immune Disord Drug Targets. 2015;15:105–118. doi: 10.2174/1871530315666150316120920. [DOI] [PubMed] [Google Scholar]
- 3.Lim SY, et al. Targeting the CCL2-CCR2 signaling axis in cancer metastasis. Oncotarget. 2016;7:28697–28710. doi: 10.18632/oncotarget.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solari R, Pease JE, Begg M. Chemokine receptors as therapeutic targets: Why aren’t there more drugs? European Journal of Pharmacology. 2015;746:363–367. doi: 10.1016/j.ejphar.2014.06.060. [DOI] [PubMed] [Google Scholar]
- 5.Cooke RM, Brown AJ, Marshall FH, Mason JS. Structures of G protein-coupled receptors reveal new opportunities for drug discovery. Drug Discov Today. 2015;20:1355–1364. doi: 10.1016/j.drudis.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Carter PH, et al. Discovery of a Potent and Orally Bioavailable Dual Antagonist of CC Chemokine Receptors 2 and 5. ACS medicinal chemistry letters. 2015;6:439–444. doi: 10.1021/ml500505q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dasse O, et al. Novel, acidic CCR2 receptor antagonists: lead optimization. Letters in Drug Design & Discovery. 2007;4:263–271. http://dx.doi.org/10.2174/157018007784619989. [Google Scholar]
- 8.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat Protoc. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu B, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan Q, et al. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science. 2013;341:1387–1390. doi: 10.1126/science.1241475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin L, et al. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine. Science. 2015;347:1117–1122. doi: 10.1126/science.1261064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burg JS, et al. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor. Science. 2015;347:1113–1117. doi: 10.1126/science.aaa5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monteclaro FS, Charo IF. The Amino-terminal Domain of CCR2 Is Both Necessary and Sufficient for High Affinity Binding of Monocyte Chemoattractant Protein 1: RECEPTOR ACTIVATION BY A PSEUDO-TETHERED LIGAND. J Biol Chem. 1997;272:23186–23190. doi: 10.1074/jbc.272.37.23186. [DOI] [PubMed] [Google Scholar]
- 14.Zhang K, et al. Structure of the human P2Y12 receptor in complex with an antithrombotic drug. Nature. 2014;509:115–118. doi: 10.1038/nature13083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weik M, et al. Specific chemical and structural damage to proteins produced by synchrotron radiation. Proc Natl Acad Sci U S A. 2000;97:623–628. doi: 10.1073/pnas.97.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cherney RJ, et al. Discovery of disubstituted cyclohexanes as a new class of CC chemokine receptor 2 antagonists. J Med Chem. 2008;51:721–724. doi: 10.1021/jm701488f. [DOI] [PubMed] [Google Scholar]
- 17.Berkhout TA, et al. CCR2: characterization of the antagonist binding site from a combined receptor modeling/mutagenesis approach. J Med Chem. 2003;46:4070–4086. doi: 10.1021/jm030862l. [DOI] [PubMed] [Google Scholar]
- 18.Hall SE, et al. Elucidation of binding sites of dual antagonists in the human chemokine receptors CCR2 and CCR5. Mol Pharmacol. 2009;75:1325–1336. doi: 10.1124/mol.108.053470. [DOI] [PubMed] [Google Scholar]
- 19.Cherney RJ, et al. Synthesis and evaluation of cis-3,4-disubstituted piperidines as potent CC chemokine receptor 2 (CCR2) antagonists. Bioorg Med Chem Lett. 2008;18:5063–5065. doi: 10.1016/j.bmcl.2008.07.123. [DOI] [PubMed] [Google Scholar]
- 20.Zweemer AJ, et al. Discovery and mapping of an intracellular antagonist binding site at the chemokine receptor CCR2. Mol Pharmacol. 2014;86:358–368. doi: 10.1124/mol.114.093328. [DOI] [PubMed] [Google Scholar]
- 21.Blankenship E, Vahedi-Faridi A, Lodowski David T. The High-Resolution Structure of Activated Opsin Reveals a Conserved Solvent Network in the Transmembrane Region Essential for Activation. Structure. 2015;23:2358–2364. doi: 10.1016/j.str.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rasmussen SG, et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zweemer AJ, et al. Multiple binding sites for small-molecule antagonists at the CC chemokine receptor 2. Mol Pharmacol. 2013;84:551–561. doi: 10.1124/mol.113.086850. [DOI] [PubMed] [Google Scholar]
- 24.Hino T, et al. G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody. Nature. 2012;482:237–240. doi: 10.1038/nature10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staus DP, et al. Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. Nature. 2016;535:448–452. doi: 10.1038/nature18636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katritch V, Cherezov V, Stevens RC. Structure-function of the G protein-coupled receptor superfamily. Annual review of pharmacology and toxicology. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manglik A, et al. Structural Insights into the Dynamic Process of β2-Adrenergic Receptor Signaling. Cell. 2015;161:1101–1111. doi: 10.1016/j.cell.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 29.Rasmussen SG, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 30.Pease J, Horuk R. Chemokine Receptor Antagonists. Journal of Medicinal Chemistry. 2012;55:9363–9392. doi: 10.1021/jm300682j. [DOI] [PubMed] [Google Scholar]
- 31.Andrews G, Jones C, Wreggett KA. An intracellular allosteric site for a specific class of antagonists of the CC chemokine G protein-coupled receptors CCR4 and CCR5. Mol Pharmacol. 2008;73:855–867. doi: 10.1124/mol.107.039321. [DOI] [PubMed] [Google Scholar]
- 32.Nicholls DJ, et al. Identification of a putative intracellular allosteric antagonist binding-site in the CXC chemokine receptors 1 and 2. Mol Pharmacol. 2008;74:1193–1202. doi: 10.1124/mol.107.044610. [DOI] [PubMed] [Google Scholar]
- 33.Alexandrov AI, Mileni M, Chien EYT, Hanson MA, Stevens RC. Microscale fluorescent thermal stability assay for membrane proteins. Structure. 2008;16:351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Cherezov V, et al. Rastering strategy for screening and centring of microcrystal samples of human membrane proteins with a sub-10 μm size X-ray synchrotron beam. Journal of The Royal Society Interface. 2009;6:S587–S597. doi: 10.1098/rsif.2009.0142.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabsch W. Xds. Acta Crystallographica Section D: Biological Crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/s0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCoy AJ, et al. Phaser crystallographic software. Journal of applied crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/s0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/s0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doyon J, et al. Discovery of potent, orally bioavailable small-molecule inhibitors of the human CCR2 receptor. Chem Med Chem. 2008;3:660–669. doi: 10.1002/cmdc.200700276. [DOI] [PubMed] [Google Scholar]
- 41.Brodmerkel CM, et al. Discovery and pharmacological characterization of a novel rodent-active CCR2 antagonist, INCB3344. J Immunol. 2005;175:5370–5378. doi: 10.4049/jimmunol.175.8.5370. [DOI] [PubMed] [Google Scholar]
- 42.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]