Highlights
-
•
Engineered live attenuated vaccines may improve the control porcine CoVs infection.
-
•
Porcine CoVs affect many host-cell pathways modulating pathogenesis.
-
•
CoV genes acting as virulence factors should be modified for virus attenuation.
-
•
To use attenuated CoVs as vaccine candidates several safety guards should be included.
Keywords: Coronavirus, Virus-host interaction, Vaccines, Pathogenesis, Innate immune response
Abstract
Porcine enteric coronaviruses (CoVs) cause severe disease in the porcine herds worldwide, leading to important economic losses. Despite the knowledge of these viruses since the 1970s, vaccination strategies have not been implemented, leading to continuous re-emergence of novel virulent strains. Live attenuated vaccines historically have been the most efficient. We consider that the new trend is the development of recombinant vaccines by using reverse genetics systems to engineer attenuated viruses, which could be used as effective and safe modified live vaccine candidates. To this end, host cell signaling pathways influencing porcine CoV virulence should be identified. Similarly, the identity of viral proteins involved in the modulation of host cell pathways influencing CoV pathogenesis should be analyzed. With this information, and using reverse genetics systems, it is possible to design viruses with modifications in the viral proteins acting as virulence factors, which may lead to attenuated viruses and, therefore, vaccine candidates. In addition, novel antiviral drugs may be developed once the host cell pathways and the molecular mechanism affecting porcine CoV replication and virulence are known. This review is focused in the host cell responses to enteric porcine CoV infection and the viral proteins involved in pathogenesis.
1. Introduction
Coronaviruses (CoVs) are enveloped viruses with a positive-sense RNA genome that belong to the Coronaviridae family within the Nidovirales order (de Groot et al., 2012). CoVs are the causative agents of a variety of human and animal diseases. In animals, CoVs cause life-threatening diseases, such as severe enteric and respiratory tract infections, and are economically relevant pathogens (Perlman and Netland, 2009). Acute infectious diarrhea is a major cause of high morbidity and mortality in piglets worldwide. Enteric infections in animals are frequently associated with viruses, including rotaviruses and CoVs (Chattha et al., 2015). A metagenomics analysis of diarrheic and healthy samples from China in 2012 found porcine CoVs in 77% of the diarrheic samples, and only in around 7% of the healthy samples, highlighting the potential importance of CoVs as enteric porcine pathogens (Zhang et al., 2014a).
There are six different porcine CoVs described so far: alphacoronaviruses transmissible gastroenteritis virus (TGEV), with porcine respiratory coronavirus (PRCV) as a variant, and porcine epidemic diarrhea virus (PEDV), the neurotropic betacoronavirus porcine hemagglutinating encephalomyelitis virus (PHEV), and porcine deltacoronavirus (PDCoV) (de Groot et al., 2012). Recently, a swine enteric coronavirus (SeCoV) that is a recombinant between TGEV and PEDV has been described (Akimkin et al., 2016, Boniotti et al., 2016).
This review will focus on porcine enteric CoVs, as emergent and re-emergent pathogens causing enteric disease in swine population worldwide.
1.1. Genome structure of porcine enteric and respiratory CoVs
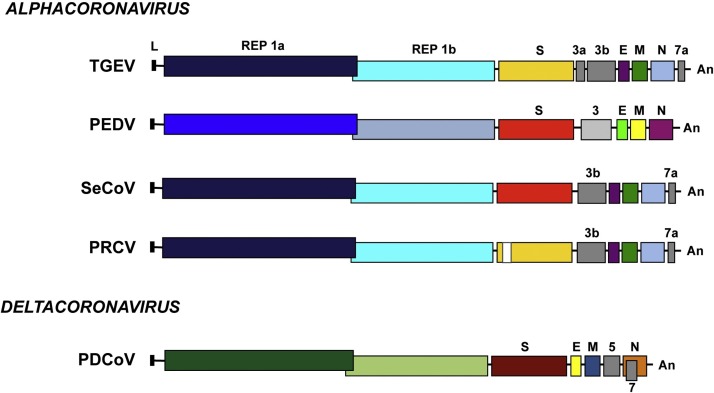
Coronaviridae members, including swine CoVs contain the largest RNA genome known among RNA viruses, consisting in a positive-sense RNA molecule of 25–30 Kb in length (Enjuanes et al., 2008). This RNA is similar to cellular mRNAs, as it contains 5′-capped and 3′ polyadenylated ends. The first two thirds of the genome contain two overlapping open reading frames (ORFs) ORF1a and ORF1b (Fig. 1 ). Translation of ORF1a yields polyprotein 1a (pp1a), and −1 ribosomal frameshifting allows translation of ORF1b to yield polyprotein pp1ab (Ziebuhr, 2005). These polyproteins are co- and post-translationally processed into 16 non-structural proteins (nsps), most of them driving viral genome replication and subgenomic mRNA (sgmRNA) synthesis. PDCoV replicase pp1ab is processed in 15 nsps, as Deltacoronavirus genomes lack nsp1 gene (Woo et al., 2012). The 3′ third of the porcine CoV genome encodes the structural proteins in the order 5′-S-E-M-N-3′. In addition, it includes up to three genus-specific genes different among CoV members (Fig. 1).
Fig. 1.
Porcine enteric and respiratory CoV genome organization. Schematic representation of the genome structure of porcine CoVs: transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV), swine enteric coronavirus (SeCoV), porcine respiratory coronavirus (PRCV), and porcine deltacoronavirus (PDCoV). The genus in which each CoV is included is indicated. Letters above genomes indicate replicase genes (REP1a and REP1b), structural genes (spike S, envelope E, membrane M and nucleocapsid N) and the different genus-specific genes (in gray). L, leader sequence; An, poly(A) tail.
1.2. Pathogenesis of porcine enteric CoVs
The three enteric CoVs (TGEV, PEDV and PDCoV) infect mainly small intestinal villous enterocytes, causing acute necrosis that leads to villi atrophy (Chattha et al., 2015, Jung et al., 2015b, Jung and Saif, 2015, Jung et al., 2014). This could produce a severe diarrhea as a consequence of malabsorption. Often, diarrhea is accompanied by vomiting, which increases severe dehydration, anorexia and appetite loss. These clinical symptoms could lead to animal death. In general, PEDV and TGEV are considered more virulent than PDCoV (Chen et al., 2015), although after experimental inoculation of gnotobiotic piglets, PDCoV caused a severe disease (Jung et al., 2015b). Further analyses will be needed to determine whether the difference in viral strain, animal age, or other factors influence PDCoV pathogenesis.
The severity of clinical signs caused by both PEDV and TGEV is inversely related to the age of animals. Symptoms are very severe in nursing piglets, under 2-weeks of age, with a mortality of up to 95%. In weaned to finisher pigs and pregnant sows the clinical signs are milder and self-limiting within 5–10 days after the onset of disease (Jung and Saif, 2015). The higher susceptibility of younger piglets has been associated with the slower turnover of enterocytes (up to one week) in neonatal piglets compared with older animals (2–3 days). Enterocytes turnover correlated with the presence of LGR5+ stem cells in the intestinal crypt, which is reduced in nursing piglets (Jung et al., 2015a). In addition, a correlation between the severity of the infection and the innate immune responses has been observed. In young piglets natural killer (NK) cell activity and interferon (IFN)-γ production were lower than those in weaned pigs (Annamalai et al., 2015).
1.3. Vaccines available for porcine enteric CoVs
The most promising vaccines to protect against porcine enteric viruses are those that elicit mucosal immunity in the gut (Chattha et al., 2015). These vaccines may lead to a protection higher than 90% of a litter from a vaccinated sow. In general, there are different types of vaccines that could prevent infections by enteric CoVs: (i) subunit vaccines, (ii) inactivated vaccines, (iii) vectored vaccines, (iv) live attenuated vaccines based on viruses that have been passaged in cell cultures, and (v) engineered modified live attenuated vaccines.
Both inactivated and live attenuated TGEV and PEDV vaccines are manufactured and have been extensively used in Asia (http://www.cfsph.iastate.edu/Vaccines/). The emergence of virulent PEDV strains in China during 2010, even in previously vaccinated farms, and in 2013 in the USA points to the necessity for more effective vaccines. Subunit or killed vaccines are only partially effective inducing mucosal immunity, although they could be suitable for boosting immunity in sows prior to farrowing (Oh et al., 2014, Paudel et al., 2014). Live vaccines are more effective inducing long-lasting immunity, although current live attenuated vaccines did not prevent virus shedding. This could represent a biosafety problem as the vaccine strain can revert to virulence by recombination with circulating strains. In fact, such recombinant PEDV isolates have been identified in Asia (Li et al., 2016).
The engineering of well-defined recombinant enteric porcine CoVs may represent a major progress towards the development of more effective and biosafe vaccines. To this end, a combination of reverse genetics systems, using infectious cDNA clones, and analysis of virus-host interactions is required. First, viral genes modulating the innate immune response should be identified, as their deletion frequently leads to attenuated viruses and, therefore to potential vaccine candidates. The first CoV infectious cDNA clone was obtained for TGEV, as a bacterial artificial chromosome (BAC) (Almazan et al., 2000). Since then, additional cDNA clones were obtained for TGEV and PEDV, based either on BACs or in vitro ligation (Almazan et al., 2014, Beall et al., 2016, Jengarn et al., 2015). This review will focus on the study of enteric CoVs interaction with the host to determine virulence genes, since the modification or deletion of these genes may lead to virus attenuation, and to the generation of vaccine candidates to prevent infection by porcine CoVs. Alternatively, analysis of virus-host interactions may teach us the signaling pathways required by the virus for its replication or to cause pathogenesis. Once this has been accomplished, the selection of inhibitors of these signaling pathways could result in the identification of antivirals.
2. Host cell pathways involved in pathogenesis
CoVs affect many host-cell pathways that may have a positive or negative impact on viral replication and virulence. There is an extensive crosstalk between the cellular pathways altered after CoV infection. Modulation of a cell-signaling pathway affecting CoV replication may also have an impact on the host, contributing to CoV virulence. Thus, an increase of the innate immune response to decrease viral replication may enhance deleterious pro-inflammatory responses exacerbating pathogenesis. Moreover, as all vaccine types would also interact with the same host-cell pathways as the pathogenic CoV, knowledge on the signaling pathways altered by porcine enteric CoVs may have implications for vaccine development. Cell pathways affected by porcine CoV infection are revised below, to clarify their effect on viral replication and pathogenesis.
2.1. Apoptosis
TGEV and PEDV infection induce programmed cell death in cell cultures and in vivo (Cruz et al., 2011, Eleouet et al., 2000, Kim and Lee, 2014). In contrast, apoptosis produced by PDCoV has only been detected in cell cultures but not in vivo (Jung et al., 2016). The mechanism of apoptosis induction, and its effect on infection is different for TGEV and PEDV. TGEV-induced cell death is caspase-dependent, and apoptosis inhibition has no effect on viral replication (Eleouet et al., 1998). In contrast, PEDV induces a caspase-independent cyclophilin D-dependent pathway, and apoptosis inhibition decreases viral replication (Kim and Lee, 2014). Independently of the mechanism involved, in the infection by both CoVs, apoptosis has been associated with the promotion of virus shedding and pathogenesis (Cruz et al., 2011, Kim and Lee, 2014). It is worth noting that apoptosis is also related with innate immune response. For TGEV it has been demonstrated that apoptosis induction correlates with IFN and pro-inflammatory cytokines production (Becares et al., 2016, Cruz et al., 2011). Therefore, apoptosis may contribute to enteric CoV pathogenesis not only by killing infected cells, but also by facilitating viral dissemination and inflammation in the infected tissues.
2.2. Endoplasmic reticulum (ER) stress
Virus infection often induces ER stress, specifically the unfolded protein response (UPR). UPR activates three main signaling pathways to avoid the accumulation of proteins with folding alterations in the ER. Sometimes ER stress is irreversible, and cells undergo apoptosis. ER stress is also interconnected with the innate immune response and other host-cell responses (Hetz, 2012). The infection with many CoVs induces ER stress, and a role of UPR in pathogenesis has been described (DeDiego et al., 2011). In the case of enteric porcine CoVs, the induction of the UPR response has been reported for PEDV, with a negative effect on viral replication (Wang et al., 2014b). In contrast, TGEV infection does not induce ER stress (Cruz et al., 2011), despite the increased expression of several UPR mediators such as ATF4 or GADD34 after infection [(Cruz et al., 2011), S. Zuñiga and L. Enjuanes, unpublished results]. ER stress may then be an additional factor contributing to PEDV pathogenesis.
2.3. Mitogen-activated protein kinase (MAPK) signaling pathways
MAPK pathways are involved in the regulation of key host-cell processes, including innate immune response. The three main MAPK subfamilies: extracellular-signal regulated kinase (ERK), p38 kinase, and Jun-N-terminal kinase (JNK) regulated the expression of many host-cell genes that together modulate the inflammatory response (Arthur and Ley, 2013). Consequently, pathogens have evolved mechanisms to affect MAPK pathways to ensure their survival.
Both PEDV and TGEV induced the activation of MAPK pathways, positively affecting viral replication. After infection of polarized porcine intestinal epithelial cells (IPEC-J2), TGEV and PEDV induced MAPK activation to affect cell microfilaments and alter epithelial barrier integrity (Zhao et al., 2014). In both TGEV and PEDV infection, ERK activation is required for efficient virus replication. Interestingly, PEDV replication is not needed for ERK signaling pathway induction, as shown after infection of Vero cells (Kim and Lee, 2015).
PEDV and TGEV infection cause p38 kinase activation in polarized IPEC-J2 cells (Zhao et al., 2014). In addition, TGEV infection of porcine PK-15 cells also induces p38 kinase activation (Huang et al., 2013). Interestingly, inhibition of p38 kinase activation using a specific inhibitor (SB203580) does not affect PEDV or TGEV replication (Huang et al., 2013, Zhao et al., 2014). p38 kinase has an important role in the modulation of pro-inflammatory responses and, in fact, there are many anti-inflammatory drugs in different phases of clinical trials that inhibit p38 kinase activation (Arthur and Ley, 2013). In addition, it has been recently demonstrated that p38 kinase activation has a role in human severe and acute respiratory syndrome (SARS)-CoV pathogenesis, and that p38 kinase inhibition promotes survival of infected animals (Jimenez-Guardeño et al., 2014). Therefore, most likely p38 kinase plays a role in porcine CoV pathogenesis.
2.4. Cell cycle regulation
Although CoV replication occurs in the cytoplasm, they usually alter host cell cycle progression to create an optimum environment for their replication. TGEV arrests cell cycle at S and G2/M phases to favor its replication (Ding et al., 2013). It has been shown that TGEV cell cycle arrest was due to the activation of p53 transcription factor, which may also cause apoptosis (Ding et al., 2013, Huang et al., 2013). Transcription factor p53 is a key hub connecting many host signaling pathways responding to different stimuli. In this sense, there is a crosstalk between p53, which is also stimulated by IFN, and the antiviral response (Sato and Tsurumi, 2013, Takaoka et al., 2003). Altogether, these data suggest that cell cycle arrest and p53 activation may have a role on the pathogenesis produced by porcine CoVs.
2.5. Antiviral dsRNA pathway
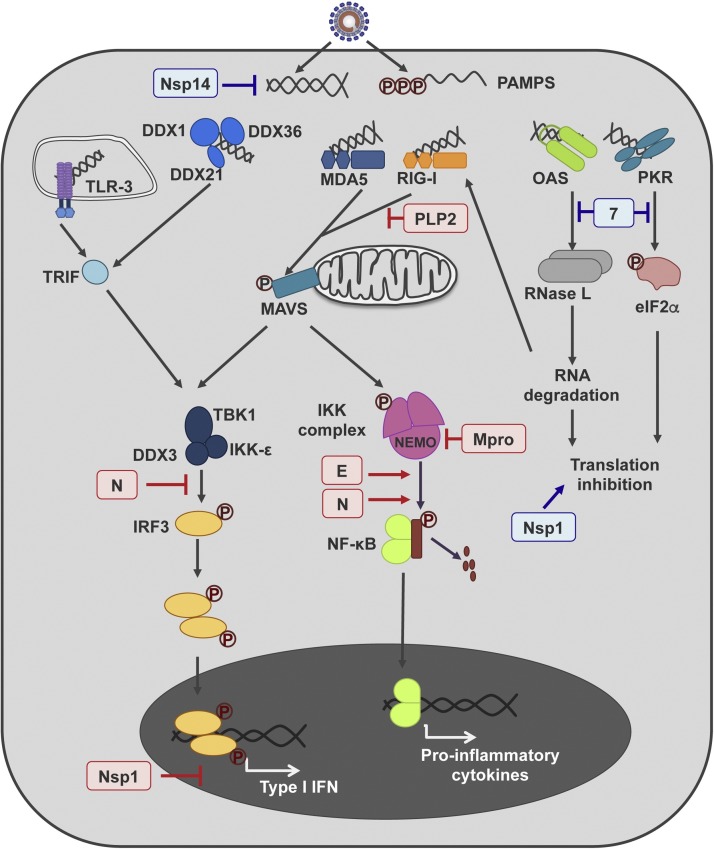
CoV infection triggers the activation of dsRNA-induced antiviral response (Fig. 2 ), which leads to the production of IFN and pro-inflammatory cytokines, modulating CoV pathogenesis (Cruz et al., 2011, Totura and Baric, 2012, Zust et al., 2011).
Fig. 2.
Porcine enteric CoV genes modulating innate immune response. A simplified representation of the main cell pathways activated after CoV infection leading to innate immune response, is shown in the figure. dsRNA is recognized as a pathogen-associated molecular pattern (PAMP) by a set of cellular sensors (TLR-3, DDX1 complex, MDA5, RIG-I, OAS and PKR). The binding of these sensors to dsRNA triggers several signaling pathways leading to type I IFN and pro-inflammatory cytokines production. The PEDV (in red) and TGEV (in blue) proteins affecting this response are indicated in the figure.
TGEV infection induces IFN and pro-inflammatory cytokines production in cell culture and in vivo (Cruz et al., 2013). A direct correlation between dsRNA antiviral response induction and TGEV virulence has been demonstrated (Cruz et al., 2011). In contrast, similarly to highly virulent human CoVs, PEDV infection inhibits IFN production in several cell types, such as monkey MARC-145 or porcine IEC cells (Cao et al., 2015a, Zhang et al., 2016). On the other hand, PEDV induced NF-κB activation both in monkey Vero and porcine IEC cells (Cao et al., 2015a, Guo et al., 2016). The modulation of these innate immunity pathways may have an effect on PEDV pathogenesis. In fact, it has been shown that virulent PEDV induces an increased NF-κB activation and pro-inflammatory cytokine production compared with an attenuated vaccine-like strain (Guo et al., 2016). Similarly to PEDV, PDCoV infection has been recently reported to inhibit IFN production in cell culture (Luo et al., 2016).
In addition, dsRNA is the molecule that also induces the host-cell RNA interference pathway (RNAi). Innate immune response and RNAi are highly related and there is a crosstalk between these pathways modulating virus pathogenesis (Pedersen et al., 2007). It has been recently shown that TGEV infection causes an extensive alteration of host cell RNAi response, changing the expression of multiple cellular microRNAs (Liu et al., 2015). There are preliminary reports suggesting that some of these microRNAs may be involved in the regulation of TGEV virulence, although these observations require further confirmation.
3. Viral proteins as virulence factors
CoVs have evolved different mechanisms affecting the host-pathways described above in order to modulate their pathogenesis. There is limited information on the viral proteins modulating enteric CoV virulence (Fig. 2), as many results are derived from overexpression studies, out of the context of the infection. Additional information could also be extracted from sequence comparison of virulent and attenuated strains, differing in the presence or absence of virulence factors, respectively. In this sense, the full-length genomes of virulent DR13 PEDV strain and its attenuated vaccine counterpart were sequenced (Park et al., 2012). Genome sequence comparison between these two strains, and also with other reference PEDV strains from Asia, EU and USA, showed that the attenuated DR13 virus accumulated a total of 536 nucleotide (nt) changes and several deletions after 100 passages in Vero cells leading to attenuation (Table 1 ). Only 25% of the nt changes were non-silent mutations, and just 35% of these (12% of the total nucleotide changes) were unique in the attenuated virus and were not present in other virulent PEDV circulating strains (Table 1). Most of the non-silent mutations in the attenuated virus were accumulated in the nsp3 and S genes (Table 1). In addition, ORF3 in the attenuated virus contains a 48 nt deletion that led to protein 3 truncation (Table 1). Similar results were obtained when full-length genomes of parental CV777 strain and the CV777 attenuated vaccine strain (GenBank accession number KT323979) were compared (S. Zuñiga and L. Enjuanes, unpublished results). The attenuated CV777 strain accumulated a total of 577 nt changes, 16% of them were non-silent and unique in that PEDV strain. Most of these mutations were accumulated in the nsp3 and S genes. Interestingly, the same deletions in nsp3 and ORF3 appeared in vaccine DR13 and CV777 strains. These data suggested that nsp3, S and ORF3 genes may be important targets for virus attenuation. This hypothesis could be tested with the availability of infectious cDNA clones.
Table 1.
Mutations present in attenuated DR13 strain. Full length genomes of virulent (GenBank JQ023161) and attenuated DR13 (GenBank JQ023162) viruses were compared with those of reference PEDV strains AH2012 (GenBank KC210145), CV777 (GenBank AF353511) and CO-2013 (GenBank KF272920).
| Point Mutations |
Deletions |
||||
|---|---|---|---|---|---|
| Gene | Totala | Non-silent | Uniqueb | nt | Resultc |
| 5′ UTR | 6 | – | – | ||
| Nsp1 | 4 | 1 | – | ||
| Nsp2 | 64 | 30 | 6 | ||
| Nsp3 | 14 | 30 | 12 | 23 | Δ1036–1043 |
| Nsp4 | 27 | 6 | 5 | ||
| Nsp5 | 24 | 6 | 2 | ||
| Nsp6 | 22 | 2 | 1 | ||
| Nsp7 | 4 | – | – | ||
| Nsp8 | 6 | 3 | – | ||
| Nsp9 | 9 | – | – | ||
| Nsp10 | 2 | – | – | ||
| Nsp12 | 39 | 2 | 2 | ||
| Nsp13 | 39 | 3 | 1 | ||
| Nsp14 | 39 | 2 | 2 | ||
| Nsp15 | 24 | 1 | – | ||
| Nsp16 | 19 | 3 | 1 | ||
| S | 67 | 25 | 25 | 3 | Δ152 |
| ORF3 | 7 | 1 | – | 48 | Δ92–224 |
| E | 5 | 2 | 2 | 21 | Δ23–29 |
| M | 13 | 4 | 2 | ||
| N | 40 | 13 | 3 | ||
| 3′UTR | 2 | – | – | ||
All mutations in attenuated DR13 compared with parental DR13 are included.
Non-silent mutations present exclusively in attenuated DR13 genome.
Aminoacid position in the replicase polyprotein is indicated for Nsp3.
3.1. Non-structural proteins
Many of the CoV replicase-encoded nsps have been described as IFN antagonists (Totura and Baric, 2012). Similarly, PEDV proteins nsp1, nsp3, nsp5, nsp7, nsp14, nsp15 and nsp16 have been identified as IFN antagonists in overexpression studies (Wang et al., 2015, Zhang et al., 2016). It would be interesting to study the effect of either the presence of deletion of these nsp genes on the innate immune response to PEDV, their role in virus attenuation and, therefore, their relevance in the generation of a live attenuated vaccine.
3.1.1. Nsp1
The most N-terminal nsp in CoV replicase polyproteins is only encoded by alpha- and betacoronaviruses. Despite the low conservation in sequence, the overall nsp1 structure is highly conserved in the two CoV genera (Jansson, 2013), and mutations in nsp1 could lead to attenuated viruses (Jauregui et al., 2013, Jimenez-Guardeño et al., 2015). Overexpression studies indicated that TGEV nsp1 protein suppresses protein translation, similarly to other CoV nsp1 proteins (Huang et al., 2011). Reduction of host protein translation has been suggested as a mechanism for IFN antagonism, although mutational analyses have indicated that these two nsp1 functions may be separated (Jauregui et al., 2013). Overexpression of PEDV nsp1 protein also led to inhibition of the innate immune response (Zhang et al., 2016). The nsp1 protein induced proteasomal-dependent CREB-binding protein (CBP) degradation, and suppression of IFN-stimulated genes (ISGs) expression. Interestingly, CBP degradation also occurs during PEDV infection (Zhang et al., 2016). Therefore, mutations affecting Alphacoronavirus nsp1 could lead to attenuated viruses to be used as vaccine candidates.
3.1.2. Nsp3
Nsp3 is a large transmembrane protein containing several functional domains that are conserved in porcine CoVs: one or two papain-like protease (PLP) domains, ubiquitin-like domains, and an ADP-ribose-phosphatase (ADRP) domain. PLP and de-ubiquitinase activities have been demonstrated for TGEV and PEDV nps3 proteins (Wojdyla et al., 2010, Xing et al., 2013). Also, ADRP activity has been reported for TGEV nsp3 protein (Putics et al., 2006). These activities have been associated with modulation of CoV pathogenesis (Fehr et al., 2015, Mielech et al., 2015). Overexpressed PEDV PLP2 de-ubiquitinates cellular nucleic acid sensors RIG-I and STING, decreasing IFN production (Xing et al., 2013). Interestingly, attenuated DR13 and CV777 vaccine viruses contain 12 and 21 non-silent point mutations, respectively, and an 8 amino acid deletion in nsp3 protein (Table 1). Altogether, these data strongly suggest a role of porcine CoV nsp3 protein in the modulation of virus pathogenesis. Therefore, its modification may lead to attenuated viruses useful as vaccine candidates.
3.1.3. Nsp5
Nsp5 gene encodes CoV main proteinase (Mpro) that is essential for replicase polyprotein processing (Ziebuhr, 2005). It has been recently reported that overexpression of PEDV nsp5 cleaves NF-κB essential modulator (NEMO), antagonizing IFN production (Wang et al., 2015). Further studies are required to know whether this also occurs during infection and the possible role of nsp5 protein in pathogenesis, as the extent of NF-κB activation has been related with PEDV virulence (Guo et al., 2016).
3.1.4. Nsp14
Nsp14 is a bifunctional protein encoding 3′-5′ exoribonuclease (ExoN) and N7-methyltransferase (N7-MTase) activities (Smith et al., 2014, Subissi et al., 2014). Therefore, this protein has essential functions in viral RNA synthesis, as a component of the unique CoV proofreading system, and the viral mRNA capping machinery. A role of nsp14 in the modulation of the innate immune response was recently hypothesized (Kindler and Thiel, 2014). Indeed, PEDV nsp14 overexpression antagonizes IFN production (Zhang et al., 2016). The first demonstration of nsp14 role in the modulation of the innate immune response in the viral context comes from recent findings with TGEV (Becares et al., 2016). A TGEV virus mutant in the first zinc-finger motif included in the ExoN domain of nsp14 protein was generated (TGEV-ZF-C). This mutant virus caused a decreased accumulation of dsRNA in the infected cells and, as a consequence, a reduced IFN, pro-inflammatory cytokines and ISGs production (Becares et al., 2016). It remains to be determined whether this mutant virus will be attenuated in vivo and could be used as a vaccine candidate.
3.2. Structural proteins
Overexpression studies have indicated that PEDV E, M and N proteins antagonize IFN induction (Zhang et al., 2016). It has been proposed that PEDV E protein induces UPR response, which leads to an increase in NF-κB activation (Xu et al., 2013a). This information is in agreement with previously reported data for SARS-CoV E protein, which has an important role in pathogenesis (DeDiego et al., 2011, DeDiego et al., 2014).
N protein is a multifunctional phosphoprotein essential for CoV RNA synthesis and with key roles in virus-host interaction (Surjit and Lal, 2008, Zuñiga et al., 2010). In fact, the interaction of TGEV N protein with several host cell proteins has been shown (Zhang et al., 2015b, Zhang et al., 2014c), and overexpression of PDCoV N protein in porcine cells affected the accumulation of many host cell proteins (Lee and Lee, 2015). In addition, both TGEV and PEDV N protein overexpression induces cell cycle arrest (Ding et al., 2014a, Xu et al., 2013b).
The role that N protein may have in porcine CoV pathogenesis has not been analyzed, in part due to the essential and multifunctional characteristics of N protein. Therefore N protein modification, required for the study of N protein functions in the infection context, represents a challenge. Nevertheless, there is accumulating evidence indicating that N protein could be a virulence factor. PEDV N protein expression has been correlated with the activation of NF-κB and increased production of pro-inflammatory cytokines (Xu et al., 2013b) possibly by the activation of Toll-like receptor (TLR) pathways (Cao et al., 2015b). On the other hand, PEDV N protein overexpression prevents TBK1-IRF3 interaction, avoiding IRF3 activation and IFN production (Ding et al., 2014b). Finally, the alteration of porcine RNAi response has been related with pathogenesis (Liu et al., 2015). In this sense, both TGEV and PEDV N proteins have RNA silencing suppressor activity, which may modulate host cell RNAi response (Cui et al., 2015). Interestingly, RNA silencing suppressor activity of TGEV N protein was stronger than that of PEDV N protein (Cui et al., 2015), although the implications of this observation in virulence require further analyses.
3.3. Genus-specific proteins
Porcine enteric CoVs contain several genus-specific genes that differ in number and genome location (Fig. 1). With the exception of ORF3 encoded proteins, these genes have no common features (see below). In general, genus-specific genes have been related with the modulation of pathogenesis, although there is limited knowledge on their function.
PDCoV encodes two genus-specific proteins of unknown function, one encoded by gene 5, located between M and N genes, and another one encoded by ORF7, overlapping with N gene.
3.3.1. ORF3 encoded proteins
Both TGEV and PEDV contain one or two genus-specific genes located between S and E genes (Fig. 1): TGEV ORFs 3a and 3b, and PEDV ORF3.
Although there is very limited sequence homology, the predicted structure of TGEV 3b, PEDV 3, HCoV-229E 4, SARS-CoV 3a and HCoV-NL63 3 proteins is similar. All include three transmembrane domains and human CoVs proteins are located in the ERGIC during infection or when overexpressed (Muller et al., 2010, Zhang et al., 2014b). All human CoV ORF3-encoded proteins have ion channel activity (Zhang et al., 2015a), most of them have been involved in morphogenesis (Donaldson et al., 2008, Zhang et al., 2014b), and can be incorporated into the viral envelope (Ito et al., 2005, Muller et al., 2010). In fact, ion channel activity of PEDV protein 3 has been reported (Wang et al., 2012). Therefore, most likely TGEV protein 3b may share this characteristic with PEDV and their human counterparts.
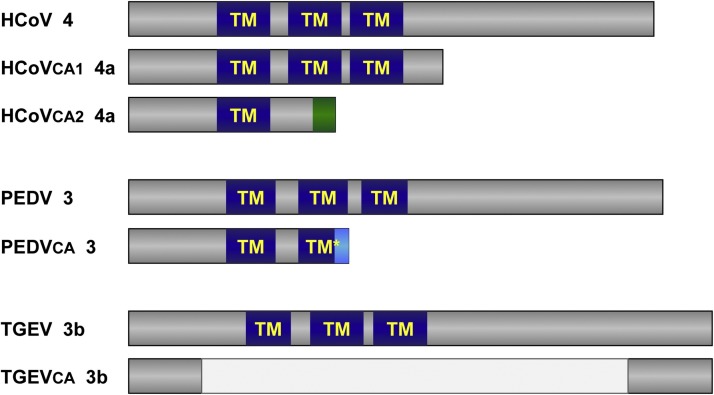
ORF3 encoded genes are not required for CoV growth in cell culture, although virus yield could be increased in their presence (Ye et al., 2015, Zhang et al., 2014b). A common feature between HCoV-229E, TGEV and PEDV is that cell-adapted strains tend to modify ORF3 (Fig. 3 ). In the case of HCoV-229E, a full length ORF is present in patient samples, while in cell-adapted strains this gene is either divided in two independent ORFs, 4a and 4b, or replaced by an ORF encoding for a truncated protein containing just the first transmembrane domain (Dijkman et al., 2006). PEDV DR13 strain, attenuated after 100 passages in Vero cells, encoded a truncated protein 3 (Park et al., 2012), and TGEV attenuated M60 strain, passed 64 times in ST cells, encodes a truncated 3b protein in which all transmembrane domains have been eliminated (Zhang et al., 2007) (Fig. 3).
Fig. 3.
Scheme of ORF3 encoded protein in different cell-adapted CoVs. Genus-specific ORF3-encoded protein is shown for: (i) HCoV-229E clinical isolate J0304 (HCoV, GenBank JX503061), and for cell-adapted viruses VR-740 (HCoVca1, AF304460) and HC-LP (HCoVca2, EF198671), upper scheme; (ii) PEDV virulent DR13 (JQ023161) and attenuated DR13 (PEDVca, JQ023162), middle scheme; and (iii) TGEV virulent Miller (DQ811785) and attenuated M60 (TGEVca, DQ811786), lower scheme. Predicted transmembrane domains (TM) are indicated in all cases. Mutations leading to a reduction in sequence identity are indicated in green and light blue.
TGEV virus lacking proteins 3a and 3b is as enteropathogenic as the parental virus, although its growth in enteric tract is reduced (Sola et al., 2003). Similarly, PEDV lacking ORF3 causes lethal disease in gnotobiotic piglets, although the diarrheic scores suggest that this mutant could be attenuated (Beall et al., 2016). Interestingly, a novel SeCoV has been recently discovered from diarrheic samples, which is a recombinant virus with a TGEV genomic background and S gene from PEDV (Boniotti et al., 2016). This SeCoV does not encode protein 3a, as it contains extensive deletions in the coding sequence and in transcription regulating sequence (TRS) including conserved core sequence (CS) essential for sgmRNA expression (Sola et al., 2015). Similarly, most PRCV isolates contain genome deletions in ORF 3a region, resulting either in a truncated 3a protein or in the complete absence of protein 3a expression. These data suggest that ORF3 encoded proteins may play a role in vivo.
3.3.2. TGEV protein 7
TGEV gene 7 is located at the 3′end of the genome and is only present in Alphacoronavirus 1 species (de Groot et al., 2012). Protein 7 antagonizes host antiviral response through its binding to cellular phosphatase PP1, decreasing translation initiation factor eIF2α phosphorylation and RNase L activation (Cruz et al., 2011). Mutant viruses lacking protein 7 expression produce an enhanced pathogenesis compared with parental viruses, both for enteric and respiratory tropism viruses (Cruz et al., 2011). The increased virulence of the mutant virus is due to an exacerbated pro-inflammatory response, both in cell culture and in vivo (Cruz et al., 2013). These data highlight the key role of protein 7 in modulating TGEV virulence.
4. Attenuated viruses as vaccine candidates
Theoretically, the modification of any of the viral genes involved in virulence may lead to attenuated phenotypes. These attenuated viruses could be the basis for the development of vaccine candidates. To guarantee the safety of vaccine candidates the introduction of several attenuating modifications, mapping at distal locations of CoV genome, would be required. Some of the already studied vaccine candidates are discussed below.
4.1. S protein mutants
In contrast with PEDV and PDCoV, TGEV has both enteric and respiratory tropism, and an increase in the respiratory tropism correlates with virus attenuation (Ballesteros et al., 1997, Sanchez et al., 1999). Mutations in TGEV S protein have been associated with the virus respiratory or enteric tropism. Thus, changes in the coding sequence, including small deletions and insertions (1 or 2 aa), into the first 390 aa of TGEV S protein correlate with changes in tropism [(Ballesteros et al., 1997), C.M. Sanchez and L. Enjuanes, unpublished results]. The emergence of the PRCV with a large S protein deletion in the N-terminus, ranging from aa 23 to aa 249 depending on the PRCV strain, led to the maintenance of the respiratory tropism and the loss of enteric tropism (Callebaut et al., 1988, Pensaert et al., 1986). This virus became prevalent in the porcine population worldwide, which may have elicited a protective immune response against TGEV (Bernard et al., 1989, Saif and Sestak, 2006). In fact, pigs seropositive for PRCV could be infected with TGEV, although they develop mild enteritis and usually recover form the infection.
The growth of TGEV and PEDV in cell cultures usually leads to the accumulation of point mutations and deletions in the half amino-terminus of S protein (first 400 aa), leading to cell adaptation of these CoVs. Frequently, these changes are associated with virus attenuation. Comparison of the genome sequence of the parental viruses and Vero-passaged DR13 and 83P-5 Korean and Japanese PEDV vaccine strains showed an increase in the accumulation of S protein changes (Fig. 4 ). Several of these mutations were shared between the two attenuated viruses, passed 100 times in cell culture, and most of these changes clustered in the N-terminal domain of S protein (Fig. 4) (S. Zuñiga, A. Pascual-Iglesias, L. Enjuanes, unpublished results).
Fig. 4.
Mutations accumulated in PEDV S protein with passages in cell cultures. The grey bars indicate the overall structure of PEDV S protein, indicating S1 and S2 domains. Different functional motifs are also represented, such as the signal peptide (SP), heptad-repeats domains (HR1 and HR2), and transmembrane domain (TM). In addition, the described domains inducing neutralizing antibodies are also represented (NE1, NE2 and NE3). Comparison was performed between virulent (GenBank JQ023161) and attenuated DR13 (JQ023162) strains, and 83P-5 parental strain (P0, AB548618) and virus passaged 34 (P34, AB548619), 61 (P61, AB548620) and 100 times (P100, AB548621) in Vero cells. Vertical bars into the dark blue rectangles represent non-silent point mutations that were identical (yellow bars) or different (red bars) between DR13 and 83P-5 P100 vaccine strains. The common deletion in the N-terminal is represented by the vertical white bar.
Similarly to TGEV, PEDV S-INDEL isolates also contain small deletions or insertions (1–4 aa) in the first 390 aa of S protein. In addition, two PEDV strains with a large deletion (up to 197 aa) in the N-terminus of S protein have also been isolated, one in the USA (PC177) and the other one in Japan (TTR-2) (Masuda et al., 2015, Oka et al., 2014). This deletion was similar, in location and size, to that found in PRCV that appeared in the field. The data indicate that PC177 virus deletion was acquired during cell culture passage (Oka et al., 2014) and preliminary data indicate that PC177 strain retains the ability to grow in vivo (Lin et al., 2015b). In contrast, TTR-2 virus deletion appeared in the field (Masuda et al., 2015). The virulence of TTR-2 strain in colostrum-deprived piglets has been recently analyzed. The results indicate that this PEDV strain is attenuated, as all piglets developed clinical signs although all of them eventually recovered from the infection (Suzuki et al., 2016).
The S-INDEL strains are considered as mild-strains, as they caused a reduced pathogenesis and faster return to baseline conditions in the farms were they were isolated, compared with virulent PEDV strains (Wang et al., 2014a). Similar results were observed after an experimental inoculation of 4-day-old conventional suckling piglets with an S-INDEL strain that led to a delay in diarrhea appearance, shorter symptom duration, and a reduction in the number of deaths compared with a virulent PEDV strain (Lin et al., 2015a). This data have been reinforced by a recently published study showing a reduced pathogenicity of S-INDEL strains in 5-day-old piglets (Chen et al., 2016). Interestingly, inoculation with S-INDEL strains confers cross-protection against a challenge with virulent PEDV strains, indicating the potential of S-INDEL mild strains as bases for the development of novel PEDV vaccines (Goede et al., 2015, Lin et al., 2015a).
4.2. Replication-competent propagation-defective
A TGEV virus lacking E gene (TGEV-ΔE) is replication-competent propagation-defective, and requires E protein in trans to grow in cell cultures (Ortego et al., 2002). TGEV-ΔE virus produced in packaging cell lines expressing E protein could be a good vaccine candidate in terms of safety, as it could not disseminate to neighboring cells. In addition, TGEV-ΔE virus particles contain an S protein with the native conformation for the induction of neutralizing antibodies. Moreover, as this deletion mutant virus will target mucosal tissues it could induce secretory immunity.
To date, the role of E protein from PEDV or PDCoV in virus growth has not been analyzed. Nevertheless, it could be a good target for the engineering of vaccine candidates due to its role as virulence factor.
5. Conclusions
Improvement of current vaccination strategies against enteric porcine CoVs is required, as novel strains of these viruses re-emerge continuously and are a threat for the porcine herds worldwide. The best vaccination results, in terms of protection and vaccine safety, should in principle be obtained using engineered live attenuated vaccines. To this end, a deeper knowledge of the porcine CoVs-host interactions is needed, to identify which host cell pathways are affected during porcine enteric CoV infection that influence the modulation of porcine CoV pathogenesis. The analysis of the viral genes involved in porcine CoV virulence will be valuable for the rational design of attenuated viruses that may serve as effective vaccine candidates.
Acknowledgements
This work was supported by grants from the Government of Spain (BIO2013-42869-R), a U.S. National Institutes of Health (NIH) project (2P01AI060699-06A1), and financial support of IMI and European Commission and in-kind contributions from EFPIA partners (ZAPI project, IMI Grant Agreement n°115760). SZ received a contract from NIH. We thank Marga González for her technical assistance.
References
- Akimkin V., Beer M., Blome S., Hanke D., Höper D., Jenckel M., Pohlmann A. New chimeric porcine coronavirus strain found in swine feces collected in Germany 2012. Emerg. Infect. Dis. 2016;22:1314–1315. doi: 10.3201/eid2207.160179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazan F., Gonzalez J.M., Penzes Z., Izeta A., Calvo E., Plana-Duran J., Enjuanes L. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5516–5521. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazan F., Sola I., Zuñiga S., Marquez-Jurado S., Morales L., Becares M., Enjuanes L. Coronavirus reverse genetic systems: infectious clones and replicons. Virus Res. 2014;189:262–270. doi: 10.1016/j.virusres.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annamalai T., Saif L.J., Lu Z., Jung K. Age-dependent variation in innate immune responses to porcine epidemic diarrhea virus infection in suckling versus weaned pigs. Vet. Immunol. Immunopathol. 2015;168:193–202. doi: 10.1016/j.vetimm.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur J.S., Ley S.C. Mitogen-activated protein kinases in innate immunity. Nat. Rev. Immunol. 2013;13:679–692. doi: 10.1038/nri3495. [DOI] [PubMed] [Google Scholar]
- Ballesteros M.L., Sanchez C.M., Enjuanes L. Two amino acid changes at the N-terminus of transmissible gastroenteritis coronavirus spike protein result in the loss of enteric tropism. Virology. 1997;227:378–388. doi: 10.1006/viro.1996.8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall A., Yount B., Lin C.M., Hou Y., Wang Q., Saif L., Baric R. Characterization of a pathogenic full-length cDNA clone and transmission model for porcine epidemic diarrhea virus strain PC22A. MBio. 2016;7:e01451–15. doi: 10.1128/mBio.01451-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becares M., Pascual-Iglesias A., Nogales A., Sola I., Enjuanes L., Zuniga S. Mutagenesis of coronavirus Nsp14 reveals its potential role in modulation of the innate immune response. J. Virol. 2016;90:5399–5414. doi: 10.1128/JVI.03259-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard S., Bottreau E., Aynaud J.M., Have P., Szymansky J. Natural infection with the porcine respiratory coronavirus induces protective lactogenic immunity against transmissible gastroenteritis. Vet. Microbiol. 1989;21:1–8. doi: 10.1016/0378-1135(89)90013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniotti M.B., Papetti A., Lavazza A., Alborali G., Sozzi E., Chiapponi C., Faccini S., Bonilauri P., Cordioli P., Marthaler D. Porcine epidemic diarrhea virus and discovery of a recombinant swine enteric coronavirus, Italy. Emerg. Infect. Dis. 2016;22:83–87. doi: 10.3201/eid2201.150544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut P., Correa I., Pensaert M., Jiménez G., Enjuanes L. Antigenic differentiation between transmissible gastroenteritis virus of swine and a related porcine respiratory coronavirus. J. Gen. Virol. 1988;69:1725–1730. doi: 10.1099/0022-1317-69-7-1725. [DOI] [PubMed] [Google Scholar]
- Cao L., Ge X., Gao Y., Herrler G., Ren Y., Ren X., Li G. Porcine epidemic diarrhea virus inhibits dsRNA-induced interferon-beta production in porcine intestinal epithelial cells by blockade of the RIG-I-mediated pathway. Virol. J. 2015;12:127. doi: 10.1186/s12985-015-0345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Ge X., Gao Y., Ren Y., Ren X., Li G. Porcine epidemic diarrhea virus infection induces NF-kappaB activation through the TLR2, TLR3 and TLR9 pathways in porcine intestinal epithelial cells. J. Gen. Virol. 2015;96:1757–1767. doi: 10.1099/vir.0.000133. [DOI] [PubMed] [Google Scholar]
- Chattha K.S., Roth J.A., Saif L.J. Strategies for design and application of enteric viral vaccines. Annu. Rev. Anim. Biosci. 2015;3:375–395. doi: 10.1146/annurev-animal-022114-111038. [DOI] [PubMed] [Google Scholar]
- Chen Q., Gauger P., Stafne M., Thomas J., Arruda P., Burrough E., Madson D., Brodie J., Magstadt D., Derscheid R., Welch M., Zhang J. Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology. 2015;482:51–59. doi: 10.1016/j.virol.2015.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Gauger P.C., Stafne M.R., Thomas J.T., Madson D.M., Huang H., Zheng Y., Li G., Zhang J. Pathogenesis comparison between the United States porcine epidemic diarrhea virus prototype and S-INDEL-variant strains in conventional neonatal piglets. J. Gen. Virol. 2016;97:1107–1121. doi: 10.1099/jgv.0.000419. [DOI] [PubMed] [Google Scholar]
- Cruz J.L.G., Sola I., Becares M., Alberca B., Plana J., Enjuanes L., Zuñiga S. Coronavirus gene 7 counteracts host defenses and modulates virus virulence. PLoS Pathog. 2011;7:e1002090. doi: 10.1371/journal.ppat.1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz J.L.G., Becares M., Sola I., Oliveros J.C., Enjuanes L., Zuñiga S. Alphacoronavirus protein 7 modulates host innate immune response. J. Virol. 2013;87:9754–9767. doi: 10.1128/JVI.01032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L., Wang H., Ji Y., Yang J., Xu S., Huang X., Wang Z., Qin L., Tien P., Zhou X., Guo D., Chen Y. The nucleocapsid protein of coronaviruses acts as a viral suppressor of RNA silencing in mammalian cells. J. Virol. 2015;89:9029–9043. doi: 10.1128/JVI.01331-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R., Enjuanes L., Gorbalenya A.E., Holmes K.V., Perlman S., Poon L., Rottier P.J.M., Talbot P.J., Woo P.C.Y., Ziebuhr J. Coronaviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; San Diego: 2012. pp. 774–796. [Google Scholar]
- DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeño J.M., Regla-Nava J.A., Alvarez E., Oliveros J.C., Zhao J., Fett C., Perlman S., Enjuanes L. Severe acute respiratory syndrome coronavirus envelope protein regulates cell stress response and apoptosis. PLoS Pathog. 2011;7:e1002315. doi: 10.1371/journal.ppat.1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeño J.M., Regla-Nava J.A., Castaño-Rodriguez C., Fernandez-Delgado R., Usera F., Enjuanes L. Coronavirus virulence genes with main focus on SARS-CoV envelope gene. Virus Res. 2014;194:124–137. doi: 10.1016/j.virusres.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkman R., Jebbink M.F., Wilbrink B., Pyrc K., Zaaijer H.L., Minor P.D., Franklin S., Berkhout B., Thiel V., van der Hoek L. Human coronavirus 229E encodes a single ORF4 protein between the spike and the envelope genes. Virol. J. 2006;3:106. doi: 10.1186/1743-422X-3-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Huang Y., Dai M., Zhao X., Du Q., Dong F., Wang L., Huo R., Zhang W., Xu X., Tong D. Transmissible gastroenteritis virus infection induces cell cycle arrest at S and G2/M phases via p53-dependent pathway. Virus Res. 2013;178:241–251. doi: 10.1016/j.virusres.2013.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Huang Y., Du Q., Dong F., Zhao X., Zhang W., Xu X., Tong D. TGEV nucleocapsid protein induces cell cycle arrest and apoptosis through activation of p53 signaling. Biochem. Biophys. Res. Commun. 2014;445:497–503. doi: 10.1016/j.bbrc.2014.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Fang L., Jing H., Zeng S., Wang D., Liu L., Zhang H., Luo R., Chen H., Xiao S. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J. Virol. 2014;88:8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson E.F., Yount B., Sims A.C., Burkett S., Pickles R.J., Baric R.S. Systematic assembly of a full-length infectious clone of human coronavirus NL63. J. Virol. 2008;82:11948–11957. doi: 10.1128/JVI.01804-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleouet J.F., Chilmonczyk S., Besnardeau L., Laude H. Transmissible gastroenteritis coronavirus induces programmed cell death in infected cells through a caspase-dependent pathway. J. Virol. 1998;72:4918–4924. doi: 10.1128/jvi.72.6.4918-4924.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleouet J.F., Slee E.A., Saurini F., Castagné N., Poncet D., Garrido C., Solary E., Martin S.J. The viral nucleocapsid protein of transmissible gastroenteritis coronavirus (TGEV) is cleaved by caspase-6 and −7 during TGEV-induced apoptosis. J. Virol. 2000;74:3975–3983. doi: 10.1128/jvi.74.9.3975-3983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes L., Gorbalenya A.E., de Groot R.J., Cowley J.A., Ziebuhr J., Snijder E.J. The nidovirales. In: Mahy B.W.J., Van Regenmortel M., Walker P., Majumder-Russell D., editors. Third Edition. vol. 5. Elsevier Ltd.; Oxford: 2008. pp. 419–430. (Encyclopedia of Virology). [Google Scholar]
- Fehr A.R., Athmer J., Channappanavar R., Phillips J.M., Meyerholz D.K., Perlman S. The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J. Virol. 2015;89:1523–1536. doi: 10.1128/JVI.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goede D., Murtaugh M.P., Nerem J., Yeske P., Rossow K., Morrison R. Previous infection of sows with a mild strain of porcine epidemic diarrhea virus confers protection against infection with a severe strain. Vet. Microbiol. 2015;176:161–164. doi: 10.1016/j.vetmic.2014.12.019. [DOI] [PubMed] [Google Scholar]
- Guo X., Hu H., Chen F., Li Z., Ye S., Cheng S., Zhang M., He Q. iTRAQ-based comparative proteomic analysis of Vero cells infected with virulent and CV777 vaccine strain-like strains of porcine epidemic diarrhea virus. J. Proteom. 2016;130:65–75. doi: 10.1016/j.jprot.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- Huang C., Lokugamage K.G., Rozovics J.M., Narayanan K., Semler B.L., Makino S. Alphacoronavirus transmissible gastroenteritis virus nsp1 protein suppresses protein translation in mammalian cells and in cell-free HeLa cell extracts but not in rabbit reticulocyte lysate. J. Virol. 2011;85:638–643. doi: 10.1128/JVI.01806-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Ding L., Li Z., Dai M., Zhao X., Li W., Du Q., Xu X., Tong D. Transmissible gastroenteritis virus infection induces cell apoptosis via activation of p53 signalling. J. Gen. Virol. 2013;94:1807–1817. doi: 10.1099/vir.0.051557-0. [DOI] [PubMed] [Google Scholar]
- Ito N., Mossel E.C., Narayanan K., Popov V.L., Huang C., Inoue T., Peters C.J., Makino S. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J. Virol. 2005;79:3182–3186. doi: 10.1128/JVI.79.5.3182-3186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson A.M. Structure of alphacoronavirus transmissible gastroenteritis virus nsp1 has implications for coronavirus nsp1 function and evolution. J. Virol. 2013;87:2949–2955. doi: 10.1128/JVI.03163-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauregui A.R., Savalia D., Lowry V.K., Farrell C.M., Wathelet M.G. Identification of residues of SARS-CoV nsp1 that differentially affect inhibition of gene expression and antiviral signaling. PLoS One. 2013;8:e62416. doi: 10.1371/journal.pone.0062416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jengarn J., Wongthida P., Wanasen N., Frantz P.N., Wanitchang A., Jongkaewwattana A. Genetic manipulation of porcine epidemic diarrhoea virus recovered from a full-length infectious cDNA clone. J. Gen. Virol. 2015;96:2206–2218. doi: 10.1099/vir.0.000184. [DOI] [PubMed] [Google Scholar]
- Jimenez-Guardeño J.M., Nieto-Torres J.L., DeDiego M.L., Regla-Nava J.A., Fernandez-Delgado R., Castaño-Rodriguez C., Enjuanes L. The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis. PLoS Pathog. 2014;10:e1004320. doi: 10.1371/journal.ppat.1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Guardeño J.M., Regla-Nava J.A., Nieto-Torres J.L., DeDiego M.L., Castano-Rodriguez C., Fernandez-Delgado R., Perlman S., Enjuanes L. Identification of the mechanisms causing reversion to virulence in an attenuated SARS-CoV for the design of a genetically stable vaccine. PLoS Pathog. 2015;11:e1005215. doi: 10.1371/journal.ppat.1005215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Saif L.J. Porcine epidemic diarrhea virus infection etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet. J. 2015;204:134–143. doi: 10.1016/j.tvjl.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Wang Q., Scheuer K.A., Lu Z., Zhang Y., Saif L.J. Pathology of US porcine epidemic diarrhea virus strain PC21A in gnotobiotic pigs. Emerg. Infect. Dis. 2014;20:662–665. doi: 10.3201/eid2004.131685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Annamalai T., Lu Z., Saif L.J. Comparative pathogenesis of US porcine epidemic diarrhea virus (PEDV) strain PC21A in conventional 9-day-old nursing piglets vs. 26-day-old weaned pigs. Vet. Microbiol. 2015;178:31–40. doi: 10.1016/j.vetmic.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Eyerly B., Lu Z., Chepngeno J., Saif L.J. Pathogenicity of 2 porcine deltacoronavirus strains in gnotobiotic pigs. Emerg. Infect. Dis. 2015;21:650–654. doi: 10.3201/eid2104.141859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Saif L.J. Porcine deltacoronavirus induces apoptosis in swine testicular and LLC porcine kidney cell lines in vitro but not in infected intestinal enterocytes in vivo. Vet. Microbiol. 2016;182:57–63. doi: 10.1016/j.vetmic.2015.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Lee C. Porcine epidemic diarrhea virus induces caspase-independent apoptosis through activation of mitochondrial apoptosis-inducing factor. Virology. 2014;460–461:180–193. doi: 10.1016/j.virol.2014.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Lee C. Extracellular signal-regulated kinase (ERK) activation is required for porcine epidemic diarrhea virus replication. Virology. 2015;484:181–193. doi: 10.1016/j.virol.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler E., Thiel V. To sense or not to sense viral RNA-essentials of coronavirus innate immune evasion. Curr. Opin. Microbiol. 2014;20C:69–75. doi: 10.1016/j.mib.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee C. Functional characterization and proteomic analysis of the nucleocapsid protein of porcine deltacoronavirus. Virus Res. 2015;208:136–145. doi: 10.1016/j.virusres.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Qiao S., Yang Y., Guo J., Xie S., Zhou E., Zhang G. Genome sequencing and analysis of a novel recombinant porcine epidemic diarrhea virus strain from Henan, China. Virus Genes. 2016;52:91–98. doi: 10.1007/s11262-015-1254-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.M., Annamalai T., Liu X., Gao X., Lu Z., El-Tholoth M., Hu H., Saif L.J., Wang Q. Experimental infection of a US spike-insertion deletion porcine epidemic diarrhea virus in conventional nursing piglets and cross-protection to the original US PEDV infection. Vet. Res. 2015;46:134. doi: 10.1186/s13567-015-0278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.M., Gao X., Oka T., Vlasova A.N., Esseili M.A., Wang Q., Saif L.J. Antigenic relationships among porcine epidemic diarrhea virus and transmissible gastroenteritis virus strains. J .Virol. 2015;89:3332–3342. doi: 10.1128/JVI.03196-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhu L., Liao S., Xu Z., Zhou Y. The porcine microRNA transcriptome response to transmissible gastroenteritis virus infection. PLoS One. 2015;10:e0120377. doi: 10.1371/journal.pone.0120377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Fang L., Dong N., Fang P., Ding Z., Wnag D., Chen H., Xiao S. Porcine deltacoronavirus (PDCoV) infection suppresses RIG-I-mediated interferon-ß production. Virology. 2016;495:10–17. doi: 10.1016/j.virol.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T., Murakami S., Takahashi O., Miyazaki A., Ohashi S., Yamasato H., Suzuki T. New porcine epidemic diarrhoea virus variant with a large deletion in the spike gene identified in domestic pigs. Arch. Virol. 2015;160:2565–2568. doi: 10.1007/s00705-015-2522-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielech A.M., Deng X., Chen Y., Kindler E., Wheeler D.L., Mesecar A.D., Thiel V., Perlman S., Baker S.C. Murine coronavirus ubiquitin-like domain is important for papain-like protease stability and viral pathogenesis. J. Virol. 2015;89:4907–4917. doi: 10.1128/JVI.00338-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M.A., van der Hoek L., Voss D., Bader O., Lehmann D., Schulz A.R., Kallies S., Suliman T., Fielding B.C., Drosten C., Niedrig M. Human coronavirus NL63 open reading frame 3 encodes a virion-incorporated N-glycosylated membrane protein. Virol. J. 2010;7:6. doi: 10.1186/1743-422X-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J., Lee K.W., Choi H.W., Lee C. Immunogenicity and protective efficacy of recombinant S1 domain of the porcine epidemic diarrhea virus spike protein. Arch. Virol. 2014;159:2977–2987. doi: 10.1007/s00705-014-2163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T., Saif L.J., Marthaler D., Esseili M.A., Meulia T., Lin C.M., Vlasova A.N., Jung K., Zhang Y., Wang Q. Cell culture isolation and sequence analysis of genetically diverse US porcine epidemic diarrhea virus strains including a novel strain with a large deletion in the spike gene. Vet. Microbiol. 2014;173:258–269. doi: 10.1016/j.vetmic.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortego J., Escors D., Laude H., Enjuanes L. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 2002;76:11518–11529. doi: 10.1128/JVI.76.22.11518-11529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Kim H.K., Song D.S., An D.J., Park B.K. Complete genome sequences of a Korean virulent porcine epidemic diarrhea virus and its attenuated counterpart. J. Virol. 2012;86:5964. doi: 10.1128/JVI.00557-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paudel S., Park J.E., Jang H., Hyun B.H., Yang D.G., Shin H.J. Evaluation of antibody response of killed and live vaccines against porcine epidemic diarrhea virus in a field study. Vet. Q. 2014;34:194–200. doi: 10.1080/01652176.2014.973999. [DOI] [PubMed] [Google Scholar]
- Pedersen I.M., Cheng G., Wieland S., Volinia S., Croce C.M., Chisari F.V., David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M., Callebaut P., Vergote J. Isolation of a porcine respiratory, non-enteric coronavirus related to transmissible gastroenteritis. Vet. Q. 1986;8:257–260. doi: 10.1080/01652176.1986.9694050. [DOI] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putics A., Gorbalenya A.E., Ziebuhr J. Identification of protease and ADP-ribose 1-monophosphatase activities associated with transmissible gastroenteritis virus non-structural protein 3. J. Gen. Virol. 2006;87:651–656. doi: 10.1099/vir.0.81596-0. [DOI] [PubMed] [Google Scholar]
- Saif L., Sestak K. Transmissible gastroenteritis and porcine respiratory coronavirus. In: Straw B., Zimmerman J., D’Allaire S.D.J.T., editors. Diseases of Swine. Blackwell Publishing; Ames, Ames, IA: 2006. pp. 489–516. [Google Scholar]
- Sanchez C.M., Izeta A., Sanchez-Morgado J.M., Alonso S., Sola I., Balasch M., Plana-Duran J., Enjuanes L. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 1999;73:7607–7618. doi: 10.1128/jvi.73.9.7607-7618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Tsurumi T. Genome guardian p53 and viral infections. Rev. Med. Virol. 2013;23:213–220. doi: 10.1002/rmv.1738. [DOI] [PubMed] [Google Scholar]
- Smith E.C., Sexton N.R., Denison M.R. Thinking outside the triangle: replication fidelity of the largest RNA viruses. Ann. Rev. Virol. 2014;1:111–132. doi: 10.1146/annurev-virology-031413-085507. [DOI] [PubMed] [Google Scholar]
- Sola I., Alonso S., Zuñiga S., Balasch M., Plana-Duran J., Enjuanes L. Engineering the transmissible gastroenteritis virus genome as an expression vector inducing lactogenic immunity. J. Virol. 2003;77:4357–4369. doi: 10.1128/JVI.77.7.4357-4369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Almazán F., Zúñiga S., Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subissi L., Imbert I., Ferron F., Collet A., Coutard B., Decroly E., Canard B. SARS-CoV ORF1b-encoded nonstructural proteins 12–16: replicative enzymes as antiviral targets. Antivir. Res. 2014;101:122–130. doi: 10.1016/j.antiviral.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M., Lal S.K. The SARS-CoV nucleocapsid protein: a protein with multifarious activities. Infect. Genet. Evol. 2008;8:397–405. doi: 10.1016/j.meegid.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Shibahara T., Yamaguchi R., Nakade K., Yamamoto T., Miyazaki A., Ohashi S. Pig epidemic diarrhoea virus S gene variant with a large deletion nonlethal to colostrum-deprived newborn piglets. J. Gen. Virol. 2016 doi: 10.1099/jgv.1090.000513. [DOI] [PubMed] [Google Scholar]
- Takaoka A., Hayakawa S., Yanai H., Stoiber D., Negishi H., Kikuchi H., Sasaki S., Imai K., Shibue T., Honda K., Taniguchi T. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature. 2003;424:516–523. doi: 10.1038/nature01850. [DOI] [PubMed] [Google Scholar]
- Totura A.L., Baric R.S. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2012;2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Lu W., Chen J., Xie S., Shi H., Hsu H., Yu W., Xu K., Bian C., Fischer W.B., Schwarz W., Feng L., Sun B. PEDV ORF3 encodes an ion channel protein and regulates virus production. FEBS Lett. 2012;586:384–391. doi: 10.1016/j.febslet.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg. Infect. Dis. 2014;20:917–919. doi: 10.3201/eid2005.140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li J.R., Sun M.X., Ni B., Huan C., Huang L., Li C., Fan H.J., Ren X.F., Mao X. Triggering unfolded protein response by 2-deoxy-d-glucose inhibits porcine epidemic diarrhea virus propagation. Antivir. Res. 2014;106:33–41. doi: 10.1016/j.antiviral.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Fang L., Shi Y., Zhang H., Gao L., Peng G., Chen H., Li K., Xiao S. Porcine epidemic diarrhea virus 3C-like protease regulates Its interferon antagonism by cleaving NEMO. J. Virol. 2015;90:2090–2101. doi: 10.1128/JVI.02514-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojdyla J.A., Manolaridis I., van Kasteren P.B., Kikkert M., Snijder E.J., Gorbalenya A.E., Tucker P.A. Papain-like protease 1 from transmissible gastroenteritis virus: crystal structure and enzymatic activity toward viral and cellular substrates. J. Virol. 2010;84:10063–10073. doi: 10.1128/JVI.00898-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Chen J., Tu J., Zhang B., Chen X., Shi H., Baker S.C., Feng L., Chen Z. The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase. J. Gen. Virol. 2013;94:1554–1567. doi: 10.1099/vir.0.051169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhang H., Zhang Q., Dong J., Liang Y., Huang Y., Liu H.J., Tong D. Porcine epidemic diarrhea virus E protein causes endoplasmic reticulum stress and up-regulates interleukin-8 expression. Virol. J. 2013;10:26. doi: 10.1186/1743-422X-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhang H., Zhang Q., Huang Y., Dong J., Liang Y., Liu H.J., Tong D. Porcine epidemic diarrhea virus N protein prolongs S-phase cell cycle, induces endoplasmic reticulum stress, and up-regulates interleukin-8 expression. Vet. Microbiol. 2013;164:212–221. doi: 10.1016/j.vetmic.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S., Li Z., Chen F., Li W., Guo X., Hu H., He Q. Porcine epidemic diarrhea virus ORF3 gene prolongs S-phase, facilitates formation of vesicles and promotes the proliferation of attenuated PEDV. Virus Genes. 2015;51:385–392. doi: 10.1007/s11262-015-1257-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Hasoksuz M., Spiro D., Halpin R., Wang S., Stollar S., Janies D., Hadya N., Tang Y., Ghedin E., Saif L. Complete genomic sequences, a key residue in the spike protein and deletions in nonstructural protein 3b of US strains of the virulent and attenuated coronaviruses, transmissible gastroenteritis virus and porcine respiratory coronavirus. Virology. 2007;358:424–435. doi: 10.1016/j.virol.2006.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Tang C., Yue H., Ren Y., Song Z. Viral metagenomics analysis demonstrates the diversity of viral flora in piglet diarrhoeic faeces in China. J. Gen. Virol. 2014;95:1603–1611. doi: 10.1099/vir.0.063743-0. [DOI] [PubMed] [Google Scholar]
- Zhang R., Wang K., Lv W., Yu W., Xie S., Xu K., Schwarz W., Xiong S., Sun B. The ORF4a protein of human coronavirus 229E functions as a viroporin that regulates viral production. Biochim. Biophys. Acta. 2014;1838:1088–1095. doi: 10.1016/j.bbamem.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Shi H., Chen J., Shi D., Li C., Feng L. EF1A interacting with nucleocapsid protein of transmissible gastroenteritis coronavirus and plays a role in virus replication. Vet. Microbiol. 2014;172:443–448. doi: 10.1016/j.vetmic.2014.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Wang K., Ping X., Yu W., Qian Z., Xiong S., Sun B. The ns12. 9 accessory protein of human coronavirus OC43 is a viroporin involved in virion morphogenesis and pathogenesis. J. Virol. 2015;89:11383–11395. doi: 10.1128/JVI.01986-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Shi H., Chen J., Shi D., Dong H., Feng L. Identification of the interaction between vimentin and nucleocapsid protein of transmissible gastroenteritis virus. Virus Res. 2015;200:56–63. doi: 10.1016/j.virusres.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Shi K., Yoo D. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology. 2016;489:252–268. doi: 10.1016/j.virol.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Gao J., Zhu L., Yang Q. Transmissible gastroenteritis virus and porcine epidemic diarrhoea virus infection induces dramatic changes in the tight junctions and microfilaments of polarized IPEC-J2 cells. Virus Res. 2014;192:34–45. doi: 10.1016/j.virusres.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. The coronavirus replicase. In: Enjuanes L., editor. vol. 287. Springer-Verlag; Berlin-Heidelberg: 2005. pp. 57–94. (Coronavirus Replication and Reverse Genetics). [Google Scholar]
- Zuñiga S., Cruz J.L., Sola I., Mateos-Gomez P.A., Palacio L., Enjuanes L. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J. Virol. 2010;84:2169–2175. doi: 10.1128/JVI.02011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S., Siddell S.G., Ludewig B., Thiel V. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]