Abstract
The CD1 proteins are a family of non-polymorphic, MHC class I-related molecules that present lipid antigens to subsets of T lymphocytes with innate- or adaptive-like immune functions. Recent studies have provided new insight into the identity of immunogenic CD1 antigens and the mechanisms that control the generation and loading of these antigens onto CD1 molecules. Furthermore, substantial progress has been made in identifying CD1-restricted T cells and decoding the diverse immunological functions of distinct CD1-restricted T cell subsets. These findings shed new light on the contributions of the CD1 antigen presentation pathway to normal health and a diverse array of pathologies, and provide a new impetus for exploiting this fascinating recognition system for the development of vaccines and immunotherapies.
Antigen Presentation Systems
Products encoded by the major histocompatibility complex (MHC) region of the vertebrate genome bind peptide fragments from pathogens and display them at the surface of antigen-presenting cells (APCs) for appraisal by T lymphocytes [1]. A hallmark of the classical MHC class I and class II proteins is their extensive polymorphism, which determines histocompatibility, controls host resistance to infection, and influences susceptibility to autoimmunity. In addition to the classical MHC class I products, many jawed vertebrates express non-polymorphic, MHC-related proteins with diverse immune functions [2]. Members of the CD1 family of MHC class I-related proteins present self- and foreign lipid antigens to T lymphocyte subsets whose functions are less well understood than conventional MHC-restricted T cells. Nevertheless, the CD1 antigen presentation system provides new targets for the development of vaccines and immunotherapies against a variety of diseases. To accomplish this goal, it is critically important to identify the antigens that are recognized by CD1-restricted T cells, to understand the pathways that control the generation and loading of these antigens onto CD1 molecules, and to clarify the molecular basis for lipid antigen recognition by CD1-restricted T cell receptors (TCRs). Recent studies have provided important insight into the mechanisms involved in the generation of immunogenic CD1 antigens, which is invaluable for understanding the functions of this antigen presentation system in health and disease, and for exploiting this system for vaccine development and therapeutic purposes.
General Themes in the CD1 Antigen Presentation System
CD1 Genes, Proteins and Evolution
CD1 proteins were originally identified as β2-microglobulin- (β2m; see Glossary) associated heavy chains encoded in a locus on human chromosome 1 [3, 4]. This region encodes five CD1 isoforms (CD1a–e) that, based on sequence homology, were classified into group 1 (CD1a–c) and group 2 (CD1d) proteins, whereas CD1e was considered an intermediate isoform, sometimes referred to as group 3 [5]. Group 1 and 2 CD1 proteins are expressed at the cell surface and function as antigen-presenting molecules, whereas CD1e is only expressed intracellularly and is involved in processing and editing lipids for presentation by the other human CD1 isoforms. Another distinguishing feature is that group 1 CD1 proteins are expressed predominantly on professional APCs, whereas group 2 CD1 proteins are expressed more widely. Additionally, expression of group 1 but not group 2 CD1 proteins is highly inducible by microbial products and cytokines. Each of the CD1 proteins is constitutively expressed on cortical thymocytes, which is required for the intrathymic development of CD1d-restricted T cells [6] and most likely for selection of group 1 CD1-restricted T cells as well [7].
The CD1 antigen presentation system predates the evolutionary split between mammals and reptiles [8]. The ancient origin of CD1, together with its evolutionary conservation among all mammalian species examined [9], suggests important functions of this antigen presentation system during an immune response. The number of CD1 genes in mammals differs widely among species, with some mammals expressing over ten CD1 genes. Like humans, several other mammals such as dogs, horses and guinea pigs contain genes for all five CD1 isotypes, whereas mice only encode CD1d protein. The absence of group 1 CD1 genes in mice has complicated the functional analysis of this group of CD1 proteins, which has been partially overcome by studying humanized mice [10, 11].
CD1 Structure, Antigens and TCR Interaction
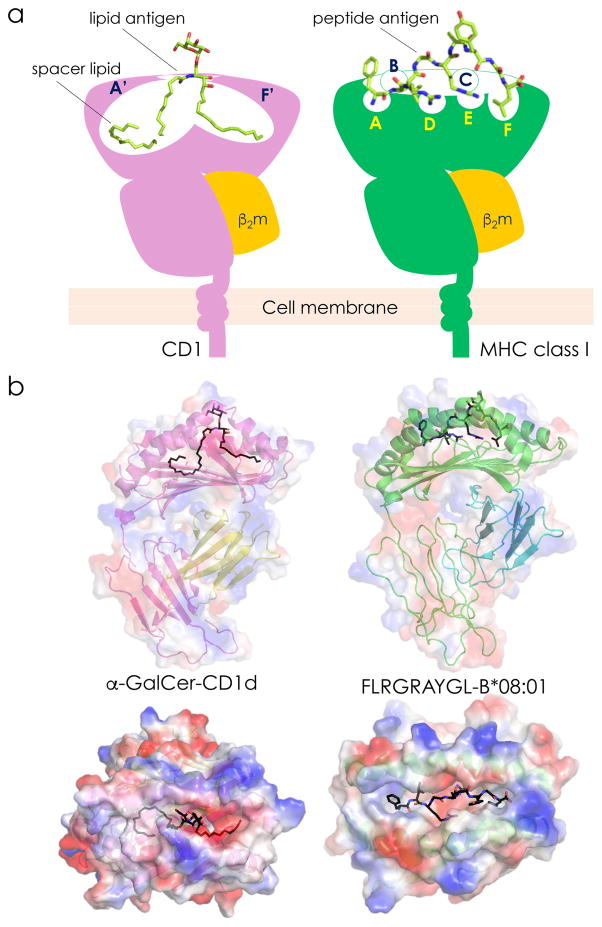
Crystal structures of CD1 molecules have revealed an overall resemblance to MHC class I but with two key differences [1, 8, 12]: (1) the CD1 inner surface is lined with hydrophobic residues, and (2) the α-helices of CD1 are extended further away from the floor of the cleft, resulting in a deeper antigen-binding groove (Figure 1a). The size of the antigen-binding groove differs substantially among distinct CD1 isoforms in the following order: CD1a<CD1d<CD1c<CD1e<CD1b (Table 1). Similar to the specificity pockets (labeled A–F) of classical MHC class I molecules, all CD1 molecules contain two antigen-binding pockets, called A’ and F’ (Figure 1). Additionally, CD1b contains a C’ pocket similar to the C pocket of MHC class I, as well as an additional T’ or “tunnel” pocket.
Figure 1. Comparative Anatomy of the α-GalCer-CD1d and Peptide-MHC Class I Ternary Structures.
a. A schematic view showing the key features of CD1 and MHC class I proteins. The specificity pockets are labeled A’ and F’ for CD1 and A–F for MHC class I molecules. Note that CD1b contains additional pockets (C’ and T’; not depicted). The CD1 groove is occupied by a lipid antigen and the MHC class I groove is occupied by a peptide antigen. In addition to an antigenic lipid, the CD1 groove may also contain a spacer lipid that fills up extra space in the antigen-binding pockets to stabilize the CD1 structure. b. The structures of human α-GalCer-CD1d (top left) and peptide- (Epstein Barr virus-derived FLRGRAYGL) HLA–B*08:01 (top right) complexes showing the electrostatic surfaces of the heavy chain and β2m. Note the depth of the CD1d antigen-binding groove that is accessed by the ligand through a narrow portal. In contrast, the peptide antigen-binding groove is relatively shallow and is accessed by a larger opening. These same structures were turned 90° toward the reader to show the solvent exposed polar galactose head group of α-GalCer (bottom left) and the roughly alternating amino acid side chains of the MHC-bound peptide (bottom right). Structures were made by PyMol using the protein database IDs 3HUJ (left) [119] and 3SJV (right) [120]. Abbreviation: β2m, β2-microglobulin.
Table 1.
Salient Features of the CD1 Antigen Presentation System
| CD1 isoform | Groove volume | TCR | TCR variability: T cell subset designation | Antigens recognized | References |
|---|---|---|---|---|---|
| CD1a | 1.35 nm3 | αβ | diverse | sulfatide, PE, PI, PC, lyso-PC, fatty acids, squalene, wax esters, dideoxymycobactin | [27, 28, 30, 107] |
| CD1b | 2.20 nm3 | αβ | diverse | sulfatide, mycolic acid, glycerol monomycolate, diacylated sulphoglycolipids | [107–110] |
| αβ | semi-invariant: GEM T | glucose monomycolate | [35] | ||
| αβ | biased: LDN5-like T | glucose monomycolate | [36] | ||
| CD1c | 1.78 nm3 | αβ | diverse | phosphomycoketide, mannosyl-phosphomycoketide, lipopeptides | [41, 111] |
| γδ | diverse | sulfatide, lyso-PC, phosphomycoketide, mannosyl-phosphomycoketide | [43, 107] | ||
| CD1d | 1.65 nm3 | Type I NKT or iNKT cells: | |||
| αβ | semi-invariant: Vα14 (mouse) or Vα24 (human) NKT | α-GalCer, β-GalCer, α-GlcCer, α-GalACer, α-GalDAG, α-GlcDAG, cholesteryl α-glucoside, asperamide B, GD3, iGb3, PE, PI, PC, lyso-PC, lysopeptidophosphoglycan | [50–59, 74, 77, 78, 112–116] | ||
| αβ | semi-invariant: Vα10 NKT | α-GalCer, α-GlcCer, α-GlcADAG | [81] | ||
| Type II NKT, dNKT or vNKT cells: | |||||
| αβ | biased: sulfatide-reactive dNKT | sulfatide, β-GlcCer, β-GalCer | [62, 107, 117] | ||
| αβ | diverse: “atypical” dNKT | α-GalCer | [97] | ||
| αβ | diverse: non-sulfatide-reactive dNKT | PG, PI, cardiolipin, lyso-PE non-lipid small molecules (e.g., PPBF), peptides (synthetic, ovalbumin-derived, collagen-derived) | [60, 61, 63, 98, 99, 102] | ||
| TCRγδ+ NKT cells: | |||||
| γδ | biased | cardiolipin | [118] | ||
| CD1e | 2.00 nm3 | - | - | sulfatide, PI, dimannosylated PI, diacylated sulphoglycolipids, hemi-bis(monoacylglycero)-phosphate | [105, 106] |
Abbreviations: dNKT, diverse NKT; GalCer, galactosylceramide; GalACer, galacturonosylceramide; GalADAG, galacturonosyldiacylglycerol; GalDAG, galactosyldiacylglycerol; GD3, ganglioside D3; GEM, germline-encoded mycolyl-specific; GlcCer, glucosylceramide; iGb3, isoglobotrihexosylceramide; iNKT, invariant NKT; NKT, natural killer T; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; vNKT, variant NKT; PPBF, phenyl 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonate.
CD1 molecules can bind with a variety of self- and foreign lipid antigens, but only a fraction of these activate T cells [8, 13, 14]. Examples of CD1 antigens are shown in Figure 2. Certain lipids such as sulfatide (3′-sulfated β1-D-galactosylceramide), an endogenous glycolipid highly expressed in neuronal cells, can promiscuously bind with all CD1 isotypes [15]. Sulfatide-reactive T cell lines restricted by all group 1 and 2 CD1 isotypes have been identified (Table 1). The diversity of CD1 antigens includes lipids, glycolipids, phospholipids, lipopeptides, oils and even non-lipid molecules. Crystal structures have revealed that lipids are oriented so that hydrophobic alkyl chains are buried deep within the antigen-binding pockets and that the hydrophilic head groups are solvent-exposed (Figure 1) [1, 8, 12]. Because of their differing volumes and shapes, the antigen-binding grooves of distinct CD1 isoforms can accommodate lipids containing alkyl chains of variable lengths. Interestingly, CD1b and CD1c can even bind lipids with alkyl chains that are longer than the calculated size of their respective antigen-binding groove. To accommodate long alkyl chains, one end of the lipid protrudes out of the groove via accessory portals. Such a detour ensures that the long alkyl chain does not interfere with TCR interactions. Conversely, CD1 molecules often present lipids with alkyl chains that are much shorter than the predicted size of the antigen-binding groove, which would leave extra space in the pockets, possibly causing structural problems. Analyses of CD1 complexes bound with short-chain lipids revealed that the unoccupied pockets were filled with spacer lipids that provide structural stability to the groove (Figure 1a, left) [16, 17]. Thus, with the assistance of multiple antigen-binding pockets, accessory portals and spacer lipids, CD1 molecules can diversify the size and range of lipids they present to T cells.
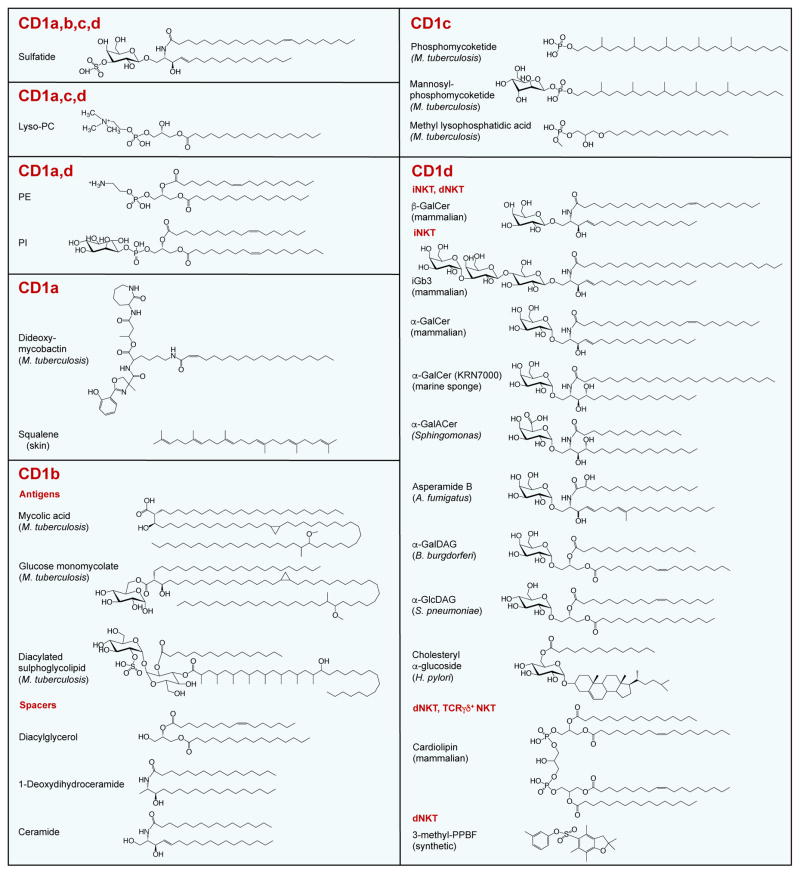
Figure 2. Select Antigens and Spacer Lipids of the CD1 Antigen Presentation System.
Structures are shown of select endogenous and exogenous antigens presented by distinct CD1 isotypes. Spacer lipids bound with CD1b, the only CD1 isotype where such lipids have been molecularly characterized thus far, are also depicted. For CD1d, antigens recognized by key subsets of CD1d-restricted T cells are shown. Note that endogenous, ER-derived phospholipids such as PE and PI, while recognized by some CD1-restricted T cells, also function as chaperone lipids to stabilize the CD1 groove and facilitate egress to the cell surface. Abbreviations: A. fumigatus, Aspergillus fumigatus; B. burgdorferi, Borrelia burgdorferi; dNKT, diverse NKT; GalCer, galactosylceramide; GalACer, galacturonosylceramide; GalDAG, galactosyldiacylglycerol; GlcDAG, glucosyldiacylglycerol; H. pylori, Helicobacter pylori; iGb3, isoglobotrihexosylceramide; iNKT, invariant NKT; M. tuberculosis, Mycobacterium tuberculosis; NKT, natural killer T; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; S. pneumoniae, Streptococcus pneumoniae; PPBF, phenyl 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonate.
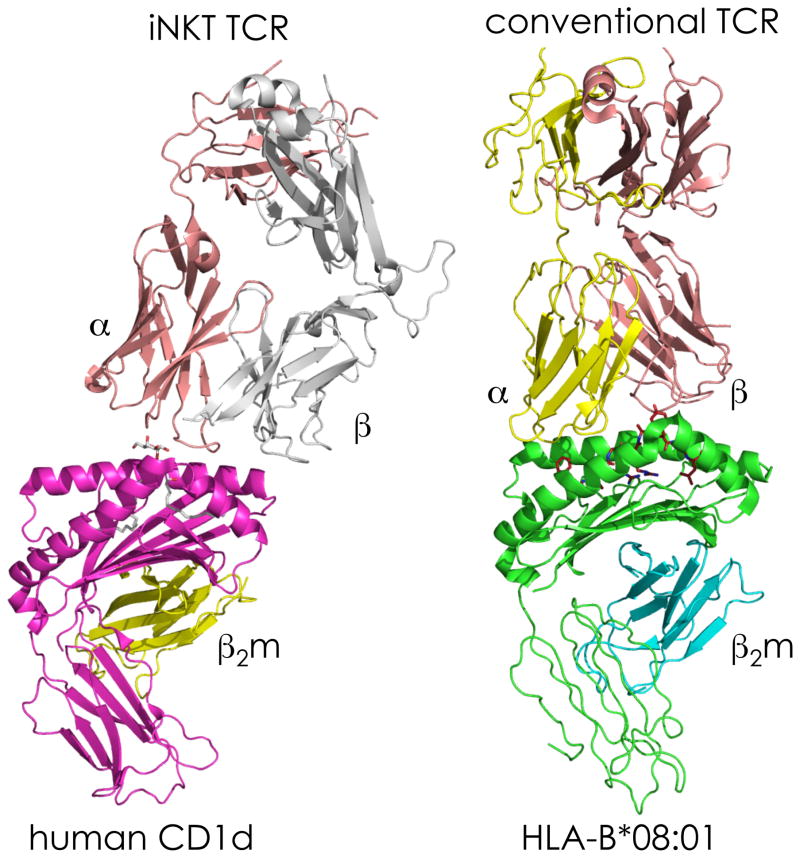
The crystal structures of CD1-lipid complexes have revealed striking similarities and important differences with MHC-peptide complexes that have critical implications for TCR recognition [1, 8, 12]. Whereas amino acid side chains of peptide are solvent-exposed and available for TCR interactions along the MHC groove, the region above the A’ pocket at the left side of the CD1 groove (referred to as the A’ roof) is closed, leaving only the polar moiety of the lipid to protrude out of CD1 via the F’ portal at the right side of the groove, thereby making the head group available for interactions with the TCR (Figure 1). Crystallographic studies of TCR/MHC-peptide complexes have revealed a diagonal or sometimes near-orthogonal binding mode with a common docking topology along the center of the groove in which the TCRα chain is positioned over one α-helix (from the α2 domain of MHC class I or the β1 domain of MHC class II) and the TCRβ chain is positioned over the second α-helix (from the α1 domain of MHC class I or the α1 domain of MHC class II). A similar topology has been observed in several TCR/CD1-lipid complexes, although the semi-invariant TCR expressed by a subset of CD1d-restricted T cells is oriented parallel to the axis of the groove (Figure 3) [1, 8]. Additionally, due to the asymmetric nature of the CD1 groove, TCRs may shift either to the left side or to the right side of the groove, resulting in wide variation in the extent of interactions with the protruding lipid head group.
Figure 3. Comparative Anatomy of the Semi-invariant NKTCR/α-GalCer-CD1d and Conventional TCR/peptide-MHC Class I Ternary Structures.
The interactions between human iNKTCR and its ligand, α-GalCer-CD1d (left), as well as the interactions between a conventional human TCR and its antigen, peptide- (as in Figure 1b) HLA–B*08:01 (right), are shown. Structures were made by PyMol using the protein database IDs 3HUJ (left) [119] and 3SJV (right) [120]. Abbreviations: α, TCRα; β, TCRβ; β2m, β2-microglobulin; HLA, human leukocyte antigen; TCR, T cell receptor.
CD1 Antigen Processing Pathways
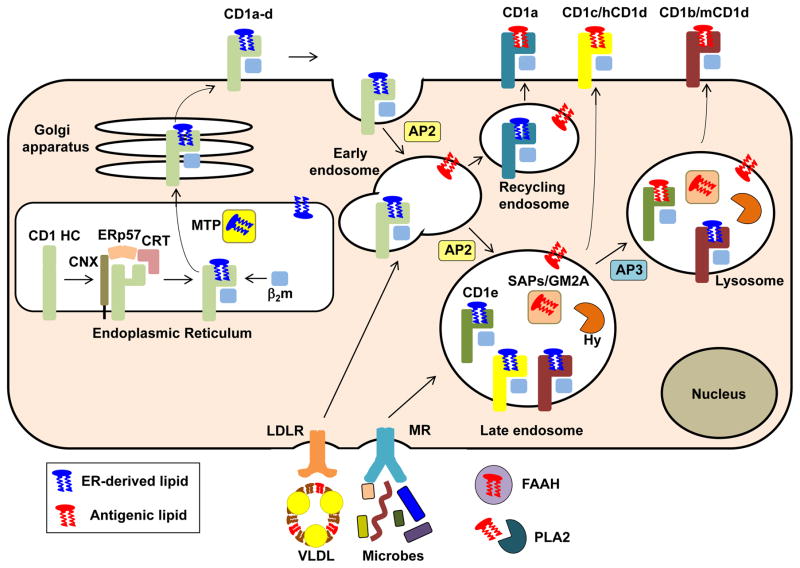
The assembly of CD1-lipid antigen complexes is initiated in the lumen of the endoplasmic reticulum (ER) by association of CD1 heavy chains with a variety of chaperones such as calnexin, calreticulin and the thiol oxidoreductase ERp57 (Figure 4), which assist in folding and assembly [5, 18, 19]. CD1 heavy chains then bind with β2m as well as with a variety of endogenous ER lipids, possibly with the assistance of spacer lipids, that together function as chaperones to stabilize the CD1 molecule. This association is reminiscent of the binding of newly synthesized MHC class II molecules with the MHC class II-associated invariant chain in the ER. Loading of such ER-resident lipids onto newly synthesized CD1 molecules is facilitated by microsomal transfer protein (MTP), an ER-resident lipid-transfer protein (LTP) that is also known for its capacity to facilitate the assembly of very low-density lipoproteins (VLDL) and chylomicrons. The CD1-lipid complexes subsequently egress to the plasma membrane, followed by internalization and entry into endosomes. CD1b–d proteins contain a tyrosine-based sorting motif that permits their binding with the adaptor protein complex 2 (AP2), which facilitates entry into a variety of endosomal compartments. Human CD1b and mouse CD1d (but not human CD1d) also contain AP3 sorting motifs that facilitate entry of these CD1 isoforms into lysosomes. CD1a, which lacks such sorting motifs, exhibits AP-independent recycling to early endosomes. In this manner, different CD1 isoforms can sample the lipid antigens that may be present in distinct intracellular compartments (Figure 4), potentially eliciting specific and non-redundant T cell responses. The ER-derived lipid cargo on these internalized and differentially sorted CD1 proteins may then be replaced with other endogenous or exogenous lipids. Lipid transport to these compartments is facilitated by lipoproteins, lipoprotein receptors and lectin receptors [20] (Figure 4). Additionally, some CD1 isoforms, especially CD1a, can exchange lipids at the cell surface. While many lipids do not require any additional processing for their association with CD1 proteins, some antigens require trimming by intracellular or extracellular carbohydrate hydrolases or phospholipases. Lipid exchange in endosomal compartments, which may or may not involve removal of spacer lipids, is facilitated by CD1e (in human but not mouse) or a variety of LTPs, including several saposins and the GM2 activator protein [21].
Figure 4. CD1-restricted Antigen Processing Pathways.
Newly synthesized CD1 heavy chains (generic CD1 heavy chains are indicated in light green whereas individual heavy chains are indicated in other colors) in the ER are stabilized by a variety of chaperones (CNX, ERp57 and CRT) and assemble with β2m and ER-derived chaperone lipids that are loaded onto CD1 with the assistance of the lipid-transfer protein (LTP) MTP. CD1-lipid complexes then transit to the cell surface, except for CD1e, which is directly transported to endosomal compartments, where it is cleaved into a soluble form. Following their arrival at the cell surface, CD1 proteins are internalized via their cytoplasmic, tyrosine-based sorting motifs that interact with AP2 (CD1b–d) and AP3 (CD1b and mCD1d) complexes, which permit CD1 proteins access to distinct intracellular compartments. CD1a, which lacks AP2 and AP3 sorting motifs, only accesses early endosomes in an AP-independent manner. ER-resident CD1d proteins may also gain access to endolysosomal compartments via an auxiliary pathway that involves the MHC class II-associated invariant chain (not depicted). In the endocytic system, CD1 proteins are loaded with endogenous or exogenous (e.g., microbial) lipid antigens, with the assistance of a variety of LTPs such as CD1e, SAPs and GM2A. CD1 antigens may be delivered to these intracellular compartments via extracellular lipid-binding proteins such as VLDL and FAAH, followed by receptor-mediated uptake via a variety of receptors such as the mannose and LDL receptors. Some products also require processing into antigenic ligands via extracellular factors such as PLA2 or intracellular factors such as lipid or carbohydrate Hy. Following loading with lipid in intracellular compartments CD1-lipid complexes recycle back to the cell surface. Note that most antigen-presenting CD1 isoforms can also be loaded with some antigens at the cell surface (not depicted). Abbreviations: AP, adaptor protein; β2m, β2-microglobulin; CNX, calnexin; CRT, calreticulin; ER, endoplasmic reticulum; FAAH, fatty acid amide hydrolase; GM2A, GM2 activator; HC, heavy chain; hCD1d, human CD1d; Hy, hydrolase; LDLR, low-density lipoprotein receptor; mCD1d, mouse CD1d; MR, mannose receptor; MTP, microsomal triglyceride protein; PLA2, phospholipase A2; SAP, saposin; VLDL, very low-density lipoprotein.
CD1-restricted T Cells
The analysis of CD1-restricted T cells has been greatly facilitated by the generation of CD1-lipid tetramers, which are now available for all group 1 and 2 CD1 isoforms [13]. A general theme in CD1-restricted T cell responses is a propensity for autoreactivity, which has been observed for each of the CD1 isoforms [5, 13]. CD1-restricted T cells may express αβ or γδ TCRs (Table 1), and include CD4+, CD8+ and double-negative cells [5, 13, 22]. Most studies on group 1 CD1-restricted T cells have focused on TCRs reactive with self- or mycobacterial antigens [5, 13]. T cells with reactivity against mycobacterial antigens are enriched in tuberculosis patients, and many of these cells have cytotoxic properties. With the exception of some CD1b-restricted T cells, group 1 CD1-restricted TCRαβ+ T cells express diverse TCRs and exhibit adaptive-like effector functions similar to conventional MHC-restricted T cells [5, 13]. The majority of CD1d-restricted T cells express natural killer (NK) cell surface markers such as NK1.1 (in mice) and are referred to as natural killer T (NKT) cells [23, 24] (Table 1). One subset of NKT cells, called Type I NKT cells or invariant NKT (iNKT) cells, expresses TCRα chains that are germ-line encoded, whereas a second subset of NKT cells, called Type II NKT cells, diverse NKT (dNKT) cells or variant NKT cells, expresses relatively diverse TCRs. In this review article we will refer to these subsets as iNKT and dNKT cells. NKT cells recognize a variety of endogenous and exogenous lipid antigens and exhibit innate-like effector functions, with complementary although sometimes opposing roles. One common feature of the CD1 antigen recognition system is that, in addition to TCRs with exquisite antigen-specificity, TCRs with limited or no antigen-specificity and a heavy bias towards recognition of CD1 alone are frequently observed. The latter property is consistent with the finding that this recognition system frequently involves reactivity of individual TCRs against both self- and foreign antigens. In addition to TCR-mediated activation, CD1-restricted T cells are also highly responsive to innate cytokine signals, even in the absence of TCR engagement [24–26], in this respect behaving much like innate lymphoid cells.
CD1a
Like the other CD1 isotypes, CD1a is expressed by myeloid APCs, but is especially abundant on epidermal Langerhans cells, and is employed as a characteristic cell marker for this cell type in humans [5, 13]. CD1a-restricted T cells have been identified that react with antigens derived from mycobacteria, pollen and endogenous skin oils [27–29]. Although autoreactivity is a common feature of CD1-restricted T cells, that is particularly the case for CD1a-restricted T cells [30]. The latter finding is consistent with the capacity of the CD1a groove to accommodate a large variety of antigens [8]. CD1a has the smallest antigen-binding groove of all CD1 proteins and can accommodate both lipids with polar head groups such as sulfatide, as well as “head-less” hydrophobic lipids such as skin-derived squalene, wax esters and triacylglycerides [8, 12, 31]. CD1a-restricted T cells reactive with both types of lipid antigen have been identified. Because the TCRs reactive with lipids containing polar head groups are usually very sensitive to head group modifications, it has been suggested that such TCRs interact with both CD1a and lipid, consistent with the classical concept of dual recognition of both MHC and antigen by TCRs. This mode of recognition contrasts sharply with that of CD1a-restricted TCRs to lipids that lack polar head groups. Crystallographic studies of an autoreactive TCR showed that it interacted with the A’ roof of CD1a in an asymmetric, left-sided manner but did not contact the lipid antigen in the groove [30]. It was proposed that in those cases the lipid largely plays a CD1a-stabilizing role without directly contributing to TCR interactions and that it is therefore not an antigen in the strict sense but rather a permissive ligand. Such a recognition mode, which is likely common among autoreactive CD1a-restricted T cells, might be able to accommodate a variety of lipids that can stabilize the CD1a groove and activate the TCR, as long as their head groups do not interfere with the capacity of the TCR to dock onto the A’ roof of the CD1a groove. Indeed, permissive lipids for this autoreactive TCR include phospholipids, lysophosphatidylcholine (lyso-PC) and fatty acids that lack polar head groups, whereas non-permissive lipids include sphingomyelin and sulfatide that contain polar head groups [8, 12, 31].
CD1a is the only antigen-presenting CD1 isoform that lacks a tyrosine-based sorting motif for internalization into endosomes [5]. Its localization is therefore restricted to the cell surface and early endosomal compartments with a close to neutral pH. CD1a is relatively stable in the absence of bound lipid and can exchange lipids at the cell surface, without the need for additional accessory factors. Recent studies have provided evidence that the permissive free fatty acids that stabilize the CD1a groove for recognition by autoreactive CD1a-restricted TCRs are generated from common skin phospholipids via endogenous or exogenous phospholipase A2 (PLA2) action [28, 31–33].
Among the CD1a-restricted T cells, the autoreactive T cells in the skin have been characterized most extensively [31]. These cells are present in the dermis of the skin and are thus physically separated from the CD1a-expressing Langerhans cells in the epidermis. These T cells exhibit autoreactivity against hydrophobic skin lipids such as squalene, fatty acids and wax esters. These lipids are contained within skin sebum, which is produced by sebaceous glands and exits the skin via hair follicles to coat the outer layer of the epidermis. Thus, access of these lipids to CD1a-expressing Langerhans cells and CD1a-restricted T cells requires a skin breach, via infection or injury. Recent studies have suggested that endogenous PLA2 secreted by skin cells may be involved in processing ubiquitous skin phospholipids into the natural oils recognized by CD1a-reactive T cells [33]. This possibility was suggested by a provocative study showing that PLA2 in insect venom, when introduced into the dermis of the skin, can convert phospholipids into CD1a-binding fatty acids that are capable of activating autoreactive CD1a-restricted T cells [32]. Consistent with this finding, CD1a-restricted T cells were expanded in individuals allergic to bee and wasp venom [34]. Autoreactive CD1a-restricted T cells produce the cytokines IFN-γ and IL-22 [28], which contribute to antimicrobial defenses, keratinocyte proliferation and a variety of skin inflammatory diseases. In this manner, CD1a-reactive T cells might be able to respond quickly to tissue injury, infectious agents and venoms by promoting antimicrobial and inflammatory responses. Consequently, this pathway may be exploited for therapeutic purposes against a variety of diseases. In this context it is interesting that several skin pathobionts, including the fungal organism Candida albicans and the bacterial organism Staphylococcus aureus, secrete phospholipases, which may thus contribute to the generation of stimulatory ligands for CD1a-reactive T cells. Additionally, some vaccine adjuvants contain squalene or other oils that may be able to activate CD1a-restricted T cells, thus potentially contributing to adjuvanticity. In addition to skin-derived self-lipids, CD1a-restricted T cells reactive with mycobacterial and pollen-derived lipids have also been identified [29], although their physiological function requires further study.
CD1b
Apart from thymocytes, CD1b is nearly exclusively expressed by dendritic cells (DCs). Autoreactive and pathogen-reactive CD1b-restricted T cells have been identified [5, 13]. CD1b has the largest binding groove of all CD1 proteins and contains four interconnected pockets (A’, C’, F’ and T’). This permits CD1b to bind with lipids containing long alkyl chains such as mycobacterial mycolic acid, glucose monomycolate and glycerol monomycolate [8, 12]. Additionally, shorter lipids such as mycobacterial sulfoglycolipids and a short-chain form of glucose monomycolate can be loaded onto CD1b in the presence of a spacer lipid, a diacylglycerol or a deoxyceramide (see chemical structures in Figure 2) [16]. Such natural spacer lipids in CD1b push the groove upwards to enhance antigen recognition and thus function as scaffolds to support the TCR/CD1b-lipid interface. This concept is consistent with the finding that long-chain glucose monomycolate requires the low pH environment of lysosomes for lipid exchange, whereas short-chain monomycolate can be loaded at the cell surface [8, 12]. Presumably, the former type of lipid exchange involves release of either a large ER-derived lipid, or a short ER-derived lipid together with a spacer lipid, whereas the latter type of lipid exchange may involve release of only the short ER-derived lipid while the spacer lipid remains bound with CD1b.
Although most CD1b-restricted T cells express diverse TCRs, subsets of glucose monomycolate-reactive T cells expressing semi-invariant (termed germ-line encoded mycolyl [GEM] lipid-reactive T cells) [35] or biased (termed LDN5-like T cells) [36] TCRs have been identified (Table 1). Among all CD1-restricted T cells that recognize mycobacterial antigens, CD1b-restricted T cells are most abundant [5]. Such cells are enriched in peripheral blood and at sites of infection in tuberculosis patients [37]. These cells exhibit cytotoxic activities and produce a variety of cytokines such as IL-2 and IFN-γ [37]. Recent studies with humanized transgenic animals expressing all human group 1 CD1 proteins together with a mycolic acid-specific CD1b-restricted TCR have provided the first convincing evidence for rapid and protective host immune responses mediated by the CD1 antigen presentation system [38]. This rapid response to Mycobacterium tuberculosis infection suggests that this system can be targeted for the development of tuberculosis vaccines.
The recent analysis of a set of autoreactive CD1b-restricted T cells revealed reactivity against phosphatidylglycerol (PG) produced by mitochondria as well as by several different bacterial pathogens such as Salmonella and Staphylococcus species [39]. Thus, these cells exhibit mixed reactivity against self- and foreign antigens. These findings also provide evidence for a much broader reactivity of CD1-reactive T cells to bacterial pathogens and suggest a role for these cells in settings such as metabolic diseases that cause mitochondrial stress.
CD1c
CD1c is abundantly expressed by myeloid DCs and B cells, especially marginal zone B cells. CD1c-restricted αβ and γδ T cells that react with endogenous sulfatides or cholesteryl esters, or with mycobacterial cell wall antigens have been identified [5, 13, 40, 41]. Each of the exogenous CD1c antigens identified, phosphomycoketide, mannosyl-phosphomycoketide and a synthetic lipopeptide (acyl-12) contain a single alkyl chain, which is embedded in the A’ pocket [17, 42]. In one of the CD1c-lipid co-crystals, the F’ pocket contained a C12 hydrocarbon chain likely derived from a detergent in the solution used during crystallization [17]. This finding suggests that spacer lipids are involved in the CD1c-mediated presentation of lipid antigens containing a single alkyl chain.
CD1c-reactive T cells have been identified in human blood, expand in patients with tuberculosis, and infiltrate organs affected by autoimmunity [5]. Although CD1c-restricted γδ T cells were identified over 25 years ago, natural ligands of Vδ1+ TCRs have only recently been identified as mycobacterial phosphomycoketide [43]. Strikingly, these γδ T cells also reacted with diverse lipids, including lyso-PC, sulfatide, and mannosyl-phosphomycoketide, which is consistent with a dominant role of TCR interactions with CD1c and the concept of mixed self- and foreign antigen reactivity often observed in the CD1 system. Although the available evidence is highly suggestive of functions in infections and autoimmunity, this has not yet been conclusively demonstrated.
CD1d
In contrast with group 1 CD1 proteins, whose expression is limited to hematopoietic cells, CD1d is expressed more widely by both hematopoietic and non-hematopoietic cells, including thymocytes, professional APCs, hepatocytes, hepatic stellate cells, intestinal epithelial cells and adipocytes, and is particularly abundant on marginal zone B cells [6, 44]. CD1d can present a variety of glycosphingolipids, diacylglycerols, phospholipids, lipopeptides, ether lipids, non-lipid small molecules and possibly even peptides to CD1d-restricted cells [8, 13, 24, 31, 45–48]. Exogenous antigens recognized by iNKT cells include lipids derived from ubiquitous environmental bacteria, pathogenic bacteria and fungi, commensal bacteria, and pollen. Much of our understanding of iNKT cell biology has been gleaned from studies with KRN7000, a synthetically optimized version of an α-galactosylceramide (α-GalCer), agelasphin 9b, derived from the marine sponge Agelas mauritianus [49, 50]. Several microbes, including the commensal bacterium Bacteroides fragilis [51, 52] and the fungal pathogen Aspergillus fumigatus [53], a common cause of airway hypersensitivity, contain iNKT cell-stimulating α-GalCers. These findings also suggest that agelasphin 9b is derived from commensal organisms associated with the marine sponge, rather than from the sponge itself. Other microbial iNKT cell agonists include α-glycuronosylceramides from Sphingomonas bacteria [54–56], diacylglycerols from Streptococcus pneumoniae [57] and the Lyme disease agent Borrelia burgdorferi [58], and cholesteryl α-glucosides from Helicobacter pylori [59], a common agent of stomach ulcers and gastric cancer. Endogenous antigens recognized by iNKT cells include α- and β-linked GalCers and glucosylceramides (GlcCers), isoglobotrihexosylceramide (iGb3), ganglioside D3 (GD3), ether-bonded lipids, and glycerophospholipids such as phosphatidylinositol (PI), phosphatidylethanolamine (PE) and lyso-PC [24]. Antigens recognized by subsets of dNKT cells include endogenous sulfatide, lysophospholipids, lyso-PC, β-glycosylceramides (β-GlyCers), and bacteria-derived PG, di-PG (cardiolipin) and PI [60–62]. In addition to lipid-reactivity, CD1d-restricted T cells have been identified that react with synthetic, non-lipidic phenyl pentamethyldihydrobenzofuran sulfonates (PPBF) [63]. While provocative, the functional significance of this type of reactivity remains unknown.
Crystal structures of human or mouse CD1d complexed with a variety of ligands have been analyzed [8, 24]. The fatty acyl chain of α-GalCer binds with the large A’ pocket of CD1d whereas the sphingosine chain fits within the F’ pocket, with the galactosyl head extending out of the groove [64, 65] (Figure 1b, left). Other NKT cell antigens, including β-GlyCer, sulfatide, iGb3, phosphoglycerolipids and microbial diacylglycerols bind with CD1d in the same conserved manner. In the case of diacylglycerols, however, the acyl chains can bind with CD1d in two different orientations, with the individual chains positioned within either the A’ or F’ pockets, which has a major impact on TCR recognition. The sugar head groups of α-linked glycolipids that extend out of the CD1d groove adopt a similar orientation so that they are easily accessible by the TCR. In sharp contrast, the head groups of the β-linked glycolipids project up and away from the CD1d groove. Co-crystals of TCR/CD1d-lipid complexes have revealed that, in sharp contrast with MHC- and group 1 CD1-restricted TCRs, the iNKT TCR (iNKTCR) docks parallel onto the CD1d groove with most of the interface dominated by the germ-line encoded TCRα chain (Figure 3) [1, 8, 24, 66]. To maintain this conserved footprint, the TCR was able to induce structural changes in both CD1d and the orientation of the ligand. For example, the iNKTCR interacted with CD1d-β-GlcCer and CD1d-iGb3 complexes by flattening the sugars that protrude out of the antigen-binding groove [67, 68]. Interestingly, the structure of a dNKTCR complexed with CD1d-lysosulfatide revealed a diagonal footprint similar to TCR/MHC-peptide complexes, with lysosulfatide recognized exclusively by the TCRβ chain [69], thus revealing features of recognition similar to iNKT cells and conventional peptide-reactive T cells.
Like the group 1 CD1 proteins, CD1d assembles in the ER where it is stabilized by chaperone lipids that have been identified as phospholipids, which are rarely recognized by NKT cells [19, 24, 60]. Association with ER-derived chaperone lipids is catalyzed by MTP. Following display at the cell surface, CD1d is internalized via its tyrosine-based sorting motif and enters endosomal and lysosomal compartments. While some autoreactive CD1d-restricted T cells recognize antigens that do not require CD1d internalization, and α-GalCer can be loaded directly onto CD1d at the cell surface, most CD1d antigens are acquired in endocytic compartments. An alternative, auxiliary pathway for CD1d to arrive in these intracellular structures is via association with the MHC class II-associated invariant chain [70]. In these intracellular compartments, CD1d binds lipids with the assistance of LTPs, including saposins A–D, GM2 activator, Niemann-Pick type C2 protein, and thymocyte-derived cathepsin L. Delivery of antigens to these compartments may involve extracellular lipid-binding proteins such as apolipoprotein E-containing VLDL and fatty acid amide hydrolase (FAAH), and receptor-mediated entry via lipid receptors such as the low-density lipoprotein receptor (LDLR) or lectin receptors such as the mannose receptor [20]. Several precursors to CD1d-presented antigens require processing by carbohydrate hydrolases or phospholipases. For example, in the case of CD1d-restricted T cells in the liver that respond to hepatitis B virus infection, antigenic ER lipids such as lyso-PE were generated from ubiquitous phospholipids via secretory PLA2 [71]. Additionally, lysosomal PLA2 was shown to be required for the intrathymic development of iNKT cells and for the presentation of endogenous antigens by CD1d [72].
A topic of controversy in the NKT cell field that remains to be fully resolved is the identity of the natural self-antigen(s) that mediate the intrathymic development and peripheral functions of iNKT cells [24]. For many years it was thought that iNKT cells selectively react with α-linked glycosphingolipids, which was unanticipated in the context of the common belief that mammalian cells only produce β-linked glycosphingolipids. A first surprise was that cells deficient in β-GlcCer synthase were unable to activate autoreactive mouse iNKT cell hybridomas [73]. Subsequent studies provided evidence that β-GlyCer may be recognized by iNKT cells, which was supported by X-ray crystallographic studies [68], yet the synthetic preparations used in these studies appeared to contain minute amounts of α-linked GlyCers. A second surprise was the identification of the endogenous, lysosomal β-linked glycosphingolipid iGb3 as a weak iNKT cell agonist [74]. However, human cells do not produce iGb3 and the lack of an iNKT cell phenotype in iGb3 synthase-deficient mice [75] raised doubt about the physiological relevance of iGb3 to iNKT cell function. The final surprise came when two research groups performed a variety of elegant biochemical and structural analyses of lipid preparations that were able to stimulate autoreactive iNKT cells [76–78]. These studies provided strong evidence for α-GlcCers and α-GalCers – not thought to be present in mammalian cells – as natural iNKT cell ligands. The minute amounts of these antigens produced by mammals were likely overlooked in previous studies, but their low prevalence is consistent with the physiological functions of iNKT cells. The nature of the enzymatic activities involved in the generation of mammalian α-linked GlyCers and how these activities are controlled during normal and pathological conditions remain to be elucidated. While current evidence indicates that these α-GlyCers control the physiological functions of iNKT cells in the periphery, they do not appear to be required for the intrathymic development of these cells. Instead, one study provided evidence for the involvement of peroxisome-derived, ether-bonded lipids in the intrathymic development of iNKT cells [79]. If and how the requirement of lysosomal PLA2 activity for iNKT cell development [72] aligns with these findings remains to be determined.
As already noted, NKT cells are CD1d-restricted T cells that co-express lipid-reactive αβ (or γδ) TCRs together with NK cell markers [6, 23, 44, 47, 80]. iNKT cells express TCRs composed of an invariant TCRα chain (Vα14-Jα18 in mouse or Vα24-Jα18 in human) and a restricted set of TCRβ chains that react with α-GalCer. These cells are substantially more abundant in mice than humans and their numbers vary widely among different human subjects. iNKT cells are most prevalent in liver and are also abundant in spleen, peripheral blood, bone marrow, thymus and mucosal tissues in gut and lung. Apart from Vα14+ NKT cells, an additional subset of α-GalCer-reactive NKT cells has been identified that expresses a unique Vα10-Jα50 TCRα chain in mice [81]. Strikingly, the latter cells exhibited greater reactivity to α-GlcCer and also reacted with mycobacterial α-glucuronosyldiacylglycerol (α-GlcADAG), suggesting important functions. iNKT cells express the innate master transcription factor promyelocytic leukemia zinc finger (PLZF), exhibit cytotoxic activities and can produce a wide variety of cytokines. Subsets of iNKT cells with specialized effector functions similar to the diverse properties of adaptive CD4+ helper T cell subsets have been identified and include NKT1 cells producing IFN-γ (and IL-4), NKT2 cells producing IL-4 and IL-13, NKT10 cells producing the immunosuppressive cytokine IL-10, NKT17 cells producing IL-17a, and follicular helper NKT cells producing IL-21 [24, 82, 83]. These iNKT cell subsets are selectively enriched in distinct organs and tissues. Consequently, iNKT cells can influence the functions of a variety of innate and adaptive immune cells. These cells rapidly elicit their broad effector functions following stimulation with α-GalCer and, instead of developing immune memory, become unresponsive to antigen restimulation as they acquire an anergic and regulatory phenotype [23, 84, 85]. The functions of iNKT cells span the entire range of the immune response, including host defense against pathogens, autoimmunity, tissue graft rejection, hypersensitivities, tumor immunity, and metabolic disease [5, 6, 23, 24, 44]. The role of iNKT cells in immune responses against pathogens is not limited to organisms containing natural iNKT cell antigens but extends to many organisms such as viruses that lack cognate iNKT cell antigens. For such pathogens, iNKT cells are activated in response to innate cytokine signals, in the presence or absence of TCR signaling via autoantigens [26]. In fact, one study provided evidence that the response of iNKT cells to pathogens is dominated by innate cytokines, even for those pathogens containing cognate iNKT cell antigens [86]. The role of CD1d-restricted T cell responses against pathogenic organisms is underscored by evasion mechanisms employed by multiple pathogens to subvert CD1d-restricted antigen presentation [87]. Studies over the past several years have further shown that iNKT cells influence the composition of the natural gut microbiota and conversely, that members of the microbiota such as Bacteroides fragilis that contain iNKT cell antigens shape iNKT cell effector functions [51, 88]. This dynamic interaction between iNKT cells and the microbiota may have major consequences on human health, raising the possibility to prevent the development of diseases such as asthma and inflammatory bowel disease by administering neonates with probiotics that influence iNKT cell functions [89]. iNKT cell agonists such as α-GalCer have been extensively employed to explore the adjuvant and therapeutic activities of iNKT cells, with promising preclinical studies for some tumors, infectious agents and autoimmune diseases, and some encouraging findings with cancer patients [49, 90, 91]. Nevertheless, inducing biological responses of iNKT cells in humans has proven challenging, and the potential for generating adverse effects rather than protect against disease remains an important concern.
In contrast with iNKT cells, dNKT cells do not typically react with α-GalCer and most of these cells express diverse TCRs [13, 47, 80]. Nevertheless, the subset of sulfatide-reactive dNKT cells contains populations with biased TCRs. Importantly, dNKT cells are more prevalent in humans than in mice. Like iNKT cells, dNKT cells express PLZF [92], exhibit innate-like functions and can influence a wide variety of immune responses [47, 93, 94]. An interesting phenomenon is that dNKT cells often oppose the functions of iNKT cells [95]. For example, while iNKT cells exhibit natural immunity against some metastatic cancers, dNKT cells have a propensity for promoting cancer growth [96]. Preclinical studies with sulfatide have provided promising results with several autoimmune diseases, raising the possibility of developing dNKT cell-based immunotherapies for human diseases.
Recently, a subset of human dNKT cells termed “atypical NKT cells” expressing diverse α-GalCer-reactive TCRs has been identified [97]. While the functions of these cells remain unclear, crystal structures of two of these TCRs bound with CD1d-α-GalCer complexes revealed orthogonal binding over the A’ pocket of CD1d, contrasting sharply with the docking mode of iNKTCRs [97].
A recent provocative report [98] has renewed interest into the possibility that CD1d-restricted T cells can react with peptides, a notion that has been entertained for over 20 years. Early studies provided evidence that mouse CD1d can present a variety of hydrophobic synthetic peptides with the common motif [FW]-X-X-[ILM]-X-X-W to CD1d-restricted T cells [99]. Subsequent studies identified CD1d-restricted CD8+ T cells reactive with peptides from the model antigen ovalbumin [100, 101]. However, due to difficulties in understanding the molecular basis of peptide recognition and the preponderance of lipid-reactivity among CD1d-restricted T cells, the issue of peptide-reactivity was largely put to rest. A more recent study reported CD1d-restricted recognition of a collagen-derived peptide by CD4+ T cells [102], but its molecular basis was unclear. Until the recent study by Girardi et al. [98], the exact location of peptide binding to CD1d has remained elusive. These investigators determined the crystal structure of the complex between mouse CD1d and the first peptide identified to bind CD1d, synthetic p99. This peptide adopts an α-helical conformation in the CD1d groove that orients the motif residues towards the bottom of the groove, in a manner that is consistent with its presentation to TCRs. Although the functions of peptide-reactive dNKT cells remain to be determined, it is striking that the CD1d peptide motif is contained within peptides derived from several viruses such as HIV [103], suggesting potential anti-viral functions. Additionally, studies with the CD1d-binding, collagen-derived peptide have provided evidence for potent immune modulatory activities of peptide-reactive, CD1d-restricted T cells [102].
CD1e
CD1e is expressed by thymocytes and DCs. As already noted, CD1e is only expressed intracellularly and does not function as an antigen-presenting molecule [5]. Instead, membrane-anchored CD1e is transported to endolysosomal compartments where it is cleaved into soluble proteins (Figure 4). CD1e is a lipid-binding protein with a wide, solvent-exposed antigen-binding groove that includes contiguous A’ and F’ pockets [104]. CD1e modulates the presentation of endogenous and exogenous lipids by human CD1b, CD1c and CD1d, which has been proposed to be due to its capacity to accelerate the generation and dissociation of CD1-lipid complexes [105]. Additionally, CD1e has been suggested to assist lysosomal α-mannosidase in the processing of mycobacterial lipids into antigenic CD1b ligands [106]. In this manner, CD1e might influence both lipid availability and the generation and persistence of CD1-lipid complexes.
Concluding Remarks and Future Perspectives
The studies reviewed here highlight the emergence of the CD1 antigen presentation system as an important complement to the classical MHC antigen presentation system in health and disease. Recent studies have provided new insight into this system by identifying novel cognate antigens and the factors involved in the generation and processing of antigens. These studies have also revived the idea that CD1d can present peptide antigens to T cells. We now understand in some depth how distinct antigens bind with individual CD1 isoforms, which does not only involve antigenic lipids but also lipids that function as chaperones, spacers and scaffolds. TCRs engage CD1-antigen complexes with footprints that exhibit a surprising amount of diversity. While significant progress has been made regarding the effector functions and immunological properties of CD1-restricted T cells, many questions remain to be addressed (see Outstanding Questions) before this fascinating antigen presentation system can be fully exploited for the development of vaccines and immune therapies.
Outstanding Questions Box.
From a structural perspective, how do CD1 proteins bind chemically diverse ligands, including lipids, non-lipidic small molecules and even peptides? How can CD1-restricted TCRs engage such diverse CD1-antigen complexes?
What are the mechanisms that control the activities and functions of CD1-restricted T cells that exhibit mixed reactivity against self- and foreign antigens?
Does the range of microbial products recognized by group 1 CD1-restricted T cells extend much beyond mycobacterial cell wall products?
Is the prevalence and function of group 1 CD1-restricted T cells influenced by environmental mycobacteria or commensal microorganisms?
How can one effectively target group 1 CD1-restricted T cells for vaccine development and immunotherapies? Studies with humanized mice and with guinea pigs that express all 5 CD1 isoforms would be revealing.
What are the physiological ligands that control the intrathymic development of CD1d-restricted NKT cells?
What are the mechanisms that control the enzymatic activities responsible for the generation of the natural, endogenous α-linked glycosylceramide ligands of iNKT cells?
What is the significance of peptide presentation by CD1d?
How can one selectively activate distinct iNKT cell subsets for therapeutic purposes?
Can one manipulate iNKT cell functions by administering probiotics to human neonates so as to lower disease susceptibility later in life?
How can the preclinical vaccine and therapeutic studies with NKT cell antigens be effectively translated to human subjects?
Trends Box.
The CD1-lipid presentation system allows the immune system to sense alterations in lipid homeostasis and complements the classical MHC-peptide presentation system. There are remarkable similarities and surprising differences in the way that TCRs engage CD1-lipid versus MHC-peptide complexes.
Group 1 CD1 proteins (CD1a–c) present a variety of endogenous, mycobacterial and potentially other bacterial lipids to T cells. CD1b-restricted T cells include subsets expressing germline-encoded TCRs.
Group 2 CD1 proteins (CD1d) present diverse endogenous and exogenous lipid antigens to subsets of natural killer T (NKT) cells expressing semi-invariant, biased or diverse TCRs. α-linked glycosylceramides have emerged as major endogenous ligands that control the functions of invariant NKT cells.
Significant progress has been made towards the development of lipid-based vaccines and immunotherapies.
Acknowledgments
We thank Dr. Benjamin Spiller (Vanderbilt University) for help with PyMol to generate protein structures. Work of the authors is supported by grants from the National Institutes of Health, the Department of Veterans Affairs, the National Multiple Sclerosis Society, and the Crohn’s and Colitis Foundation of America. We apologize to colleagues whose work we did not cite due to space constraints.
Glossary
- adjuvant
Chemical substances used to induce innate immune response, which then enhance and direct adaptive immune responses to the vaccine antigen.
- anergic
An immune cell property characterized by unresponsiveness to antigen resulting from cell-intrinsic tolerance induction.
- β2-microglobulin
The invariant soluble component of all MHC class I and many MHC class I-related molecules such as the CD1 proteins. It provides stability to MHC class I and class I-like structures.
- chaperone
In antigen processing, chaperones facilitate the assembly and transport of MHC proteins. They include products involved in general ER quality-control and those with specific functions in MHC class I-, MHC class II- or CD1-restricted antigen presentation.
- double-negative cells
T cells that lack CD4 and CD8 co-receptor expression.
- hepatic stellate cells
These cells, also called perisinusoidal or Ito cells, store vitamin A in the liver, become activated during liver injury, and play a major role in liver fibrosis.
- humanized mice
Mice carrying functional human genes, cells, tissues or organs.
- Langerhans cells
Dendritic cells that uniquely express langerin (CD207) and reside in the epidermis of the skin and some mucosal epithelia.
- lectin
A protein that binds with certain sugars.
- marginal zone B cells
A subset of non-circulating B cells with innate-like properties that reside within the marginal zone – the region between the white (lymphoid) and the red (non-lymphoid) pulp – of the spleen.
- microbiota
The variety of symbiotic, commensal and parasitic microorganisms that are normally associated with metazoans.
- MHC class II-associated invariant chain
This transmembrane protein, also known as the MHC class II γ chain or CD74, is a chaperone that binds with the groove of MHC class II molecules, facilitates class II transit to endolysosomal compartments, and prevents peptide binding until its release and exchange with antigenic peptides.
- pathobiont
Any organism that under normal circumstances lives as a symbiont with its host but under certain situations can cause disease.
- phospholipase
Any of four enzymes that hydrolyze specific ester bonds in phospholipids.
- portal
For CD1 proteins, portals are open areas of the antigen-binding groove where portions of the antigen can project out of the groove.
- professional antigen-presenting cells
A group of antigen-presenting cells that include dendritic cells, macrophages and B cells, which can activate naïve T cells.
- specificity pockets
For the classical MHC class I and II proteins, specificity pockets in the antigen-binding groove interact with conserved residues of the bound peptide. CD1 proteins similarly contain specificity pockets that interact with conserved portions of the bound lipid.
- tetramers
In T cell biology, fluorescently-labeled, tetrameric forms of MHC or CD1 proteins loaded with specific antigens are employed to identify antigen-specific T cells.
- cortical thymocytes
Thymocytes are hematopoietic precursor cells to the T cell lineage in the thymus. Thymocytes in the cortical area of the thymus, the outer layer of this organ, are most immature and express CD1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rossjohn J, et al. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol. 2015;33:169–200. doi: 10.1146/annurev-immunol-032414-112334. [DOI] [PubMed] [Google Scholar]
- 2.Rodgers JR, Cook RG. MHC class Ib molecules bridge innate and acquired immunity. Nat Rev Immunol. 2005;5:459–471. doi: 10.1038/nri1635. [DOI] [PubMed] [Google Scholar]
- 3.Calabi F, Milstein C. A novel family of human major histocompatibility complex-related genes not mapping to chromosome 6. Nature. 1986;323:540–543. doi: 10.1038/323540a0. [DOI] [PubMed] [Google Scholar]
- 4.McMichael AJ, et al. A human thymocyte antigen defined by a hybrid myeloma monoclonal antibody. Eur J Immunol. 1979;9:205–210. doi: 10.1002/eji.1830090307. [DOI] [PubMed] [Google Scholar]
- 5.Brigl M, Brenner MB. CD1: antigen presentation and T cell function. Annu Rev Immunol. 2004;22:817–890. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- 6.Bendelac A, et al. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 7.Li S, et al. Autoreactive CD1b-restricted T cells: a new innate-like T-cell population that contributes to immunity against infection. Blood. 2011;118:3870–3878. doi: 10.1182/blood-2011-03-341941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Rhijn I, et al. Lipid and small-molecule display by CD1 and MR1. Nat Rev Immunol. 2015;15:643–654. doi: 10.1038/nri3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogers SL, Kaufman J. Location, location, location: the evolutionary history of CD1 genes and the NKR-P1/ligand systems. Immunogenetics. 2016 doi: 10.1007/s00251-016-0938-6. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felio K, et al. CD1-restricted adaptive immune responses to Mycobacteria in human group 1 CD1 transgenic mice. J Exp Med. 2009;206:2497–2509. doi: 10.1084/jem.20090898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lockridge JL, et al. Analysis of the CD1 antigen presenting system in humanized SCID mice. PLoS One. 2011;6:e21701. doi: 10.1371/journal.pone.0021701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ly D, Moody DB. The CD1 size problem: lipid antigens, ligands, and scaffolds. Cell Mol Life Sci. 2014;71:3069–3079. doi: 10.1007/s00018-014-1603-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godfrey DI, et al. The burgeoning family of unconventional T cells. Nat Immunol. 2015;16:1114–1123. doi: 10.1038/ni.3298. [DOI] [PubMed] [Google Scholar]
- 14.Mori L, et al. The Immunology of CD1- and MR1-Restricted T Cells. Annu Rev Immunol. 2016;34:479–510. doi: 10.1146/annurev-immunol-032414-112008. [DOI] [PubMed] [Google Scholar]
- 15.Shamshiev A, et al. Self glycolipids as T-cell autoantigens. Eur J Immunol. 1999;29:1667–1675. doi: 10.1002/(SICI)1521-4141(199905)29:05<1667::AID-IMMU1667>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 16.Huang S, et al. Discovery of deoxyceramides and diacylglycerols as CD1b scaffold lipids among diverse groove-blocking lipids of the human CD1 system. Proc Natl Acad Sci U S A. 2011;108:19335–19340. doi: 10.1073/pnas.1112969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scharf L, et al. The 2.5 A structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation. Immunity. 2010;33:853–862. doi: 10.1016/j.immuni.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joyce S, Van Kaer L. CD1-restricted antigen presentation: an oily matter. Curr Opin Immunol. 2003;15:95–104. doi: 10.1016/s0952-7915(02)00012-2. [DOI] [PubMed] [Google Scholar]
- 19.De Libero G, Mori L. Novel insights into lipid antigen presentation. Trends Immunol. 2012;33:103–111. doi: 10.1016/j.it.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Freigang S, et al. Transport and uptake of immunogenic lipids. Mol Immunol. 2013;55:179–181. doi: 10.1016/j.molimm.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darmoise A, et al. The immunological functions of saposins. Adv Immunol. 2010;105:25–62. doi: 10.1016/S0065-2776(10)05002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salio M, et al. Biology of CD1- and MR1-restricted T cells. Annu Rev Immunol. 2014;32:323–366. doi: 10.1146/annurev-immunol-032713-120243. [DOI] [PubMed] [Google Scholar]
- 23.Van Kaer L, et al. Invariant natural killer T cells: bridging innate and adaptive immunity. Cell Tissue Res. 2011;343:43–55. doi: 10.1007/s00441-010-1023-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill TM, et al. CD1d-restricted natural killer T cells. Encycl Life Sci. 2016:a0020180. [Google Scholar]
- 25.Van Kaer L, et al. iNKT cells as sensors and managers of inflammation. Trends Immunol. 2013;34:50–58. doi: 10.1016/j.it.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brigl M, et al. Mechanism of CD1d-restricted natural killer T cell activation during microbial infection. Nat Immunol. 2003;4:1230–1237. doi: 10.1038/ni1002. [DOI] [PubMed] [Google Scholar]
- 27.Moody DB, et al. T cell activation by lipopeptide antigens. Science. 2004;303:527–531. doi: 10.1126/science.1089353. [DOI] [PubMed] [Google Scholar]
- 28.de Jong A, et al. CD1a-autoreactive T cells recognize natural skin oils that function as headless antigens. Nat Immunol. 2014;15:177–185. doi: 10.1038/ni.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agea E, et al. Human CD1-restricted T cell recognition of lipids from pollens. J Exp Med. 2005;202:295–308. doi: 10.1084/jem.20050773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Birkinshaw RW, et al. alphabeta T cell antigen receptor recognition of CD1a presenting self lipid ligands. Nat Immunol. 2015;16:258–266. doi: 10.1038/ni.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Jong A. Activation of human T cells by CD1 and self-lipids. Immunol Rev. 2015;267:16–29. doi: 10.1111/imr.12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bourgeois EA, et al. Bee venom processes human skin lipids for presentation by CD1a. J Exp Med. 2015;212:149–163. doi: 10.1084/jem.20141505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Kaer L. Bee venom stirs up buzz in antigen presentation. J Exp Med. 2015;212:126. doi: 10.1084/jem.2122insight1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramaniam S, et al. Elevated and cross-responsive CD1a-reactive T cells in bee and wasp venom allergic individuals. Eur J Immunol. 2016;46:242–252. doi: 10.1002/eji.201545869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Rhijn I, et al. A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nat Immunol. 2013;14:706–713. doi: 10.1038/ni.2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Rhijn I, et al. TCR bias and affinity define two compartments of the CD1b-glycolipid-specific T Cell repertoire. J Immunol. 2014;192:4054–4060. doi: 10.4049/jimmunol.1400158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montamat-Sicotte DJ, et al. A mycolic acid-specific CD1-restricted T cell population contributes to acute and memory immune responses in human tuberculosis infection. J Clin Invest. 2011;121:2493–2503. doi: 10.1172/JCI46216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao J, et al. Mycolic acid-specific T cells protect against Mycobacterium tuberculosis infection in a humanized transgenic mouse model. Elife. 2015:4. doi: 10.7554/eLife.08525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Rhijn I, et al. Human autoreactive T cells recognize CD1b and phospholipids. Proc Natl Acad Sci U S A. 2016;113:380–385. doi: 10.1073/pnas.1520947112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moody DB, et al. CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature. 2000;404:884–888. doi: 10.1038/35009119. [DOI] [PubMed] [Google Scholar]
- 41.Matsunaga I, et al. Mycobacterium tuberculosis pks12 produces a novel polyketide presented by CD1c to T cells. J Exp Med. 2004;200:1559–1569. doi: 10.1084/jem.20041429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roy S, et al. Molecular basis of mycobacterial lipid antigen presentation by CD1c and its recognition by alphabeta T cells. Proc Natl Acad Sci U S A. 2014;111:E4648–4657. doi: 10.1073/pnas.1408549111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roy S, et al. Molecular Analysis of Lipid-Reactive Vdelta1 gammadelta T Cells Identified by CD1c Tetramers. J Immunol. 2016;196:1933–1942. doi: 10.4049/jimmunol.1502202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kronenberg M. Toward an understanding of NKT cell biology: progress and paradoxes. Annu Rev Immunol. 2005;26:877–900. doi: 10.1146/annurev.immunol.23.021704.115742. [DOI] [PubMed] [Google Scholar]
- 45.Joyce S, et al. NKT cell ligand recognition logic: molecular basis for a synaptic duet and transmission of inflammatory effectors. J Immunol. 2011;187:1081–1089. doi: 10.4049/jimmunol.1001910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McEwen-Smith RM, et al. CD1d-dependent endogenous and exogenous lipid antigen presentation. Curr Opin Immunol. 2015;34:116–125. doi: 10.1016/j.coi.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 47.Kumar V, Delovitch TL. Different subsets of natural killer T cells may vary in their roles in health and disease. Immunology. 2014;142:321–336. doi: 10.1111/imm.12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joyce S, et al. Natural ligand of mouse CD1d1: cellular glycosylphosphatidylinositol. Science. 1998;279:1541–1544. doi: 10.1126/science.279.5356.1541. [DOI] [PubMed] [Google Scholar]
- 49.Van Kaer L. a-Galactosylceramide therapy for autoimmune diseases: prospects and obstacles. Nat Rev Immunol. 2005;5:31–42. doi: 10.1038/nri1531. [DOI] [PubMed] [Google Scholar]
- 50.Kawano T, et al. CD1d-restricted and TCR-mediated activation of Va14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 51.An D, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wieland Brown LC, et al. Production of alpha-galactosylceramide by a prominent member of the human gut microbiota. PLoS Biol. 2013;11:e1001610. doi: 10.1371/journal.pbio.1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Albacker LA, et al. Invariant natural killer T cells recognize a fungal glycosphingolipid that can induce airway hyperreactivity. Nat Med. 2013;19:1297–1304. doi: 10.1038/nm.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mattner J, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 55.Kinjo Y, et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434:520–525. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- 56.Sriram V, et al. Cell wall glycosphingolipids of Sphingomonas paucimobilis are CD1d-specific ligand for NKT cells. Eur J Immunol. 2005;35:1692–1701. doi: 10.1002/eji.200526157. [DOI] [PubMed] [Google Scholar]
- 57.Kinjo Y, et al. Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria. Nat Immunol. 2011;12:966–974. doi: 10.1038/ni.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kinjo Y, et al. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol. 2006;7:978–986. doi: 10.1038/ni1380. [DOI] [PubMed] [Google Scholar]
- 59.Chang YJ, et al. Influenza infection in suckling mice expands an NKT cell subset that protects against airway hyperreactivity. J Clin Invest. 2011;121:57–69. doi: 10.1172/JCI44845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolf BJ, et al. Identification of a Potent Microbial Lipid Antigen for Diverse NKT Cells. J Immunol. 2015;195:2540–2551. doi: 10.4049/jimmunol.1501019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tatituri RV, et al. Recognition of microbial and mammalian phospholipid antigens by NKT cells with diverse TCRs. Proc Natl Acad Sci U S A. 2013;110:1827–1832. doi: 10.1073/pnas.1220601110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rhost S, et al. Identification of novel glycolipid ligands activating a sulfatide-reactive, CD1d-restricted, type II natural killer T lymphocyte. Eur J Immunol. 2012;42:2851–2860. doi: 10.1002/eji.201142350. [DOI] [PubMed] [Google Scholar]
- 63.Van Rhijn I, et al. CD1d-restricted T cell activation by nonlipidic small molecules. Proc Natl Acad Sci USA. 2004;101:13578–13583. doi: 10.1073/pnas.0402838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zajonc DM, et al. Structure and function of a potent agonist for the semi-invariant natural killer T cell receptor. Nat Immunol. 2005;6:810–818. doi: 10.1038/ni1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koch M, et al. The crystal structure of human CD1d with and without a-galactosylceramide. Nat Immunol. 2005;6:819–826. doi: 10.1038/ni1225. [DOI] [PubMed] [Google Scholar]
- 66.Borg NA, et al. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- 67.Zajonc DM, et al. Crystal structures of mouse CD1d-iGb3 complex and its cognate Valpha14 T cell receptor suggest a model for dual recognition of foreign and self glycolipids. J Mol Biol. 2008;377:1104–1116. doi: 10.1016/j.jmb.2008.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pellicci DG, et al. Recognition of beta-linked self glycolipids mediated by natural killer T cell antigen receptors. Nat Immunol. 2011;12:827–833. doi: 10.1038/ni.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Patel O, et al. Recognition of CD1d-sulfatide mediated by a type II natural killer T cell antigen receptor. Nat Immunol. 2012;13:857–863. doi: 10.1038/ni.2372. [DOI] [PubMed] [Google Scholar]
- 70.Jayawardena-Wolf J, et al. CD1d endosomal trafficking is independently regulated by an intrinsic CD1d-encoded tyrosine motif and by the invariant chain. Immunity. 2001;15:897–908. doi: 10.1016/s1074-7613(01)00240-0. [DOI] [PubMed] [Google Scholar]
- 71.Zeissig S, et al. Hepatitis B virus-induced lipid alterations contribute to natural killer T cell-dependent protective immunity. Nat Med. 2012;18:1060–1068. doi: 10.1038/nm.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Paduraru C, et al. Role for lysosomal phospholipase A2 in iNKT cell-mediated CD1d recognition. Proc Natl Acad Sci U S A. 2013;110:5097–5102. doi: 10.1073/pnas.1302923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stanic AK, et al. Defective presentation of the CD1d1-restricted natural Va14Ja18 NKT lymphocyte antigen caused by b-D-glucosylceramide synthase deficiency. Proc Natl Acad Sci USA. 2003;100:1849–1854. doi: 10.1073/pnas.0430327100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou D, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306:1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
- 75.Porubsky S, et al. Normal development and function of invariant natural killer T cells in mice with isoglobotrihexosylceramide (iGb3) deficiency. Proc Natl Acad Sci USA. 2007;104:5977–5982. doi: 10.1073/pnas.0611139104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kain L, et al. Endogenous ligands of natural killer T cells are alpha-linked glycosylceramides. Mol Immunol. 2015;68:94–97. doi: 10.1016/j.molimm.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brennan PJ, et al. Activation of iNKT cells by a distinct constituent of the endogenous glucosylceramide fraction. Proc Natl Acad Sci U S A. 2014;111:13433–13438. doi: 10.1073/pnas.1415357111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kain L, et al. The identification of the endogenous ligands of natural killer T cells reveals the presence of mammalian alpha-linked glycosylceramides. Immunity. 2014;41:543–554. doi: 10.1016/j.immuni.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Facciotti F, et al. Peroxisome-derived lipids are self antigens that stimulate invariant natural killer T cells in the thymus. Nat Immunol. 2012;13:474–480. doi: 10.1038/ni.2245. [DOI] [PubMed] [Google Scholar]
- 80.Godfrey DI, et al. NKT cells: what’s in a name? Nat Rev Immunol. 2004;4:231–237. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- 81.Uldrich AP, et al. A semi-invariant Valpha10+ T cell antigen receptor defines a population of natural killer T cells with distinct glycolipid antigenrecognition properties. Nat Immunol. 2011;12:616–623. doi: 10.1038/ni.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lynch L, et al. Regulatory iNKT cells lack expression of the transcription factor PLZF and control the homeostasis of T(reg) cells and macrophages in adipose tissue. Nat Immunol. 2015;16:85–95. doi: 10.1038/ni.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Constantinides MG, Bendelac A. Transcriptional regulation of the NKT cell lineage. Curr Opin Immunol. 2013;25:161–167. doi: 10.1016/j.coi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Parekh VV, et al. Glycolipid antigen induces long-term natural killer T cell anergy in mice. J Clin Invest. 2005;115:2572–2583. doi: 10.1172/JCI24762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sag D, et al. IL-10-producing NKT10 cells are a distinct regulatory invariant NKT cell subset. J Clin Invest. 2014;124:3725–3740. doi: 10.1172/JCI72308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brigl M, et al. Innate and cytokine-driven signals, rather than microbial antigens, dominate in natural killer T cell activation during microbial infection. J Exp Med. 2011;208:1163–1177. doi: 10.1084/jem.20102555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Van Kaer L, Joyce S. Viral evasion of antigen presentation: not just for peptides anymore. Nat Immunol. 2006;7:795–797. doi: 10.1038/ni0806-795. [DOI] [PubMed] [Google Scholar]
- 88.Gensollen T, et al. How colonization by microbiota in early life shapes the immune system. Science. 2016;352:539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zeissig S, Blumberg RS. Commensal microbial regulation of natural killer T cells at the frontiers of the mucosal immune system. FEBS Lett. 2014;588:4188–4194. doi: 10.1016/j.febslet.2014.06.042. [DOI] [PubMed] [Google Scholar]
- 90.Van Kaer L, et al. Invariant NK T cells: potential for immunotherapeutic targeting with glycolipid antigens. Immunotherapy. 2011;3:59–75. doi: 10.2217/imt.10.85. [DOI] [PubMed] [Google Scholar]
- 91.Carreno LJ, et al. Synthetic glycolipid activators of natural killer T cells as immunotherapeutic agents. Clin Transl Immunology. 2016;5:e69. doi: 10.1038/cti.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao J, et al. Polyclonal type II natural killer T cells require PLZF and SAP for their development and contribute to CpG-mediated antitumor response. Proc Natl Acad Sci U S A. 2014;111:2674–2679. doi: 10.1073/pnas.1323845111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Marrero I, et al. Type II NKT Cells in Inflammation, Autoimmunity, Microbial Immunity, and Cancer. Front Immunol. 2015;6:316. doi: 10.3389/fimmu.2015.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rhost S, et al. Immunomodulatory type II natural killer T lymphocytes in health and disease. Scand J Immunol. 2012;76:246–255. doi: 10.1111/j.1365-3083.2012.02750.x. [DOI] [PubMed] [Google Scholar]
- 95.Dasgupta S, Kumar V. Type II NKT cells: a distinct CD1d-restricted immune regulatory NKT cell subset. Immunogenetics. 2016 doi: 10.1007/s00251-016-0930-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Terabe M, et al. A nonclassical non-Va14Ja18 CD1d-restricted (type II) NKT cell is sufficient for down-regulation of tumor immunosurveillance. J Exp Med. 2005;202:1627–1633. doi: 10.1084/jem.20051381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Le Nours J, et al. Atypical natural killer T-cell receptor recognition of CD1d-lipid antigens. Nat Commun. 2016;7:10570. doi: 10.1038/ncomms10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Girardi E, et al. Structure of an alpha-Helical Peptide and Lipopeptide Bound to the Nonclassical Major Histocompatibility Complex (MHC) Class I Molecule CD1d. J Biol Chem. 2016;291:10677–10683. doi: 10.1074/jbc.M115.702118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Castano AR, et al. Peptide binding and presentation by mouse CD1. Science. 1995;269:223–226. doi: 10.1126/science.7542403. [DOI] [PubMed] [Google Scholar]
- 100.Lee DJ, et al. Induction of an antigen-specific, CD1-restricted cytotoxic T lymphocyte response In vivo. J Exp Med. 1998;187:433–438. doi: 10.1084/jem.187.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tangri S, et al. Presentation of peptide antigens by mouse CD1 requires endosomal localization and protein antigen processing. Proc Natl Acad Sci U S A. 1998;95:14314–14319. doi: 10.1073/pnas.95.24.14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu Y, et al. Endogenous collagen peptide activation of CD1d-restricted NKT cells ameliorates tissue-specific inflammation in mice. J Clin Invest. 2011;121:249–264. doi: 10.1172/JCI43964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brossay L, et al. Antigen-presenting function of mouse CD1: one molecule with two different kinds of antigenic ligands. Immunol Rev. 1998;163:139–150. doi: 10.1111/j.1600-065x.1998.tb01193.x. [DOI] [PubMed] [Google Scholar]
- 104.Garcia-Alles LF, et al. Crystal structure of human CD1e reveals a groove suited for lipid-exchange processes. Proc Natl Acad Sci U S A. 2011;108:13230–13235. doi: 10.1073/pnas.1105627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Facciotti F, et al. Fine tuning by human CD1e of lipid-specific immune responses. Proc Natl Acad Sci U S A. 2011;108:14228–14233. doi: 10.1073/pnas.1108809108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cala-De Paepe D, et al. Deciphering the role of CD1e protein in mycobacterial phosphatidyl-myo-inositol mannosides (PIM) processing for presentation by CD1b to T lymphocytes. J Biol Chem. 2012;287:31494–31502. doi: 10.1074/jbc.M112.386300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shamshiev A, et al. Presentation of the same glycolipid by different CD1 molecules. J Exp Med. 2002;195:1013–1021. doi: 10.1084/jem.20011963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Beckman EM, et al. Recognition of a lipid antigen by CD1-restricted alpha beta+ T cells. Nature. 1994;372:691–694. doi: 10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- 109.Sieling PA, et al. CD1-restricted T cell recognition of microbial lipoglycan antigens. Science. 1995;269:227–230. doi: 10.1126/science.7542404. [DOI] [PubMed] [Google Scholar]
- 110.Gilleron M, et al. Diacylated sulfoglycolipids are novel mycobacterial antigens stimulating CD1-restricted T cells during infection with Mycobacterium tuberculosis. J Exp Med. 2004;199:649–659. doi: 10.1084/jem.20031097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Moody DB, et al. CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature. 2000;404:884–888. doi: 10.1038/35009119. [DOI] [PubMed] [Google Scholar]
- 112.Natori T, et al. Agelasphins, novel a-galactosylceramides from the marine sponge Agelas mauritianus. Tetrahedron Lett. 1993;34:5591–5592. [Google Scholar]
- 113.Brennan PJ, et al. Invariant natural killer T cells recognize lipid self antigen induced by microbial danger signals. Nat Immunol. 2011;12:1202–1211. doi: 10.1038/ni.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gumperz JE, et al. Murine CD1d-restricted T cell recognition of cellular lipids. Immunity. 2000;12:211–221. doi: 10.1016/s1074-7613(00)80174-0. [DOI] [PubMed] [Google Scholar]
- 115.Fox LM, et al. Recognition of lyso-phospholipids by human natural killer T lymphocytes. PLoS Biol. 2009;7:e1000228. doi: 10.1371/journal.pbio.1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wu DY, et al. Cross-presentation of disialoganglioside GD3 to natural killer T cells. J Exp Med. 2003;198:173–181. doi: 10.1084/jem.20030446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jahng A, et al. Prevention of autoimmunity by targeting a distinct, noninvariant CD1d-reactive T cell population reactive to sulfatide. J Exp Med. 2004;199:947–957. doi: 10.1084/jem.20031389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dieude M, et al. Cardiolipin binds to CD1d and stimulates CD1d-restricted gammadelta T cells in the normal murine repertoire. J Immunol. 2011;186:4771–4781. doi: 10.4049/jimmunol.1000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pellicci DG, et al. Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors. Immunity. 2009;31:47–59. doi: 10.1016/j.immuni.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gras S, et al. A structural basis for varied alphabeta TCR usage against an immunodominant EBV antigen restricted to a HLA-B8 molecule. J Immunol. 2012;188:311–321. doi: 10.4049/jimmunol.1102686. [DOI] [PubMed] [Google Scholar]