Summary
Renal disease is growing in prevalence and has striking co-morbidities with metabolic and cardiovascular disease. Indoxyl sulfate (IS) is a toxin that accumulates in plasma when the kidney function declines and contributes to the progression of chronic kidney disease. IS derives exclusively from the gut microbiota. Bacterial tryptophanases convert tryptophan to indole, which is absorbed and modified by the host to produce IS. Here, we identify a widely distributed family of tryptophanases in the gut commensal Bacteroides and find that deleting this gene eliminates the production of indole in vitro. By altering the status or abundance of the Bacteroides tryptophanase, we can modulate IS levels in gnotobiotic mice and in the background of a conventional murine gut community. Our results demonstrate that it is possible to control host IS levels by targeting the microbiota and suggest a possible strategy for treating renal disease.
Keywords: indoxyl sulfate, human microbiome, chronic kidney disease, tryptophanase, Bacteroides, genetic engineering
Graphical abstract
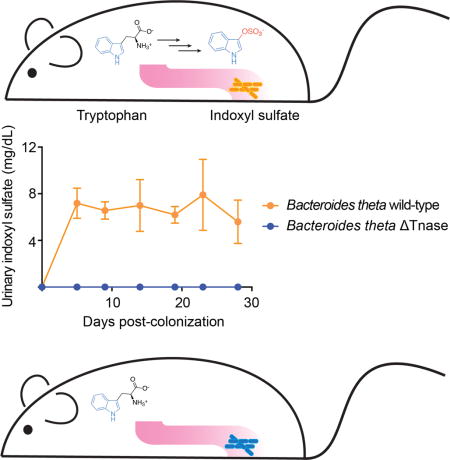
Gut bacteria convert diet-derived molecules into hundreds of diffusible organic compounds, some of which are toxic and must be cleared from the body. Many of these molecules are normally excreted in urine. In people with chronic kidney disease (CKD), however, kidney function remains diminished for a long period, and these compounds accumulate in serum and contribute to disease progression (Schepers et al., 2010; Meyer and Hostetter, 2012; Poesen et al., 2013).
One such metabolite is indoxyl sulfate (IS), a uremic toxin that is produced in the healthy human host at concentrations that range from 10 to 130 mg/day (Patel et al., 2012). In some contexts, IS and its precursor indole can be beneficial to the host, most notably by suppressing CNS inflammation (Rothhammer et al., 2016) or enhancing gut barrier function (Shimada et al., 2013). However, in CKD patients, inadequate renal clearance leads to highly elevated IS levels in the blood, causing deleterious effects (Niwa and Shimizu, 2012). Patients with end stage renal disease who are maintained on dialysis have IS levels that are up 20-fold greater than normal (Duranton et al., 2012; Sirich et al., 2013), and accumulating evidence suggests that IS contributes to the progression of CKD. Its levels are inversely correlated with renal function in CKD patients (Barreto et al., 2009; Wu et al., 2011), and it accelerates tubular cell injury and induces interstitial fibrosis and glomerular sclerosis in animal models of disease (Niwa and Ise, 1994; Niwa et al., 1994).
In addition to acting as a nephrotoxin, IS may play a crucial role in vascular dysfunction and death in CKD patients (Niwa, 2010). Cardiovascular disease (CVD) mortality is significantly higher in CKD patients compared to the general population and is responsible for 40–50% of deaths among patients receiving dialysis (Johnson et al., 2007; Parfrey and Foley, 1999). IS stimulates severe aortic calcification in vivo (Adijiang et al., 2008), and its levels correlate with aortic calcification and arterial stiffness in CKD patients (Barreto et al., 2009). In addition, CKD patients suffer from high rates of thrombosis. IS functions as a pro-thrombotic metabolite by activating the aryl hydrocarbon receptor (AHR), which then stabilizes tissue factor (TF) in vascular smooth muscle cells, thereby promoting thrombosis (Shivanna et al., 2016). Taken together, these data suggest that IS contributes to negative cardiovascular outcomes in CKD patients (Niwa, 2010). Indeed, the highest tertile of IS level is a powerful predictor of overall and cardiovascular mortality in CKD patients (Barreto et al., 2009), and the level of IS is associated with the first heart failure event in hemodialysis patients (Cao et al., 2014).
IS derives exclusively from the action of the microbiota (Carter et al., 1966; Einheber and Carter, 1966; Wikoff et al., 2009). Gut bacteria convert tryptophan to indole (Evenepoel et al., 2009), which is 3-hydroxylated to indoxyl by hepatic cytochrome P450 enzymes, including CYP 2E1, (Banoglu et al., 2001; Gillam et al., 2000) and then O-sulfated by sulfotransferase (SULT) 1A1, producing IS (Figure 1A) (Banoglu and King, 2002). Manipulation of the microbiota to lower the amount of indole produced, thereby reducing circulating IS levels, could provide a new means to limit the adverse effects of kidney disease. However, it was previously unknown which gut bacterial strains produce indole and what genes are responsible. This knowledge would allow us to better understand, predict, and reprogram IS levels in vivo.
Figure 1. Identification of a family of Bacteroides tryptophanases responsible for producing indole, the precursor to the uremic toxin indoxyl sulfate.
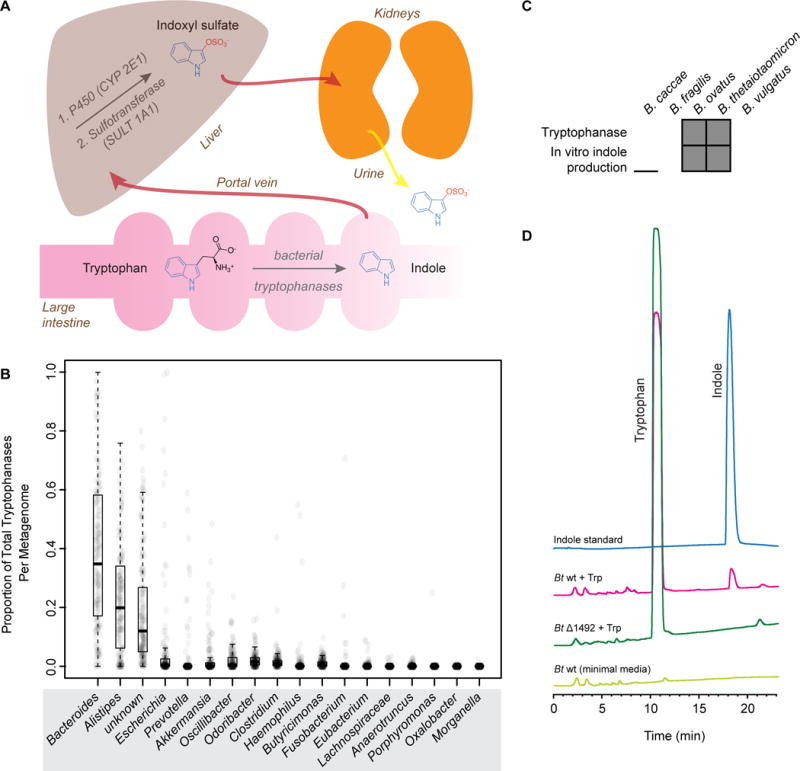
(A) Schematic representation of indoxyl sulfate (IS) production and excretion in vivo. (B) Known and putative tryptophanases are widely distributed across human gut metagenomes. At the genus level, tryptophanases from Bacteroides are the most abundant. Tryptophanase homolog abundances were aggregated based on their taxonomic annotation at the genus level and normalized to sum to 1.0 per sample. The distribution of these values for 1,267 gut metagenomes is shown for 18 different genera represented as a box plot. A value of 0.25 for a sample-genus pair indicates that the tryptophanases from that genus comprised 25% of the total pool of tryptophanases in the sample. For a given genus, 150 random data points are plotted as transparent circles in order to show the distribution across samples. (C) Bacteroides species that produced indole from tryptophan (Trp) in vitro also contained putative tryptophanase BT1492 or an ortholog thereof. A gray box indicates that the strain contains BT1492 or an ortholog (top row) or is positive for indole production in vitro (bottom row). (D) Representative HPLC traces showing that wild-type Bt produces indole in vitro whereas Bt Δ1492 does not, demonstrating that BT1492 is responsible for indole production by Bt. Light green: Bt wild-type plus minimal media only. Green: Bt Δ1492 plus 5 mg/mL Trp. Pink: Bt wild-type plus 5 mg/mL Trp. Blue: indole standard. See also Figures S1 and S2 and Tables S1A–D.
The production of indole from tryptophan has long been used as a phenotypic test in microbiology, and to date ~85 indole-producing bacterial species have been identified (Lee and Lee, 2010). Tryptophanases (EC 4.1.99.1, Tnase) are the only enzymes identified thus far that are capable of performing this transformation. These pyridoxal 5′-phosphate (PLP)-dependent enzymes are present in bacteria but not eukaryotes and convert L-tryptophan to indole, pyruvate, and ammonia (Newton and Snell, 1964). The majority of human-associated bacterial species that are known to produce indole are low-abundance gut colonists or pathogens (Kraal et al., 2014; Lee and Lee, 2010).
Because healthy humans accumulate large amounts of indoxyl sulfate (10–130 mg/day), we hypothesized that additional more abundant gut commensal genera may produce indole from tryptophan. We performed a computational search for putative tryptophanases in the genome sequences of gut-associated reference strains in the NCBI database. This search revealed a putative tryptophanase from Bacteroides, a much more abundant gut genus than had been previously identified to harbor indole-producing members. This gene possesses two distinguishing characteristics that prompted us to focus our experimental efforts on the enzyme it encodes: First, it is present in some of the most common species in this genus, including B. thetaiotaomicron (Bt) and B. ovatus (BT1492 and BACOVA_04222, respectively, 93.4% sequence similarity), but not in others, including B. vulgatus, B. caccae, and B. fragilis. Second, those species containing the gene have been reported to produce indole, whereas the other species have been reported to be non-producers (Cato and Johnson, 1976; Johnson et al., 1986).
Using a recently developed computational method (Nayfach et al., 2015) and a set of query sequences consisting of both experimentally characterized and putative tryptophanases (Table S1A), we quantified the abundance of known and putative tryptophanase genes across 1,267 gut metagenomes from American, European, and Chinese subjects. We found evidence for the presence of tryptophanases in all metagenomes surveyed (minimum abundance = 1 copy/1000 cells), suggesting that this may be a core function in the human gut (Table S1A). On average we estimated that tryptophanases are found once per 6 microbial cells or once per 22,000 genes. Strikingly, the level of these genes spanned over two orders of magnitude across individuals (Figure S1A–C, Table S1B), indicating that tryptophanase abundance varies widely among healthy gut metagenomes.
At the genus level, BT1492 and its orthologs were the most abundant and comprised nearly 40% of the total tryptophanases across samples (Figure 1B, Table S1C). We also found a number of diverse tryptophanases from Bacteroidetes and Firmicutes that were unannotated at the genus level based on a comparison to currently sequenced reference genomes (Figure 1B and Table S1C). As an alternative approach, we compared tryptophanase gene abundance to the relative abundance of different taxonomic groups across the 1,267 metagenomes (Table S1D) and found that both tryptophanase copy number per cell (R2=0.22, p<1e–16) and relative abundance (R2=0.12, p<1e–16) were most strongly associated with the relative abundance of Bacteroides (Figure S1D and E). In several individuals with high tryptophanase levels but low levels of Bacteroides species, the abundance of several other genera including Alistipes and Escherichia were correlated with tryptophanase levels (Figure S1F and G), suggesting that in a small minority of people, non-Bacteroides tryptophanases may be responsible for indole production. The data indicate that in the majority of individuals, however, the most abundant tryptophanases in their gut are harbored by Bacteroides species.
To determine whether the putative tryptophanase BT1492 and its orthologs were responsible for the production of indole, we cultured Bacteroides species in both rich medium (TYG) containing L-tryptophan (Trp) (Holdeman et al., 1977) and minimal medium (Kotarski and Salyers, 1984) to which 5 mg/mL Trp had been added and assayed for indole production using Ehrlich’s test (Lombard and Dowell, 1983) and HPLC, respectively. Consistent with literature reports (Cato and Johnson, 1976; Lombard and Dowell, 1983), Bt and B. ovatus produced indole in both media conditions, whereas B. fragilis, B. vulgatus, and B. caccae did not (Figure 1C and 1D, Figure S2A–D). To determine whether the putative tryptophanase in Bt was necessary and sufficient for indole production, we constructed an unmarked deletion using allelic exchange and assayed the resultant strain (Bt Δ1492) in the same in vitro assays. In contrast to wild-type Bt, the BtΔ1492 mutant did not produce indole from tryptophan (Figure 1D, Figure S2C and D), suggesting that BT1492 is a tryptophanase and is solely responsible for indole production by Bt.
Healthy subjects vary substantially in terms of which Bacteroides species are present and at what level (Kraal et al., 2014). These data suggest that communities dominated by a tryptophanase-positive Bacteroides species could differ predictably in host IS load from those composed largely of tryptophanase-negative Bacteroides.
We next sought to test the hypothesis that the status of a single bacterial gene (BT1492) in Bt-mono-colonized mice could determine the presence or absence of IS in the host. Two groups of germ-free Swiss-Webster mice were colonized with either Bt wild-type (WT) or Δ1492. As expected, IS accumulated in the urine of mice colonized with WT Bt (Figure 2A); excreted levels of this metabolite remained consistent throughout the course of the four-week experiment (~7 mg/dl), suggesting that Bt is a robust and stable indole producer in vivo. Strikingly, the Δ1492-colonized mice had no detectable urinary IS.
Figure 2. Modulating indoxyl sulfate production by controlling the level of BT1492 and its orthologs.
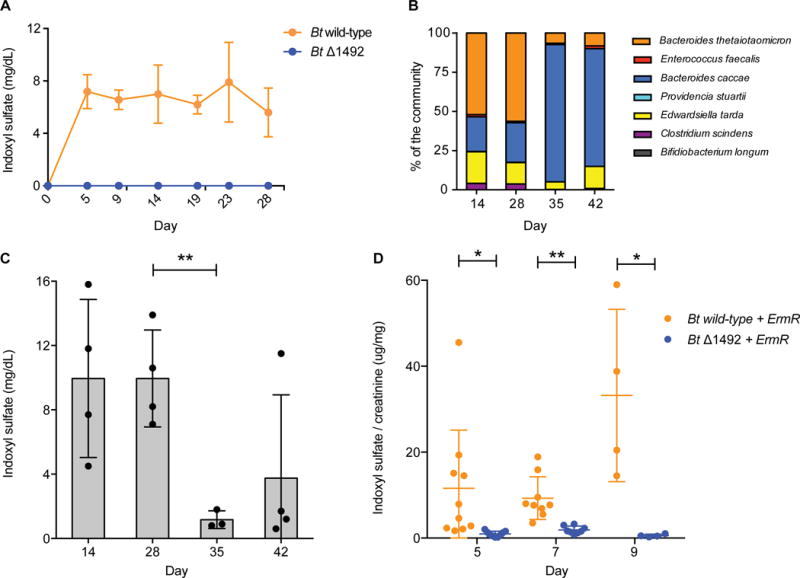
(A) Germ-free mice were mono-colonized with either Bt wild-type or Bt Δ1492 and the level of circulating IS was determined by analyzing host urine and serum (Figure S3) samples. In contrast to Bt wild-type-colonized animals, Bt Δ1492-colonized mice had no detectable IS in urine or serum (Figure S3C) (limit of detection in serum 0.003 mg/dL). Data shown are means from five biological replicates. Error bars represent standard deviation. (B) When gnotobiotic mice colonized with a defined bacterial community were switched from a standard diet to a fructo-oligosaccharide (FOS) diet 28 days post-inoculation, a shift from a Bt (indole producer)-dominant to a B. caccae (indole non-producer)-dominant community was observed. Data shown are from five biological replicates. (C) Concomitant with this shift in community composition, the level of urinary IS decreased significantly from day 28 to day 35 (P = 0.0085). Asterisks represent a significant difference between groups, Welch’s t test, n = at least 3 per group. (D) Conventional mice were reprogrammed to accumulate near-zero levels of IS. Following three days of treatment with an antibiotic cocktail, mice were treated with erythromycin and colonized with either Bt wild-type + ErmR or Bt Δ1492 + ErmR (days 3–9). Colonization with Bt wild-type + ErmR resulted in higher IS levels (Day 5, P = 0.035, at least 9 biological replicates per group; Day 7, P = 0.002, at least 7 biological replicates per group; Day 9, P = 0.047, 4 biological replicates per group). Error bars represent standard deviation. Asterisks represent a significant difference between groups, Welch’s t test. IS levels in (A) and (B) were not normalized to creatinine because a diet change in these experiments ablated the effectiveness of using creatinine normalization to account for changes in hydration state of the animals.
In order to ascertain whether urinary IS excretion corresponded directly to circulating IS levels, we repeated this experiment and collected both urine and serum on day 7 post-colonization. As expected, the relative serum IS levels mirrored those in urine (Figure S3). IS accumulated in the serum of WT Bt-colonized mice but was nearly undetectable in Δ1492-colonized mice, suggesting that differences observed in urinary IS excretion were due to increased circulating IS, not differences in kidney excretion rate. Taken together, these data show that BT1492 is necessary and sufficient for IS production in Bt-colonized mice, and they demonstrate that by modifying a single bacterial gene, the production of a high-abundance circulating metabolite can be toggled on and off.
To address whether IS levels could be controlled in the context of a more complex community, we used a synthetic community that consists of six bacterial species (Bt, Bacteroides caccae, Enterococcus faecalis, Providencia stuartii, Edwardsiella tarda, and Clostridium scindens) and includes two key features: First, Bt was the only indole producer in the community, so its level should have solely determined the total quantity of indole synthesized in the gut. Second, B. caccae – an indole-non-producing Bacteroides species – is well-adapted to use β-2,1-linked fructans such as fructo-oligosaccharides (FOS), whereas Bt is not (Sonnenburg et al., 2010). This enabled us to boost the level of B. caccae at the expense of Bt by shifting from a standard diet to a FOS-based diet.
After colonizing germ-free mice with the synthetic community, we fed the mice a standard diet for four weeks and then shifted to a fructo-oligosaccharide (FOS) diet for two weeks; bacterial counts and urinary IS levels were monitored at one-week intervals. As expected, on day 35 (seven days after switching to the FOS diet), we observed a significant decrease in Bt abundance and a concomitant increase in B. caccae abundance (Figure 2B). Importantly, host IS levels decreased markedly at the same timepoint; this lower level was maintained one week later, indicating a stable decrease in the amount of toxin produced (Figure 2C). These results suggest that we can use a dietary shift to rationally alter the level of IS in vivo.
Finally, we sought to determine whether the gut community of a conventional animal could be reprogrammed in order to modulate its IS level. In order to alter microbial composition in the background of a conventional gut community, we used antibiotics to facilitate the stable colonization of wild-type Bt or the indole-deficient mutant Δ1492. We treated conventional mice with an antibiotic cocktail that included erythromycin in drinking water for three days. Since the vast majority of Bacteroides species, including B. thetaiotaomicron and B. ovatus, are erythromycin-sensitive (Cato and Johnson, 1976), we reasoned that native indole-producing Bacteroides strains would be unlikely to grow in the presence of erythromycin. Mice were then switched to water with only erythromycin on day 4 and were colonized with either Bt-ermR or Δ1492-ermR, both of which had been modified to possess the erythromycin resistance gene. Bacteria were administered by gavage daily for six days, and urinary IS levels were monitored every two days.
Mice colonized with Δ1492-ermR excreted very low levels of IS on days 5, 7, and 9 (Figure 2D); in contrast, Bt-ermR-colonized mice exhibited significantly higher levels of urinary IS. The level of IS in Bt-ermR-colonized mice is lower by an order of magnitude than that of conventional mice, potentially due to a reduction in bacterial cell number as a consequence of antibiotic treatment or the absence of a second species that helps to liberate tryptophan from peptides, or stimulates Bt to convert tryptophan to indole. Nonetheless, these data prove the concept that in the background of a conventional gut community, a rational manipulation of the gut bacterial roster can modulate the level of excreted IS. Importantly, antibiotics have been used in clinical trials as an ameliorative treatment for dialysis patients (Poesen et al., 2013), so a therapeutic regimen that consists of antibiotic treatment plus re-colonization with an indole-deficient Bacteroides strain is plausible in the setting of kidney disease.
There are currently ~500,000 patients in the United States with end stage renal disease; their care consumes an estimated $50 billion annually, including ~7% of the Medicare budget (USRDS Annual Data Report, 2015). An additional 8 million people in the US have renal filtration that is reduced to below 50% of the normal rate and suffer ill effects (Coresh et al., 2005). Outcomes with current treatment are poor. Hemodialysis provides low quality of life and an average 5-year survival rate under 50% (Meyer and Hostetter, 2007). Certain solutes, including IS, are not efficiently removed by hemodialysis because they are highly bound to plasma protein (Niwa et al., 1988). Successful renal transplantation represents the best treatment option for patients with end-state kidney disease (Wolfe et al., 1999), but its utility is limited by the shortage of donor kidneys, the risk of death associated with surgery, and the subsequent life-long immunosuppression required to prevent organ rejection (Poesen et al., 2013).
Given the limitations of current therapies, reducing the production of uremic solutes, including IS, presents an increasingly attractive means to slow disease progression and increase quality and duration of life for CKD patients. Along these lines, clinical studies involving limiting dietary protein intake, ingesting adsorbent, or increasing dietary fiber intake have in some cases resulted in the reduction of circulating uremic toxins, including IS, and the amelioration of uremic symptoms (Kopple et al., 2000; Shoji et al., 2007; Ueda et al., 2008). As we explore here, a further means to limit solute production is to manipulate the composition of the gut microbiota.
In this work, we have shown that we can modulate indoxyl sulfate levels in vivo. In both a mono-colonized germ-free mouse experiment and a synthetic community experiment, we observed that by deleting or altering the abundance of a single gene (BT1492) and its orthologs, we could eliminate or dramatically reduce the level of urinary IS. We also showed that by rationally altering diet, we could favor the growth of a tryptophanase-negative Bacteroides species within a model gut community, demonstrating the feasibility of using a dietary manipulation to decrease the indole production capacity of the microbiota.
We then showed that we could reprogram a conventional murine gut community to control its indole-producing status, dictating the level of IS that accumulates in host circulation. Further optimization is necessary in order to achieve a more pronounced ON/OFF effect in a conventional animal. Despite the remaining challenges, the markedly low level of IS observed when we colonized with the tryptophanase-deficient B. theta strain is particularly striking, considering the challenges associated with controlling the level of a bacterially derived metabolite in a conventionally colonized animal. The use of antibiotics alone to suppress indole-producing strains is not an optimal strategy, due to potential complications from microbiota ablation such as antibiotic-associated pathogen invasion (Kamada et al., 2013). However, the judicious use of antibiotics to enable or enforce colonization of strains selected for particular metabolic traits may provide a strategy that accomplishes a reduction in toxin production and allows for the retention of many of the microbiota’s health benefits.
IS is one of a group of uremic solutes that are elevated in dialysis patients (Sirich et al., 2013). Until now, it has not been possible to deconvolute the specific effects of these compounds by selectively removing one molecule from circulation. We have developed a strategy by which we can study the effect of one of these solutes by toggling its levels up and down in the host. In doing so, our study opens the door to the possibility of treating renal disease by manipulating the gut microbiota.
Supplementary Material
Acknowledgments
We are indebted to members of the Fischbach and Sonnenburg Groups for helpful discussions. This work was supported by a research award from BASF (M.A.F.), a Fellowship for Science and Engineering from the David and Lucile Packard Foundation (M.A.F.), DARPA award HR0011-15-C-0084 (M.A.F.), the Program for Breakthrough Biomedical Research (M.A.F.), an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund (M.A.F.), and NIH grant DK101674 (J.L.S., T.W.M., and M.A.F.). M.A.F. is a scientific advisor to NGM Biopharmaceuticals and Revolution Medicines, and a Director of Achaogen.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental Information includes Supplemental Experimental Procedures, three Supplemental Figures, and one Excel Spreadsheet and can be found with this article online at:
Author Contributions
A.S.D, D.D., A.M., S.N., T.M., J.L.S., and M.A.F. designed the experiments. A.S.D., D.D., A.M., S.N., and N.P. performed the experiments and analyzed the data. A.S.D., S.N., D.D., A.M., J.L.S., and M.A.F. wrote the paper, and K.P. and T.M. provided edits and comments.
References
- Adijiang A, Goto S, Uramoto S, Nishijima F, Niwa T. Indoxyl sulphate promotes aortic calcification with expression of osteoblast-specific proteins in hypertensive rats. Nephrol Dial Transplant. 2008;23:1892–1901. doi: 10.1093/ndt/gfm861. [DOI] [PubMed] [Google Scholar]
- US Renal Data System. USRDS Annual Data Report. Volume 2: End-Stage Renal Disease in the United States 2015 [Google Scholar]
- Banoglu E, King RS. Sulfation of indoxyl by human and rat aryl (phenol) sulfotransferases to form indoxyl sulfate. Eur J Drug Metab Ph. 2002;27:135–140. doi: 10.1007/BF03190428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banoglu E, Jha GG, King RS. Hepatic microsomal metabolism of indole to indoxyl, a precursor of indoxyl sulfate. Eur J Drug Metab Ph. 2001;26:235–240. doi: 10.1007/BF03226377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto FC, Barreto DV, Liabeuf S, Meert N, Glorieux G, Temmar M, Choukroun G, Vanholder R, Massy ZA, European Uremic Toxin Work Group (EUTox) Serum indoxyl sulfate is associated with vascular disease and mortality in chronic kidney disease patients. Clin J Am Soc Nephrol. 2009;4:1551–1558. doi: 10.2215/CJN.03980609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao XS, Chen J, Zou JZ, Zhong YH, Teng J, Ji J, Chen ZW, Liu ZH, Shen B, Nie YX, et al. Association of indoxyl sulfate with heart failure among patients on hemodialysis. Clin J Am Soc Nephrol. 2014;10:111–119. doi: 10.2215/CJN.04730514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter D, Einheber A, Bauer H, Rosen H, Burns WF. The role of the microbial flora in uremia. II. Uremic colitis, cardiovascular lesions, and biochemical observations. J Exp Med. 1966;123:251–266. doi: 10.1084/jem.123.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cato EP, Johnson JL. Reinstatement of species rank for Bacteroides fragilis, B. ovatus, B. distasonis, B. thetaiotaomicron, and B. vulgatus: Designation of neotype strains for Bacteroides fragilis (Veillon and Zuber) Castellani and Chalmers and Bacteroides thetaiotaomicron (Distaso) Castellani and Chalmers. Int J Syst Bacteriol. 1976;26:230–237. [Google Scholar]
- Coresh J, Byrd-Holt D, Astor BC, Briggs JP, Eggers PW, Lacher DA, Hostetter TH. Chronic kidney disease awareness, prevalence, and trends among U.S. adults, 1999 to 2000. J Am Soc Nephrol. 2005;16:180–188. doi: 10.1681/ASN.2004070539. [DOI] [PubMed] [Google Scholar]
- Duranton F, Cohen G, De Smet R, Rodriguez M, Jankowski J, Vanholder R, Argilés A, European Uremic Toxin Work Group Normal and pathologic concentrations of uremic toxins. J Am Soc Nephrol. 2012;23:1258–1270. doi: 10.1681/ASN.2011121175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einheber A, Carter D. The role of the microbial flora in uremia. I. Survival times of germfree, limited-flora, and conventionalized rats after bilateral nephrectomy and fasting. J Exp Med. 1966;123:239–250. doi: 10.1084/jem.123.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evenepoel P, Meijers BKI, Bammens BRM, Verbeke K. Uremic toxins originating from colonic microbial metabolism. Kidney Int Suppl. 2009:S12–S19. doi: 10.1038/ki.2009.402. [DOI] [PubMed] [Google Scholar]
- Gillam EMJ, Notley LM, Cai H, De Voss JJ, Guengerich FP. Oxidation of Indole by Cytochrome P450 Enzymes. Biochem. 2000;39:13817–13824. doi: 10.1021/bi001229u. [DOI] [PubMed] [Google Scholar]
- Holdeman LV, Cato EP, Moore W. Anaerobe Laboratory Manual. Blacksburg, VA: Anaerobe Laboratory Virginia Polytechnic Institute and State University; 1977. [Google Scholar]
- Johnson DW, Craven AM, Isbel NM. Modification of cardiovascular risk in hemodialysis patients: An evidence-based review. Hemodial Int. 2007;11:1–14. doi: 10.1111/j.1542-4758.2007.00146.x. [DOI] [PubMed] [Google Scholar]
- Johnson JL, Moore WEC, Moore LVH. Bacteroides caccae sp nov., Bacteroides merdae sp nov., and Bacteroides stercoris sp nov Isolated from Human Feces. Int J Syst Bacteriol. 1986;36:499–501. [Google Scholar]
- Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotarski SF, Salyers AA. Isolation and characterization of outer membranes of Bacteroides thetaiotaomicron grown on different carbohydrates. J Bacteriol. 1984;158:102–109. doi: 10.1128/jb.158.1.102-109.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopple JD, Greene T, Chumlea WC, Hollinger D, Maroni BJ, Merrill D, Scherch LK, Schulman G, Wang SR, Zimmer GS. Relationship between nutritional status and the glomerular filtration rate: Results from the MDRD Study. Kidney Int. 2000;57:1688–1703. doi: 10.1046/j.1523-1755.2000.00014.x. [DOI] [PubMed] [Google Scholar]
- Kraal L, Abubucker S, Kota K, Fischbach MA, Mitreva M. The prevalence of species and strains in the human microbiome: A resource for experimental efforts. Plos One. 2014;9:e97279. doi: 10.1371/journal.pone.0097279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev. 2010;34:426–444. doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- Lombard GL, Dowell VR. Comparison of three reagents for detecting indole production by anaerobic bacteria in microtest systems. J Clin Microbiol. 1983;18:609–613. doi: 10.1128/jcm.18.3.609-613.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer TW, Hostetter TH. Uremia. N Engl J Med. 2007;357:1316–1325. doi: 10.1056/NEJMra071313. [DOI] [PubMed] [Google Scholar]
- Meyer TW, Hostetter TH. Uremic solutes from colon microbes. Kidney Int. 2012;81:949–954. doi: 10.1038/ki.2011.504. [DOI] [PubMed] [Google Scholar]
- Nayfach S, Fischbach MA, Pollard KS. MetaQuery: a web server for rapid annotation and quantitative analysis of specific genes in the human gut microbiome. Bioinformatics. 2015;31:3368–3370. doi: 10.1093/bioinformatics/btv382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton WA, Snell EE. Catalytic properties of tryptophanase, a multifunctional pyridoxal phosphate enzyme. Proc Natl Acad Sci USA. 1964;51:382–389. doi: 10.1073/pnas.51.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa T, Ise M. Indoxyl sulfate, a circulating uremic toxin, stimulates the progression of glomerular sclerosis. J Lab Clin Med. 1994;124:96–104. [PubMed] [Google Scholar]
- Niwa T, Ise M, Miyazaki T. Progression of glomerular sclerosis in experimental uremic rats by administration of indole, a precursor of indoxyl sulfate. Am J Nephrol. 1994;14:207–212. doi: 10.1159/000168716. [DOI] [PubMed] [Google Scholar]
- Niwa T, Shimizu H. Indoxyl sulfate induces nephrovascular senescence. J Ren Nutr. 2012;22:102–106. doi: 10.1053/j.jrn.2011.10.032. [DOI] [PubMed] [Google Scholar]
- Niwa T, Takeda N, Tatematsu A, Maeda K. Accumulation of indoxyl sulfate, an inhibitor of drug-binding, in uremic serum as demonstrated by internal-surface reversed-phase liquid chromatography. Clin Chem. 1988;34:2264–2267. [PubMed] [Google Scholar]
- Niwa T. Uremic toxicity of indoxyl sulfate. Nagoya J Med Sci. 2010;72:1–11. [PMC free article] [PubMed] [Google Scholar]
- Parfrey PS, Foley RN. The clinical epidemiology of cardiac disease in chronic renal failure. J Am Soc Nephrol. 1999;10:1606–1615. doi: 10.1681/ASN.V1071606. [DOI] [PubMed] [Google Scholar]
- Poesen R, Meijers B, Evenepoel P. The colon: an overlooked site for therapeutics in dialysis patients. Semin Dial. 2013;26:323–332. doi: 10.1111/sdi.12082. [DOI] [PubMed] [Google Scholar]
- Rothhammer V, Mascanfroni ID, Bunse L, Takenaka MC, Kenison JE, Mayo L, Chao CC, Patel B, Yan R, Blain M, et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat Med. 2016 doi: 10.1038/nm.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers E, Glorieux G, Vanholder R. The gut: the forgotten organ in uremia? Blood Purif. 2010;29:130–136. doi: 10.1159/000245639. [DOI] [PubMed] [Google Scholar]
- Shimada Y, Kinoshita M, Harada K, Mizutani M, Masahata K, Kayama H, Takeda K. Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. Plos One. 2013;8:e80604. doi: 10.1371/journal.pone.0080604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivanna S, Kolandaivelu K, Shashar M, Belghasim M, Al-Rabadi L, Balcells M, Zhang A, Weinberg J, Francis J, Pollastri MP, et al. The aryl hydrocarbon receptor is a critical regulator of tissue factor stability and an antithrombotic target in uremia. J Am Soc Nephrol. 2016;27:189–201. doi: 10.1681/ASN.2014121241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T, Wada A, Inoue K, Hayashi D, Tomida K, Furumatsu Y, Kaneko T, Okada N, Fukuhara Y, Imai E, et al. Prospective randomized study evaluating the efficacy of the spherical adsorptive carbon AST-120 in chronic kidney disease patients with moderate decrease in renal function. Nephron Clin Pract. 2007;105:c99–c107. doi: 10.1159/000097985. [DOI] [PubMed] [Google Scholar]
- Sirich TL, Funk BA, Plummer NS, Hostetter TH, Meyer TW. Prominent accumulation in hemodialysis patients of solutes normally cleared by tubular secretion. J Am Soc Nephrol. 2013;25:615–622. doi: 10.1681/ASN.2013060597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Ueda H, Shibahara N, Takagi S, Inoue T, Katsuoka Y. AST-120 treatment in pre-dialysis period affects the prognosis in patients on hemodialysis. Renal Failure. 2008;30:856–860. doi: 10.1080/08860220802356531. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe RA, Ashby VB, Milford EL, Ojo AO, Ettenger RE, Agodoa LYC, Held PJ, Port FK. Comparison of mortality in all patients on dialysis, patients on dialysis awaiting transplantation, and recipients of a first cadaveric transplant. N Engl J Med. 1999;341:1725–1730. doi: 10.1056/NEJM199912023412303. [DOI] [PubMed] [Google Scholar]
- Wu IW, Hsu KH, Lee CC, Sun CY, Hsu HJ, Tsai CJ, Tzen CY, Wang YC, Lin CY, Wu MS. p-Cresyl sulphate and indoxyl sulphate predict progression of chronic kidney disease. Nephrol Dial Transplant. 2011;26:938–947. doi: 10.1093/ndt/gfq580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


