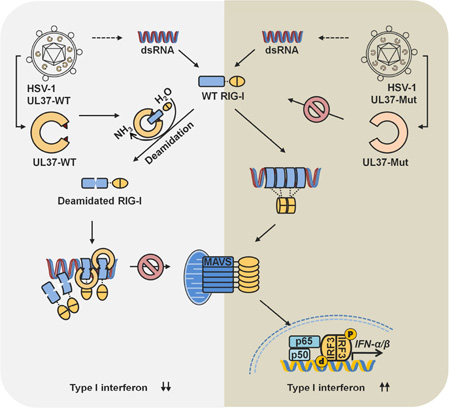
SUMMARY
RIG-I detects double-stranded RNA (dsRNA) to trigger antiviral cytokine production. Protein deamidation is emerging as a post-translational modification that chiefly regulates protein function. We report here that UL37 of herpes simplex virus 1 (HSV-1) is a protein deamidase that targets RIG-I to block RNA-induced activation. Mass spectrometry analysis identified two asparagine residues in the helicase 2i domain of RIG-I that were deamidated upon UL37 expression or HSV-1 infection. Deamidation rendered RIG-I unable to sense viral dsRNA, thus blocking its ability to trigger antiviral immune responses and restrict viral replication. Purified full-length UL37 and its carboxyl-terminal fragment were sufficient to deamidate RIG-I in vitro. Uncoupling RIG-I deamidation from HSV-1 infection, by engineering deamidation-resistant RIG-I or introducing deamidase-deficient UL37 into the HSV-1 genome, restored RIG-I activation and antiviral immune signaling. Our work identifies a viral deamidase and extends the paradigm of deamidation-mediated suppression of innate immunity by microbial pathogens.
Graphical Abstract
INTRODUCTION
Innate immunity is the first line of host defense. In response to invading pathogens, pattern recognition receptors (PRRs) sense pathogen-associated molecular patterns (PAMPs) that are structural components or replication intermediates (Medzhitov, 2007; Takeuchi and Akira, 2010; Ting et al., 2010). PRRs include the cytosolic receptors (e.g., cGAS, IFI16, RIG-I-like and NOD-like receptors) and the membrane-anchored Toll-like receptors (TLRs) and C-type lectins. Upon binding to PAMPs, PRRs recruit cognate adaptor molecules that signal to activate two closely-related kinase complexes, IKKα/β and TBK-1/IKKε. IKKα/β phosphorylates and induces the degradation of the inhibitor of NF-κB (IkBs), leading to the nuclear translocation of NF-κB (Chen et al., 1996; Zandi et al., 1997). TBK-1/IKKε can directly phosphorylate interferon regulatory factors (IRFs) to induce its dimerization and translocation into the nucleus (Fitzgerald et al., 2003; Sharma et al., 2003). Along with other transcription factors, nuclear NF-κB and IRFs coordinate to up-regulate the expression of many immune genes to engender an antiviral state (Bhatt and Ghosh, 2014). The cytosolic RIG-I receptor is a genuine RNA sensor that, in response to viral infection, activates NF-κB and IRFs through the mitochondrion antiviral signaling (MAVS) protein (Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005). Studies entailing gene knockout mice demonstrate that loss of RIG-I or MAVS severely impairs host innate immune response and greatly increases viral replication (Kato et al., 2006; Sun et al., 2006). Not surprisingly, viruses have evolved diverse strategies to halt or hijack antiviral signaling downstream of RIG-I and MAVS (Chan and Gack, 2015; Feng et al., 2013).
Post-translational modification (PTM) is a major means to regulate protein function and underpins diverse fundamental biological processes. First reported more than five decades ago (Mycek and Waelsch, 1960), deamidation of asparagine/glutamine in protein has long been regarded as a non-specific process associated with protein “aging”. Early protein deamidation research surveyed the overall deamidation of the cellular proteome, and led to the postulate that non-enzymatic protein deamidation serves as a biological clock for protein “aging” (Robinson and Robinson, 2001; Weintraub and Deverman, 2007). As such, research in protein deamidation is scarce and accordingly our understanding is rudimentary at best. A few proteins (e.g., Bcl-xL and 4EBP2) were shown to be regulated by deamidation in mammalian cells, which was postulated to be the consequence of an increase in cellular pH (Bidinosti et al., 2010; Deverman et al., 2002; Dho et al., 2013). Recent studies demonstrate that pathogenic bacteria secrete effectors to deamidate key signaling molecules to evade host immune defenses (Cui et al., 2010; Sanada et al., 2012) and manipulate cellular signaling (Flatau et al., 1997; Schmidt et al., 1997), indicating that protein deamidation can be catalyzed by bacterial enzymes and is highly regulated. The roles of protein deamidation in metazoan remain largely unclear.
We previously reported that gamma herpesviruses, including human Kaposi’s sarcoma-associated herpesvirus (KSHV) and murine gamma herpesvirus 68 (γHV68), deploy vGAT pseudo-enzymes to induce RIG-I deamidation (He et al., 2015; Kolakofsky and Garcin, 2015). Though lacking intrinsic enzyme activity, vGAT proteins recruited cellular phosphoribosylformyglycinamidine synthetase (PFAS, also known as FGARAT) to deamidate and concomitantly activate RIG-I. Activated RIG-I was harnessed by γHV68 to evade antiviral cytokine production (Dong and Feng, 2011; Dong et al., 2012). We report here that herpes simplex virus 1 (HSV-1) induces RIG-I deamidation to prevent RIG-I activation by viral dsRNA. The UL37 tegument protein was sufficient to deamidate RIG-I in cells and in vitro, revealing a role for a viral protein deamidase in immune regulation. Site-specific deamidation within the helicase 2i domain impaired the RNA detection and ATP hydrolysis of RIG-I. Uncoupling RIG-I deamidation from HSV-1 infection restored RIG-I activation and anti-viral cytokine production, thereby reducing HSV-1 replication. Our work delineates a pivotal role of protein deamidation in sensing nucleic acid by a PRR and demonstrates that HSV-1 exploits protein deamidation to evade innate immune defense.
RESULTS
HSV-1 Evades RNA-induced RIG-I Activation
We previously reported that the vGAT proteins of human KSHV and murine γHV68 recruit PFAS to deamidate RIG-I. In addition to these gamma herpesviruses, HSV-1 infection also increased negative charge of RIG-I as analyzed by two-dimensional gel electrophoresis (2-DGE), indicative of deamidation (Figure 1A). However, genomes of HSV-1 and other alpha- herpesviruses contain no homologue of gamma herpesvirus vGAT proteins, suggesting a distinct mechanism of RIG-I deamidation. Additionally, the antiviral roles of RIG-I against DNA viruses, such as herpesviruses, are not well defined. Thus, we investigated whether HSV-1 infection induces RIG-I deamidation and determined the functional consequence of RIG-I deamidation on host immune responses.
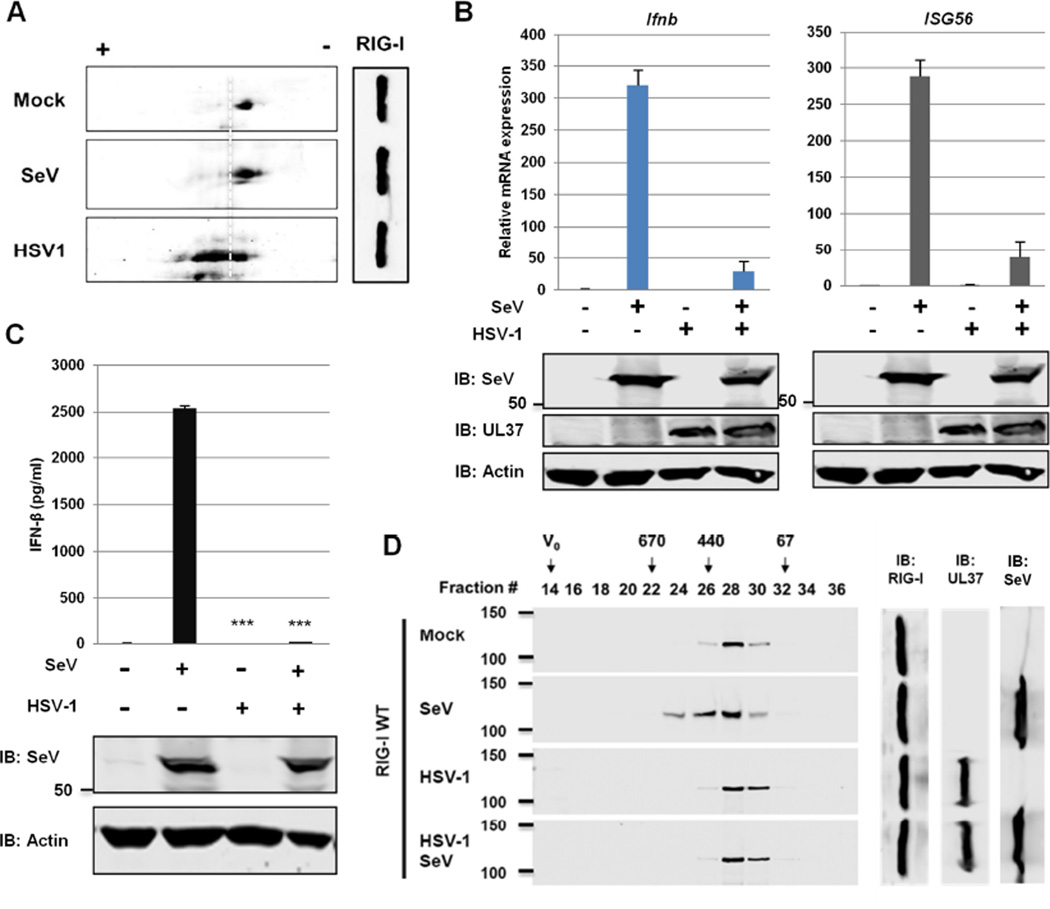
Figure 1. HSV-1 evades RNA-induced activation of RIG-I.
(A) HEK293/Flag-RIG-I cells were mock-infected or infected with HSV-1 (MOI=2) or Sendai virus (SeV, 100 HAU/ml) for 4 hours. Whole cell lysates (WCLs) were analyzed by two-dimensional gel electrophoresis.
(B and C) HEK293 cells were mock-infected or infected with HSV-1 (MOI=2) for 1 h and super-infected with SeV (100 HAU/ml) for 8 (B) or 16 h (C). The expression of the indicated antiviral genes was analyzed by real-time PCR using total RNA (B). Supernatant was collected to determine IFN-β by ELISA (C).
(D) HEK293/Flag-RIG-I were mock-infected or infected with HSV-1 for 1 h, followed by SeV infection (100 HAU/ml) for 4 h. RIG-I was purified and analyzed by gel filtration and immunoblotting. Numbers indicate the size of RIG-I in kDa and V0 denotes void volume.
For B-D, WCLs were analyzed by immunoblotting with antibodies against SeV, HSV-1 UL37 and β-actin.
***, p<0.001 was calculated in reference to cells infected with SeV. For B and C, data are presented as mean ± SD.
See also Figure S1.
To probe the roles of RIG-I in host defense against HSV-1, we depleted RIG-I expression and assessed the IFN-β mRNA in primary human foreskin fibroblasts (HFF). We found that knockdown of RIG-I reduced IFN-β mRNA at 24 h induced by HSV-1 infection (Figure S1A). A similar effect was observed with knockdown of IFI16, a DNA sensor implicated in detecting herpesviruses (Kerur et al., 2011; Unterholzner et al., 2010). In 293T and HeLa cells that HSV-1 replication is very robust, we observed that RIG-I depletion impaired IFN and ISG56 induction at multiplicity of infection (MOI) of 0.1 and 0.5 (Figure S1B,S1C). No difference in IFN and ISG56 induction was observed in RIG-I knockdown cells upon high MOI (=5) HSV-1 infection, suggesting that HSV-1 can blunt IFN induction. Moreover, RIG-I depletion in human THP-1 macrophages also reduced IFN-β and ISG56 mRNA induced by HSV-1 infection (Figure S1D). These results indicate that RIG-I senses dsRNA produced by HSV-1-infected cells and contributes to the IFN induction by HSV-1 infection.
To determine whether HSV-1 infection inhibits RIG-I activation, we sequentially infected 293T cells with HSV-1 and SeV, and determined IFN-β and ISG56 expression. We found that HSV-1 infection significantly reduced IFN-β and ISG56 mRNA induced by SeV (Figure 1B), which correlated with minimal IFN-β secretion (Figure 1C). Importantly, HSV-1 infection did not significantly reduce SeV replication as evidenced by expression of the major protein of SeV (Figure 1B and 1C). Furthermore, SeV infection induced the oligomerization of RIG-I that eluted in fractions corresponding to protein sizes between ~440 kDa and ~670 kDa, while RIG-I eluted in fraction corresponding to ~230 kDa in mock-infected cells or cells that were infected with HSV-1 (Figure 1D). Strikingly, RIG-I purified from HSV-1- and SeV-infected cells had an elution pattern identical to that of mock-infected cells. Finally, HSV-1 infection completely blunted the induction of IFN and ISG56 mRNA in 293T cells transfected with LMW poly[I:C] (Figure S1E). These results show that HSV-1 inhibits RIG-I activation triggered by SeV, a prototype RIG-I activator.
HSV-1 UL37 Interacts with RIG-I
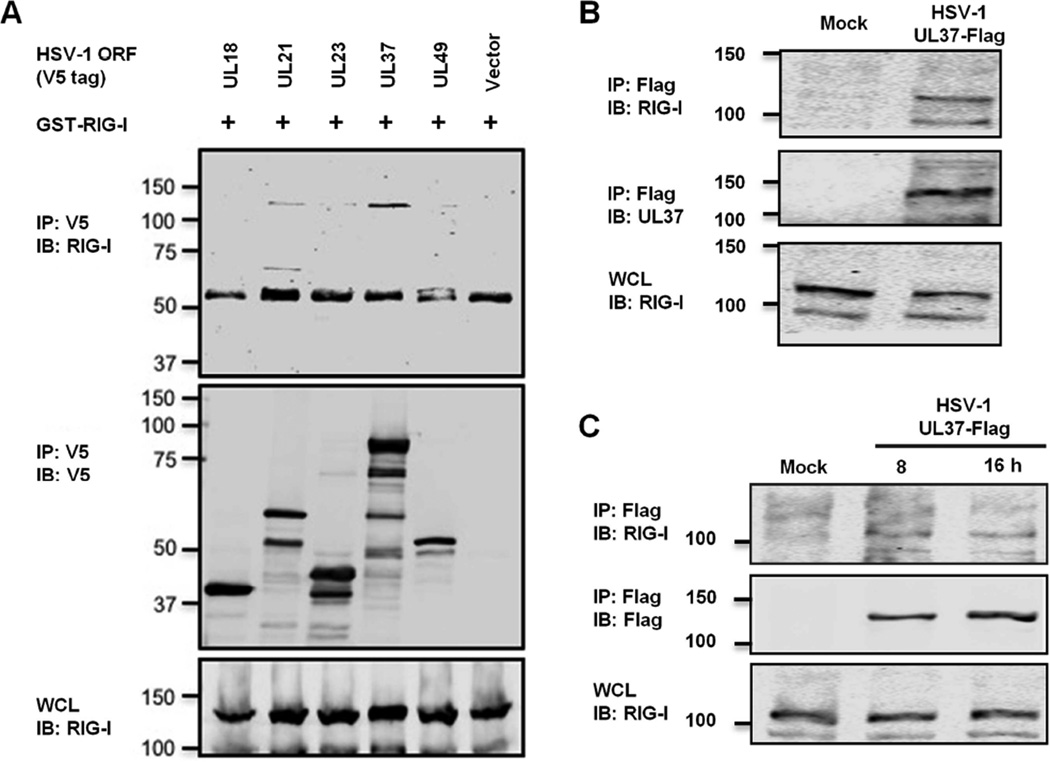
To delineate the mechanism by which HSV-1 abrogates RIG-I activation, we screened for RIG-I-binding proteins by co-immunoprecipitation (Co-IP) using a HSV-1 expression library, with a particular focus on gene products that operate in the early phase of infection. Co-IP assays identified open reading frames UL21 and UL37 as RIG-I-interacting proteins (Figure 2A and S2A). Although UL37 has no sequence homology with gamma herpesvirus vGAT proteins, it shares multiple functions with the vGAT proteins, e.g., activating NF-κB and promoting viral replication (Desai et al., 2001; Full et al., 2014; Gaspar et al., 2008; Liu et al., 2008). Thus, we examined whether UL37 evades RIG-I-dependent immune defense. Indeed, UL37 was readily detected in protein complexes precipitated by antibody against RIG-I in HSV-1-infected 293T cells as early as 1 hour post-infection (hpi) at high MOI (=30) (Figure 2B) and during late lytic replication at lower MOI (=1) (Figure 2C). When expressed in 293T cells, UL37 co-precipitated with RIG-I (Figure S2B), indicating that UL37 interacts with RIG-I in the absence of any other viral proteins. Interestingly, UL37 also interacted with MDA5 in transfected 293T cells (Figure S2B). Gel filtration analyses further showed that UL37 co-eluted with RIG-I in fractions corresponding to ~220–440 kDa, supporting that these proteins form a complex in HSV-1-infected cells (Figure S2C). UL37 also partly co-eluted with its interacting partner TRAF6 by gel filtration analysis. Thus, UL37 interacts with RIG-I in HSV-1-infected or transfected cells.
Figure 2. HSV-1 UL37 interacts with RIG-I.
(A) 293T cells were transfected with plasmids containing GST-RIG-I and the indicated open reading frames of HSV-1. WCLs were precipitated with the indicated antibody. WCLs and precipitated proteins were analyzed by immunoblotting.
(B and C) 293T cells were infected with recombinant HSV-1 UL37-Flag at MOI of 30 for 1 h (B) or MOI of 1 for 8 and 16 h (C). WCLs were precipitated with anti-Flag (M2) antibody. RIG-I and WCLs were analyzed by immunoblotting with indicated antibodies.
See also Figure S2.
HSV-1 UL37 Inhibits RIG-I Activation
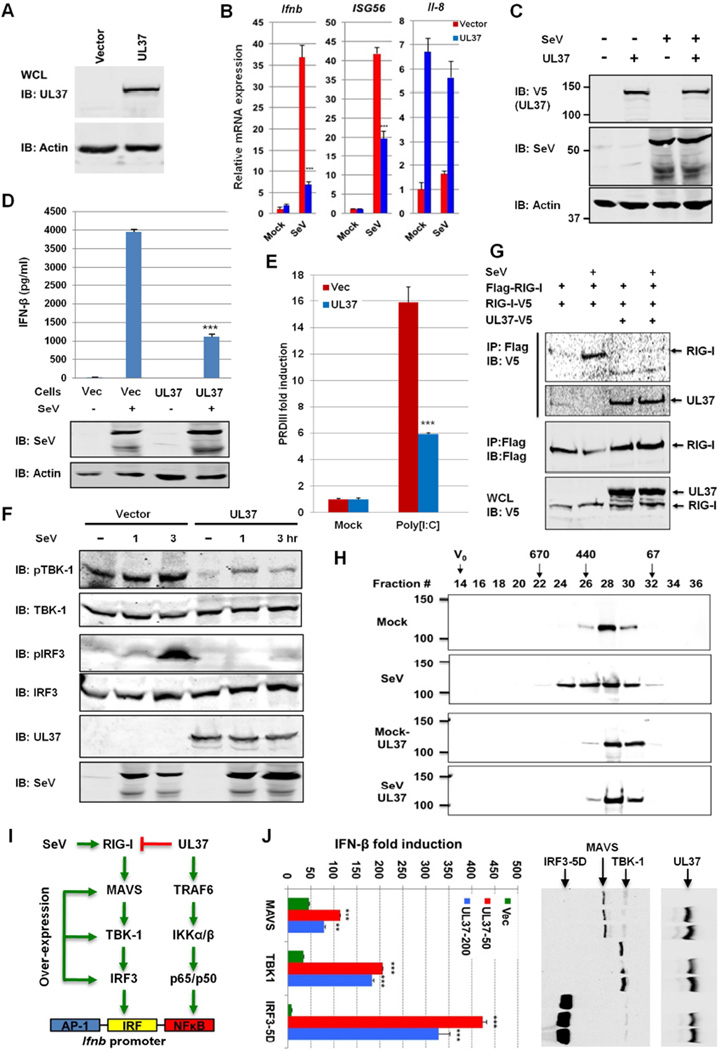
We established 293T cells stably expressing UL37 (Figure 3A). Upon SeV infection, UL37 expression significantly reduced IFN-β and ISG56 expression by real-time PCR analysis (Figure 3B) and reporter assays (Figure S3A, S3B). UL37 did not reduce SeV protein expression (Figure 3C). UL37 significantly up-regulated IL-8 and CxCL2 expression (Figure S3C), likely due to the NF-κB activation by UL37 (Liu et al., 2008), while had a marginal effect on IL-8 expression upon SeV infection (Figure 3B). ELISA further confirmed that UL37 expression reduced IFN-β secretion by ~75% in response to SeV infection (Figure 3D). Moreover, UL37 expression inhibited IFN induction upon LMW poly[I:C] transfection (Figure 3E). Over-expression of UL37 was sufficient to activate NF-κB (Figure S3D), but had no detectable effect on PRDIII, an IRF-responsive element of the IFN-β promoter (Figure S3E). Loss of RIG-I in mouse embryonic fibroblasts (MEFs) had no effect on UL37-induced NF-κB activation (Figure S3F). UL37 expression did not alter the transcription of the PRDIII promoter induced by MDA5 over-expression (Figure S3F). These results collectively show that UL37 specifically inhibits RIG-I-dependent IFN-β induction.
Figure 3. UL37 inhibits RIG-I activation.
(A–F) Whole cell lysates (WCLs) of 293T cells stably expressing UL37 were analyzed by immunoblotting with anti-V5 (UL37) and anti-β-actin antibodies (A). Cells were infected with Sendai virus (SeV) (100 HAU, 8 h) and total RNA was analyzed by real-time PCR with primers specific for the indicated genes (B). WCLs were analyzed by immunoblotting with antibodies against V5 (UL37), SeV and β-actin (C). Supernatant was harvested for cytokines determined by ELISA at 16 hpi, and WCLs were analyzed by immunoblotting with antibodies against SeV and β-actin (D). Cells were transfected with poly[I:C] and the PRDIII-luc reporter. Activation of the PRDIII promoter was determined by luciferase reporter assay (E). 293T stable cells were infected with SeV (100 HAU/ml) for 1 and 3 h, and WCLs were analyzed for the phosphorylation of TBK-1 and IRF3 by immunoblotting (F).
(G and H) 293T cells stably expressing Flag-RIG-I and RIG-I-V5 were transfected with an empty or UL37-containing plasmid. At 30 h post-transfection, cells were infected with SeV (100 HAU/ml) for 4 h. WCLs were precipitated with anti-Flag. Precipitated proteins and WCLs were analyzed by immunoblotting with the indicated antibodies (G). 293T/Flag-RIG-I cells, without or with UL37-V5 expression (by lentivirus), were mock-infected or infected with SeV (100 HAU/ml) for 4 h. Purified RIG-I was analyzed by gel filtration and immunoblotting. Numbers at the top indicate the size of RIG-I in kDa and V0 denotes void volume (H).
(I) Diagram of key components of the RIG-I-mediated IFN induction pathway.
(J) 293T cells were transfected with plasmids containing MAVS, TBK1 and the constitutively active IRF3-5D, along with the IFN-β reporter plasmid and a UL37-containing plasmid. Activation of the IFN-β promoter was determined by luciferase assay. WCLs were analyzed by immunoblotting with anti-Flag (M2) (MAVS, TBK-1 and IRF3-5D) and anti-V5 (UL37) antibodies (right panels).
***, p<0.001. For B, D, E and J, data are presented as mean ± SD.
See also Figure S3.
To probe the effect of UL37 on signaling events downstream of RIG-I, we analyzed the phosphorylation of TBK-1 (Ser172) and IRF3 (Ser396), markers of activated TBK-1 and IRF3, respectively. As shown in Figure 3F, UL37 expression inhibited the phosphorylation of TBK-1 and IRF3 upon SeV infection. Moreover, UL37 expression reduced the dimerization and nuclear translocation of IRF3 (Figure S3G and S3H). Using 293T cells stably expressing Flag-RIG-I and RIG-I-V5, we found that UL37 expression abolished RIG-I dimerization upon SeV infection by Co-IP assay (Figure 3G). Furthermore, UL37 diminished the SeV-induced oligomerization of RIG-I as analyzed by gel filtration (Figure 3H). To test whether UL37 inhibits key components of the IRF-IFN pathway downstream of RIG-I, we over-expressed MAVS, TBK-1 and the constitutively active IRF3–5D mutant (Figure 3I) and examined the activation of the IFN-β reporter. Consistent with NF-κB activation by UL37 (Figure S3D), we found that UL37 enhanced, rather than inhibited, the transcription of the IFN-β reporter in a dose-dependent manner with all three components (Figure 3J). UL37 did not alter the protein level of MAVS, TBK-1 and IRF3–5D. These results conclude that UL37 specifically targets RIG-I to block IFN induction by viral dsRNA.
UL37 deamidates RIG-I
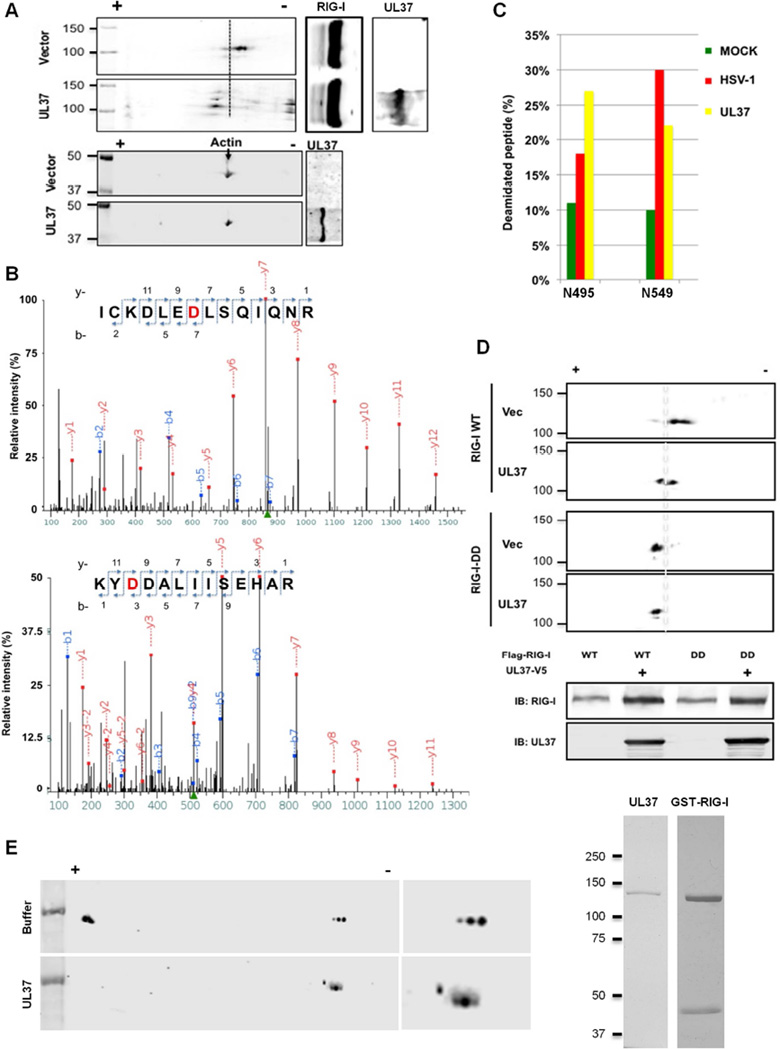
HSV-1 infection reduced the charge of RIG-I, suggesting that HSV-1 induces RIG-I deamidation. We found that UL37 expression was sufficient to reduce the charge of RIG-I, but not that of β-actin (Figure 4A). Furthermore, UL37 expression did not alter the charge of MDA5, an RNA sensor akin to RIG-I (Figure S4A). We thus purified RIG-I 293T stable cells upon HSV-1 infection or UL37 expression (Figure S4B). Tandem mass spectrometry analyses of both samples identified two peptides that contained aspartates at residue 495 and 549, indicative of deamidation of N495 and N549 (Figure 4B). HSV-1 infection and UL37 expression had similar effect on the deamidation of N495 and N549 (Figure 4C), suggesting that UL37 is responsible for RIG-I deamidation during HSV-1 infection. When N495D and N549D were introduced into RIG-I, designated RIG-I-DD, we found that RIG-I-DD migrated toward the positive end of the strip, to a position identical to that of RIG-I-WT when UL37 was expressed (Figure 4D). Moreover, UL37 expression did not further shift RIG-I-DD by 2-DGE analysis, indicating that N495 and N549 are the two sites of deamidation by UL37.
Figure 4. UL37 deamidates RIG-I in cells and in vitro.
(A) HEK293/Flag-RIG-I cells were transfected with an empty or UL37-containing plasmid. Whole cell lysates (WCLs) were analyzed by two-dimensional gel electrophoresis and immunoblotting with the indicated antibodies.
(B and C) HEK293/Flag-RIG-I cells were transfected with a UL37-expressing plasmid or infected with HSV-1 (MOI=1) for 12 h. RIG-I was purified and analyzed by tandem mass spectrometry. Two peptides containing deamidated asparagines were identified. D495 and D549 (in red) were shown (B). Deamidated peptides were quantitatively determined by tandem mass spectrometry analysis and data represents one of two independent experiments (C).
(D) HEK293/Flag-RIG-I or HEK293/Flag-RIG-I-DD cells were transfected with an empty or UL37-containing plasmid. WCLs were analyzed by two-dimensional gel electrophoresis and immunoblotting.
(E) GST-RIG-I and UL37 were purified from transfected 293T cells and E.coli, respectively, and analyzed by silver staining (right panels). Deamidation reaction was analyzed by two-dimensional gel electrophoresis and immunoblotting with anti-RIG-I antibody.
See also Figure S4.
To probe the mechanism of UL37-induced deamidation, we first determined whether a specific inhibitor of glutamine amidotransferase, 6-diazo-5-oxo-L-norleucine (DON), can block UL37-induced RIG-I deamidation. Indeed, DON inhibited RIG-I deamidation in cells expressing UL37 (Figure S4C). This result suggests that UL37-mediated deamidation of RIG-I depends on an enzymatic activity akin to glutamine amidotransferase. Thus, we sought to determine whether UL37 is intrinsically a protein deamidase. We purified UL37 full-length from E. coli to homogeneity and examined RIG-I deamidation in vitro. Analysis by 2-DGE indicated that UL37 was sufficient to reduce RIG-I charge, suggestive of deamidation (Figure 4E). These results indicate that UL37 deamidates RIG-I in cells and in vitro.
Deamidated RIG-I Fails to Sense RNA and Hydrolyze ATP
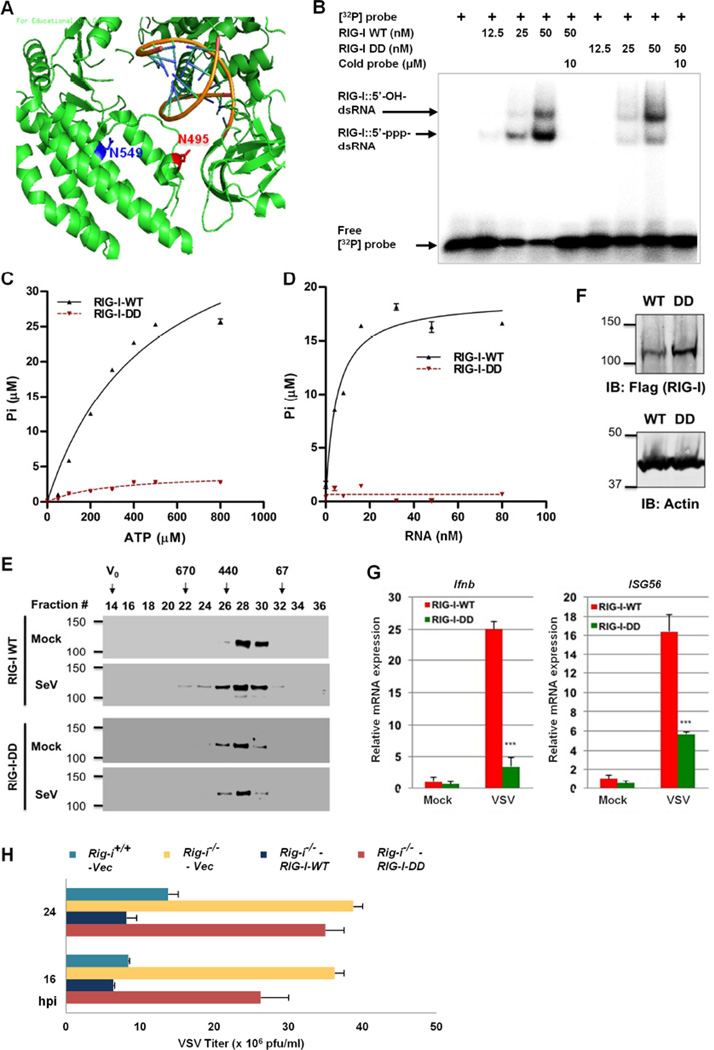
We previously showed that m-vGAT induced deamidation and concomitant activation of RIG-I. However, the RIG-I-DD mutant failed to activate NF-κB and IFN-β reporters (Figure S5A, S5B). N495 and N549 reside in the helicase 2i (Hel2i) domain that specializes in duplex RNA recognition (Luo et al., 2013). Previous structural analyses of RNA-bound RIG-I showed that these two residues flank the dsRNA-binding α-helix, α23 (Figure 5A) (Kowalinski et al., 2011; Luo et al., 2011). Residues of the α-helix (α23), specifically K508 and Q511, make direct contact with dsRNA. While N495 precedes the α23 helix, N549 is located in the middle of a spatially adjacent α-helix (α24). Thus, we opted to determine whether deamidation of N495 and N549 affects the RNA-binding ability of RIG-I, an important function for RNA detection by RIG-I. We purified RIG-I-WT and its mutants to homogeneity from transfected 293T cells (Figure S5C) and performed electrophoresis mobility shift assay (EMSA). We found two distinct RIG-I:RNA complexes that correlated with increasing doses of RIG-I (Figure 5B and S5D). The RIG-I-DD mutant was significantly impaired in forming the fast migrating RIG-I:RNA complex, while formed higher levels of the more slowly migrating RIG-I:RNA complex compared to RIG-I-WT (Figure 5B). EMSA also showed that the deamidation of N549 had a major effect on the RNA-binding ability of RIG-I, while RIG-I-N495D demonstrated comparable RNA-binding affinity to RIG-I-WT (Figure S5D). Using a control dsRNA 19 mer lacking the 5’-triphosphate, we found that the more slowly migrating RIG-I:RNA complex consisted of RIG-I and dsRNA without 5’-triphophate (Figure S5E). Interestingly, the K270A mutant previously shown to have impaired ATPase activity, bound to the 5’-triphosphate 19mer dsRNA (5’ppp-RNA) with affinity similar to RIG-I-WT (Figure S5F). Although ATP hydrolysis is not required for RIG-I signaling, it has been proposed that ATPase activity is necessary for recycling of RIG-I from RNA-bound complexes and critical for RIG-I-mediated innate immune signaling against nonself RNA (Anchisi et al., 2015; Lassig et al., 2015; Luo et al., 2013). We thus examined the ATP hydrolysis activity using purified RIG-I proteins. An in vitro ATPase assay showed that RIG-I-DD completely lost its ability to hydrolyze ATP (Figure 5C). RIG-I-WT and RIG-I-DD demonstrated ATPase activity with kcat of 944 and 32.6 sec−1 at physiological ATP concentrations, respectively. Furthermore, RIG-I-DD failed to hydrolyze ATP upon 5’ppp-RNA stimulation (Figure 5D). RIG-I-N549D and RIG-I-N495D demonstrated basal or no ATPase activity similar to RIG-I-DD, with or without 5’ppp-RNA (Figure S5G, S5H). These results indicate that deamidation of N495 and N549 abolishes RIG-I activity to bind RNA and hydrolyze ATP.
Figure 5. The deamidated RIG-I-DD mutant fails to sense viral dsRNA.
(A) N495 and N549 are located in the helicase 2i domain of the RIG-I structure (PDB ID: 3TMI). dsRNA is shown as helices in dark yellow.
(B) Purified RIG-I and RIG-I-DD were incubated with [32P]-labeled 5’-triphosphate 19mer dsRNA and analyzed by electrophoresis mobility shift assay.
(C and D) Purified RIG-I and RIG-I-DD were used for in vitro ATP hydrolysis with increasing amount of ATP (C) or 5’-triphosphate 19 mer dsRNA (D).
(E) 293T cells stably expressing RIG-I-WT and RIG-I-DD were mock-infected or infected with Sendai virus (SeV, 100 HAU/ml) for 4 h. Purified RIG-I was analyzed by gel filtration and immunoblotting. Numbers at the top indicate the size of RIG-I in kDa and V0 denotes void volume.
(F–H) Rig-i−/− MEFs “reconstituted” with RIG-I-WT or RIG-I-DD were analyzed by immunoblotting with the indicated antibodies (F). Cells were infected with recombinant eGFP VSV (Indiana Strain, MOI=20) for 8 h, and total RNA was analyzed by real-time PCR (G). Cells were infected with VSV (MOI=0.05) and viral titer in the supernatant was determined by plaque assay (H). hpi, hours post-infection.
For C, D, G and H, data are presented as mean ± SD.
See also Figure S5.
To assess the functional consequence of RIG-I deamidation, we examined RIG-I activation by gel filtration. SeV infection induced oligomerization of RIG-I-WT as evidenced by fractions corresponding to protein complexes of ~440–670 kDa sizes, while RIG-I-WT in mock-infected cells eluted in fractions corresponding to ~130–230 kDa (Figure 5E). However, SeV infection failed to induce the oligomerization of RIG-I-DD. Notably, a low level of RIG-I-DD was detected in fractions corresponding to protein sizes of ~440 kDa regardless of SeV infection. We then “reconstituted” RIG-I expression in Rig-i−/− MEF with RIG-I-WT or RIG-I-DD (Figure 5F), and examined host immune responses and viral infection. Compared to RIG-I-WT, RIG-I-DD induced basal or lower expression of IFN-β and ISG56 upon vesicular stomatitis virus (VSV) infection (Figure 5G). Similar results were observed in SeV-infected cells (Figure S5I). Consequently, RIG-I-WT, but not RIG-I-DD, reduced VSV replication in Rig-i−/− MEFs (Figure 5H). These results demonstrate that deamidation of N495 and N549 eleminates RIG-I detection of viral RNA and restriction of viral replication.
A Deamidation-resistant RIG-I-QQ Mutant Restores Antiviral Immune Responses Against HSV-1 Infection
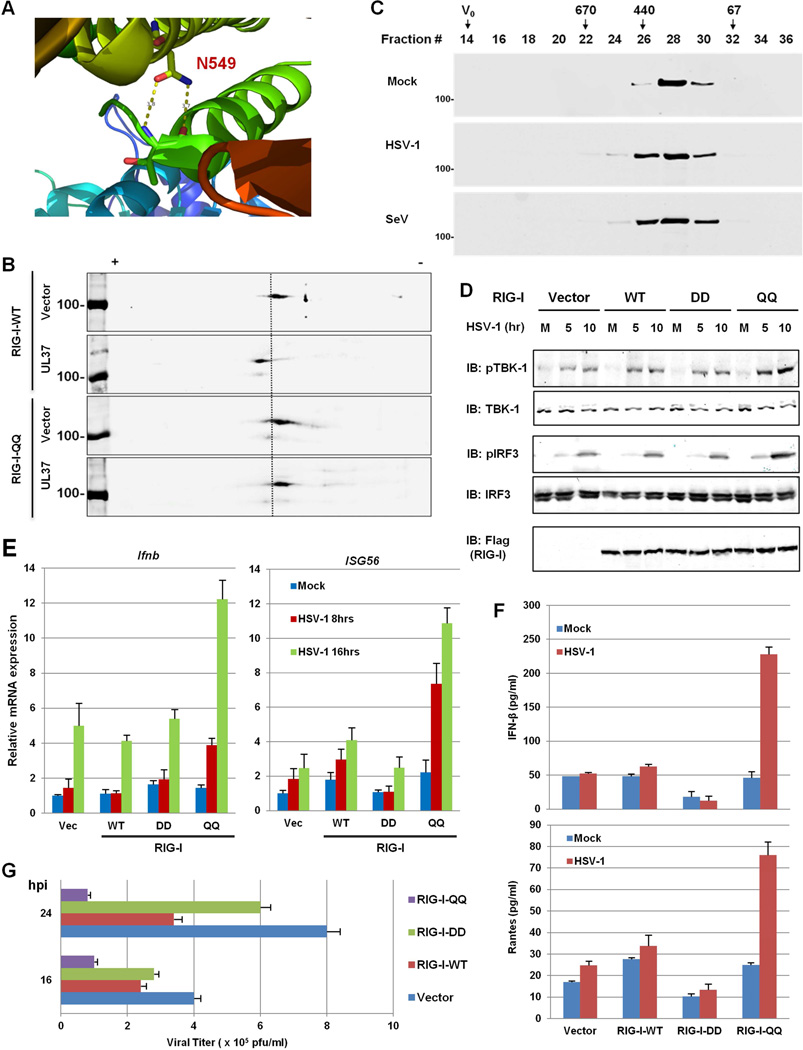
Our mutational analysis indicates that N549 is critical for the RNA-binding and ATPase activities of RIG-I. Previously solved crystal structure of RIG-I showed that the amide group of N549 (within α24) forms two hydrogen bonds with the backbone of threonine 504 of the RNA-binding α-helix (α23) (Figure 6A)(Kowalinski et al., 2011; Luo et al., 2011). The RIG-I-N549A mutant failed to trigger IFN induction by SeV infection (data not shown), suggesting that the hydrogen bonds between N549 and T504 of the two neighboring helices are critical for RIG-I immune signaling. The side chain of glutamine contains a primary amide functional group as asparagine does. We hypothesized that a glutamine residue at position 549 might conserve hydrogen bonds with T504, which translates to a predicted ~1 angstrom short difference in hydrogen bonds formed by Q549 than N549, thereby potentially resisting deamidation. We then generated a RIG-I mutant containing Q495 and Q549, designated RIG-I-QQ. In 293T cells stably expressing RIG-I-WT or the RIG-I-QQ mutant, UL37 expression shifted RIG-I-WT, but not RIG-I-QQ, toward the positive end of the strip, indicating that RIG-I-QQ is deamidation-resistant (Figure 6B). Furthermore, RIG-I-QQ was eluted in fractions corresponding to sizes of ~440–670 kDa in cells infected with HSV-1, demonstrating similar levels of oligomerization as RIG-I-WT (Figure 1D) and RIG-I-QQ (Figure 6C) induced by SeV infection. These results indicate that RIG-I-QQ is refractory to deamidation, and therefore, restores RIG-I activation induced by HSV-1 infection.
Figure 6. The deamidation-resistant RIG-I-QQ restores antiviral cytokine production in response to HSV-1 infection.
(A) N549 forms two hydrogen bonds with the backbone of T504 within the helicase 2i domain of the RIG-I structure (PDB ID: 4A36).
(B) HEK293/Flag-RIG-I-WT or HEK293/Flag-RIG-I-QQ cells were transfected with an empty or UL37-containing plasmid. Whole cell lysates (WCLs) were analyzed by two-dimensional gel electrophoresis.
(C) HEK293/Flag-RIG-I-QQ cells were mock-infected or infected with HSV-1 (MOI=5) or Sendai virus (SeV, 100 HAU/ml) for 4 h. Purified RIG-I was resolved by gel filtration and analyzed by immunoblotting.
(D–F) Control HEK293 (Vec) or HEK293 cells stably expressing RIG-I-WT, RIG-I-DD or RIG-I-QQ were were infected with HSV-1 (MOI=5) for 4h and WCLs were analyzed by immunoblotting with the indicated antibodies (D). Total RNA was analyzed by real-time PCR (E). Supernatant was collected at 16 hpi and cytokines (IFN-β and RANTES) were quantified by ELISA (F). M, mock-infected; numbers on the top indicate hours post-infection in (D).
(G) Stable HEK293 cell lines as above were infected with HSV-1 at MOI=0.5 and viral titer was determined by plaque assay.
For E, F and G, data are presented as mean ± SD.
We reasoned that only the deamidation-resistant RIG-I-QQ mutant will confer gain-of-function in RIG-I-mediated innate immune response, thus we used wild-type HEK293 to establish stable cell lines expressing RIG-I wild-type and mutants. In resting cells, the level of phosphorylated TBK-1 (Ser172) was below detection in all four cell lines. HSV-1 infection increased the phosphorylation of TBK-1 to similar levels in control cells and cells expressing RIG-I-WT or RIG-I-DD (Figure 6D). Remarkably, HSV-1 infection induced TBK-1 phosphorylation to much more pronounced levels in cells expressing RIG-I-QQ than the other three cell lines. Similar results were observed for phosphorylated IRF3. Consistent with this, HSV-1 infection also more significantly up-regulated IFN-β and ISG56 expression in RIG-I-QQ cells than control cells and cells expressing RIG-I-WT or RIG-I-DD (Figure 6E). The low levels of IFN-β and ISG56 induction in the other three cell lines are likely due to activation of innate sensors other than RIG-I. Increased IFN-β and RANTES expression were detected only in the supernatant of HSV-1-infected 293T cells expressing RIG-I-QQ, but not the other three cell lines (Figure 6F). To determine the antiviral activities of RIG-I wild-type and these mutants, we examined viral replication in HEK293 stable cells. As shown in Figure 6G, RIG-I-WT reduced HSV-1 replication by ~50%, while RIG-I-DD had a marginal effect on HSV-1 replication. Consistent with the robust antiviral response induced by RIG-I-QQ, RIG-I-QQ reduced HSV-1 titer by ~75–90% in HEK293 cells. These results show that the deamidation-resistant RIG-I-QQ restores RIG-I antiviral activity against HSV-1 and efficiently restricts HSV-1 replication.
The Carboxyl Terminal of UL37 Contains a Deamidase Domain
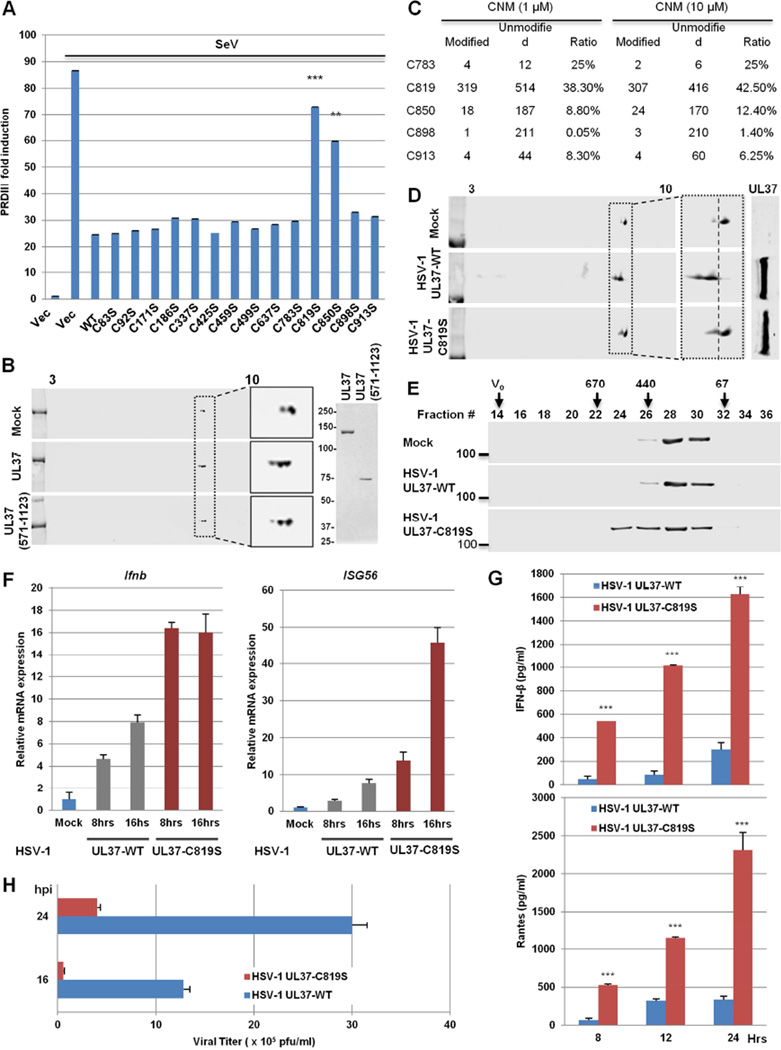
UL37 purified from E.coli is sufficient to deamidate RIG-I, implying that UL37 is a bona fide protein deamidase. Because all known protein deamidases (e.g., PFAS) are cysteine hydrolases (Zhao et al., 2016), we suspect that UL37 also contains a catalytic cysteine residue. Thus, we mutated all 14 cysteines of UL37 individually to serines and screened for the loss of inhibition of RIG-I-mediated activation of the PRDIII promoter upon SeV infection. The C819S and C850S mutants were identified to have greatly impaired blockade of PRDIII induction by SeV (Figure 7A and Figure S6A). Analysis by 2-DGE also showed that the C819S and C850S mutants of UL37 failed to induce RIG-I deamidation in transfected cells (Figure S6B), indicating that these cysteines are required for the deamidase activity of UL37. Previous crystallography analysis showed that the N-terminus of UL37 adopts a helical bundle structure similar to multisubunit tethering complexes involved in intracellular trafficking (Pitts et al., 2014). Coupled with the observation that C819 and C850 are required for UL37 to deamidate RIG-I, we reasoned that the C-terminal half (571–1123, designated UL37C) contains a protein deamidase domain. We first determined whether UL37C was sufficient to block RIG-I-dependent IFN induction. Indeed, UL37C expression inhibited the SeV-induced transcription of PRDIII (Figure S6C). We then expressed and purified UL37C from E.coli to homogeneity for RIG-I deamidation studies. Consistent with results from transfected cells, UL37C was sufficient to deamidate RIG-I in vitro, demonstrating that UL37C contains intrinsic protein deamidase activity (Figure 7B).
Figure 7. A cysteine is required for UL37 deamidase activity.
(A) 293T cells were transfected with plasmids containing UL37 wild-type or mutants, along with the PRDIII-luciferase reporter. Cells were infected with Sendai virus (SeV, 100 HAU/mL) at 30 h later for 15 hours. Activation of the PRDIII promoter was determined by luciferase reporter assay.
(B) UL37 and UL37 (571-1123) were purified from E.coli and analyzed by silver staining (right panel). In vitro RIG-I deamidation reaction was analyzed by two-dimensional gel electrophoresis and immunoblotting.
(C) UL37C (571-1123) was reacted with CNM (1 and 10 µM) for 45 minutes and analyzed by tandem mass spectrometry. The ratio of peptide containing the indicated cysteines is shown as the percentage of the CNM-modified peptide to total peptide. Data represents one of two independent experiments.
(D and E) HEK293/Flag-RIG-I cells were infected with recombinant HSV-1 UL37-WT or HSV-1 UL37-C819S (MOI=5) for 4 h. WCLs were analyzed by two-dimensional gel electrophoresis and immunoblotting (D). RIG-I was purified and analyzed by gel filtration and immunoblotting (E). Numbers indicate the size of RIG-I in kDa and V0 denotes void volume.
(F and G) THP-1 cells were harvested at 8 h after HSV-1 infection (MOI=2) and total RNA was analyzed by real-time PCR (F). Supernatant was collected at 16 hpi to quantify cytokines by ELISA (G).
(H) HFF cells were infected with HSV-1 UL37-WT and HSV-1 UL37-C819S at MOI of 0.1. Viral replication was determined by plaque assays.
**, p<0.01 and ***, p<0.001. For A, F, G and H, data are presented as mean ± SD.
See also Figure S6 and S7.
To pinpoint the cysteine residue of the active site, we employed a small molecule electrophile for mass spectrometry analysis, an approach that was successfully used to quantitatively profile functional cysteines in proteomes (Weerapana et al., 2010). The rationale is that functional cysteines, such as those in enzymatic active sites, are hyper-reactive and react with small molecule electrophiles independent of concentration. As such, a ratio of the percentage of labeled peptides at high concentration to that at low concentration near 1 predicts functional cysteines. After reacting with 2-Chloro-N-(hydroxymethyl)acetamide (CNM), mass spectrometry analysis identified that C819 was primarily labeled by CNM within UL37C. Specifically, 38.3% and 42.5% of C819 were labeled by CNM at 1 and 10 µM, respectively (Figure 7C). C850 was labeled at minimal level (<10%) by CNM, suggesting that C850 is largely inaccessible. Taken together, these results support the conclusion that C819 is the active site of the catalytic triad.
To probe the roles of UL37-mediated deamidation in viral infection, we introduced UL37 wild-type (UL37-WT) and UL37-C819S into the HSV-1 genome (designated HSV-1 UL37-WT and HSV-1 UL37-C819S) and examined RIG-I-mediated innate immune signaling. Gel electrophoresis of viral genomic DNA after BamHI digestion revealed identical pattern of migration, indicative of lack of large chromosome rearrangement (Figure S6D). Immunoblotting analysis showed that UL37-WT and UL37-C819S were expressed at similar levels in 293T cells (Figure S6E). Compared to HSV-1 UL37-WT, HSV-1 UL37-C819S failed to deamidate RIG-I by 2-DGE analysis (Figure 7D). Infection of HSV-1 UL37-C819S, but not HSV-1 UL37-WT, induced RIG-I oligomerization corresponding to protein sizes of ~440–670 kDa analyzed by gel filtration (Figure 7E). Moreover, HSV-1 UL37-C819S induced higher levels of IFN-β and ISG56 expression (Figure 7F), and IFN-β and RANTES secretion (Figure 7G) in THP-1 macrophages. Similar results were obtained in HSV-1-infected HeLa, HFF and 293T cells (Figure S7A–S7C). These results show that the deamidase activity of UL37 is critical for HSV-1 to evade RIG-I-mediated immune response.
We then analyzed HSV-1 lytic replication and found that HSV-1 UL37-C819S produced ~10% of virion progeny of HSV-1 UL37-WT in HFF (Figure 7H). In HeLa cells, HSV-1 UL37-C819S was more impaired at 36 than at 24 hpi compared to HSV-1 UL37-WT with the MOI of 1, whereas the impaired replication phenotype of HSV-1 UL37-C819S was more pronounced at 12 than 24 hpi with the MOI of 0.1 (Figure S7D). To determine whether the reduced replication of HSV-1 UL37-C819S is due to the elevated IFN response, we characterized HSV-1 replication in Vero cells that are deficient in IFN induction. Compared to HSV-1 UL37-WT, HSV-1 UL37-C819S showed identical viral replication at 12 and 24 hpi (Figure S7E). However, HSV-1 UL37-C819S produced ~50% and 35% as many virion progeny as HSV-1 UL37-WT at 36 and 48 hpi, respectively. To further corroborate the roles of RIG-I in inhibiting HSV-1 replication, we knocked down RIG-I and examined HSV-1 replication. As shown in Figure S7F, RIG-I depletion restored the lytic replication HSV-1 UL37-C819S to levels of HSV-1 UL37-WT, at 12 and 24 hpi. However, RIG-I knockdown had no effect on the difference in lytic replication between HSV-1 UL37-WT and HSV-1 UL37-C819S, at 36 and 48 hpi. These results show that RIG-I-mediated antiviral activity suppresses HSV-1 lytic replication during early infection and that UL37 deamidase activity is important to antagonize RIG-I-mediated antiviral defense. Furthermore, UL37 deamidase activity is important for late stages of HSV-1 lytic replication.
DISCUSSION
We previously reported that vGAT pseudo-enzymes of human KSHV and murine γHV68 recruited cellular PFAS to deamidate RIG-I and evade antiviral cytokine production (He et al., 2015). Interestingly, HSV-1 infection also induced RIG-I deamidation, despite the fact that genomes of alpha herpesviruses do not contain sequence homologues of vGAT proteins. Herein, we identified UL37 as a viral deamidase that targets RIG-I for deamidation and inactivation, thereby preventing RIG-I from sensing viral dsRNA. Previously reported protein deamidases contain either a cysteine-protease fold or a GAT domain (Cui et al., 2010; He et al., 2015; Sanada et al., 2012; Wang et al., 2009). UL37-mediated deamidation of RIG-I disarms downstream innate immune signaling, suggesting the critical, and likely more ubiquitous, roles of protein deamidation in signal transduction. UL37 is a large tegument protein that is implicated in viral trafficking, egress and innate immune regulation (Desai et al., 2001; Liu et al., 2008; Pitts et al., 2014). Taken together, UL37 inhibits the IRF-IFN branch of innate immune signaling through deamidation of RIG-I, while activating the NF-κB cascade, sharing functions similar to the gamma herpesvirus vGAT proteins.
Our biochemical analyses show that UL37 is intrinsically a protein deamidase. UL37 and its carboxyl terminal fragment (571–1123) purified from E.coli were sufficient to deamidate RIG-I in vitro. Mutational analysis and electrophile reaction profiling of hyper-reactive cysteines identified C819 as the single residue critical for the deamidase activity, implying that C819 is the active cysteine of the catalytic triad of UL37. Interestingly, C850 is more conserved in alpha herpesviruses than C819 (data not shown). The fact that C850 is largely inaccessible suggests that it may be required for the structural integrity of the deamidase domain. It is unclear whether other UL37 homologs are deamidases. Future structural studies of the UL37 deamidase domain may define a new fold catalyzing protein deamidation and “visualize” the catalytic cysteine.
Although previous studies implicated RIG-I in sensing dsRNA produced by herpesviruses (da Silva and Jones, 2013; Jacquemont and Roizman, 1975; Rasmussen et al., 2009; Weber et al., 2006), our work provides further credence concerning the RIG-I-mediated immune defense against a model DNA virus and viral immune evasion thereof. HSV-1 infection prevents RIG-I activation and innate immune responses triggered by subsequent SeV infection. These phenotypes were recapitulated by UL37 expression, pointing to the key roles of UL37 in evading RIG-I activation by viral dsRNA. The deamidated RIG-I-DD (D495 and D549) mutant, failed to sense 5’ppp-RNA and SeV, which correlated with its inability to initiate host immune signaling and control VSV replication. Comparing HSV-1 replication kinetics in IFN-competent 293T and HeLa cells to that in IFN-deficient Vero cells, we found that the deamidase activity of UL37 is critical in negating RIG-I-mediated inhibition of the early steps of HSV-1 lytic replication. The mutation abolishing UL37 deamidase activity, notably, also impaired HSV-1 replication during late stages of replication in an RIG-I- independent manner, implying the existence of other viral and cellular targets in addition to RIG-I. Nevertheless, uncoupling RIG-I deamidation from UL37, via either introducing the deamidation-resistant RIG-I-QQ into cells or engineering the C819S mutation of UL37 into the HSV-1 genome, restored RIG-I activation and downstream innate immune signaling, thereby reducing HSV-1 productive infection. These results unambiguously demonstrate the antiviral activity of RIG-I against a DNA herpesvirus and elucidate a mechanism of viral immune evasion.
N495 and N549 reside in two α-helices that constitute the RNA-binding interface of the Hel2i domain. Interestingly, N549 forms hydrogen bonds with the backbone of T504 that ends the N495-containing α23 helix, providing a physical link between these two neighboring helices that are located immediately proximal to the RIG-I-bound dsRNA. These observations suggest that the two α-helices constitute a region responsible for regulating RNA-binding/sensing by RIG-I. The susceptibility of the hydrogen bonds between N549 and T504 to the deamidase activity of UL37 underpins the inactivation of RIG-I by HSV-1 infection. Remarkably, the N549Q mutation appears to conserve hydrogen bonds, and confers resistance to UL37-mediated deamidation, demonstrating the exquisite specificity of UL37-mediated deamidation. Deamidation of N495 and N549 within the Hel2i domain, unexpectedly, abolishes 5’ppp-RNA-binding and ATP hydrolysis of RIG-I, uncovering a simple but powerful mechanism to switch off RIG-I. Although the CTD of RIG-I is responsible for sensing viral dsRNA, emerging studies support the regulatory role of helicase domains in RNA-sensing by RIG-I. It was previously reported that Hel2i “measures” the length of dsRNA stem during RNA-binding by RIG-I (Kohlway et al., 2013). Structural analysis also highlighted the direct contact between Hel2i and dsRNA (Kowalinski et al., 2011; Luo et al., 2011). Moreover, mutations within a helicase domain reduced the ATPase activity of RIG-I, increased its association with cellular dsRNA and activated downstream signaling (Lassig et al., 2015). Together with these observations, our work further lends credence to the pivotal roles of Hel2i of RIG-I and site-specific deamidation thereof in interacting with and sensing viral dsRNA, suggesting more ubiquitous roles of protein deamidation in fundamental biological processes.
EXPERIMENTAL PROCEDURES
Two-dimensional Gel Electrophoresis
Cells (1×106) were lysed in 150 µl rehydration buffer (8 M Urea, 2% CHAPS, 0.5% IPG Buffer, 0.002% bromophenol blue) by three pulses of sonication and whole cell lysates were centrifuged at 20,000 g for 15 min. Supernatants were loaded to IEF strips for focusing with a program comprising: 20 V, 10 h (rehydration); 100 V, 1 h; 500 V, 1 h; 1000 V, 1 h; 2000 V, 1 h; 4000 V, 1 h; 8000 V, 4h. After IEF, strips were incubated with SDS equilibration buffer (50 mM Tris-HCl [pH8.8], 6 M urea, 30% glycerol, 2% SDS, 0.001% Bromophenol Blue) containing 10 mg/ml DTT for 15 min and then SDS equilibration buffer containing 2-iodoacetamide for 15 min. Strips were washed with SDS-PAGE buffer, resolved by SDS-PAGE, and analyzed by immunoblotting.
In vitro Deamidation Assay
GST-RIG-I was purified from transfected 293T cells to homogeneity as determined by silver staining. In vitro on-column deamidation of RIG-I was performed as previously reported (He et al., 2015). Briefly, ~0.2 µg of His-tagged UL37/UL37(571–1123) expressed and purified from E.coli, and 0.6 µg of GST-RIG-I (bound to glutathione-conjugated agarose) were added to a total volume of 30 µl. The reaction was carried out at 30°C for 45 min in deamidation buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM MgCl2). Protein-bound GST beads were washed with deamidation buffer and GST-RIG-I was eluted with rehydration buffer (6 M Urea, 2 M Thio-urea, 2% CHAPS, 0.5% IPG Buffer, 0.002% bromophenol blue) at room temperature. Samples were then analyzed by two-dimensional gel electrophoresis and immunoblotting.
Constructing Recombinant HSV-1
Recombinant HSV-1 was engineered as previously described (Dong et al., 2010). Briefly, DNA fragments containing UL37 WT and C819S were amplified using overlapping primers. First round PCR products of ~500 bp fragment upstream of UL37, UL37 open reading frames (WT and C819S) and ~500 bp fragments downstream of UL37 were used as the template for second round PCR amplification. Purified PCR products of the second round, along with HSV-1 ΔUL37 (KOS) Bacmid, were transfected into 293T cells to generate recombinant HSV-1. The revertant (containing wild-type UL37, designated wild-type) and UL37-C819S mutant were plaque purified and validated by restriction digestion of viral genomic DNA and sequencing of the UL37 open reading frame.
RNA Electrophoresis Mobility Shift Assay (EMSA)
RNA EMSA was performed as previously described (Takahasi et al., 2008). 5’-ppp-dsRNA (5’-triphosphate dsRNA) and control dsRNA were purchased from Invivogen and bottom strands were labeled with γ-[P32] ATP by T4 polynucleotide kinase (NEB). Purified RIG-I and RIG-I mutants were incubated with dsRNA at room temperature for 15 min. Binding buffer contains 20 mM Tris-HCl (pH=8.0), 1.5 mM MgCl2 and 1.5 mM DTT. Unlabeled ppp-dsRNA was used as competitor at 100-fold in excess. The reaction mixtures were run on 5% native polyacrylamide gels at a constant voltage of 200 V. Gels were dried and subjected to phosphorimaging.
| Labeled 5’-ppp- | Top Strand | 5’-ppp-GCAUGCGACCUCUGUUUGA-3’ |
| dsRNA | Bottom Strand | 3’-CGUACGCUGGAGACAAACU-5’-32P |
| Labeled 5’ control | Top Strand | 5’-GCAUGCGACCUCUGUUUGA-3’ |
| dsRNA | Bottom Strand | 3’-CGUACGCUGGAGACAAACU-5’-32P |
In Vitro ATPase Activity Assay
Purified RIG-I or RIG-I mutants were incubated with 5’-ppp-dsRNA (Invivogen) at 37°C for 20 min in ATPase reaction buffer (50 mM Tr is-HCl, pH 7.5, 2.5 mM MgCl2, and ATP). Released phosphates were measured using a PiColorLockTM phosphate detection reagent (Innova Biosciences). For reactions with varying concentrations of ATP, the concentrations of RIG-I proteins and RNA were 20 nM and 80 nM, respectively. For reactions with varying concentrations of the RNA, the concentrations of RIG-I proteins and ATP were 20 nM and 500 µM, respectively.
Additional experimental procedures, including reagents, lentivirus, viral infection, luciferase reporter assay, DNA and RNA transfection, confocal immunofluorescence microscropy, protein expression and purification, immunoprecipitation and immunoblotting, knockdown, gel filtration and real-time PCR, can be found in Supplemental Information.
Supplementary Material
Acknowledgments
We thank Dr. Steve Gygi and Ross Tamarin for mass spectrometry analysis, Thomas Mettenleiter, Katya Heldwein and Greg Smith for HSV-1 and PRV reagents. We thank Ms. Stacy Lee for editing this manuscript. This work is supported by grants from NIDCR (DE021445, DE026003), Joint Research Fund for Oversea Chinese, Hong Kong and Macao Scholars (81528011) to P. Feng, core services performed through Norris Cancer Center grant P30CA014089-34 and the National Key Research and Development Program of China (2016YFD0500300 and 2016YFC1200200) to Z. Xia. C.Y is supported by a fellowship from Gilead Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes seven figures and Extended Experimental Procedures.
CONTRIBUTIONS
J.Z., Y.Z., G.S., S.X. and P.F. performed experiments. J.C., C.Y., D.K., W.Y., J.P., W.X., C.Z. and Z.X. contributed reagents. J.Z. and P.F. designed experiments, analyzed data and wrote the paper.
REFERENCES
- Anchisi S, Guerra J, Garcin D. RIG-I ATPase activity and discrimination of self-RNA versus non-self-RNA. MBio. 2015;6:e02349. doi: 10.1128/mBio.02349-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt D, Ghosh S. Regulation of the NF-kappaB-Mediated Transcription of Inflammatory Genes. Front Immunol. 2014;5:71. doi: 10.3389/fimmu.2014.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidinosti M, Ran I, Sanchez-Carbente MR, Martineau Y, Gingras AC, Gkogkas C, Raught B, Bramham CR, Sossin WS, Costa-Mattioli M, et al. Postnatal Deamidation of 4E–BP2 in Brain Enhances Its Association with Raptor and Alters Kinetics of Excitatory Synaptic Transmission. Molecular cell. 2010;37:797–808. doi: 10.1016/j.molcel.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YK, Gack MU. RIG-I-like receptor regulation in virus infection and immunity. Curr Opin Virol. 2015;12:7–14. doi: 10.1016/j.coviro.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Parent L, Maniatis T. Site-specific phosphorylation of kappaBalpha by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- Cui J, Yao Q, Li S, Ding X, Lu Q, Mao H, Liu L, Zheng N, Chen S, Shao F. Glutamine deamidation and dysfunction of ubiquitin/NEDD8 induced by a bacterial effector family. Science. 2010;329:1215–1218. doi: 10.1126/science.1193844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva LF, Jones C. Small non-coding RNAs encoded within the herpes simplex virus type 1 latency associated transcript (LAT) cooperate with the retinoic acid inducible gene I (RIG-I) to induce beta-interferon promoter activity and promote cell survival. Virus research. 2013;175:101–109. doi: 10.1016/j.virusres.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai P, Sexton GL, McCaffery JM, Person S. A null mutation in the gene encoding the herpes simplex virus type 1 UL37 polypeptide abrogates virus maturation. J Virol. 2001;75:10259–10271. doi: 10.1128/JVI.75.21.10259-10271.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deverman BE, Cook BL, Manson SR, Niederhoff RA, Langer EM, Rosova I, Kulans LA, Fu X, Weinberg JS, Heinecke JW, et al. Bcl-xL deamidation is a critical switch in the regulation of the response to DNA damage. Cell. 2002;111:51–62. doi: 10.1016/s0092-8674(02)00972-8. [DOI] [PubMed] [Google Scholar]
- Dho SH, Deverman BE, Lapid C, Manson SR, Gan L, Riehm JJ, Aurora R, Kwon KS, Weintraub SJ. Control of cellular Bcl-xL levels by deamidation-regulated degradation. PLoS Biol. 2013;11:e1001588. doi: 10.1371/journal.pbio.1001588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Feng H, Sun Q, Li H, Wu TT, Sun R, Tibbetts SA, Chen ZJ, Feng P. Murine gamma-herpesvirus 68 hijacks MAVS and IKKbeta to initiate lytic replication. PLoS pathogens. 2010;6:e1001001. doi: 10.1371/journal.ppat.1001001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Feng P. Murine gamma herpesvirus 68 hijacks MAVS and IKKbeta to abrogate NFkappaB activation and antiviral cytokine production. PLoS pathogens. 2011;7:e1002336. doi: 10.1371/journal.ppat.1002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, He Z, Durakoglugil D, Arneson L, Shen Y, Feng P. Murine gammaherpesvirus 68 evades host cytokine production via replication transactivator-induced RelA degradation. Journal of virology. 2012;86:1930–1941. doi: 10.1128/JVI.06127-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng P, Moses A, Fruh K. Evasion of adaptive and innate immune response mechanisms by gamma-herpesviruses. Curr Opin Virol. 2013;3:285–295. doi: 10.1016/j.coviro.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nature immunology. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- Flatau G, Lemichez E, Gauthier M, Chardin P, Paris S, Fiorentini C, Boquet P. Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature. 1997;387:729–733. doi: 10.1038/42743. [DOI] [PubMed] [Google Scholar]
- Full F, Jungnickl D, Reuter N, Bogner E, Brulois K, Scholz B, Sturzl M, Myoung J, Jung JU, Stamminger T, et al. Kaposi’s sarcoma associated herpesvirus tegument protein ORF75 is essential for viral lytic replication and plays a critical role in the antagonization of ND10-instituted intrinsic immunity. PLoS pathogens. 2014;10:e1003863. doi: 10.1371/journal.ppat.1003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar M, Gill MB, Losing JB, May JS, Stevenson PG. Multiple functions for ORF75c in murid herpesvirus-4 infection. PLoS One. 2008;3:e2781. doi: 10.1371/journal.pone.0002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Zhao J, Song S, He X, Minassian A, Zhou Y, Zhang J, Brulois K, Wang Y, Cabo J, et al. Viral pseudo-enzymes activate RIG-I via deamidation to evade cytokine production. Mol Cell. 2015;58:134–146. doi: 10.1016/j.molcel.2015.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont B, Roizman B. Rna-Synthesis in Cells Infected with Herpes-Simplex Virus .10. Properties of Viral Symmetric Transcripts and of Double-Stranded-Rna Prepared from Them. Journal of virology. 1975;15:707–713. doi: 10.1128/jvi.15.4.707-713.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nature immunology. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlway A, Luo D, Rawling DC, Ding SC, Pyle AM. Defining the functional determinants for RNA surveillance by RIG-I. EMBO Rep. 2013;14:772–779. doi: 10.1038/embor.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolakofsky D, Garcin D. gammaHV68 vGAT: a viral pseudoenzyme pimping for PAMPs. Mol Cell. 2015;58:3–4. doi: 10.1016/j.molcel.2015.03.021. [DOI] [PubMed] [Google Scholar]
- Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- Lassig C, Matheisl S, Sparrer KM, de Oliveira Mann CC, Moldt M, Patel JR, Goldeck M, Hartmann G, Garcia-Sastre A, Hornung V, et al. ATP hydrolysis by the viral RNA sensor RIG-I prevents unintentional recognition of self-RNA. Elife. 2015:4. doi: 10.7554/eLife.10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Fitzgerald K, Kurt-Jones E, Finberg R, Knipe DM. Herpesvirus tegument protein activates NF-kappaB signaling through the TRAF6 adaptor protein. Proc Natl Acad Sci U S A. 2008;105:11335–11339. doi: 10.1073/pnas.0801617105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Kohlway A, Pyle AM. Duplex RNA activated ATPases (DRAs): platforms for RNA sensing, signaling and processing. RNA Biol. 2013;10:111–120. doi: 10.4161/rna.22706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Mycek MJ, Waelsch H. The enzymatic deamidation of proteins. J Biol Chem. 1960;235:3513–3517. [PubMed] [Google Scholar]
- Pitts JD, Klabis J, Richards AL, Smith GA, Heldwein EE. Crystal structure of the herpesvirus inner tegument protein UL37 supports its essential role in control of viral trafficking. J Virol. 2014;88:5462–5473. doi: 10.1128/JVI.00163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SB, Jensen SB, Nielsen C, Quartin E, Kato H, Chen ZJ, Silverman RH, Akira S, Paludan SR. Herpes simplex virus infection is sensed by both Toll-like receptors and retinoic acid-inducible gene-like receptors, which synergize to induce type I interferon production. J Gen Virol. 2009;90:74–78. doi: 10.1099/vir.0.005389-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NE, Robinson AB. Molecular clocks. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:944–949. doi: 10.1073/pnas.98.3.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada T, Kim M, Mimuro H, Suzuki M, Ogawa M, Oyama A, Ashida H, Kobayashi T, Koyama T, Nagai S, et al. The Shigella flexneri effector OspI deamidates UBC13 to dampen the inflammatory response. Nature. 2012;483:623–U149. doi: 10.1038/nature10894. [DOI] [PubMed] [Google Scholar]
- Schmidt G, Sehr P, Wilm M, Selzer J, Mann M, Aktories K. Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature. 1997;387:725–729. doi: 10.1038/42735. [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, Gale M, Jr, Inagaki F, Fujita T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Molecular cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Ting JP, Duncan JA, Lei Y. How the noninflammasome NLRs function in the innate immune system. Science. 2010;327:286–290. doi: 10.1126/science.1184004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nature immunology. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Piatkov KI, Brower CS, Varshavsky A. Glutamine-specific N-terminal amidase, a component of the N-end rule pathway. Mol Cell. 2009;34:686–695. doi: 10.1016/j.molcel.2009.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. Journal of virology. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub SJ, Deverman BE. Chronoregulation by asparagine deamidation. Science’s STKE : signal transduction knowledge environment. 2007;2007:re7. doi: 10.1126/stke.4092007re7. [DOI] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Molecular cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- Zhao J, Li J, Xu S, Feng P. Emerging roles of regulated protein deamidation in innate immune signaling. J Virol. 2016 doi: 10.1128/JVI.01980-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.