SUMMARY
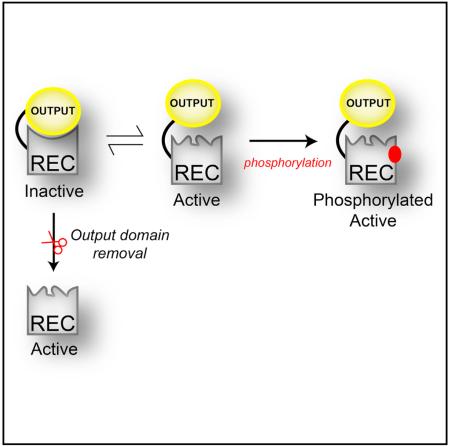
Information transmission in biological signaling networks is commonly considered to be a unidirectional flow of information between protein partners. According to this view, many bacterial response regulator proteins utilize input receiver (REC) domains to “switch” functional outputs, using REC phosphorylation to shift pre-existing equilibria between inactive and active conformations. However, recent data indicate that output domains themselves also shift such equilibria, implying a “mutual inhibition” model. Here we use solution nuclear magnetic resonance to provide a mechanistic basis for such control in a PhyR-type response regulator. Our structure of the isolated, non-phosphorylated REC domain surprisingly reveals a fully active conformation, letting us identify structural and dynamic changes imparted by the output domain to inactivate the full-length protein. Additional data reveal transient structural changes within the full-length protein, facilitating activation. Our data provide a basis for understanding the changes that REC and output domains undergo to set a default “inactive” state.
Graphical abstract
In Brief Many protein switches use mutual inhibition between sensory and output domains to control spurious activation. Correa and Gardner elucidated the molecular mechanisms utilized for such control in a bacterial response regulator, identifying structural and dynamic changes important for such regulation.
INTRODUCTION
Intracellular signal transduction often relies on cascades of interactions among multidomain proteins that operate as molecular switches, coupling changes in environmental stimuli with adaptive responses. Many principles of such cascades are incorporated, and arguably best understood, in two-component systems (TCS), a prototypical signaling mechanism found chiefly in bacteria (Stock et al., 2000). Most simply composed of a sensory histidine kinase (HK) and a response regulator (RR), TCS signal transduction is achieved via a phosphotransfer reaction from a phosphohistidine residue in the HK to an aspartyl residue in the receiver (REC) domain of the RR. Upon this phosphorylation, the RR undergoes a conformational change that regulates a catalytic or signaling activity, making the REC domain a molecular switch that controls the output response (Stock et al., 2000; Gao and Stock, 2009). Similar to other natural protein-based switches (Nussinov et al., 2014), several RECs exhibit a pre-existent equilibrium between major inactive and minor active conformations in the absence of phosphorylation, with formation of the phosphor-Asp adduct shifting the balance between the two (Volkman et al., 2001; Stock and Guhaniyogi, 2006; Gardino et al., 2009).
However, while most RRs are multidomain proteins with both REC and output domains (Galperin, 2006), many of the data supporting this signaling model have come from experiments utilizing simpler natural REC-only proteins (Gardino et al., 2003; McDonald et al., 2012) or artificial truncations of more complex ones (Volkman et al., 2001; Gardino et al., 2009). While informative, such studies have yet to address a critical point: how the output domains themselves influence the activation properties of the REC domains that are tethered nearby. Biochemical data suggest that such an effect is present, as small molecule phosphodonors can phosphorylate full-length RRs more slowly than their truncated REC counterparts, despite the fact that output domains do not directly block phosphorylation sites (Barbieri et al., 2010). These findings suggest a “mutual inhibition” model where both sensor and effector domains influence each other's activities in the absence of triggering stimuli. Left unanswered are multiple mechanistic questions about this process, not only in this important group of bacterial proteins but also more generally in the very large number of protein switches composed of regulatory and effector domains: How does an effector domain modulate the properties of its own regulatory components? What allosteric events are propagated between and within domains to achieve this control? What conformational fluctuations between domains allow the protein to be activated? How are internal motions redistributed when proteins interconvert between states?
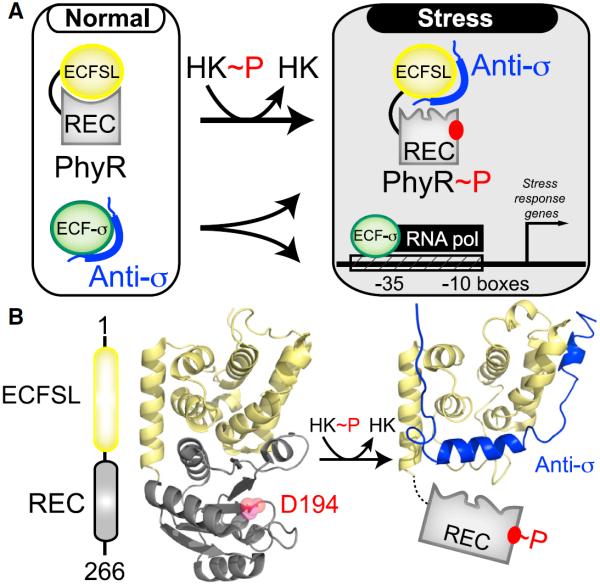
To address these questions, we combined solution nuclear magnetic resonance (NMR) and biochemical approaches to compare structural, dynamic, and functional aspects of an REC domain both in isolation and within in its native full-length protein context with a tightly bound output domain. For this work, we chose the Erythrobacter litoralis PhyR (EL_PhyR) RR protein (Francez-Charlot et al., 2009; Herrou et al., 2010; Correa et al., 2013), which contains an N-terminal output domain (called here ECFSL, extracytoplasmic function sigma like), followed by a C-terminal REC domain (Figure 1A). PhyR plays an important role in the alphaproteobacterial general stress response through a “partner-switching mechanism” (Francez-Charlot et al., 2009) that regulates its affinity for an anti-σ factor transcriptional inhibitor. Under normal growth conditions, PhyR adopts an inactive unphosphorylated state, leaving an anti-σ factor free to interact with its cognate σ factor (ECF-σ, extracytoplasmic function sigma factor). With stress-induced phosphorylation, PhyR undergoes a conformational change that allows it to bind the anti-σ factor, increasing free ECF-σ levels and promoting the transcription of general stress-response genes (Francez-Charlot et al., 2009) (Figure 1A). Biochemical and genetic studies with parallel light-sensitive systems in E. litoralis (Correa et al., 2013), Brucella abortus (Kim et al., 2014), and Caulobacter crescentus (Foreman et al., 2012) support this regulatory model and indicate that PhyR homologs play similar roles in these organisms. Complementary structural work on unphosphorylated C. crescentus PhyR demonstrated that the REC and ECFSL domains directly interact, burying about 1,000 Å2 of surface area (Figure 1B) (Herrou et al., 2010). Notably, the ECFSL domain uses the same surface to bind the REC domain and its cognate anti-σ factor (Campagne et al., 2012; Herrou et al., 2012), necessitating that phosphorylation-induced changes in the REC domain somehow lead to its undocking from the output domain for anti-σ binding to occur.
Figure 1. PhyR Regulates General Stress Response in Bacteria by the Regulated “Partner Switching” of an Anti-σ Factor.
(A) Overview of PhyR-based partner switching. Under standard conditions, PhyR adopts a closed conformation with the ECFSL and REC domains, burying surfaces of both domains. Upon phosphorylation by an HK, PhyR changes conformation to expose a binding surface for an anti-σ factor, titrating it away from ECF sigma factors (ECF-σ) and allowing it to enhance the transcription of target genes.
(B) An EL_PhyR domain architecture and homology model, showing how the ECFSL and REC domains bind each other in the unphosphorylated state while not directly occluding the D194 site of phosphorylation. In comparison, a structural model of ECFSL bound to anti-σ suggests that phospho-induced conformational changes will undock the REC and output domains, exposing regions required for anti-σ factor binding (Campagne et al., 2012; Herrou et al., 2012). See also Figure S1.
Here, we demonstrate that EL_PhyR uses mutual inhibition between its regulatory and output domains, with the ECFSL (output) domain stabilizing the inactive conformation of the REC (regulatory) domain just as the REC domain inhibits the anti-σ binding activity of the output. We support this conclusion with several lines of evidence, including solution NMR data comparing the structure and dynamics of the PhyR REC domain in full-length and REC-only contexts. These data show that truncation leads to marked changes in the structure and dynamics of the REC domain, not just at the α4-β5-α5 interface used to bind the ECFSL domain but also at distant regions including the α3 helix, over 20Å away. Additional experiments with an engineered PhyR containing an extended linker between REC and output domains showed that this subtler way of destabilizing interdomain interactions also favors the active conformation, as demonstrated by increased affinities for the phosphomimic beryllium fluoride (BeF3−) (Yan et al., 1999). We conclude that, by providing paramagnetic relaxation enhancement (PRE) measurements, part of the REC domain may transiently disrupt its interactions with the output domain in the full-length context, allowing the REC domain to prime the active site for subsequent phosphorylation. Here, we demonstrate a range of allosteric changes required for activation and phosphorylation in multidomain protein switches from structural and dynamics perspectives.
RESULTS
REC Conformation Is Affected by Intramolecular Interactions with an Output Domain
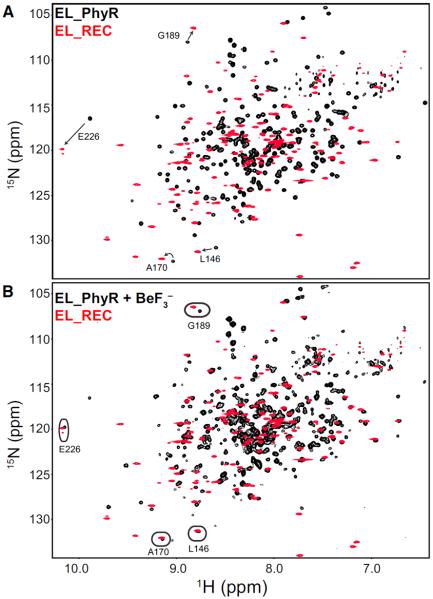
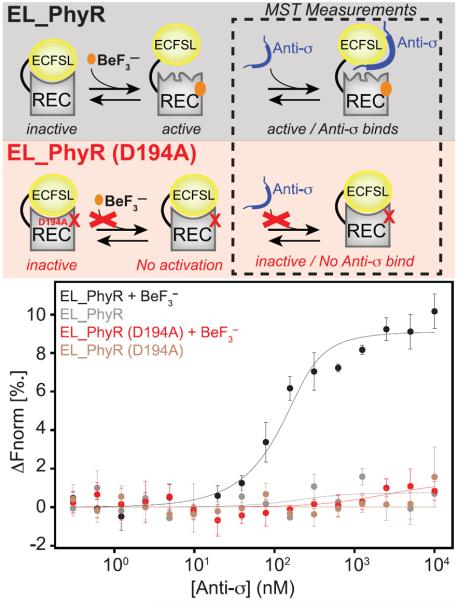
Relaxation dispersion NMR studies have previously demonstrated that several isolated REC domains undergo millisecond-timescale dynamic equilibria between abundant inactive and rare active conformations (Volkman et al., 2001; Stock and Guhaniyogi, 2006; Gardino et al., 2009). To complement these studies by examining how an output domain affects the structure and dynamics in a multicomponent RR, we performed a series of solution NMR studies with EL_PhyR, a member of the PhyR family of RR proteins (Gourion et al., 2008). This RR is ideal for such studies as it (1) is monomeric and does not change oligomerization state upon phosphorylation (Francez-Charlot et al., 2009; Correa et al., 2013), (2) has a closely related homolog (CC_PhyR) (58.4% identity, Figure S1) with a high-resolution crystal structure (Figure 1B) (Herrou et al., 2010), and (3) has a well-characterized biochemical signaling pathway (Correa et al., 2013). The CC_PhyR crystal structure demonstrates a large buried surface area between the REC and ECFSL domains (approximately 1,000 Å2), suggesting that the output domain could substantially influence the REC domain conformation, as suggested for OmpR/PhoB-type RRs with smaller interacting surfaces (Barbieri et al., 2010). To directly examine the effect of interdomain interaction on the REC conformation, we compared 2D 15N-1H NMR spectra of full-length EL_PhyR and a truncated REC-only construct (EL_REC, residues 142–266; Figure 2A). The relatively poor overlap between these spectra strongly suggests that the REC domain adopts different conformations in the native full-length and isolated domain contexts. Furthermore, the EL_REC spectrum had more peaks than expected from the sequence, suggesting that the protein underwent slow exchange on the millisecond to second timescale (or slower) among two conformations in a 65%:35% ratio. Titrations with MgCl2, supplying the important Mg2+ cation needed to activate RRs (McDonald et al., 2012; Ocasio et al., 2015), shifted the equilibrium in favor of the less populated of these slow-exchanging states (Figures S2A–S2C). In comparison, 2D 15N-1H NMR spectra of full-length EL_PhyR did not display comparable peaks indicative of conformational exchange and addition of MgCl2 caused only minor chemical shift displacements. We suspected that such binding to Mg2+ might be correlated to REC domain activation triggered by output domain removal, since perturbations at the active site due to protein activation play an important role in how Mg2+ associates to the protein (Bellsolell et al., 1994; Kern et al., 1999). To evaluate this hypothesis, we obtained 15N-1H transverse relaxation-optimized spectroscopy (TROSY)-heteronuclear single-quantum correlation (HSQC) spectra of EL_PhyR in the presence of Mg2+ and phosphomimic BeF3−. Addition of both compounds markedly changed the EL_PhyR spectrum, indicating a potential stabilization of the active state of the molecule. Remarkably, under these conditions (5 mM MgCl2, 11 mM BeF3−) we observed substantial improvement in the overlap between EL_PhyR and EL_REC spectra (Figure 2B), suggesting that in the absence of the output domain, the isolated REC domain adopts an active conformation. Even though we did not obtain assignments of EL_PhyR in the presence of BeF3−, chemical shift displacements at EL_PhyR + BeF3− could still be assigned due to the frequency proximity among some chemical shifts in EL_PhyR and EL_REC spectra (Figure 2), which further confirms the structural association between EL_REC and active BeF3−-bound EL_PhyR. On the other hand, addition of BeF3− to a sample of EL_REC containing MgCl2 caused minor changes in the overall spectrum, likely due to the already active-like conformation of the truncated protein (Figure S2D). To further confirm that BeF3− binding to EL_PhyR was indeed mimicking the effect of protein phosphorylation, we monitored the binding of the cognate anti-σ factor to fluorescently labeled EL_PhyR (wild-type and D194A mutant) in the presence of 5 mM BeF3− using microscale thermophoresis (MST; Seidel et al., 2013; Scheuermann et al., 2016) (Figure 3). Our data show that EL_PhyR strongly binds to its anti-σ factor (Kd = 34 ± 17 nM) in an activation-dependent manner, comparable with homologous systems (Campagne et al., 2012; Herrou et al., 2012). Mutation of the D194 phosphoacceptor residue disrupted EL_PhyR binding to anti-σ regardless of activation (Figure 3), which we attribute to the inability of the D194A mutant to bind BeF3− (Figures S3 and S4).
Figure 2. Removing the Output Domain Affects the Overall Conformation of the EL_PhyR REC Domain.
(A) Comparison of 15N-1H spectra of full-length EL_PhyR plus 5 mM MgCl2 (black) and EL_REC plus 10 mM MgCl2 (red) shows poor overlap for the majority of cross-peaks, suggesting that the removal of the output ECFL domain affected the overall conformation of the REC domain.
(B) Comparison of 15N-1H spectra of EL_PhyR plus 11 mM BeF3−, 5 mM MgCl2 (black), and EL_REC plus 10 mM MgCl2 (red). Under these conditions, greater overlap between spectra is observed indicating that removal of output domain stabilized the active REC domain conformation. Circles highlight some examples of improved peak overlap between the isolated EL_REC and full-length EL_PhyR when the latter is loaded with BeF3−. See also Figures S2–S4 and S7.
Figure 3. BeF3− Binding to EL_PhyR Enhances its Interaction with its Cognate Anti-σ Factor.
Conformational changes induced by BeF3− binding strongly favors interaction between EL_PhyR and anti-σ factor, mimicking the effect of protein phosphorylation. Removal of the phosphoaccepting carboxylate group (D194A) disrupts BeF3− binding, locking the EL_PhyR REC domain in an inactive conformation and thus indirectly inhibiting EL_PhyR:anti-σ interactions. Microscale thermophoresis (MST) titration assays monitoring the binding of labeled EL_PhyR and EL_PhyR (D194A) to anti-σ factor in the absence and presence of 5 mM BeF3− shows a tight binding between wild-type EL_PhyR and anti-σ (Kd = 34 ± 17 nM) when a phosphomimic compound is present. Curves represent averages of titration curves measured from three to five independent replicates, with error bars at ±1 standard deviation. See also Figures S2–S4 and S7.
Solution Structure and Dynamics of the EL_PhyR REC Domain
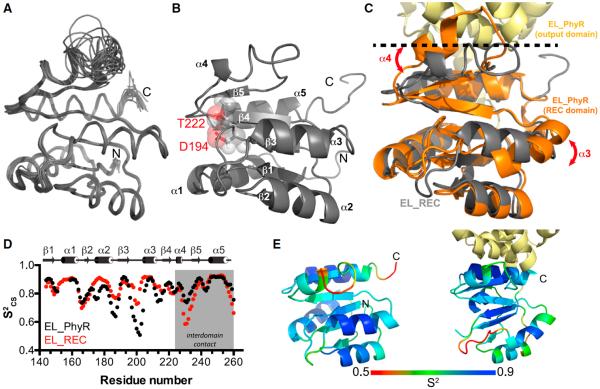
To characterize the conformational changes in the REC domain caused by the removal of the output domain, we determined the solution structure of the EL_REC protein using standard triple-resonance NMR approaches (Cavanagh et al., 2007). After measuring 2,719 distances, 195 dihedral angles and 106 hydrogen bond restraints (Table S1), we calculated an ensemble of 20 structures consistent with the experimental data (Figures 4A and 4B; Table S1). This ensemble demonstrates that the molecule adopts the typical doubly wound α/β fold of an REC domain (Gao and Stock, 2009) with a central parallel β sheet surrounded by five α helices. Analysis of our nuclear Overhauser effect spectroscopy (NOESY) data demonstrated that very similar patterns of nuclear Overhauser effects (NOEs) for the major (65%) and minor species (35%) (Figure S2E), indicating limited structural differences between conformations. Furthermore, mapping the conformationally exchanging residues onto the tertiary structure showed that they cluster in loop regions (Figure S2F), limiting their impact on the protein topology overall. Therefore, we removed restraints from all of the identified low-intensity resonance doublings from the dataset used to calculate the EL_REC structure.
Figure 4. Effects of Output Domain Removal on the Structure and Dynamics of the REC Domain.
(A) Solution structure of EL_REC showing an ensemble of the 20 lowest energy structures.
(B) Cartoon representation of EL_REC demonstrating the typical α5-β5 wobbly fold of an REC domain. Phosphoaccepting D194 is indicated in the structure.
(C) Overlap between the EL_PhyR homology model based on that of Herrou et al. (2010) and the representative NMR structure. Regions that underwent substantial changes (α3, α4, loop α4-β5) upon output domain removal are indicated.
(D) Comparison of backbone order parameter (S2CS) values between full-length (black) and REC only (red) domains. Results show that structural changes were followed by motion redistribution.
(E) S2CS value distribution on tertiary structure ranging from dynamic (red) to rigid (blue) in EL_REC(e) and EL_PhyR(f), respectivley. See also Figures S2, S5, and S6; Table S1.
We compared the EL_REC NMR structure with a full-length E. litoralis PhyR homology model based on the C. crescentus PhyR structure (Herrou et al., 2010), which we validated by comparing backbone 13Cα and 13Cβ chemical shifts from structure-based predictions (using the program SHIFTX2; Han et al., 2011) with our almost completely experimentally derived values (>90% assignment coverage for Cα and Cβ backbone atoms) obtained from triple-resonance-based assignments. We observed minor differences between predicted and experimental chemical shifts, comparable with those reported for the accuracy observed with high-resolution crystal structures (Figure S5), supporting that the homology model is a reasonable representation of the EL_PhyR structure. Comparing these EL_PhyR and EL_REC structures, we noted marked differences in the orientation of the α3 helix (average backbone root-mean-square deviation [RMSD] = 3.4 Å) and α4 helix plus α4-β5 loop (RMSD = 3.8 Å), when compared with other regions of the molecule (RMSD = ~ 1.2 Å) (Figures 4C and S6). In the full-length homology model, residues in the α4 helix and the following α4-β5 loop contact two helices within the output domain (Herrou et al., 2010). Removal of the output domain allows α4 to undergo conformational changes and establish interactions with α3 through van der Waals (I204-L228) and electrostatic (S202-R227, D205, and E208 with R233) interactions.
These structural rearrangements correlated with changes in fast-timescale (ps-ns) dynamics observed throughout the entire molecule, as established by comparing backbone amide order parameters derived from chemical shifts (S2CS; Berjanskii and Wishart, 2005) of the full-length and REC-only constructs (Figure 4D). This comparison showed an intriguing redistribution of dynamics upon the removal of the output domain: while the REC surface that normally interacts with the ECFSL output domain undergoes an expected increase in dynamics (lower S2CS values in the α4-β5 loop; Figure 4E), the loops abutting the β3 strand decrease their flexibility (increased S2CS values) due to the new interactions formed between α3 and α4 helices (Figure 4E), effectively conserving entropy by changes on opposite sides of the REC domain.
Conformational Rearrangements in the Isolated REC Domain Allow the Protein to Adopt a Default Active Conformation
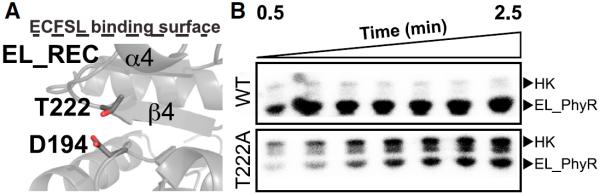
Prior structural studies on several REC domains have reported a set of common rearrangements at the active site and neighboring regions upon activation, culminating in changes to the α4-β4-α5 region that typically contacts effector domains (Gardino et al., 2003; Stock and Guhaniyogi, 2006). While not universal, these rearrangements are often accompanied by the so-called “Y-T coupling mechanism,” involving a conserved β4 strand Ser/Thr residue (T222 in EL_PhyR) that has its hydroxyl-containing side chain positioned to form a hydrogen bond with the oxygen of the active-site phosphoaccepting Asp in the inactive state. Upon activation, the Thr residue repositions itself, allowing the solvent-exposed side chain of a conserved β5 strand Phe/Tyr residue to rotate inwards (Stock and Guhaniyogi, 2006). Comparing our EL_PhyR structure with the conformations of the canonical NtrC and CheY REC domains revealed that the conserved β5 aromatic residue was replaced by a Leu in EL_PhyR (L239), which had its side chain buried and pointing toward the α4 helix in both the full-length and EL_REC structures. While there is no substantial change in the L239 location, we observed that the conserved T222 residue still moves to approach hydrogen bond distance with the D194 phosphoacceptor in the inactive EL_REC structure (Figure 5A). To evaluate the relevance of T222 for PhyR phosphorylation/activation, we examined the functional and structural impact of a T222A mutant lacking side chain hydrogen bonding capability. Functionally, in vitro phosphotransfer assays with EL_PhyR showed that this mutation impaired incorporation of phosphate by the protein compared with the wild-type (Figure 5B). In addition, BeF3− titrations monitored by 15N-1H TROSY-HSQC spectra indicated only minimal changes for the overall EL_PhyR (T222A) spectra in contrast to wild-type protein (Figure S7). Results from both the phosphotransfer assay and NMR studiers are consistent with T222 playing an important role in stabilizing the protein active state and undergoing phosphorylation (or binding a phosphomimic ligand).
Figure 5. The Isolated REC Domain Displays a Reordered Active Site.
Removal of the output domain leads the α4 helix to adopt a new position, causing reorientation of the conserved residue T222 side chain.
(A) Structure of EL_REC showing that T222 may form a hydrogen bond with the phosphoacceptor D194 mimicking events that would likely happen upon protein phosphorylation.
(B) Phosphotransfer assay showing incorporation of radiolabeled 32P by wild-type (WT) and mutant (T222A) versions of full-length EL_PhyR using HK EL368 (Correa et al., 2013) as a substrate. Reduced incorporation of phosphate by the mutant indicates that residue T222 plays an important role at the phosphotransfer reaction. The marked HK phosphorylation band due to T222A mutation at RR is a result of the reduced phosphotransfer reaction. As the T222A mutation reduces dramatically the intake of phosphate by EL_PhyR, the HK kinase activity rate overcomes the phosphotransfer rate reaction by EL_PhyR. This leads to an increase in the amount of phospho-HK, especially at the beginning of the phosphorylation assay. See also Figures S3, S4, and S7.
Increased Interdomain Linker Length Destabilizes Inactive REC Conformation
Based on our data clearly indicating that removal of the output domain destabilizes the inactive conformation of the REC domain, we hypothesized that minor perturbations to the REC/output interaction would let the REC domain begin to sample an active conformation, even within the full-length context and without mutations to either domain itself. Our EL_PhyR homology model shows that REC and output domains are normally separated by a short six-residue linker (R139-ESST-N144), suggesting that the inactive REC conformation is enforced by the high local effective concentration of the output domain (Mattison et al., 2002). To evaluate the relevance of the linker length on EL_PhyR, we generated a mutant EL_PhyR with seven additional residues inserted into the linker (140-EGNTGDGTS-141, called EL_PhyRext), and examined its binding to BeF3− by collecting 15N-1H TROSY-HSQCs in different ligand concentrations (Figure S4). We observed that the linker extension caused a substantial increase in the binding affinity of the protein to the phosphomimicking compound (wild-type Kd = 5.5 ± 1.6 mM, mutant Kd = 1.0 ± 0.8 mM). We suggest that this binding enhancement could be related to the destabilization of the inactive conformation caused by loosened interaction between REC and ECFSL, as observed with the removal of output domain.
The α4 Region of EL_PhyR Undergoes Conformational Fluctuations
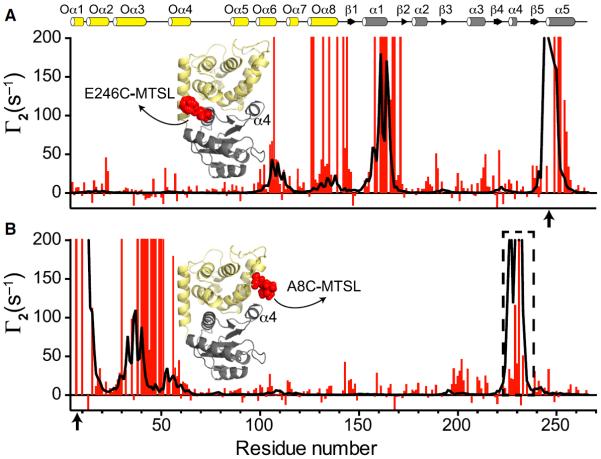
Our results demonstrate that minor or major perturbations at the interdomain interface favor the active state of the REC domain. The conformational fluctuations at the α4-β5-α5 interface could be a requirement for sampling the active state, priming the system for subsequent phosphorylation. To identify which regions undergo conformational exchange in the full-length EL_PhyR, we used PRE approaches (Clore and Iwahara, 2009). A nitroxide spin label ((1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) methanethiosulfonate [MTSL]) was attached to cysteine residues of single mutants at the output (A8C) or REC (E246C) domains. We quantitated amide proton transverse relaxation rates for spin-labeled EL_PhyR samples in the oxidized (Roxi) and reduced states (Rred). Experimental PRE rates (Γ2 = Roxi − Rred) were compared with theoretical Γ2 rates calculated from homology models with five MTSL conformers (Iwahara et al., 2004). For the probe located in the REC domain, we observed compatible experimental and theoretical PRE profiles, indicating that those regions of the molecule are well represented by the homology model (Figure 6A). On the other hand, the experimental and theoretical PRE profiles observed for A8C were inconsistent (Figure 6B), demonstrating lower-than-expected PREs for residues in the REC domain α4 helix (Figure 6B). The reduced PRE rates of residues in α4 suggest that this region of the molecule could undergo conformational changes that position it further from the output domain than portrayed in the crystal structure. These observations agree with our previous results that indicate that changes at the α4 would play an essential role in priming the active site for phosphorylation.
Figure 6. EL_PhyR Presents Conformational Fluctuations on α4 in the Inactive State.
PRE profiles comparison between experimental (red) and structure-based back-calculated values (black) for MTSL probes located at 246 (A) and 8 (B) positions (indicated by arrows). Secondary structure elements are highlighted on the top of figure (ECFSL, yellow; REC, gray). Cartoon representations indicating probe locations are indicated in the panels (ECFSL, yellow; REC, gray; MTSL, red). See also Figure S6.
DISCUSSION
An ideal protein switch has two key characteristics: (1) minimal inappropriate activation without stimuli present and (2) easy activation when these stimuli appear, necessitating a fine balance between rigidity and plasticity. These functional traits are established at the atomic level within many regulatory domains through pre-existing equilibria between favored “inactive” and disfavored “active” conformations that rapidly interconvert (Nussinov et al., 2014); by shifting such an equilibrium upon exposure to stimuli, the desired characteristics can be achieved. Studies among isolated REC domains, either from simple REC-only proteins or truncations of larger ones, support the generality of this activation model (Volkman et al., 2001; Stock and Guhaniyogi, 2006; Gardino et al., 2009). Notably, while studies often focus on the “forward” direction of this regulation (probing how a REC domain controls its cognate output domain), evidence is rapidly emerging to document a “mutual inhibition” process where the output domain controls aspects of REC structure as well (Barbieri et al., 2010). Support for this is provided by biochemical assays showing that the equilibrium between REC substates can be influenced either toward the stabilization of inactive or active conformations by the presence of either an output domain (Barbieri et al., 2010) or the binding partners to such (Ames et al., 1999; Schuster et al., 2001; Kaczmarczyk et al., 2014; Herrou et al., 2015). From a broader perspective, these principles can simply be viewed as examples of the general process of thermodynamic coupling: systems undergoing conformational change among multiple states can have the equilibria of these processes biased by the presence of binding partners that preferentially stabilize one state over the others. Considering a general process which shifts a system between almost completely active to inactive states (99% → 1%), approximately 2–3 kcal/mol of free energy would be required to achieve such a change. Within EL_PhyR, such free energy could easily be provided by the 8–10 kcal/mol of binding energy available from the ECFSL:REC interaction, as estimated from the approximately 1,000 Å2 of surface area between the two (Chen et al., 2013).
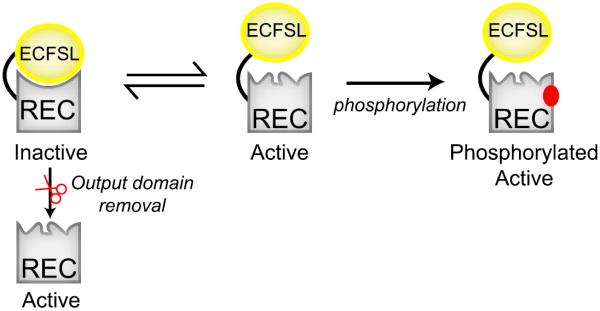
Here we clearly demonstrate the impact of the output domain on the structure, dynamics, and function of the REC domain within EL_PhyR, an interaction that has evolved to the point that even minor changes in the interdomain linker length can substantially affect these characteristics. As anticipated, the structure and dynamics of the REC domain at the interdomain (α4-β4-α5) interface are sensitive to the presence of the ECFSL, but surprisingly also at more distant regions in the molecule such as the α3 helix (20 Å away). These structural rearrangements were accompanied by the repositioning of the switch Thr residue side chain (T222) at the C-terminal end of the β4 strand, which likely forms a hydrogen bond with the nearby phosphoacceptor D194 (Figure 5A). This reorientation is typically observed when the protein is activated by phosphorylation or binding to BeF3− (Stock and Guhaniyogi, 2006), which is nicely supported by our observed overlap between EL_PhyR(+BeF3−) and EL_REC 15N-1H spectra (Figure 2B). Using rational mutagenesis, we found that T222 indeed plays an important role in the phosphorylation reaction and binding to BeF3− (Figures 5B, S4, and S7), likely by participating in the catalysis of the phosphorylation reaction (Appleby and Bourret, 1998) or by forming a hydrogen bond with one of the fluorine atoms of BeF3− (Lee et al., 2001). Collectively, our data indicate that EL_PhyR presents a case of mutual inhibition of the REC and output domains stabilized by interactions between the two, leaving the system poised for rapid activation by any perturbation that disrupts these interdomain interactions and allows the REC to adopt its preferred active conformation (Figure 7).
Figure 7. Model of the Mutual Inhibition between REC and Output Domains, as Exhibited by PhyR.
See also Figures S3, S4, S6, and S7.
Biochemical (Kaczmarczyk et al., 2014; Herrou et al., 2015) data from other PhyRs provide additional precedence that events separating ECFSLs from REC domains in these proteins, such as binding of ECFSLs to their cognate anti-σ partners, switch the REC domains into their active conformations. Data supporting this conclusion include in vitro assays showing that the addition of anti-σ factor enhances the phosphorylation rate of the Sphingomonas melonis Fr1 and C. crescentus PhyR proteins by either HKs (Kaczmarczyk et al., 2014) or small phosphodonors (Herrou et al., 2015), likely by favoring the stabilization of the phospho-accepting REC conformation.
Our data also provide important insights into the concerted conformational changes along secondary structure elements at REC (e.g., α3 and α4) that play an important role in propagating the effects of phosphorylation (as mimicked by BeF3− binding) from the REC active site to the output domain. Comparison of inactive and active structures of PhyR, represented by EL_PhyR and EL_REC, respectively, show that the most prevalent structural alterations caused by removal of the output domain are displacements of the interfacial α4 helix and its subsequent interaction with the α3 helix (Figure 4C). The α4 displacement seems to be primarily driven by the disruption of interactions between this helix and output domain. These structural rearrangements were followed by changes in ps-ns backbone motions (S2cs) for REC residues located not only at the domain interface (α4-β5 loop) but also far away (β3-α3 loop, over 10 Å distant) (Figures 4D and 4E). Given the linkage between S2 values and local entropy (Akke et al., 1993; Yang and Kay, 1996), we suggest that these changes reflect a form of entropy/enthalpy compensation whereby the loss of one kind of energetic stabilization is “compensated” by the gain of another as the REC domain switches between free and ECFSL-bound forms. For example, the loss in enthalpy caused by disruption of interactions between the output domain and the α4-β5 loop seems to be thermodynamically compensated by a gain in entropy in this region, as reflected by a decrease in S2CS values. This change is allosterically propagated to regions far from the interface (α3 helix and β3-α3 loop), where a loss in entropy (increased S2CS values) is compensated by a gain in enthalpy due to new hydrogen bonds that are potentially made between Tyr224/Arg227/Arg233 (α4) and Gln196/Ser202/Glu208 (α3 helix and α3-β3 loop) regions.
The entropy/enthalpy compensation we observed for the displacement of α4 coupled to priming of the active site for phosphorylation leads us to suggest that thermodynamically allowed conformational fluctuations for that helix occur and play an essential role in the activation of this RR. Indeed, this is indicated by our PRE measurements, where we observed a poor agreement between theoretical and experimental PRE rate values for residues located at the α4 helix when the paramagnetic MTSL probe was situated in the output domain (residue 8; Figure 6B). Based on the lower-than-expected PRE relaxation rates seen there, we suggest a heterogeneous population with conformations in which α4 is situated far apart from the output domain. In contrast, the good agreement we observed between theoretical and experimental values for amide probes near residue 246 in the REC domain strongly suggest that structural fluctuations seems to be confined to the α4 region, in agreement with low RMSD values observed between homology and NMR models for most of the protein except α3 and α4 regions when we compare the full-length protein with the truncated protein. While we were unable to simply confirm the presence of an inactive-active equilibrium in the EL_PhyR REC domain using CPMG-type relaxation dispersion measurements as in some related domains (Volkman et al., 2001), we believe that our combination of experimental data and the intrinsically dynamic nature of this type of domain (Volkman et al., 2001; Stock and Guhaniyogi, 2006; Gardino et al., 2009) strongly suggest that the REC domain likely samples several conformations. This heterogeneity generates a pool of active conformational states that can undergo phosphorylation (Figure 7), which provides a mechanism for the necessary conformational fluctuations required to allow a small fraction of anti-σ factor proteins to bind unphosphorylated PhyR, as observed indirectly in in vitro assays (Kaczmarczyk et al., 2014; Herrou et al., 2015).
In conclusion, our results demonstrate how an output domain can affect the structure and dynamics of REC switches in RRs, substantially affecting their regulatory characteristics. We suggest that this implies the potential for similar mechanisms for other protein switches which utilize intramolecular mutual inhibition between sensory and effector domains. Given interest in both understanding cellular signaling pathways and controlling them through small molecules and engineered proteins, further understanding of such principles would provide valuable insight into the natural and artificial control of protein-based switches more generally.
SIGNIFICANCE
Biological signal transduction relies on protein-based switches to convert stimuli into changes in cellular activity, requiring that individual components quickly and specifically change between “inactive” and “active” states when stimuli fluctuate. Here we examine a core aspect of such switching in the receiver domains, elements of many phosphorylation-controlled bacterial signaling pathways. While some isolated receiver domains adopt an “inactive” conformation without phosphorylation, our solution NMR and biochemical studies show that a receiver domain from a larger protein requires the presence of other domains to be similarly inactivated. The need for such interactions has implications for the co-evolution of regulatory and output domains in signaling proteins, with relevance to both natural and synthetic applications.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
DNA constructs encoding the REC-only fragment of EL_PhyR (“EL_REC,” 142–266) and a full-length variant with an extended interdomain linker (E140-GNTGDGT-S141) were generated from wild-type EL_PhyR (Correa et al., 2013) and cloned into the pHis-parallel expression vector (Sheffield et al., 1999). EL_PhyR mutants (A8C, D194A, E246C, T222A) were obtained by site-directed mutagenesis with primers containing the desired mutations. Expression in rich media was performed as described previously (Correa et al., 2013). Isotopically labeled proteins were obtained by expression in Escherichia coli BL21(DE3) cells grown in M9 minimal media containing 1 g/L of 15NH4Cl for U-15N samples and 3 g/L of [13C6]glucose for U-15N/13C-labeled samples. Cultures were grown at 37°C to an A600 of 0.6–1.0 and induced at 18°C for 12–16 hr by adding 0.5 mM isopropyl β-D-thiogalactoside. U-2H-labeled EL_PhyR was obtained similarly with the exception that protein expression was induced in media containing D2O (~99%). Protein purifications were performed as described previously (Correa et al., 2013). For details, see Supplemental Experimental Procedures.
NMR Chemical Shift Assignment and Structure Determination
NMR samples used for chemical shift assignment (full-length EL_PhyR, REC-only EL_REC) contained 0.5–1 mM U-15N/13C/2H-labeled EL_PhyR or U-15N/13C-labeled EL_REC in 10 mM Tris (pH 7.0), 50 mM NaCl, and 10% D2O. We emphasize that these samples did not contain Mg2+ to facilitate a proper comparison between them; we note that this is consistent with the conditions used to solve many REC-only structures to date. NMR data were recorded at 25°C on Varian Inova 600 and 800 MHz spectrometers, and processed and analyzed with NMRPipe (Delaglio et al., 1995) and NMRViewJ (Johnson and Blevins, 1994). EL_PhyR chemical shift assignments were obtained for over 90% of backbone positions using data from versions of 3D HNCO, CT-HNCA, CT-HN(CO)CA, CT-HN(CA)CB, and CT-HN(COCA)CB experiments optimized for perdeuterated proteins (Yamazaki et al., 1994) for Cα and Cβ assignments. Chemical shift assignments of EL_REC were obtained from 3D HNCACB, CBCA(CO)NH, HNCO, H(CCO)NH, C(CO)NH, and HCCH-TOCSY experiments (Cavanagh et al., 2007) along with distance information from a 3D 15N,13C simultaneous-edited NOESY (Pascal et al., 1994) (τm = 120 ms) spectrum.
For structure determination, the aforementioned 3D 15N,13C simultaneous-edited NOESY spectrum was used as our source of interproton distance restraints. Given that EL_REC samples two conformations in a 65%:35% ratio (Figure S2), we focused on solving the structure of the dominant species. For residues undergoing conformational exchange, we removed any NOEs arising from the minor conformer. Coupled with the similarity of NOEs between the two conformations (Figure S2E) and the preponderance of doubled residues occurring in loop regions (Figure S2F), we feel confident that any possible errors will minimally impact the overall topology of the protein. Additional structural restraints were obtained for hydrogen bonds (1.3 Å < dNH-O < 2.5 Å, 2.3 Å < dN-O < 3.5 Å) imposed on backbone amide protons that were protected from 2H exchange with factors higher than 104 (25°C, pH 7.0) and/or hydrogen bonds observed in more than 90% of structures in initial ensembles calculated without hydrogen bond restraints. ϕ and ψ torsion angle restraints were obtained from chemical shift analyses using TALOS (Cornilescu et al., 1999), with restraints set to three times the value of the calculated SD. Initial structures were calculated with automated assignments of NOESY spectra using ARIA 2.2 (Rieping et al., 2007), and subsequently refined using both automated and manual assignments. A total of 80 structures were calculated, using the 20 lowest energy models for evaluation with PROCHECK-NMR (Laskowski et al., 1996). Chemical shift assignments were deposited with BMRB (accession codes EL_PhyR: 26722, EL_REC: 25918), while coordinates for the REC_only protein were deposited with the PDB (accession code PDB: 2N9U).
Homology Modeling
We used the Modeller software suite (version 9v8; Sali and Blundell, 1993) to model the EL_PhyR atomic structure using the 1.25 Å resolution C. crescentus PhyR crystal structure as our template (PDB: 3N0R; Herrou et al., 2010). Five models were generated using automated sequence/structure alignment and default parameters; of these, we selected the model with lowest energy (DOPE) value for all analyses.
Spin Labeling and PRE Measurements
Purified U-15N-labeled samples of EL_PhyR mutants (A8C, E246C) were modified with the thiol-specific spin-label reagent (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) MTSL (Toronto Research Chemicals). Spin label was added in a 3:1 MTSL:protein ratio to 500–900 μM protein stocks and incubated overnight at room temperature in 50 mM Tris (pH 8) and 50 mM NaCl. Samples were subsequently injected over a Superdex 75 gel filtration column equilibrated with 20 mM HEPES (pH 7) and 50 mM NaCl to remove unreacted spin label and disulfide-bridged dimers of EL_PhyR. For NMR measurements, protein concentrations were used in a range of 150–250 μM in 20 mM HEPES (pH 7), 50 mM NaCl, and 5 mM MgCl2, 10% D2O. Reduced samples were generated by mixing protein samples with a 10:1 excess of sodium dithionite and incubated under anaerobic conditions for approximately 30 min at 4°C prior to NMR data collection. 1HN transverse relaxation rates (R2) were acquired on an 800 MHz Varian Inova spectrometer at 25°C using a series of five modified 15N-1H TROSY-HSQC experiments (Donaldson et al., 2001) with R2 relaxation delays (T) set between 6.5 and 16 ms. Spectral processing was performed as described above. R2 values and errors were obtained from fitting peak intensities in these spectra to exponential decay curves using NMRViewJ (Johnson and Blevins, 1994), letting us calculate PRE rates (Γ2) by subtracting R2 values between oxidized and reduced samples. Theoretical Γ2 rates were calculated by XPLOR-NIH (Schwieters et al., 2003) using the homology model of EL_PhyR generated by Modeller (Sali and Blundell, 1993) as noted above. These calculations used the SBMF mathematical model (and a rotational correlation time of 18 ns, determined from backbone 15N T1 and T2 measurements) to predict 1HN R2 relaxation rates from sites within an ensemble of ten EL_PhyR models, each of which had an MTSL probe conjugated to the protein in five different conformations.
Titration Experiments with BeF3−
Stock solutions of BeF3−, generated by mixing 400 mM NaF and 133 mM BeCl2, were freshly prepared prior to starting titrations by adding BeF3− to samples of U-15N-labeled proteins (240–300 μM) in buffer containing 20 mM HEPES (pH 7), 50 mM NaCl, 5 mM MgCl2, and 10% D2O. 15N-1H TROSY-HSQC spectra at 25°C were collected for each protein in different concentrations of BeF3− ranging from 0 to 11 mM. For EL_PhyR mutants D194A and T222A, 500 μM protein in the same buffer was titrated with 5 and 15 mM BeF3−. Spectra were processed as described above, monitoring changes in peak intensities using NMRViewJ (Johnson and Blevins, 1994).
Phosphorylation Assays
Samples of EL_PhyR (50 μM, either wild-type or T222A) were incubated with 5 μM EL368 HK (Correa et al., 2013) in 50 mM Tris (pH 8), 100 mM NaCl, 5 mM MgCl2, 1 mM ATP, and 12 μCi [γ-32P] ATP (6,000 Ci/mmol, PerkinElmer) at room temperature. Aliquots were removed at intervals of 30, 45, 60, 75, 90, 120, and 150 s, then treated and imaged for 32P as described previously (Correa et al., 2013). For details, see Supplemental Experimental Procedures.
MST
MST assays were conducted using a Monolith NT.115 instrument (Nanotemper). EL_PhyR and EL_PhyR (D194A) were labeled with Alexa Fluor 488 NHS ester (Molecular Probes) and buffer exchanged into 20 mM HEPES, 50 mM NaCl, 5 mM MgCl2, and 0.05% Tween 20). Fluorophore-labeled EL_PhyR or EL_PhyR (D194A) (200 nM) were titrated with up to 10 μM full-length anti-σ protein (Correa et al., 2013). Data points were measured from three to five times, and fit to a single-site binding equation using NanoTemper data analysis software.
Supplementary Material
Highlights.
Removing response regulator output domain stabilizes active input conformation
Protein activation correlates with extensive redistribution of backbone dynamics
NMR data reveal transient structural changes preceding phosphorylation
A rationale for mutual inhibition between sensor and effector domains is provided
ACKNOWLEDGMENTS
NMR data presented herein were collected at the UT Southwestern and at the City University of New York Advanced Science Research Center Biomolecular NMR Facilities, with the generous assistance of Qiong Wu (UT Southwestern) and James Aramini (CUNY ASRC). We thank Lewis E. Kay (University of Toronto) for providing us the pulse sequences for the acquisition of triple resonance and PRE data on large 2H-labeled proteins. We additionally thank Victor Ocasio, Giomar Rivera-Cancel, Arati Ramesh, Yirui Guo, and Doeke Hekstra for providing constructive comments for this manuscript, and all the members of Gardner laboratory for their helpful discussions. This work was supported by grants from the NIH (R01 GM106239) and the Robert A. Welch Foundation (I-1424).
Footnotes
ACCESSION NUMBERS The accession number for the EL_REC structure reported in this paper is PDB: 2N9U. The accession numbers for the EL_PhyR and EL_REC chemical shift assignments reported in this paper are BMRB: 26722 and 25918, respectively.
SUPPLEMENTAL INFORMATION Supplemental Information includes Supplemental Experimental Procedures, seven figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.chembiol.2016.07.010.
AUTHOR CONTRIBUTIONS F.C. and K.H.G. conceived the project scope, designed, performed, and analyzed the experiments, and wrote the paper.
REFERENCES
- Akke M, Bruschweiler R, Palmer AG., 3rd NMR order parameters and free energy: an analytical approach and its application to cooperative Ca2+ binding by Calbindin D9k. J. Am. Chem. Soc. 1993;115:9832–9833. [Google Scholar]
- Ames SK, Frankema N, Kenney LJ. C-terminal DNA binding stimulates N-terminal phosphorylation of the outer membrane protein regulator OmpR from Escherichia coli. Proc. Natl. Acad. Sci. USA. 1999;96:11792–11797. doi: 10.1073/pnas.96.21.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleby JL, Bourret RB. Proposed signal transduction role for conserved CheY residue Thr87, a member of the response regulator active-site quintet. J. Bacteriol. 1998;180:3563–3569. doi: 10.1128/jb.180.14.3563-3569.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri CM, Mack TR, Robinson VL, Miller MT, Stock AM. Regulation of response regulator autophosphorylation through interdomain contacts. J. Biol. Chem. 2010;285:32325–32335. doi: 10.1074/jbc.M110.157164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellsolell L, Prieto J, Serrano L, Coll M. Magnesium binding to the bacterial chemotaxis protein CheY results in large conformational changes involving its functional surface. J. Mol. Biol. 1994;238:489–495. doi: 10.1006/jmbi.1994.1308. [DOI] [PubMed] [Google Scholar]
- Berjanskii MV, Wishart DS. A simple method to predict protein flexibility using secondary chemical shifts. J. Am. Chem. Soc. 2005;127:14970–14971. doi: 10.1021/ja054842f. [DOI] [PubMed] [Google Scholar]
- Campagne S, Damberger FF, Kaczmarczyk A, Francez-Charlot A, Allain FH, Vorholt JA. Structural basis for sigma factor mimicry in the general stress response of Alphaproteobacteria. Proc. Natl. Acad. Sci. USA. 2012;109:E1405–E1414. doi: 10.1073/pnas.1117003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh J, Fairbrother WJ, Palmer AG, Rance M, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. 2nd edition Academic Press; 2007. [Google Scholar]
- Chen J, Sawyer N, Regan L. Protein-protein interactions: general trends in the relationship between binding affinity and interfacial buried surface area. Protein Sci. 2013;22:510–515. doi: 10.1002/pro.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore GM, Iwahara J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem. Rev. 2009;109:4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Correa F, Ko WH, Ocasio V, Bogomolni RA, Gardner KH. Blue light regulated two-component systems: enzymatic and functional analyses of light-oxygen-voltage (LOV)-histidine kinases and downstream response regulators. Biochemistry. 2013;52:4656–4666. doi: 10.1021/bi400617y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Donaldson LW, Skrynnikov NR, Choy WY, Muhandiram DR, Sarkar B, Forman-Kay JD, Kay LE. Structural characterization of proteins with an attached ATCUN motif by paramagnetic relaxation enhancement NMR spectroscopy. J. Am. Chem. Soc. 2001;123:9843–9847. doi: 10.1021/ja011241p. [DOI] [PubMed] [Google Scholar]
- Foreman R, Fiebig A, Crosson S. The LovK-LovR two-component system is a regulator of the general stress pathway in Caulobacter crescentus. J. Bacteriol. 2012;194:3038–3049. doi: 10.1128/JB.00182-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francez-Charlot A, Frunzke J, Reichen C, Ebneter JZ, Gourion B, Vorholt JA. Sigma factor mimicry involved in regulation of general stress response. Proc. Natl. Acad. Sci. USA. 2009;106:3467–3472. doi: 10.1073/pnas.0810291106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 2006;188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardino AK, Volkman BF, Cho HS, Lee SY, Wemmer DE, Kern D. The NMR solution structure of BeF(3)(−)-activated Spo0F reveals the conformational switch in a phosphorelay system. J. Mol. Biol. 2003;331:245–254. doi: 10.1016/s0022-2836(03)00733-2. [DOI] [PubMed] [Google Scholar]
- Gardino AK, Villali J, Kivenson A, Lei M, Liu CF, Steindel P, Eisenmesser EZ, Labeikovsky W, Wolf-Watz M, Clarkson MW, et al. Transient non-native hydrogen bonds promote activation of a signaling protein. Cell. 2009;139:1109–1118. doi: 10.1016/j.cell.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourion B, Francez-Charlot A, Vorholt JA. PhyR is involved in the general stress response of Methylobacterium extorquens AM1. J. Bacteriol. 2008;190:1027–1035. doi: 10.1128/JB.01483-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B, Liu Y, Ginzinger SW, Wishart DS. SHIFTX2: significantly improved protein chemical shift prediction. J. Biomol. NMR. 2011;50:43–57. doi: 10.1007/s10858-011-9478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrou J, Foreman R, Fiebig A, Crosson S. A structural model of anti-anti-sigma inhibition by a two-component receiver domain: the PhyR stress response regulator. Mol. Microbiol. 2010;78:290–304. doi: 10.1111/j.1365-2958.2010.07323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrou J, Rotskoff G, Luo Y, Roux B, Crosson S. Structural basis of a protein partner switch that regulates the general stress response of alpha-proteobacteria. Proc. Natl. Acad. Sci. USA. 2012;109:E1415–E1423. doi: 10.1073/pnas.1116887109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrou J, Willett JW, Crosson S. Structured and dynamic disordered domains regulate the activity of a multifunctional anti-sigma factor. MBio. 2015;6:e00910. doi: 10.1128/mBio.00910-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahara J, Schwieters CD, Clore GM. Ensemble approach for NMR structure refinement against (1)H paramagnetic relaxation enhancement data arising from a flexible paramagnetic group attached to a macromolecule. J. Am. Chem. Soc. 2004;126:5879–5896. doi: 10.1021/ja031580d. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. NMR view - a computer-program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Kaczmarczyk A, Hochstrasser R, Vorholt JA, Francez-Charlot A. Complex two-component signaling regulates the general stress response in Alphaproteobacteria. Proc. Natl. Acad. Sci. USA. 2014;111:E5196–E5204. doi: 10.1073/pnas.1410095111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern D, Volkman BF, Luginbuhl P, Nohaile MJ, Kustu S, Wemmer DE. Structure of a transiently phosphorylated switch in bacterial signal transduction. Nature. 1999;402:894–898. doi: 10.1038/47273. [DOI] [PubMed] [Google Scholar]
- Kim HS, Willett JW, Jain-Gupta N, Fiebig A, Crosson S. The Brucella abortus virulence regulator, LovhK, is a sensor kinase in the general stress response signalling pathway. Mol. Microbiol. 2014;94:913–925. doi: 10.1111/mmi.12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Lee SY, Cho HS, Pelton JG, Yan D, Berry EA, Wemmer DE. Crystal structure of activated CheY. Comparison with other activated receiver domains. J. Biol. Chem. 2001;276:16425–16431. doi: 10.1074/jbc.M101002200. [DOI] [PubMed] [Google Scholar]
- Mattison K, Oropeza R, Kenney LJ. The linker region plays an important role in the interdomain communication of the response regulator OmpR. J. Biol. Chem. 2002;277:32714–32721. doi: 10.1074/jbc.M204122200. [DOI] [PubMed] [Google Scholar]
- McDonald LR, Boyer JA, Lee AL. Segmental motions, not a two-state concerted switch, underlie allostery in CheY. Structure. 2012;20:1363–1373. doi: 10.1016/j.str.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R, Tsai CJ, Liu J. Principles of allosteric interactions in cell signaling. J. Am. Chem. Soc. 2014;136:17692–17701. doi: 10.1021/ja510028c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocasio VJ, Correa F, Gardner KH. Ligand-induced folding of a two-component signaling receiver domain. Biochemistry. 2015;54:1353–1363. doi: 10.1021/bi501143b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal SM, Muhandiram DR, Yamazaki T, Formankay JD, Kay LE. Simultaneous acquisition of N-15-edited and C-13-edited NOE spectra of proteins dissolved in H2O. J. Magn. Reson. Ser. B. 1994;103:197–201. [Google Scholar]
- Rieping W, Habeck M, Bardiaux B, Bernard A, Malliavin TE, Nilges M. ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007;23:381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Scheuermann TH, Padrick SB, Gardner KH, Brautigam CA. On the acquisition and analysis of microscale thermophoresis data. Anal Biochem. 2016;496:79–93. doi: 10.1016/j.ab.2015.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M, Silversmith RE, Bourret RB. Conformational coupling in the chemotaxis response regulator CheY. Proc. Natl. Acad. Sci. USA. 2001;98:6003–6008. doi: 10.1073/pnas.101571298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Seidel SA, Dijkman PM, Lea WA, van den Bogaart G, Jerabek-Willemsen M, Lazic A, Joseph JS, Srinivasan P, Baaske P, Simeonov A, et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods. 2013;59:301–315. doi: 10.1016/j.ymeth.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- Stock AM, Guhaniyogi J. A new perspective on response regulator activation. J. Bacteriol. 2006;188:7328–7330. doi: 10.1128/JB.01268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Volkman BF, Lipson D, Wemmer DE, Kern D. Two-state allosteric behavior in a single-domain signaling protein. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Lee W, Arrowsmith CH, Muhandiram DR, Kay LE. A suite of triple-resonance NMR experiments for the backbone assignment of N-15, C-13, H-2 labeled proteins with high-sensitivity. J. Am. Chem. Soc. 1994;116:11655–11666. [Google Scholar]
- Yan D, Cho HS, Hastings CA, Igo MM, Lee SY, Pelton JG, Stewart V, Wemmer DE, Kustu S. Beryllofluoride mimics phosphorylation of NtrC and other bacterial response regulators. Proc. Natl. Acad. Sci. USA. 1999;96:14789–14794. doi: 10.1073/pnas.96.26.14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Kay LE. Contributions to conformational entropy arising from bond vector fluctuations measured from NMR-derived order parameters: application to protein folding. J. Mol. Biol. 1996;263:369–382. doi: 10.1006/jmbi.1996.0581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.