Summary
Chromosomal structure of nuclear DNA is usually maintained by insertion of nucleosomes into preexisting chromatin, both on newly synthesized DNA at replication forks and at sites of DNA damage. But during retrovirus infection, a histone-free DNA copy of the viral genome is synthesized that must be loaded with nucleosomes de novo. Here we show that core histones are rapidly loaded onto unintegrated Moloney murine leukemia virus DNAs. Loading of nucleosomes requires nuclear entry but does not require viral DNA integration. The histones associated with unintegrated DNAs become marked by covalent modifications, with a delay relative to the time of core histone loading. Expression from unintegrated DNA can be enhanced by modulation of the histone modifying machinery. The data show that histone loading onto unintegrated DNAs occurs very rapidly after nuclear entry and does not require prior establishment of an integrated provirus.
Keywords: Retrovirus, Nucleosome, Histone, Epigenetics
Graphical abstract
INTRODUCTION
DNA of eukaryotic chromosomes is condensed into chromatin by the association with nucleosomes. Nucleosomes are normally loaded onto chromosomal DNA during DNA replication (Ramachandran and Henikoff, 2015). As a replication fork moves along the DNA, nucleosomes are displaced from the unreplicated DNA ahead of the fork and then must reassociate onto both daughter branches behind the growing fork (Lucchini et al., 2001), likely in small clusters (Annunziato, 2013). Current models suggest that the relocalization of nucleosomes is a highly dynamic process in which “old” nucleosomes are distributed to both daughter branches, and “new” nucleosomes are rapidly added to restore normal nucleosome density. During this process, epigenetic marks on the old chromatin must be reestablished on the new DNA to ensure maintenance of the appropriate pattern of gene expression (Campos et al., 2014). New nucleosomes may also be loaded at sites of DNA damage, likely under the direction of the PCNA factor present at the sites (Mello and Almouzni, 2001; Polo et al., 2006). The loading of nucleosomes is thought to be promoted by an array of dedicated chaperones, including the trimeric CAF-1, Histone regulator A (HIRA), HJURP, ASF1, DAXX and ATRX, and others (Annunziato, 2013).
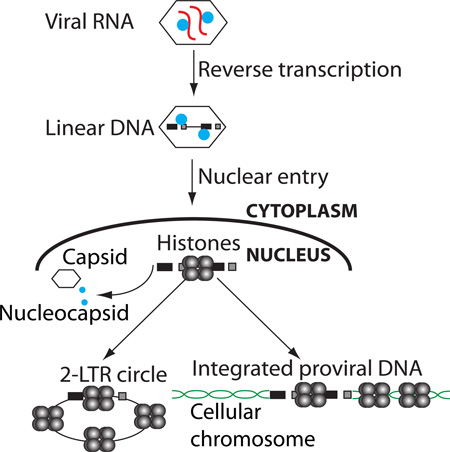
In contrast to these normal settings of chromatin maintenance, there are unusual circumstances when naked DNA, or DNA previously free of nucleosomes, enters a cell and must acquire a nucleosomal array de novo. One such circumstance occurs in the course of retroviral infection. Retroviruses initiate a complex series of events in the early stages of infection. The core of the mature virion particle consists of a protein shell made up of the viral capsid protein (CA) enclosing two copies of the RNA genome tightly associated with the viral nucleocapsid protein (NC), a small, highly basic protein. Following viral entry into the cytoplasm, the single-stranded viral RNA genome is copied by reverse transcriptase (RT) to generate a linear double-stranded DNA version of the genome (Telesnitsky and Goff, 1997). The double-stranded linear viral DNA, contained within a large complex termed the preintegration complex (PIC) (Brown et al., 1989), is trafficked into the nucleus by one of a variety of mechanisms that vary among different viral genera. The PICs of the gammaretroviruses such as Moloney murine leukemia virus (MLV) are unable to enter the intact nucleus of a nondividing cell and depend on nuclear membrane breakdown during cell division to gain access to the host DNA (Roe et al., 1993). In contrast, PICs of lentiviruses such as HIV-1 are actively transported across the nuclear membrane even in non-dividing cells (Lewis et al., 1992; Lewis and Emerman, 1994; Weinberg et al., 1991).
Once inside the nucleus, the linear DNA gives rise to a variety of DNA forms. The viral integrase (IN) directs the insertion of the linear DNA to form the integrated provirus. Homologous recombination between the two LTRs of the linear viral DNA leads to the formation of circular DNAs containing a single LTR (Kilzer et al., 2003; Yoshimura and Weinberg, 1979). Joining of the termini of the linear viral DNA by non-homologous end joining (NHEJ) DNA repair pathways leads to the formation of circles containing two LTRs (Kilzer et al., 2003; Li et al., 2001), with the LTRs joined in tandem at a unique LTR-LTR junction sequence (Shoemaker et al., 1980) that allows their levels to be readily quantified by PCR. The 2-LTR circles are thought to be generated in the nucleus, making them a useful marker of viral DNA nuclear import.
Transcriptional promoter sequences in the 5’ LTR of the proviral DNA generate full-length genomic viral mRNAs for viral gene expression and genome replication. However, certain cell types block this transcription. For example, mouse embryonic stem (ES) and embryonic carcinoma (EC) cells block transcription of the MLV provirus (Akgun et al., 1991; Teich et al., 1977). This silencing is mediated by a large protein complex containing the trans-acting repressors ZFP809, Trim28, YY1, HP1, and EBP1 (Rowe et al., 2010; Schlesinger et al., 2013; Wang et al., 2014; Wolf and Goff, 2007, 2009) recognizing DNA-binding sites on the viral LTR. This complex recruits chromatin modifiers such as ESET/SETDB1, which deposits the H3K9 trimethylation repressive histone mark (H3K9me3) (Matsui et al., 2010; Rowe et al., 2010).
Only a few studies have addressed the state of the viral DNA after reverse transcription and before integration (Fassati and Goff, 1999; Karageorgos et al., 1993). Several pressing questions are unanswered: How and when are nucleosomes loaded onto the viral DNA, which has never previously seen a nucleosome? Do unintegrated linear or circular DNAs acquire nucleosomes? When are epigenetic marks placed on the nucleosomes of the viral DNA? It is known that the unintegrated DNAs are very poorly expressed (Nakajima et al., 2001; Poon and Chen, 2003; Sakai et al., 1993; Schwartzberg et al., 1984), but the basis for this is unclear. The expression of transgenes on integration-defective retrovirus vectors is enhanced by histone deacetylase inhibitors (Pelascini et al., 2013; Schneider et al., 2012), suggesting that the unintegrated viral DNA is associated with histones lacking acetylation.
In this study we describe the kinetics of histone association with the DNA genomes of MLV. We show that unintegrated linear viral DNAs and 2-LTR circles undergo rapid core histone loading after nuclear entry and before integration. Moreover, histone loading is coordinated with the removal of viral structural proteins NC and CA from the viral DNA. The appearance of epigenetically modified histones occurs after a delay, thereby postponing host cell regulation of the viral promoter. Finally, we confirm that unintegrated viral DNAs are only weakly expressed, but that expression is significantly increased by chromatin modifiers. The data provide insights into fundamental aspects of retroviral biology.
RESULTS
Core histone loading onto retroviral DNAs occurs rapidly following infection
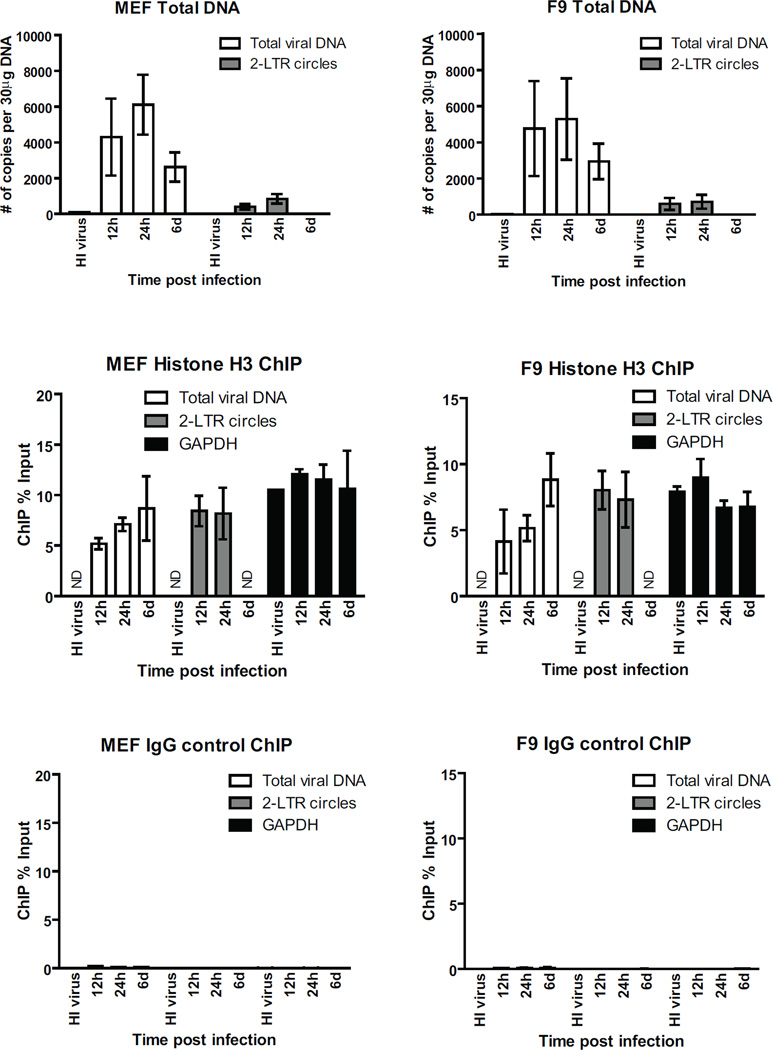
To investigate histone loading onto viral DNAs following infection, we utilized an MLV reporter genome in which viral genes were replaced with the GFP gene (MLV-GFP). Mouse embryonic fibroblasts (MEFs) were transduced with MLV-GFP reporter virus pseudotyped with the VSV-G envelope, total DNA was harvested at 12, 24h or 6d post infection, and levels of viral DNA replication intermediates were monitored by quantitative real-time PCR (qRT-PCR) using primers specific for GFP (to detect total viral DNA) or 2-LTR circles (a marker of nuclear entry). Total viral DNA, which includes linear, circular, and integrated proviral DNA, appeared at 12h post infection, peaked at 24 h, and then decreased slightly at 6d (Figure 1A). The 2-LTR circles peaked at 24h post infection, and were undetectable by 6d, indicative of their short half-life (Figure 1A). The maximum levels of 2-LTR circles at early times accounted for 5–15% of total viral DNA. MEFs transduced with heat-inactivated virus yielded no PCR signal, indicating minimal contaminating plasmid DNA carryover from the virus preparation. To determine the localization of viral DNA intermediates in the cell, MLV-GFP infected NIH-3T3 cells were subjected to cytoplasmic/nuclear fractionation at 12h post infection followed by quantitative PCR. Levels of total viral DNA were higher in the cytosol, while levels of 2-LTR circles were higher in the nuclear fraction (Figure S1); GAPDH control was highly enriched in the nuclear fraction.
FIGURE 1. Core histone loading onto retroviral DNAs occur rapidly following infection.
(A) MEF cells were infected with VSV-G pseudotyped MLV-GFP virus. To control for potential plasmid DNA carry over in the viral supernatant, heat inactivated (HI) virus were used in parallel. Total DNA from infected cells was isolated at indicated times, followed by real time quantitative PCR using primers targeting GFP (total viral DNA) and 2-LTR circles. Absolute copy number is calculated based on standard curves generated using plasmid DNA. Results shown are means ± SDs from two independent experiments performed in duplicate.
For related data, see Figure S1.
(B and C) ChIP analysis of chromatin harvested at indicated times following MLV-GFP infection of MEF cells using antibody to (B) histone H3 or (C) control rabbit IgG. ChIP data is presented as % of input DNA, calculated by dividing the ChIP copy number for each gene target by the copy number from input DNA and multiplying by 100%. Results shown are means ± SDs from two independent experiments performed in duplicate. ND, not determined.
For related data, see Figure S2.
(D, E, F) Same as (A, B, C), respectively, using F9 embryonic carcinoma cells.
We next monitored the association of nucleosomal histones with retroviral DNAs. Infected MEF cells were formaldehyde cross-linked at 12h, 24h, and 6d post infection, and chromatin immunoprecipitation (ChIP) assays were performed with anti-histone H3 or nonspecific IgG, followed by qRT-PCR using primers targeting GFP (total viral DNA) or 2-LTR circles. Primers targeting GAPDH were used to score fully chromatinized cellular DNA. Histone H3 rapidly associated with total viral DNA at 12h and reached levels comparable to that of GAPDH by 24h (Figure 1B). The ChIP signal with the GFP probe (total viral DNA) remained high at 6d, indicative of fully chromatinized integrated proviral DNA. Thus, H3 histones are rapidly loaded on the incoming viral DNAs. Histone H3 loading assayed here likely reflects the behavior of other core histones. ChIP experiments using anti-histone H2A revealed similar results (Figure S2).
Analysis of 2-LTR circles also revealed rapid histone H3 loading, with levels comparable to that of the GAPDH gene by 12h (Figure 1B). While the level of histones on circular DNA was high, the overall abundance of the circular DNA was too low to account for the entire signal of chromatin-bound DNA observed in the total DNA fraction, and thus the majority of the signal in the total DNA must derive from the linear forms.
Similar kinetics of histone loading onto viral DNAs were observed in MLV-GFP infected F9 embryonic carcinoma cells (Figure 1D and 1E). Histone H2A also associated with 2-LTR circles, though at somewhat lower levels compared to total viral DNA (Figure S2). Together, these findings suggest that both linear and circular retroviral DNAs undergo rapid chromatinization following infection.
Histone loading of retroviral DNAs is independent of viral DNA integration
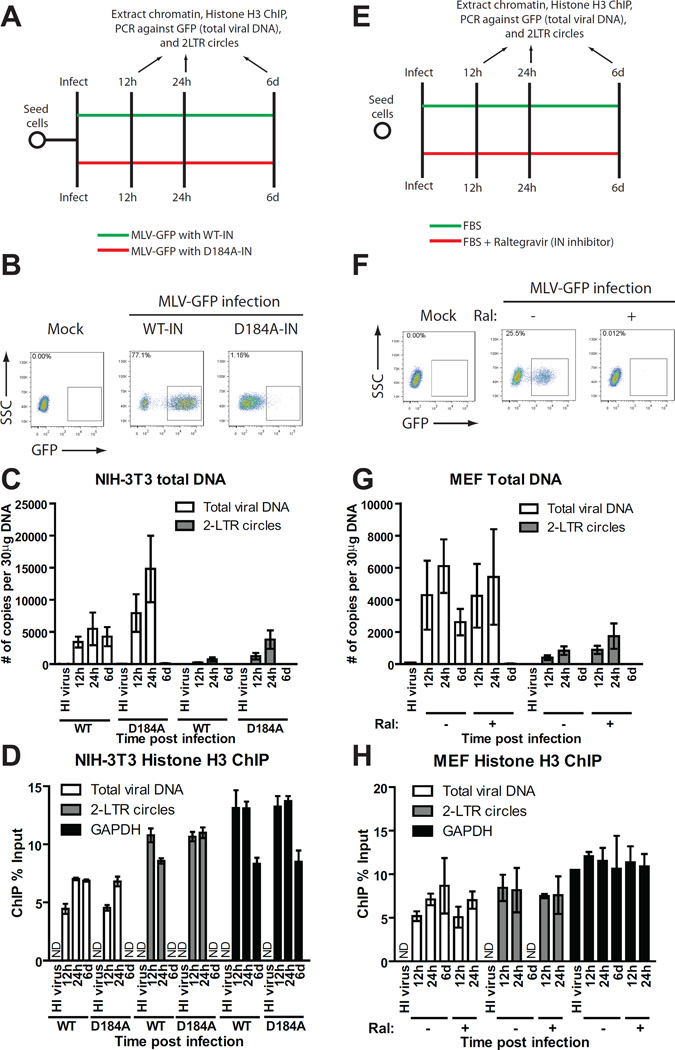
To investigate whether viral DNA integration is needed for efficient histone loading, we utilized two approaches. First, MLV-GFP reporter viruses were packaged using wild-type (WT) or a catalytically inactive MLV integrase (IN) mutant (D184A) (Lai et al., 2001). The infectivity of WT or D184A-IN virions was monitored using flow cytometry analysis of infected NIH-3T3 fibroblasts 48h post infection (Figure 2B). The infectivity of D184A-IN virus was ∼70-fold lower than that of WT virus (Figure 2B). Cells infected with D184A-IN virions contained slightly higher levels of total viral DNA (GFP) and 2-LTR circles at 12 and 24 h, but no viral DNA was detected at 6d (Figure 2C). These results confirm that the D184A mutation had no adverse effects on reverse transcription and nuclear entry of viral DNA, but efficiently blocked viral integration. Compared to WT virus, histone H3 ChIP of cells infected with D184A-IN virus showed comparable accumulation of histone H3 on both total viral DNA (GFP) and 2-LTR circles (Figure 2D). Thus, viral integration is not required for histone loading and both linear and circular forms of the preintegrative viral DNA are chromatinized even when integration is blocked.
FIGURE 2. Histone loading of retroviral DNAs occurs independently of viral integration.
(A) Schematic of experimental setup.
(B) Flow cytometry analysis of NIH-3T3 cells infected with VSV-G pseudotyped WT or integrase defective mutant (D184A) MLV-GFP viruses at 2d post infection. SSC, side scatter. One of three independent experiments is shown.
(C) NIH-3T3 cells were infected with VSV-G pseudotyped WT or integrase-defective mutant (D184A) MLV-GFP viruses. Total DNA from infected cells was isolated at indicated times post infection and number of copies of viral replication intermediates was determined (means ± SDs from two independent experiments performed in duplicate).
(D) Histone H3 ChIP analysis of chromatin harvested at indicated times following MLV-GFP infection of NIH-3T3 cells. ChIP data is presented as % of input DNA (means ± SDs from two independent experiments performed in duplicate).
(E) Schematic of experimental setup.
(F) Flow cytometry analysis of MEF cells infected with VSV-G pseudotyped WT MLV-GFP viruses in the presence or absence of raltegravir (Ral). Results are from 2d post infection. One representative of three independent experiments is shown.
(G) MEF cells were infected with VSV-G pseudotyped WT MLV-GFP viruses in the presence or absence of raltegravir (Ral). Total DNA from infected cells was isolated at indicated times post infection and the number of copies of viral replication intermediates was determined (means ± SDs from two independent experiments performed in duplicate).
(H) Histone H3 ChIP analysis of chromatin harvested at indicated times following MLV-GFP infection of MEF cells treated with or without raltegravir (Ral). ChIP data is presented as % of input DNA (means ± SDs from two independent experiments performed in duplicate).
ND, not determined.
Secondly, ChIP experiments were performed in MEF cells infected with WT virus in the presence or absence of the integrase inhibitor raltegravir (Figure 2E). Raltegravir treatment almost completely prevented transduction (Figure 2F) in accord with earlier studies (Beck-Engeser et al., 2009). Raltegravir caused a 2-fold increase in 2-LTR circles without affecting total viral DNA levels (Figure 2G). ChIP analysis of raltegravir-treated cells revealed normal accumulation of histone H3 on both total viral DNA (GFP) and 2-LTR circles (Figure 2H), again demonstrating that integration is not required for histone loading. Thus, chromatinization of viral DNAs occurs early in infection irrespective of viral DNA integration.
Histone loading of retroviral DNAs requires viral nuclear entry
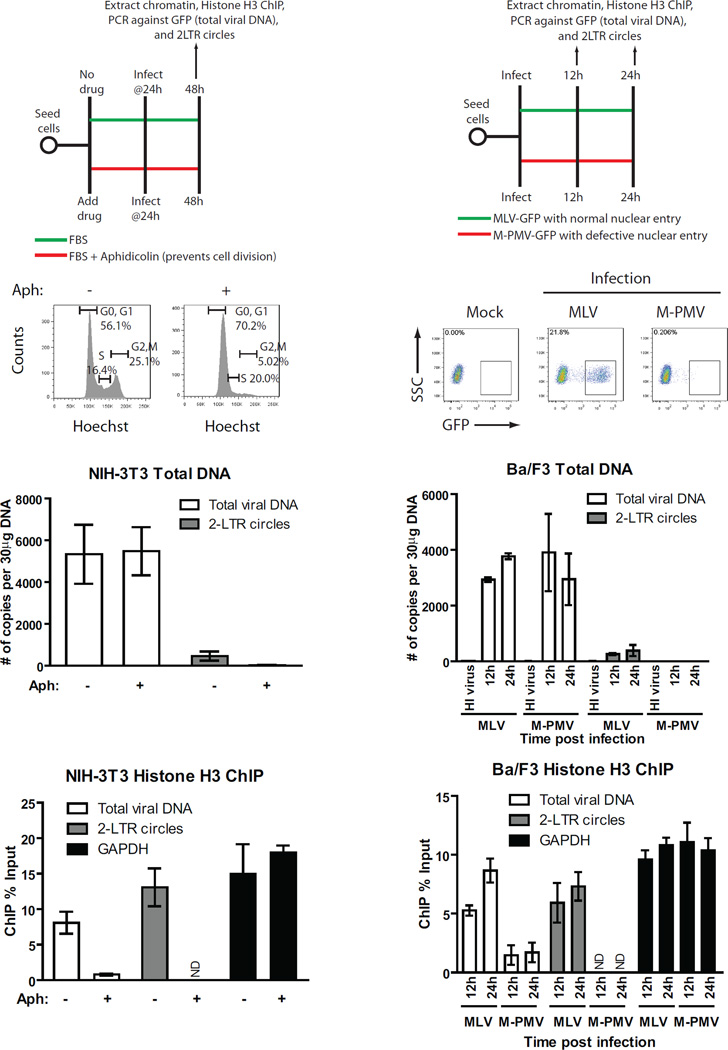
Histones are synthesized in the cytosol and subsequently imported to the nucleus (Burgess and Zhang, 2013). To test whether histone loading onto viral DNAs occurred in the cytoplasm prior to nuclear entry, we examined histone loading on viral DNA when nuclear entry was blocked. Entry of the MLV PIC into the nucleus requires breakdown of the nuclear envelope during mitosis, and growth-arrested cells that fail to undergo cell division block MLV infection at nuclear entry (Roe et al., 1993). To arrest cell division, NIH-3T3 cells were cultured for 24h in the presence of aphidicolin, an inhibitor of DNA synthesis (Figure 3A). DNA content profiling of the aphidicolin-treated cells by flow cytometry confirmed cell cycle arrest in the G1/S phase (Figure 3B). The arrested cells were transduced with MLV-GFP viruses for 24 h, and chromatin was subjected to ChIP using histone H3 antibodies. Aphidicolin treatment did not affect levels of total viral DNA, suggesting that viral entry, uncoating and reverse transcription proceeded normally (Figure 3C). The levels of 2-LTR circles, however, were reduced ∼30-fold, confirming that nuclear entry of viral DNA was blocked (Figure 3C). Strikingly, despite normal levels of total viral DNA, ChIP analysis revealed a 10-fold reduction in association of histone H3 with viral DNA in the aphidicolin-arrested cells, whereas histone H3 coverage of GAPDH was unaffected (Figure 3D). Thus, nucleosome loading on the viral DNA occurs primarily – if not exclusively - after nuclear entry.
FIGURE 3. Histone loading of retroviral DNAs requires viral nuclear entry.
(A) Schematic of experimental setup.
(B) Cell cycle analysis of NIH-3T3 cells treated with or without DNA polymerase inhibitor aphidicolin for 24h prior to infection. Y-axis, cell count, X-axis, DNA content measured by Hoechst staining. One representative of three independent experiments is shown.
(C) NIH-3T3 cells pretreated with or without aphidicolin (Aph) to induce cell cycle arrest were infected with VSV-G pseudotyped WT MLV-GFP virus. Total DNA from infected cells was isolated 24h post infection and levels of viral replication intermediates were determined (means ± SDs from two independent experiments performed in duplicate).
(D) Histone H3 ChIP analysis of chromatin harvested from NIH-3T3 cells outlined in (C) 24h post infection. ChIP data is presented as % of input DNA (means ± SDs from two independent experiments performed in duplicate).
(E) Schematic of experimental setup.
(F) Flow cytometry analysis of Ba/F3 cells infected with VSV-G pseudotyped single-round MLV or M-PMV-GFP viruses at 2d post infection. SSC, side scatter. One representative of three independent experiments is shown.
(G) Level of viral replication intermediates from Ba/F3 cells infected with MLV or M-PMV-GFP viruses. Total DNA from infected cells was isolated at indicated times post infection and number of copies of viral replication intermediates was determined (means ± SDs from two independent experiments performed in duplicate).
(H) Histone H3 ChIP analysis of chromatin harvested from infected Ba/F3 cells outlined in (G). ChIP data is presented as % of input DNA (means ± SDs from two independent experiments performed in duplicate).
ND, not determined.
To address the nuclear entry requirement for retroviral histone loading, we used Mason-Pfizer monkey virus (M-PMV), which is blocked from nuclear entry in mouse cells (Wang and Goff, 2015). Mouse Ba/F3 cells were infected with VSV-G-pseudotyped M-PMV-GFP or MLV-GFP (Figure 3E), and the efficiency of infection was monitored by flow cytometry analysis at 48h (Figure 3F). Ba/F3 cells showed a ∼100-fold reduced susceptibility to M-PMV transduction relative to MLV (Figure 3F). The cells showed normal levels of total viral DNA (GFP), but a ∼60-fold reduction in 2-LTR circles, confirming a strong block to nuclear entry (Figure 3G). Analysis of histone loading on the viral DNA by ChIP revealed a 6-fold reduction in histone H3 association with M-PMV total viral DNA relative to MLV DNA (Figure 3H). Thus, two approaches to block nuclear entry – cell-cycle arrest and species-specific restriction – demonstrated a requirement for nuclear entry for efficient histone loading onto viral DNAs.
Appearance of epigenetically modified histones on viral DNAs is delayed compared to core histones
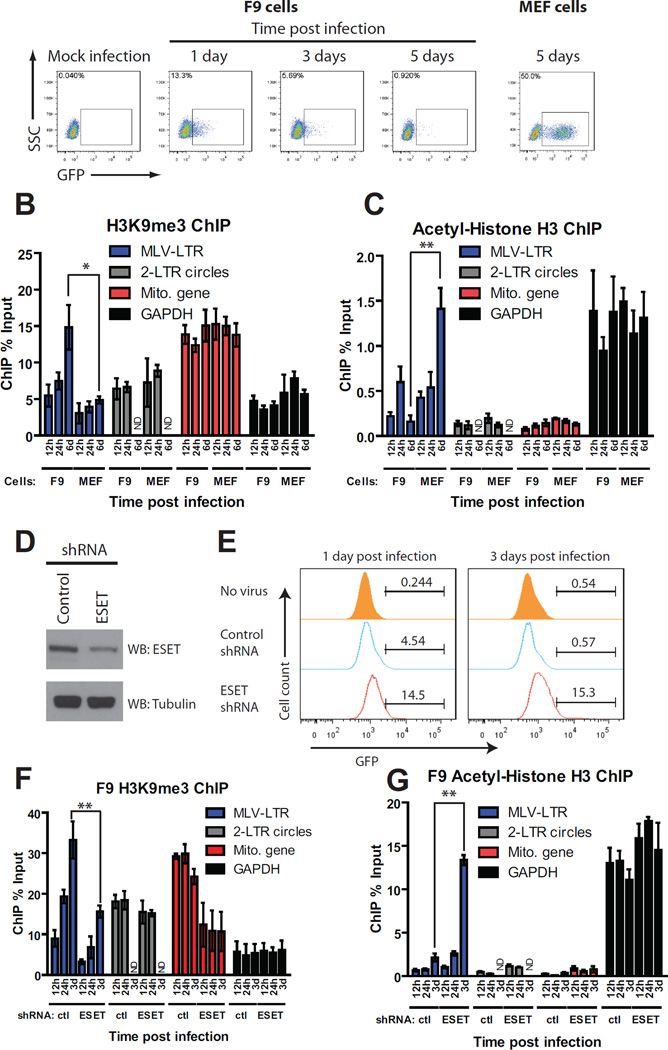
We examined the kinetics of epigenetically modified histone loading onto retroviral DNAs in the context of transcriptional silencing in F9 mouse embryonic carcinoma cells, which silence integrated proviral DNA (Schlesinger and Goff, 2013). F9 cells were exposed to high titers of MLV vectors delivering a GFP reporter. A modest fraction (13%) of cells expressed GFP at 1d post infection, and <1% of cells expressed GFP at 5d (Figure 4A). Infection of MEF cells, a differentiated cell type, resulted in 50% GFP positive cells ChIP analysis of viral DNA in F9 and MEF cells was performed using antibodies against trimethylated histone H3K9 (H3K9me3), a marker of silenced chromatin (Figure 4B), or acetylated histone H3 (H3Ac), a marker of active chromatin (Figure 4C). The specificity of these antibodies for ChIP was confirmed using positive control primers specific for endogenous promoters of the mitochondrial RNA polymerase gene (Polrmt, enriched in H3K9me3, hereafter called Mito gene) and GAPDH (enriched in H3Ac). At early times after infection of F9 cells, levels of H3K9me3 marks on viral DNA were low (Figure 4B), consistent with transient GFP expression. In contrast, H3K9me3 was highly enriched on viral DNA by 6d (Figure 4B), consistent with complete silencing of the viral promoter (Figure 4A). No enrichment of H3Ac marks of activated chromatin on viral DNAs was observed at any time in F9 cells (Figure 4C). Together, these results suggest that histone loading occurs first and that modified histones appear later, coordinately with the MLV silencing (Figure 4A). The opposite chromatin modification pattern was seen in permissive differentiated MEF cells, in which enrichment of H3Ac (Figure 4C), and not H3K9me3 (Figure 4B) marks on the viral DNA occurred at 6d, consistent with high MLV gene expression in these cells (Figure 4A). Histone H3K9me3, associated with gene silencing, is mediated by histone lysine methyltransferase ESET, which is recruited to target DNA via its interaction with TRIM28 (Schultz et al., 2002). To determine whether ESET is responsible for de novo deposition of H3K9me3, we generated F9 cells stably depleted of ESET (Figure 4D), exposed them to high titers of MLV-GFP virus and scored the fraction of cells expressing GFP at 1 and 3d (Figure 4E). In control knockdown cells, 4.5% of cells expressed GFP at 1d and <1% at 3d, whereas ESET knockdown cells showed higher transduction efficiency (14.5% expressing GFP at 1d and15.3% at 3d), reflecting loss of retroviral silencing as reported previously (Matsui et al., 2010). ESET depletion resulted in a 2-fold reduction in H3K9me3 marks on the viral LTR at both 24h and 3d (Figure 4F); the linear DNA showed the greatest loss of the H3K9me3 marks, while the circular forms were almost unaffected. ESET depletion also led to roughly a 6-fold increase in H3Ac marks (Figure 4G). Core histone H3 loading was unaffected in ESET knockdown cells at all times examined (Figure S3), further suggesting that core histone loading is independent of histone marking by ESET. Together, these data suggest that ESET is responsible for de novo deposition of H3K9me3 marks on the incoming viral DNA in mouse embryonic cells.
FIGURE 4. Loading of epigenetically modified histones onto viral DNAs is delayed compared to core histones.
(A) Flow cytometry analysis of F9 or MEF cells infected with VSV-G pseudotyped WT MLV-GFP viruses at the indicated times. SSC, side scatter. One representative of three independent experiments is shown.
(B) H3K9 trimethylation (H3K9me3) ChIP analysis of infected F9 and MEF cells at the indicated times. ChIP data is presented as % of input DNA (means ± SEMs from three independent experiments performed in duplicate). Mito gene, mitochondrial RNA polymerase (positive control), GAPDH (negative control). *, p < 0.05.
(C) Acetyl-histone H3 (H3Ac) ChIP analysis of infected F9 or MEF cells at the indicated times. ChIP data is presented as % of input DNA (means ± SEMs from three independent experiments performed in duplicate). Mito gene (negative control) and GAPDH (positive control) are shown. **, p < 0.01.
(D) Total cell lysates prepared from F9 control knockdown (KD) and F9 ESET KD cells were immunoblotted with antisera as indicated.
(E) Flow cytometry analysis of F9 control or ESET KD cells infected with VSV-G pseudotyped WT MLV-GFP viruses at the indicated times. Y-axis shows cell count, X-axis shows GFP intensity. One representative example of two independent experiments is shown.
(F and G) Similar experiments as (B and C), respectively, using MLV infected F9 control or ESET KD cells (means ± SEMs from three independent experiments performed in duplicate). **, p < 0.01.
For related data, see Figure S3.
Student’s t-test was used for statistical analysis. ND, not determined.
Expression of unintegrated MLV DNA in NIH-3T3 cells
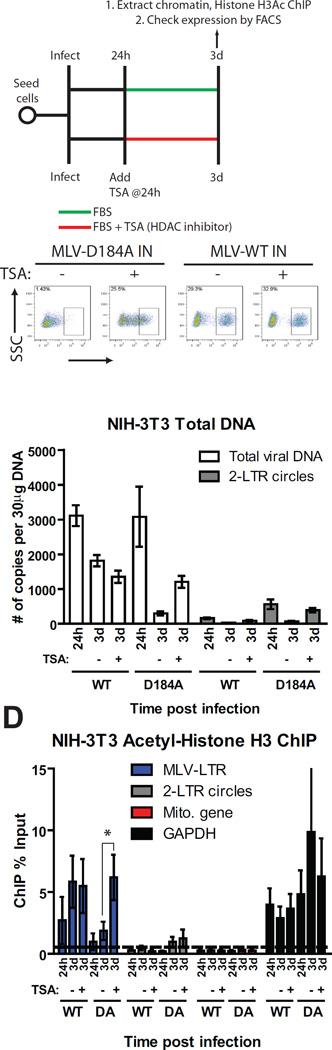
Our findings that unintegrated MLV genomes are rapidly covered with core histones (Figure 2D and 2H) together with the observation that unintegrated MLV genomes show poor gene expression in otherwise permissive cells (Figure 2B) raised the possibility that unintegrated MLV DNA lacks specific chromatin modifications such as acetylation of histone H3 and H4 that promote transcription (Grunstein, 1997; Hassig and Schreiber, 1997; Kuo and Allis, 1998). To test the possibility that unintegrated MLV DNA is poorly expressed because of a deficiency in histone acetylation, NIH-3T3 cells were infected with WT or D184A-IN viruses delivering a GFP reporter for 24h, followed by the addition of Trichostatin A (TSA), a histone deacetylase inhibitor (Figure 5A). Cells infected with D184A-IN virus showed an 18-fold increase in the percentage of GFP+ cells following TSA treatment and a 3-fold increase in the mean GFP fluorescence intensity (Figure 5B). In contrast, TSA addition only marginally increased GFP expression in cells infected with WT viruses (Figure 5B), suggesting that this drug preferentially affects expression from unintegrated viral DNAs.
FIGURE 5. Characterization of the expression of unintegrated MLV DNA in NIH-3T3 cells.
(A) Schematic of experimental setup.
(B) Flow cytometry analysis of NIH-3T3 cells infected with VSV-G pseudotyped WT or integrase defective mutant (D184A) MLV-GFP viruses with or without TSA. Results are from 2d post TSA treatment. SSC, side scatter. One representative of three independent experiments is shown.
(C) NIH-3T3 cells infected with VSV-G pseudotyped WT or integrase defective mutant (D184A) MLV-GFP virus with or without TSA. Total DNA was isolated at indicated times and levels of viral replication intermediates were determined (means ± SEMs from three independent experiments performed in duplicate).
(D) H3Ac ChIP analysis of infected NIH-3T3 cells with or without TSA. ChIP data is presented as % of input DNA (means ± SEMs from three independent experiments performed in duplicate). Student’s t-test was used for statistical analysis. *, p < 0.05. ND, not determined.
We next monitored the kinetics of formation of viral DNA replication intermediates in WT and D184A-IN infected cells with or without TSA using qRT-PCR (Figure 5C). In WT-infected cells, TSA did not affect the levels of total viral DNA or 2-LTR circles at 3d post infection. Surprisingly, TSA treatment of D184A-IN infected cells resulted in a 4-fold increase in total viral DNA and a 6-fold increase in 2-LTR circles at 3d, suggesting that the drug prolongs the half-life of unintegrated viral DNAs.
We then performed ChIP assays using acetylated histone H3 antibodies on cells infected with WT and D184A-IN viruses with and without TSA . Compared to mock treatment, TSA treatment of D184A-IN infected cells resulted in a 4-fold increase in H3 acetylation of total viral DNA, but remarkably not of the 2-LTR circles (Figure 5D). This ChIP data is expressed as a percent of input DNA, and the increase in H3 acetylation is in addition to the increased levels of viral DNA in TSA treated cells (Figure 5C). In contrast, TSA did not increase histone H3 acetylation in WT virus infected cells (Figure 5D), perhaps because integrated proviral DNA is already enriched in acetylated H3 chromatin. Together, these data suggest that gene expression from unintegrated DNAs is hampered by the lack of necessary chromatin modifications that promote RNA polymerase recruitment, and that the chromatin can be altered by inhibition of histone deacetylases. We cannot rule out the possibility that the increased gene expression from unintegrated DNAs results from indirect effects of TSA.
Kinetics of NC and CA dissociation from viral DNA following reverse transcription
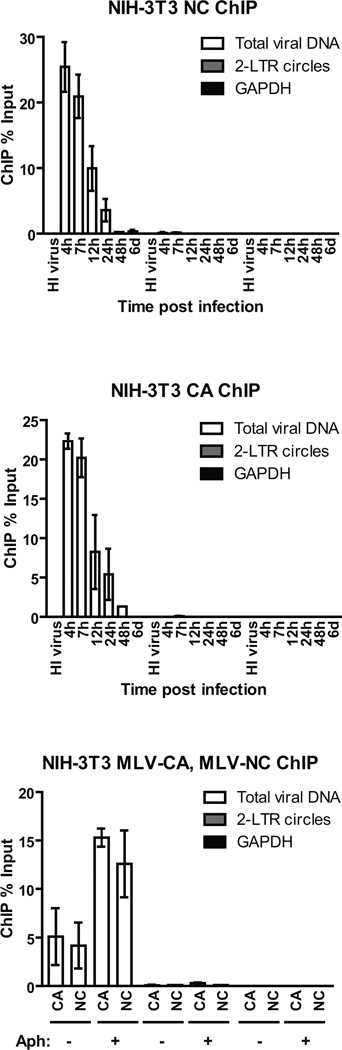
The NC proteins of retroviruses are small, highly basic nucleic acid-binding proteins that directly contact the RNA genome in the mature virion. NC is involved in many steps of viral replication including virion assembly, genomic RNA packaging, reverse transcription and integration (Darlix et al., 2014). The CA protein constitutes the major structural protein of the virion core and plays a role in early events such as nuclear entry and DNA integration. We examined the association of viral DNAs with NC and CA during the course of infection by ChIP assays with primers specific for total viral DNA (GFP) and 2-LTR circles (Figure 6A). The highest levels of viral DNA associated with NC were observed at 4h post infection, followed by a rapid decrease to very low levels by 48h (Figure 6A). Similar results were observed for CA (Figure 6B). Strikingly, no association of 2-LTR circles with NC or CA was detected at any time. The specificity of the antibodies used for ChIP was confirmed by the lack of non-specific binding to GAPDH. Cells infected with heat inactivated (HI) virus were free of viral DNA that would derive from contaminating plasmid DNAs (Figure 6A and 6B).
FIGURE 6. Characterization of the kinetics of NC and CA association with viral DNA following reverse transcription.
(A, B) ChIP analysis of chromatin harvested at indicated times following MLV-GFP infection of NIH-3T3 cells using (A) MLV-NC or (B) MLV-CA antibodies. For related data, see Figures S4 and S5.
(C) ChIP analysis of chromatin harvested from aphidicolin (Aph)-treated NIH-3T3 cells 24h post infection with MLV-NC and CA antibodies.
ChIP data is presented as % of input DNA (means ± SDs from two independent experiments performed in duplicate).
The timing of NC and CA dissociation from the total viral DNAs (Figure 6A and 6B) coincided with the rapid accumulation of core histones onto the DNA (Figure 1B and 1D), raising the possibility that core histones promote the dissociation of NC and CA from the viral DNAs by direct competition for DNA binding. If this were the case, preventing nuclear entry and histone loading might result in the stabilization of NC and CA-DNA complexes. Indeed, ChIP analysis of DNA from aphidicolin-treated NIH-3T3 cells showed a 3-fold increase in total viral DNAs associated with NC and CA (Figure 6C).
To address whether viral integration influences the kinetics of NC and CA dissociation from viral DNA, we carried out ChIP experiments in NIH-3T3 cells infected with WT or D184A-IN virus. NC and CA showed similar kinetics of dissociation from viral DNA in both cases, with highest association occurring at 12h post infection, followed by a rapid decrease to very low levels by 48h (Figure S4 and Figure S5). The data suggest that the kinetics of dissociation of NC and CA from the viral DNA are not affected by integration.
DISCUSSION
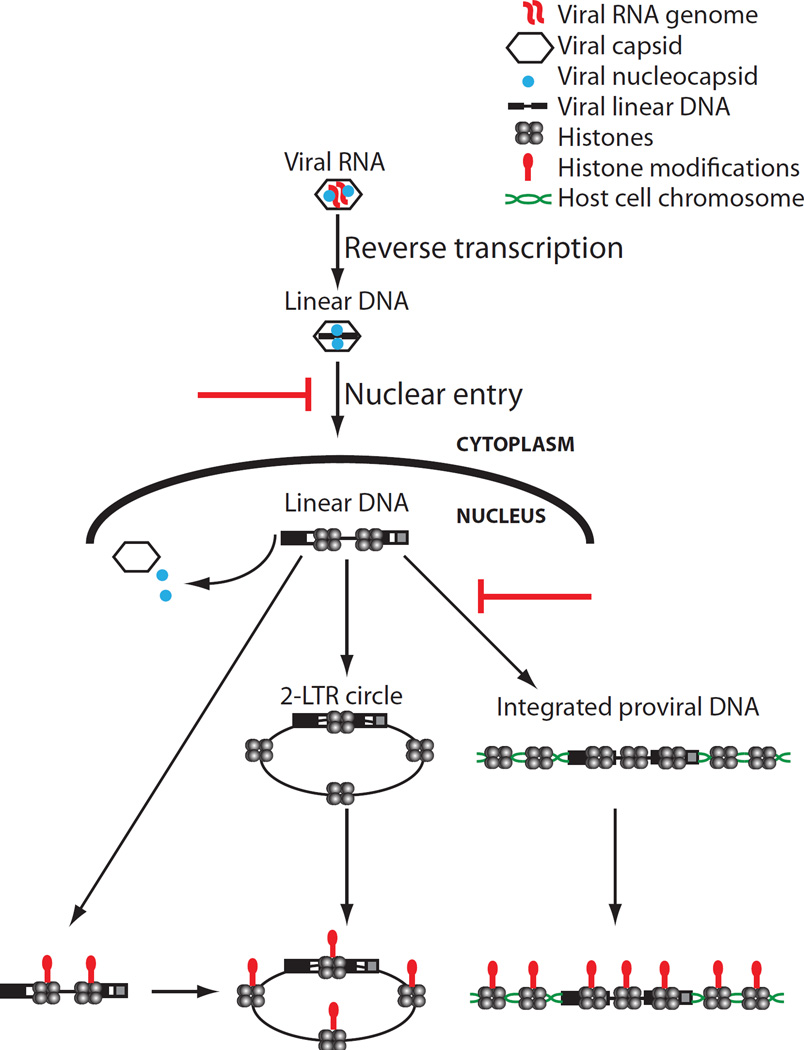
We find unintegrated viral DNAs, mainly in the form of linear DNA, rapidly associate with core histones after infection. 2-LTR circular DNAs were fully loaded with histones at all times, consistent with early observations that covalently closed circular retroviral DNAs migrate as supercoiled molecules in gel electrophoresis after deproteinization (Gianni et al., 1975). The data indicate that histones are first loaded onto the linear DNA, which is then circularized to form 2-LTR circles. The timing of loading during infections by WT virus suggests that loading occurs before viral DNA integration, and experiments using integrase-defective mutant MLV or raltegravir-treated cells demonstrate that histone loading on viral DNAs occurs even when viral DNA integration is blocked. Thus, the majority of the viral DNA in the preintegrative PIC at the time of integration is nucleosomal, and it is likely that nucleosome-covered DNA is the precursor to the provirus. Because integration is not highly efficient, we cannot rule out the remote possibility that a small fraction of viral DNA that is not histone-covered is responsible for forming some of the integrated proviruses.
The loading of histones onto viral DNA apparently requires nuclear entry. Blocking nuclear entry of viral DNA, either by infecting aphidicolin-arrested cells with MLV, or by infecting mouse cells with M-PMV, which is defective in nuclear entry in murine cells, caused a marked reduction in histone loading. We conclude that the majority, if not all of the histone loading onto viral DNAs occurs in the nucleus. Another laboratory has shown by cellular fractionation experiments that anti-histone antibodies could immunoprecipitate linear HIV-1 DNA from the cytoplasm of infected cells (Karageorgos et al., 1993). Our attempts to apply ChIP to cell lysates after separation into cytoplasmic and nuclear fractions were unsuccessful due to high backgrounds of nonspecific precipitation. We cannot rule out the possibility that a small extent of histone loading takes place in the cytoplasm.
We directly determined the timing of appearance of epigenetically modified histone association with viral DNAs. In the case of differentiated cells, where the MLV promoter is fully active, core histone loading was nearly complete at 12h, while acetylated histone H3 loading accumulated to higher levels at later times. In the case of F9 embryonic carcinoma cells, significant core histone loading again occurred as early as 12h, while H3K9me3 became evident at 6d. The delay in the appearance of modified histones on the viral DNAs correlates with the delay in transcriptional silencing of the MLV promoter. Together, these data suggest that the appearance of both active and repressive histone marks occurs after core histone loading, thereby causing a transient delay in the modulation of viral gene expression.
Expression from unintegrated viral DNAs was very low compared to integrated proviral DNAs, consistent with early studies of MLV (Schwartzberg et al., 1984) and HIV-1 (Nakajima et al., 2001; Poon and Chen, 2003; Sakai et al., 1993). However, the addition of the HDAC inhibitor TSA caused a marked increase in expression from unintegrated DNA, without causing a similar increase from integrated viral DNAs. These findings are in accord with previous studies of a variety of integration-defective vectors in a variety of cell lines (Joglekar et al., 2014; Pelascini et al., 2013; Schneider et al., 2012) and supports the notion that the unintegrated DNAs are functionally associated with nucleosomes. The increase in expression from unintegrated DNAs appears to involve two mechanisms. First, TSA caused an increase in the association of acetylated histone H3, a marker of actively transcribed genes, with the viral promoter. This increase in acetylated H3 is consistent with earlier reports, though one study observed much smaller increases in H3Ac and larger increases in a different mark, H3K4 trimethylation (Pelascini et al., 2013). Remarkably, we observed this H3Ac enrichment only on total viral DNA, but not 2-LTR circles, implying that the enhanced gene expression may be a product of increased transcription from only linear DNA templates and not circular forms. Secondly, TSA appears to increase the stability of unintegrated, but not integrated DNA forms, thereby increasing the amount of viral template available for transcription. How TSA stabilizes the viral DNA is not clear. One possibility is that histone H3 acetylation itself may somehow stabilize the viral DNA or protect it from degradation. Alternatively, TSA treatment may prolong the half-life of unintegrated DNA forms by reducing the activity of DNA degradation machinery in the nucleus. Other activities of the HDACs may also be involved. Previous reports suggested that a major mechanism by which HDACs increase expression is through modulation of the cell cycle (Joglekar et al., 2014).
The observation that TSA can significantly increase expression of unintegrated retroviral DNAs may have applications in gene therapy. Because of potentially oncogenic events associated with integration of viral DNA, many protocols have been designed to use nonintegrating vectors to deliver therapeutic genes into patients (Banasik and McCray, 2010; Philpott and Thrasher, 2007; Wanisch and Yanez-Munoz, 2009). Since the DNAs are not stably integrated, their use is limited to settings where transient expression is sufficient for a therapeutic outcome (Negri et al., 2010; Yanez-Munoz et al., 2006). These constructs typically suffer from very low levels of expression (Wanisch and Yanez-Munoz, 2009). The findings here confirm and provide mechanistic understanding for earlier work suggesting that the use of histone deacetylase inhibitors such as TSA, valproic acid, vorinostat, givinostat, and sodium butyrate increase expression and therefore the range of utility of nonintegrating retroviral vectors (Joglekar et al., 2014; Pelascini et al., 2013).
At early times after infection, high levels of viral DNA were associated with NC and CA. Retroviral NCs show high affinity for single-stranded RNA and DNA, but can also bind to double-stranded DNA with lower affinity (Priel et al., 1995; Wang et al., 2009) and promote DNA condensation (Krishnamoorthy et al., 2003), and so could be expected to persist on the DNA even after reverse transcription is complete. It has been suggested that NC is present on DNA in the nucleus (Risco et al., 1995) and plays a role in viral DNA integration (Carteau et al., 1999; Poljak et al., 2003). In contrast, purified CA does not show high affinity for nucleic acids (Gross et al., 1997), and its association with the viral DNA likely reflects its presence in the form of remnants of a cage-like structure that encapsidates the genome in the mature virion core (Ganser et al., 1999). At least some CA protein is still associated with retroviral DNAs in the nucleus (Hulme et al., 2015a), and CA can affect the timing and route of entry into the nucleus (Hulme et al., 2015b; Yamashita et al., 2007) and the distribution of proviral integration sites (Koh et al., 2013; Schaller et al., 2011). We observed that the levels of both NC and CA steadily dropped post infection, roughly corresponding to the time of nuclear entry. Excluding DNA from the nucleus by addition of aphidicolin led to increased levels of NC and CA on the viral DNA, consistent with their removal normally occurring upon nuclear entry (Figure 6C). Moreover, the circular DNAs never showed any detectable association with NC or CA, suggesting that at least the bulk of both viral proteins was removed before circularization. The timing of the loss of NC and CA also coincided with the time of histone loading, suggesting that the two events may be coordinated. One possible model is that core histones displace NC and CA from the DNA (Figure 7).
FIGURE 7.
Model of the states of retroviral DNA following infection.
The mechanism by which core histones are loaded onto retroviral DNAs remains unknown but may be similar to histone loading on naked DNAs introduced by transformation (Reeves et al., 1985). Studies of transfected DNAs by micrococcal nuclease digestion suggest that nucleosomes are loaded on non-replicating DNAs, but that the distribution of nucleosomes is not well-ordered (Jeong and Stein, 1994). Our ChIP studies do not reveal the density or distribution of the nucleosomes on the viral DNA, but simply report whether a nucleosome is crosslinked anywhere within the span of the DNA fragments produced by sonication, roughly 500 bp in length. Whether histone chaperones such as CAF1, which mediates assembly of nucleosomes on chromosomal DNA during DNA replication and repair (Annunziato, 2013), are involved in loading on retroviral DNA is not known. Cells stably depleted of CAF1 complex subunits a and b, or NAP1L and HIRA, by shRNA did not show reduced histone H3 loading onto viral DNAs (Figure S6). It is possible that these chaperones are functionally redundant, or that other known or unknown chaperones are involved.
Experimental Procedures
Detailed methods are available in Supplemental Experimental Procedures.
Cell lines
Cell lines for these experiments included human embryonic kidney (HEK)-293, F9 embryonic carcinoma, Mouse embryonic fibroblasts (MEF) and NIH-3T3 cells. All cells were cultured in Dulbecco’s modified Eagle’s Medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM glutamine, 1000 U/mL penicillin and 100 mg/mL streptomycin.
Plasmids
Plasmid pCMV-intron expresses wild-type Gag and Pol from NB-tropic MLV (Soneoka et al., 1995). pCMV-intron D184A-IN is an integrase defective mutant of pCMV-intron created by site directed mutagenesis. pMD.G expresses the vesicular stomatitis virus (VSV) envelope glycoprotein. pNCA-GFP is a replication-defective single-round MLV vector described previously (Ooi et al., 2010). pSARM-EGFP is a replication-defective single-round M-PMV vector in which the env gene has been replaced with EGFP as described (Newman et al., 2006).
Retroviral transduction assay and flow cytometry analysis
MLV-GFP reporter viruses were produced by 293T cell transfection with 8 µg of pNCA-GFP, 4 µg of pCMV-intron, and 4 µg of pMD.G DNAs using Polyethylenimine (PEI). Similarly, M-PMV-GFP reporter viruses were produced by transfection of 293T cells with 12 µg of pSARM-EGFP and 4 µg of pMD.G. All reporter viruses were harvested 48 h later, filtered (0.45 μm), DNase treated for 1 h (Turbo DNase, Thermo Scientific), and used directly for transduction assays as described previously (Wang and Goff, 2015). For some experiments, 10 µM of raltegravir (Santa Cruz Biotechnology) was added to the culture medium for the entire duration of infection. Forty-eight hours post-infection, cells were trypsinized, diluted using flow cytometry buffer (PBS with 1% BSA), and subjected to flow cytometry using automated cell analyzer (LSRII, BD Bioscience).
Chromatin immunoprecipitation
Cells were crosslinked with 1% formaldehyde, followed by quenching with 0.125 M glycine for 5 min, lysed, and sonicated to produce an average DNA fragment size of 200–800 bp. Immunoprecipitations were performed by incubating 30 µg of sonicated chromatin with 4 µg of respective antibodies overnight, and Protein A/G Dynabeads were added for an additional 4 h. Captured antibody-antigen complexes were washed repeatedly, and DNA was eluted from the beads. qPCRs were performed with specific primers.
Quantitative real-time PCR analysis of viral replication intermediates
At various time points following infection, cells were washed with PBS, and total DNA was isolated using Qiagen DNeasy kit according to the manufacturer’s instructions. For analysis of viral replication intermediates, 75 ng of total DNA was mixed with SYBR Green PCR master mix (Roche) containing 15 pmol of indicated primers (see Supplementary Table 1). PCRs were performed in 96-well plates using 7900 Fast Real-Time PCR system (Applied Biosystems) with the following reaction conditions: 10 min at 95°C, followed by 45 cycles of 30 s at 95°C, 30 s at 60°C and 30 s at 72°C. For all reactions, the number of copies of each PCR product was determined from threshold cycle number (Ct) using plasmid derived standard curves.
Aphidicolin treatment and cell cycle analysis of DNA content
NIH-3T3 cells were treated with aphidicolin (Sigma, 2 μg/ml) for 24 h prior to infection. Efficiency of aphidicolin treatment was confirmed by analyzing DNA content. Cells were stained with Hoechst 33342 (Invitrogen) at 1 μg/ml in cell culture medium for 30 min at 37°C, followed by flow cytometry using automated cell analyzer (LSRII, BD Bioscience).
Supplementary Material
Acknowledgments
This work was supported by NCI grant R01 CA 30488 from the National Cancer Institute. S.P.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental information includes Supplementary Experimental Procedures, six figures, one table and can be found with this article online.
AUTHOR CONTRIBUTIONS
GZW and SPG designed and supervised the study. GZW and YW conducted the experiments. GZW and SPG analyzed the data and wrote the manuscript.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
REFERENCES
- Akgun E, Ziegler M, Grez M. Determinants of retrovirus gene expression in embryonal carcinoma cells. J Virol. 1991;65:382–388. doi: 10.1128/jvi.65.1.382-388.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziato AT. Assembling chromatin: the long and winding road. Biochim Biophys Acta. 2013;1819:196–210. doi: 10.1016/j.bbagrm.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Banasik MB, McCray PB., Jr Integrase-defective lentiviral vectors: progress and applications. Gene Ther. 2010;17:150–157. doi: 10.1038/gt.2009.135. [DOI] [PubMed] [Google Scholar]
- Beck-Engeser GB, Eilat D, Harrer T, Jack HM, Wabl M. Early onset of autoimmune disease by the retroviral integrase inhibitor raltegravir. Proc Natl Acad Sci U S A. 2009;106:20865–20870. doi: 10.1073/pnas.0908074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PO, Bowerman B, Varmus HE, Bishop JM. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci U S A. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess RJ, Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nat Struct Mol Biol. 2013;20:14–22. doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos EI, Stafford JM, Reinberg D. Epigenetic inheritance: histone bookmarks across generations. Trends Cell Biol. 2014;24:664–674. doi: 10.1016/j.tcb.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carteau S, Gorelick RJ, Bushman FD. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J Virol. 1999;73:6670–6679. doi: 10.1128/jvi.73.8.6670-6679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlix JL, de Rocquigny H, Mauffret O, Mely Y. Retrospective on the all-in-one retroviral nucleocapsid protein. Virus Res. 2014;193:2–15. doi: 10.1016/j.virusres.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassati A, Goff SP. Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J Virol. 1999;73:8919–8925. doi: 10.1128/jvi.73.11.8919-8925.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- Gianni AM, Smotkin D, Weinberg RA. Murine leukemia virus: detection of unintegrated double-stranded DNA forms of the provirus. Proc Natl Acad Sci U S A. 1975;72:447–451. doi: 10.1073/pnas.72.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross I, Hohenberg H, Krausslich HG. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur J Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Hassig CA, Schreiber SL. Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. Curr Opin Chem Biol. 1997;1:300–308. doi: 10.1016/s1367-5931(97)80066-x. [DOI] [PubMed] [Google Scholar]
- Hulme AE, Kelley Z, Foley D, Hope TJ. Complementary Assays Reveal a Low Level of CA Associated with Viral Complexes in the Nuclei of HIV-1-Infected Cells. J Virol. 2015a;89:5350–5361. doi: 10.1128/JVI.00476-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulme AE, Kelley Z, Okocha EA, Hope TJ. Identification of capsid mutations that alter the rate of HIV-1 uncoating in infected cells. J Virol. 2015b;89:643–651. doi: 10.1128/JVI.03043-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Stein A. Micrococcal nuclease digestion of nuclei reveals extended nucleosome ladders having anomalous DNA lengths for chromatin assembled on non-replicating plasmids in transfected cells. Nucleic Acids Res. 1994;22:370–375. doi: 10.1093/nar/22.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AV, Stein L, Ho M, Hoban MD, Hollis RP, Kohn DB. Dissecting the mechanism of histone deacetylase inhibitors to enhance the activity of zinc finger nucleases delivered by integrase-defective lentiviral vectors. Hum Gene Ther. 2014;25:599–608. doi: 10.1089/hum.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karageorgos L, Li P, Burrell C. Characterization of HIV replication complexes early after cell-to-cell infection. AIDS Res Hum Retroviruses. 1993;9:817–823. doi: 10.1089/aid.1993.9.817. [DOI] [PubMed] [Google Scholar]
- Kilzer JM, Stracker T, Beitzel B, Meek K, Weitzman M, Bushman FD. Roles of host cell factors in circularization of retroviral dna. Virology. 2003;314:460–467. doi: 10.1016/s0042-6822(03)00455-0. [DOI] [PubMed] [Google Scholar]
- Koh Y, Wu X, Ferris AL, Matreyek KA, Smith SJ, Lee K, KewalRamani VN, Hughes SH, Engelman A. Differential effects of human immunodeficiency virus type 1 capsid and cellular factors nucleoporin 153 and LEDGF/p75 on the efficiency and specificity of viral DNA integration. J Virol. 2013;87:648–658. doi: 10.1128/JVI.01148-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy G, Roques B, Darlix JL, Mely Y. DNA condensation by the nucleocapsid protein of HIV-1: a mechanism ensuring DNA protection. Nucleic Acids Res. 2003;31:5425–5432. doi: 10.1093/nar/gkg738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Lai L, Liu H, Wu X, Kappes JC. Moloney murine leukemia virus integrase protein augments viral DNA synthesis in infected cells. J Virol. 2001;75:11365–11372. doi: 10.1128/JVI.75.23.11365-11372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P, Hensel M, Emerman M. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 1992;11:3053–3058. doi: 10.1002/j.1460-2075.1992.tb05376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PF, Emerman M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J Virol. 1994;68:510–516. doi: 10.1128/jvi.68.1.510-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Olvera JM, Yoder KE, Mitchell RS, Butler SL, Lieber M, Martin SL, Bushman FD. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 2001;20:3272–3281. doi: 10.1093/emboj/20.12.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini R, Wellinger RE, Sogo JM. Nucleosome positioning at the replication fork. EMBO J. 2001;20:7294–7302. doi: 10.1093/emboj/20.24.7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- Mello JA, Almouzni G. The ins and outs of nucleosome assembly. Curr Opin Genet Dev. 2001;11:136–141. doi: 10.1016/s0959-437x(00)00170-2. [DOI] [PubMed] [Google Scholar]
- Nakajima N, Lu R, Engelman A. Human immunodeficiency virus type 1 replication in the absence of integrase-mediated dna recombination: definition of permissive and nonpermissive T-cell lines. J Virol. 2001;75:7944–7955. doi: 10.1128/JVI.75.17.7944-7955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri DR, Michelini Z, Cara A. Toward integrase defective lentiviral vectors for genetic immunization. Curr HIV Res. 2010;8:274–281. doi: 10.2174/157016210791208622. [DOI] [PubMed] [Google Scholar]
- Newman RM, Hall L, Connole M, Chen GL, Sato S, Yuste E, Diehl W, Hunter E, Kaur A, Miller GM, et al. Balancing selection and the evolution of functional polymorphism in Old World monkey TRIM5alpha. Proc Natl Acad Sci U S A. 2006;103:19134–19139. doi: 10.1073/pnas.0605838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Wolf D, Hartung O, Agarwal S, Daley GQ, Goff SP, Bestor TH. Dynamic instability of genomic methylation patterns in pluripotent stem cells. Epigenetics Chromatin. 2010;3:17. doi: 10.1186/1756-8935-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelascini LP, Janssen JM, Goncalves MA. Histone deacetylase inhibition activates transgene expression from integration-defective lentiviral vectors in dividing and non-dividing cells. Hum Gene Ther. 2013;24:78–96. doi: 10.1089/hum.2012.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott NJ, Thrasher AJ. Use of nonintegrating lentiviral vectors for gene therapy. Hum Gene Ther. 2007;18:483–489. doi: 10.1089/hum.2007.013. [DOI] [PubMed] [Google Scholar]
- Poljak L, Batson SM, Ficheux D, Roques BP, Darlix JL, Kas E. Analysis of NCp7-dependent activation of HIV-1 cDNA integration and its conservation among retroviral nucleocapsid proteins. J Mol Biol. 2003;329:411–421. doi: 10.1016/s0022-2836(03)00472-8. [DOI] [PubMed] [Google Scholar]
- Polo SE, Roche D, Almouzni G. New histone incorporation marks sites of UV repair in human cells. Cell. 2006;127:481–493. doi: 10.1016/j.cell.2006.08.049. [DOI] [PubMed] [Google Scholar]
- Poon B, Chen IS. Human immunodeficiency virus type 1 (HIV-1) Vpr enhances expression from unintegrated HIV-1 DNA. J Virol. 2003;77:3962–3972. doi: 10.1128/JVI.77.7.3962-3972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priel E, Aflalo E, Seri I, Henderson LE, Arthur LO, Aboud M, Segal S, Blair DG. DNA binding properties of the zinc-bound and zinc-free HIV nucleocapsid protein: supercoiled DNA unwinding and DNA-protein cleavable complex formation. FEBS Lett. 1995;362:59–64. doi: 10.1016/0014-5793(95)00208-q. [DOI] [PubMed] [Google Scholar]
- Ramachandran S, Henikoff S. Replicating Nucleosomes. Sci Adv. 2015:1. doi: 10.1126/sciadv.1500587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R, Gorman CM, Howard B. Minichromosome assembly of non-integrated plasmid DNA transfected into mammalian cells. Nucleic Acids Res. 1985;13:3599–3615. doi: 10.1093/nar/13.10.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risco C, Menendez-Arias L, Copeland TD, Pinto da Silva P, Oroszlan S. Intracellular transport of the murine leukemia virus during acute infection of NIH 3T3 cells: nuclear import of nucleocapsid protein and integrase. J Cell Sci. 1995;108(Pt 9):3039–3050. doi: 10.1242/jcs.108.9.3039. [DOI] [PubMed] [Google Scholar]
- Roe T, Reynolds TC, Yu G, Brown PO. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Sakai H, Kawamura M, Sakuragi J, Sakuragi S, Shibata R, Ishimoto A, Ono N, Ueda S, Adachi A. Integration is essential for efficient gene expression of human immunodeficiency virus type 1. J Virol. 1993;67:1169–1174. doi: 10.1128/jvi.67.3.1169-1174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller T, Ocwieja KE, Rasaiyaah J, Price AJ, Brady TL, Roth SL, Hue S, Fletcher AJ, Lee K, KewalRamani VN, et al. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog. 2011;7:e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger S, Goff SP. Silencing of proviruses in embryonic cells: efficiency, stability and chromatin modifications. EMBO Rep. 2013;14:73–79. doi: 10.1038/embor.2012.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger S, Lee AH, Wang GZ, Green L, Goff SP. Proviral silencing in embryonic cells is regulated by Yin Yang 1. Cell Rep. 2013;4:50–58. doi: 10.1016/j.celrep.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider WM, Wu DT, Amin V, Aiyer S, Roth MJ. MuLV IN mutants responsive to HDAC inhibitors enhance transcription from unintegrated retroviral DNA. Virology. 2012;426:188–196. doi: 10.1016/j.virol.2012.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzberg P, Colicelli J, Goff SP. Construction and analysis of deletion mutations in the pol gene of Moloney murine leukemia virus: a new viral function required for productive infection. Cell. 1984;37:1043–1052. doi: 10.1016/0092-8674(84)90439-2. [DOI] [PubMed] [Google Scholar]
- Shoemaker C, Goff S, Gilboa E, Paskind M, Mitra SW, Baltimore D. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: implications for retrovirus integration. Proc Natl Acad Sci U S A. 1980;77:3932–3936. doi: 10.1073/pnas.77.7.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneoka Y, Cannon PM, Ramsdale EE, Griffiths JC, Romano G, Kingsman SM, Kingsman AJ. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teich NM, Weiss RA, Martin GR, Lowy DR. Virus infection of murine teratocarcinoma stem cell lines. Cell. 1977;12:973–982. doi: 10.1016/0092-8674(77)90162-3. [DOI] [PubMed] [Google Scholar]
- Telesnitsky A, Goff SP. Reverse Transcriptase and the Generation of Retroviral DNA. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor (NY); 1997. [PubMed] [Google Scholar]
- Wang GZ, Goff SP. Postentry restriction of Mason-Pfizer monkey virus in mouse cells. J Virol. 2015;89:2813–2819. doi: 10.1128/JVI.03051-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GZ, Wolf D, Goff SP. EBP1, a novel host factor involved in primer binding site-dependent restriction of moloney murine leukemia virus in embryonic cells. J Virol. 2014;88:1825–1829. doi: 10.1128/JVI.02578-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yeh YS, Barbara PF. HIV-1 nucleocapsid protein bends double-stranded nucleic acids. J Am Chem Soc. 2009;131:15534–15543. doi: 10.1021/ja9070046. [DOI] [PubMed] [Google Scholar]
- Wanisch K, Yanez-Munoz RJ. Integration-deficient lentiviral vectors: a slow coming of age. Mol Ther. 2009;17:1316–1332. doi: 10.1038/mt.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg JB, Matthews TJ, Cullen BR, Malim MH. Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferating human monocytes. J Exp Med. 1991;174:1477–1482. doi: 10.1084/jem.174.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Perez O, Hope TJ, Emerman M. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog. 2007;3:1502–1510. doi: 10.1371/journal.ppat.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez-Munoz RJ, Balaggan KS, MacNeil A, Howe SJ, Schmidt M, Smith AJ, Buch P, MacLaren RE, Anderson PN, Barker SE, et al. Effective gene therapy with nonintegrating lentiviral vectors. Nat Med. 2006;12:348–353. doi: 10.1038/nm1365. [DOI] [PubMed] [Google Scholar]
- Yoshimura FK, Weinberg RA. Restriction endonuclease cleavage of linear and closed circular murine leukemia viral DNAs: discovery of a smaller circular form. Cell. 1979;16:323–332. doi: 10.1016/0092-8674(79)90009-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.