Abstract
We aimed to identify new candidate genes potentially involved in early onset Alzheimer’s disease (EOAD). Exome sequencing was conducted on 45 EOAD patients with either a family history of Alzheimer’s disease (AD, <65 years) or an extremely early age at onset (≤55 years) followed by multiple variant filtering according to different modes of inheritance. We identified 29 candidate genes potentially involved in EOAD, of which the gene TYROBP, previously implicated in AD, was selected for genetic and functional follow-up. Using three patient cohorts, we observed rare coding TYROBP variants in 9 out of 1110 EOAD patients, whereas no such variants were detected in 1826 controls (p-value=0.0001), suggesting that at least some rare TYROBP variants might contribute to EOAD risk. Overexpression of the p.D50_L51ins14 TYROBP mutant led to a profound reduction of TREM2 expression, a well-established risk factor for AD. This is the first study supporting a role for genetic variation in TYROBP in EOAD, with in vitro support for a functional effect of the p.D50_L51ins14 TYROBP mutation on TREM2 expression.
Keywords: Alzheimer’s disease, TYROBP, TREM2, exome sequencing, burden test
1. Introduction
Alzheimer’s disease (AD) is the most common cause of dementia affecting over 4 million individuals in the United States. A minority of patients have early-onset AD (EOAD, arbitrarily defined as an age at disease onset before 65 years of age) and a subset of those patients carry disease-causing mutations or duplications in one of three genes: APP (MIM #104760), PSEN1 (MIM #104311) and PSEN2 (MIM #606889). In the last few years, the use of exome sequencing has provided additional insights into the genetics of AD, identifying rare variants in SORL1 (MIM #602005), NOTCH3 (MIM #600276), TTC3 (MIM #602259), PLD3 (MIM #615698) and TREM2 (MIM #605086) as potentially involved in the pathophysiology (Cruchaga, et al., 2014, Guerreiro, et al., 2013, Guerreiro, et al., 2012, Jonsson, et al., 2013, Kohli, et al., 2016, Pottier, et al., 2012). A large proportion of EOAD, however, remains unexplained due to paucity of large pedigrees for genetic screening. Although approximately 50% of EOAD patients are considered to be sporadic, we hypothesized that either variants compatible with autosomal recessive transmission or autosomal dominant transmission with reduced penetrance could be responsible for apparently sporadic EOAD patients. In order to identify candidate EOAD genes, we exome sequenced 45 EOAD patients from our Mayo Clinic EOAD cohort and applied bioinformatics filtering strategies using multiple modes of inheritance. Among the identified candidate genes, we chose to follow-up TYROBP because of its already reported upregulation in AD brains and its significant role in the pathogenesis of late-onset AD (LOAD) (Ma, et al., 2015, Zhang, et al., 2013). Moreover, TYROBP is the binding partner of TREM2 (Bouchon, et al., 2001), a well-established genetic risk factor for AD (MIM #107741) (Benitez, et al., 2013, Guerreiro, et al., 2013, Jin, et al., 2014, Jonsson, et al., 2013, Neumann and Daly, 2013, Pottier, et al., 2013, Ruiz, et al., 2014).
2. Materials and methods
2.1. Participants
Our primary patient series includes 597 EOAD patients which were ascertained from 1997 to 2015 at Mayo Clinic Jacksonville, Mayo Clinic Rochester, and the Mayo Clinic Brain Bank (Table 1). Patients were arbitrarily defined as EOAD if patients presented with a clinical diagnosis of AD at or before 65 years of age, or if patients were pathologically confirmed with AD at autopsy with an age at death before 70 years (irrespective of the onset age). Routine genetic screening of the known genes by Sanger sequencing in this EOAD cohort was performed and revealed 35 pathogenic PSEN1 mutations, 8 APP mutations and duplications, 3 PSEN2 mutations, 5 MAPT mutations, 2 PGRN mutations and 3 C9ORF72 repeat expansions (Wojtas, et al., 2012). A total of 1036 white non-Hispanic healthy subjects of similar age and gender ascertained at Mayo Clinic Jacksonville were used as a control group (Table 1).
Table 1.
Mayo Clinic EOAD and control cohort characteristics
| Mean age at onset in years |
Mean age at death in years |
Mean age last visit in years |
% females |
% White non- Hispanic |
% Hispanic |
% Black or African American |
% Asian |
% Mutation carriers |
|
|---|---|---|---|---|---|---|---|---|---|
| (range) | (range) | (range) | (n) | (n) | (n) | (n) | (n) | (n) | |
| EOAD patients (n=597) |
55.93 (25–67) |
63.95 (36–79) |
NA | 49.4 (295) |
92.8 (554) |
2.7 (16) |
4.0 (24) |
0.5 (3) |
8.7 (52) |
| Controls (n=1036) |
NA | NA | 75 (27–99) |
53.0 (549) |
100.0 (1036) |
NA | NA | NA | NA |
Key: EOAD, Early Onset Alzheimer’s Disease; n, number; NA, Not Applicable
Additional EOAD patients (n=111), negative for pathogenic mutations in APP, PSEN1 and PSEN2 genes, were recruited in the Charles F. and Joanne Knight Alzheimer’s Disease Research Center (ADRC) from Washington University, St Louis, USA (Supplementary Table 1). Additionally, 292 healthy individuals ascertained at Charles F. and Joanne Knight ADRC were used as control group (Supplementary Table 1). Finally, a cohort of French EOAD patients (n=484), negative for pathogenic mutations in APP, PSEN1 and PSEN2 genes, were recruited over a 20-year period by the French National CNR-MAJ consortium. For comparison, a total of 498 controls were recruited in 5 different French cities and were all of French ancestry (Supplementary Table 1).
All patients were diagnosed according to the NINCDS-ADRDA (National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s Disease and Related Disorders Association) criteria (McKhann, et al., 2011). A comprehensive clinical examination including personal medical and family history assessment and neurological examination was performed on each patient. Patients were considered as having a positive family history if at least one first or second degree relative presented with a neurodegenerative disorder.
2.2. Standard protocol approvals, registrations, and patient consents
Written informed consent was obtained from patients and controls individuals for genetic analyses at all sites. Each study site obtained approval from the local ethics committee or institutional research board.
2.3. Exome sequencing analysis
Exome sequencing of 23 familial EOAD patients was performed as previously described (Guerreiro, et al., 2013). Exomes of the 22 sporadic EOAD patients were captured using the Agilent SureSelect Human All Exon V5 plus UTR kit. Sequencing was performed on an Illumina Genome HiSeq 2000 by 101-base paired-end reads. Alignment of sequence reads was performed against the Human Reference Genome build GRCh37. Variants were called using default settings from the GATK tool and following the best practices (McKenna, et al., 2010). Low-quality variants that did not pass the Variant Quality Score Recalibration (VQSR) filter (DePristo, et al., 2011) were removed. Genetic data from the two pilot studies were combined and analyzed using two independent pipelines in order to maximize the chance of detecting potentially pathogenic variants. Specifically, filtering of variants was performed using the GEM.app application developed at the University of Miami (Gonzalez, et al., 2013) and using BioR developed at Mayo Clinic (Kocher, et al., 2014). All variants identified by at least one analysis pipeline were confirmed by Sanger sequencing.
2.4. Mutational analysis
For sequence validation, specific primers were designed surrounding each rare variant detected by Exome sequencing (Supplementary Table 2). In addition, all exonic regions of TYROBP were amplified by PCR using specific primers (sequences available upon request). Prior to sequencing, PCR products were purified using the AMPure system (Beckman Coulter Genomics, Brea, CA, USA). Sequence reactions were purified with CleanSEQ (Beckman Coulter Genomics, Brea, CA, USA) and then Sanger sequenced on an ABI3730xl Genetic Analyzer. Sequences were analyzed using Sequencher 4.8 software (Gene Codes Corporation, Ann Arbor, MI, USA).
2.5. Phase determination of p.G2E and p.V55L variants in TYROBP
Cerebellar RNA was extracted from patient A and a healthy subject using the RNeasy Plus Mini Kit (Qiagen, Hilden, Germany) and quality was assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), the RNA integrity number value obtained were above 8. cDNA was generated from 250ng of RNA from patient A and a healthy subject using the SuperScript® III First Strand Synthesis System kit (Invitrogen™, Carlsbad, CA, USA). A PCR was performed on the cDNA using a PCR primer set with a reverse primer specific for the mutant T allele of the p.V55L variant (NM_003332.3:c.163G>T) (primers sequences available upon request) allowing us to Sanger sequence the nucleotides surrounding the p.G2E variant.
2.6. Cell culture transfection, mRNA expression and western blotting
HeLa cells were transiently co-transfected with: 1µg of human TREM2-cMyc and 1µg of either TYROBP-eGFP wild-type (TYROBP WT) or TYROBP-D50_L51ins14-eGFP mutant (TYROBP D50_L51ins14) plasmids. Control transfections were performed using either a co-transfection with human TREM2-cMyc and peGFP plasmids, or using a single transfection with 2µg of peGFP plasmid (Supplementary Material). For mRNA expression analysis, transfected HeLa cells were harvested and total RNA was extracted using the RNeasy Plus Mini Kit (Qiagen, Hilden, Germany). Reverse transcription reaction was then performed using the Superscript III system (Life Technologies, Carlsbad, CA, USA). Real-time quantitative PCR using an ABI7900 was performed in triplicate for each sample using gene expression probes for TYROBP (Hs00182426_m1), TREM2 (Hs00219132_m1), eGFP (Mr04097229_mr), and GAPDH (Hs02758991_g1). Results were analyzed using SDS software version 2.2 (Life Technologies, Carlsbad, CA, USA) and quantifications of TYROBP and TREM2 mRNA were first normalized to GAPDH levels and were determined relative to TYROBP WT transfected cells (Supplementary Material).
For western blotting, cells were harvested using radioimmunoprecipitation assay buffer (RIPA) (Boston Bioproducts, Ashland, MA). Equal volumes of cell lysate were run on 10–20% acrylamide tris-glycine gels (Invitrogen, Carlsbad, CA, USA). After transfer, total TYROBP levels were detected using an eGFP antibody (MAB3580; Millipore, Darmstadt, Germany). TREM2 levels were assessed using a cMyc antibody (9E10; Roche, Penzberg, Germany). A GAPDH antibody was used as a protein loading control to visualize protein loading (Meridian Life Science, Memphis, ME, USA). Quantitative values from each blot were obtained using ImageJ 1.44 (National Institutes of Health, Bathesda, MD, USA). Numerical values of optical intensity were obtained by measuring the average pixel intensity per area for each band. Band intensity was then normalized to the total amount of protein using GAPDH. Protein expression level was assessed relative to the TYROBP WT condition (Supplementary Material).
2.7. Statistical analyses
Statistical analyses were performed using the Combined and Multivariate Collapsing test (CMC) using 100,000 permutation and the R software version 3.1.2 (2014-10-3, Vienna, Austria) (Li and Leal, 2008). Rare variants (minor allele frequency < 1%) affecting the TYROBP protein sequence were considered. CMC test collapses all rare variants and compares the proportions of variant carriers among cases and controls. No adjustment for age and gender were performed.
Statistical analyses for the western blot quantifications and the mRNA experiments were performed using a one-way ANOVA followed by Student t-tests for TREM2 and TYROBP levels using GraphPad Prism software (version 6.07 for Windows, GraphPad Software, La Jolla, CA, USA).
3. Results
3.1. Identification of potential candidate genes by exome sequencing
We performed two independent exome sequencing pilot studies on 45 white non-Hispanic EOAD patients selected from the overall Mayo Clinic cohort negative for mutations in the known genes: 23 patients with at least one affected relative with AD (mean age at onset: 56.7 years), and 22 patients with extreme young onset of EOAD (age at onset at or before 55, mean age at onset: 52.0 years) with no reported family history. Three different bioinformatic filtering strategies were subsequently employed to select candidate EOAD genes from our pilot sequencing dataset of 45 exomes (Table 2). First, we assessed our entire patient cohort (n=45) and selected heterozygous rare coding variants with a minor allele frequency (MAF) <0.1% in the NHLBI GO Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu/EVS/) based on the European cohort. Only variants affecting the protein sequence and shared between at least two individuals were selected (autosomal dominant model). Additionally, variants could not be present in more than 2 families in the GEM.app database. Using this strategy we were able to detect and validate 15 variants in 15 candidate genes (Table 2; Supplementary Table 2). Second, we focused on sporadic EOAD patients (n=22) and a potential recessive mode of inheritance. We selected all homozygous variants with a less stringent MAF of less than 5% in ESP European cohort that affected protein sequence, which resulted in the identification of 13 variants in 13 candidate genes (Table 2; Supplementary Table 2, homozygous model). Finally, in the same cohort of sporadic EOAD patients (n=22), we searched for compound heterozygous variants with a MAF<5% in the ESP European cohort that affected the protein sequences and were present in at least two individuals. The variants identified were filtered out if present at the homozygous state in ExAC European population. This strategy revealed 4 validated variants in 2 candidate genes (Table 2, Supplementary Table 2, compound heterozygous model). Of note, variants were considered as compound heterozygous when at least two variants were found in the same gene.
Table 2.
Strategies used to identify candidate EOAD genes in exome sequencing data
| Strategy | Autosomal Dominant | Autosomal Recessive | |
|---|---|---|---|
| Coding changes | Missense/Stop-gain/Stop- loss/Splice/Indels/Start loss and gain |
Missense/Stop-gain/Stop- loss/Splice/Indels/Start loss and gain |
Missense/Stop-gain/Stop- loss/Splice/Indels/Start loss and gain |
|
Number of EOAD patients |
n=45 | n=22 | n=22 |
| Variant types | Heterozygous variants | Homozygous variants | Compound heterozygous variants |
|
ESP European American MAF* |
<0.1% | <5% | <5% |
|
HapMap European MAF* |
<0.1% | <5% | <5% |
| Additional filters | Same variant in two unrelated individuals |
No additional filter | Same variant in at least two unrelated individuals |
| No more than 2 families in the GEM.app** database with the same variant |
No homozygotes in ExAC European*** |
||
|
Number of candidate genes |
15 | 13 | 2 |
Key: EOAD, Early Onset Alzheimer’s Disease; n, number of patients included
Minor allele frequency (MAF) from the Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu/EVS/)
Filtering was done on allele count from the GEnomes Management Application (GEM.app) database, regrouping exome sequencing from 2200 families (September 2014).
Filtering was done on Broad Institute Exome Aggregation Consortium (ExAC) genotypes.
3.2. Evaluation of TYROBP rare variants in the etiology of EOAD
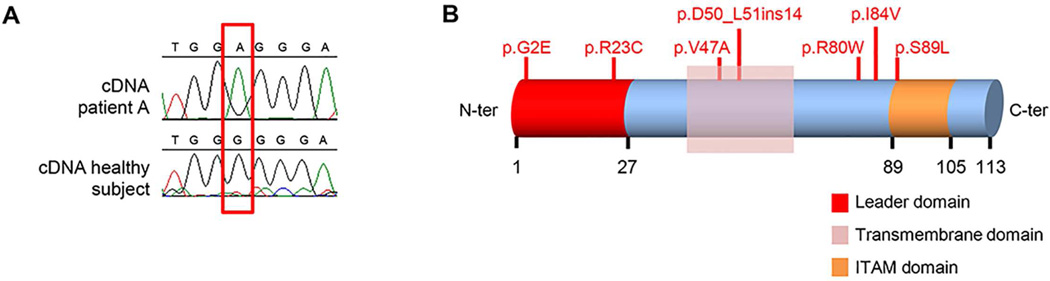
Interestingly, the gene encoding TYROBP (MIM #604142) was identified using both the autosomal dominant and the compound heterozygote disease strategy, due to presence of variants c.5G>A (p.G2E) and c.163G>T (p.V55L) in two unrelated sporadic EOAD patients (patient A and patient B). Cerebellar brain tissue, available for patient A, allowed the extraction of mRNA and subsequent cDNA analysis, which demonstrated that both variants were inherited in cis, arguing in favor of a possible autosomal dominant mode of action for TYROBP variants in EOAD (Fig. 1A; Supplementary Material). Given the strong functional link between TYROBP and AD, we expanded our genetic analysis of rare coding TYROBP variants in EOAD. We sequenced TYROBP in the entire EOAD Mayo Clinic cohort (n=597 including 43 other than white non-Hispanic patients and 56 mutation carriers) as well as in 1036 white non-Hispanic healthy subjects of similar age and gender ascertained at Mayo Clinic Jacksonville. This led to the identification of 4 additional rare variants (MAF<1%) affecting the coding region in 7 additional EOAD patients: c.67C>T (p.R23C), c.266C>T (p.S89L), c.151_152ins42 (p.Asp50_Leu51ins14), and c.238C>T (p.R80W) (Fig. 1B, Table 2, Table 3; Supplementary Table 3). No rare coding variants that affected the protein sequence were found in healthy subjects. Notably, of the 9 EOAD patients carrying rare TYROBP variants, 3 patients also carried known AD gene mutations: the p.R80W variant was found twice in two Hispanic patients also harboring the pathogenic PSEN1 c.617G>C (p.G206A) mutation; and the p.S89L variant was found in an APP duplication carrier. These patients were excluded from further statistical analysis. By collapsing rare coding variants with a MAF<1% and comparing the overall frequency of such variants between all white non-Hispanic EOAD patients without mutations in the known genes (n=6, representing 1.17% of 509 EOAD patients) and healthy subjects (n=1036; no mutation detected), we identified a significant enrichment in EOAD patients (p-value=0.002; CMC test).
Fig. 1. Molecular genetic analysis of TYROBP.
(A) Panel A includes a chromatogram of several bases surrounding the p.G2E variant (red square) of patient A and an healthy subject. (B) Schematic representation of TYROBP protein along with its different domains and the rare coding variants identified in the overall EOAD cohort. A PCR was performed on the cDNA from patient A using a PCR primer set with a reverse primer specific for the mutant T allele of the p.V55L variant (NM_003332.3:c.163G>T) allowing us to specifically amplify the chromosome carrying the c.163T allele. This PCR fragment also spanned the sequence surrounding the p.G2E (NM_003332.3:c.5G>A) variant. Sanger sequencing of the PCR product showed the mutant A allele of the p.G2E variant suggesting that the variant is in cis with the p.V55L variant.
Table 3.
Non-Hispanic whites without mutations in known AD genes included in the statistical analyses
| Site | Patients | Healthy individuals | ||
|---|---|---|---|---|
|
TYROBP mutation carriers |
Non mutation carriers |
TYROBP mutation carriers |
Non mutation carriers |
|
| Mayo Clinic* | 6 | 509 | 0 | 1036 |
| Knight ADRC | 0 | 111 | 0 | 292 |
| France | 3 | 481 | 0 | 498 |
| Total** | 9 | 1101 | 0 | 1826 |
pvalue=0.002
pvalue=0.0001
To further substantiate this finding we examined two additional independent cohorts for the presence of rare coding variants in TYROBP. We studied 111 EOAD patients without mutations in the known AD genes and 292 healthy individuals from the Knight ADRC at Washington University by Sanger sequencing and we assessed exome sequencing data available on 484 French EOAD patients and 498 French controls without mutation in the known AD genes. Interestingly, we identified and confirmed by Sanger sequencing two rare coding variants in three unrelated French patients (Nicolas, et al., 2015), whereas no rare coding variants were detected in controls (n=595 EOAD patients and n=790 healthy subjects, Table 3, Supplementary Table 3). When combining these results, we observed a significant association between rare coding variants and EOAD: 6 rare coding TYROBP variants were identified in 9 unrelated patients out of 1110 white non-Hispanic EOAD patients as compared to an absence of such variants in 1826 healthy subjects (p-value=0.0001; CMC test). Of note, none of the patients harboring rare TYROBP variants was homozygous for the APOE ε4 allele and the presence of the ε4 haplotype does not appear to have an effect on age at onset (Table 3, Supplementary Table 3).
3.3. In vitro functional analysis of TYROBP p.D50_L51ins14
To provide support for a functional effect of at least one of the identified TYROBP variants, we selected variant TYROBP p.D50_L51ins14 that overlaps with a critical amino-acid for the binding of TYROBP to TREM2 (D50). It was shown that an artificial mutation of amino-acid D50 reduced the ability of TYROBP to bind TREM2 and stabilize TREM2 C-terminal fragments resulting from γ-secretase cleavage (Wunderlich, et al., 2013,Zhong, et al., 2015). We identified the p.D50_L51ins14 mutation in a probable AD patient with an age at disease onset of 62 years. This patient showed a clear family history with a diagnosis of AD in the patient’s father.
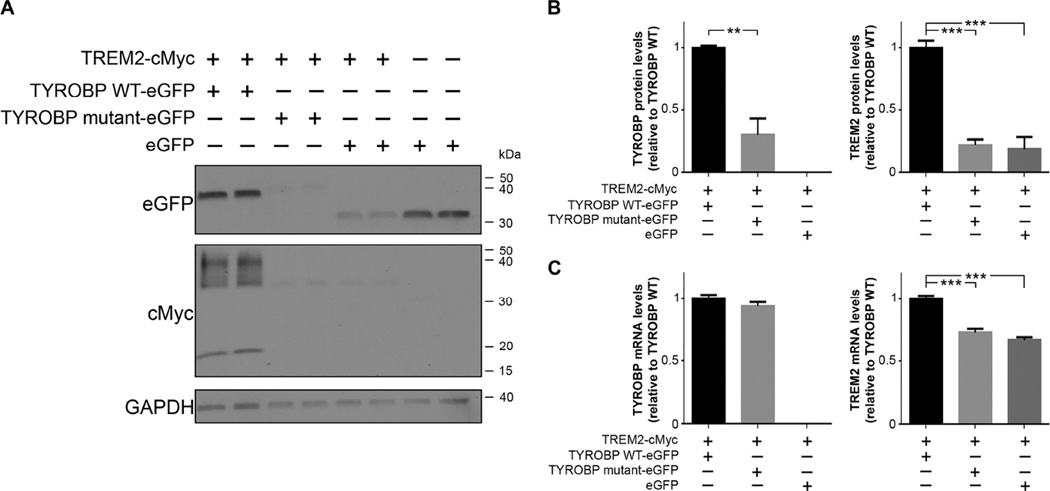
We co-expressed TYROBP p.D50_L51ins14 along with wild-type human TREM2 in HeLa cells in order to evaluate the impact of TYROBP p.D50_L51ins14 mutation on TYROBP and TREM2 levels. We found that the mutant form of TYROBP was expressed at a significantly lower protein level as compared to TYROBP WT (mean =0.30, SE=0.12; p-value=0.001, Student t-test; Fig. 2A and B), despite equal wild-type and mutant TYROBP mRNA expression (mean=0.93, SE=0.03; p-value=0.19, Student t-test; Fig. 2C). Moreover, TREM2 protein levels were significantly reduced in cells transfected with p.D50_L51ins14 mutant TYROBP as compared to cells transfected with wild-type TYROBP (mean=0.22, SE=0.02; p-value <0.0001, Student t-test; Fig. 2A and B). Interestingly, the reduction in TREM2 expression is also present at the mRNA level but to a far lesser extent than the protein level (mean=0.73, SE=0.02; p-value <0.0001, Student t-test; Fig. 2C). Our control experiment in which we co-transfected TREM2 with eGFP (in the absence of TYROBP overexpression) also showed significantly reduced levels of TREM2 mRNA (mean=0.66, SE=0.02; p-value <0.0001, Student t-test; Fig. 2C) and TREM2 protein (mean=0.18, SE=0.05; p-value <0.0001, Student t-test; Fig. 2A and B) similar to the decrease observed when co-expressed with the TYROBP mutant.
Fig. 2. Effect of TYROBP mutant p.D50_L51ins14 on TREM2 expression.
(A) Co-expression of TYROBP mutant with TREM2 decreases the level of TREM2 protein expression. Western blot of HeLa cells transfected with TREM2-Myc in combination with wild-type TYROBP-eGFP (TYROBP WT-eGFP) or with the p.D50_L51ins14 (TYROBP mutant-eGFP) or a vector control eGFP. The levels of TYROBP and TREM2 were visualized using an eGFP or cMyc antibody, respectively. GAPDH was used as loading control. (B) Quantification of TYROBP and TREM2 protein levels in HeLa cells transfected as described in (A). Quantification is assessed relative to the TYROBP WT-eGFP transfection condition after normalization to GAPDH levels. (C) Quantification of mRNA levels in HeLa cells transfected as described in (A). **: p<0.01; ***: p<0.0001, Student t-test on n≥4 replicates.
4. Discussion
In this study, we used an original bioinformatics filtering strategy to evaluate the impact of rare autosomal dominant and recessive variants in the genetics of EOAD and nominate potential candidate EOAD genes. We present a list of 29 candidate genes that could be considered in future genetics studies. We selected one gene from our list, TYROBP, based on its functional link to AD, and further characterized its potential involvement in EOAD. We first detected a nominally significant enrichment of rare coding TYROBP variants in EOAD compared to controls. Second, we identified one rare TYROBP variant with a profound effect on TREM2 expression in cell culture.
TYROBP was nominated as a candidate EOAD gene as part of our autosomal dominant and autosomal recessive (compound heterozygote) filtering strategies; however, brain mRNA and cDNA expression studies suggest that the observed variants were in cis, thereby favoring the hypothesis of an autosomal dominant inheritance pattern for TYROBP variants, possibly with reduced penetrance. TYROBP encodes a type I transmembrane protein consisting of a signal peptide sequence, a transmembrane segment and a cytoplasmic region containing an immunoreceptor tyrosine-based activation motif (ITAM; Fig. 1B). Within the central nervous system, TYROBP is preferentially expressed in microglia (Hickman, et al., 2013). Binding to its ligands, such as TREM2, leads to phosphorylation of TYROBP’s tyrosine residues (positions 91 and 102) and recruitment of downstream proteins, which regulates pro-inflammatory responses and phagocytosis (Malik, et al., 2015, Paradowska-Gorycka and Jurkowska, 2013, Zhong, et al., 2015). In addition to its major role in immune system regulation and phagocytosis, TYROBP might be involved in Aβ turnover (Zhang, et al., 2013). Similar to TREM2 mutations, loss-of-function mutations in TYROBP have already been described in the recessively inherited Nasu-Hakola disease (NHD, MIM #221770) (Paloneva, et al., 2000), a rare condition characterized by systemic bone cysts and dementia. Moreover, TYROBP regulates macrophage proliferation through CSF1R (MIM #164770), which is encoded by a gene previously implicated in hereditary diffuse leukoencephalopathy with spheroids (HDLS, MIM #221820), a white-matter disease clinically characterized by behavioral, cognitive and motor abnormalities (Rademakers, et al., 2012). The mechanism leading to cognitive decline in these diseases is still under investigation; however, partial loss of the CSF1R/TYROBP signaling pathway has been suggested to be responsible for the neurological phenotypes observed in NHD and HDLS.
In our study, we detected an enrichment of TYROBP rare coding variants in EOAD compared to controls. We acknowledge that the level of significance did not reach the threshold of exome-wide significance (2.5×10−6 for Bonferroni correction accounting for ~20,000 genes); however, we argue that our results are at least suggestive of a subset of TYROBP variants impacting EOAD disease risk. Remarkably, in line with this hypothesis, overexpression of TYROBP p.D50_L51ins14 leads to a loss of TYROBP protein and an almost total loss of TREM2 protein expression in cell culture. This reduction in TREM2 protein levels was partially explained by a 30% loss in TREM2 mRNA expression but other mechanisms such as destabilization of TREM2 protein may also be involved. In either way, the strong reduction in TREM2 levels when co-expressed with mutant TYROBP suggests that, based on the current cellular model, the p.D50_L51ins14 mutation leads to a loss of TYROBP expression and/or function and reduced TREM2 levels. In this respect it is important to note that a decrease in soluble TREM2 expression has been reported in cerebrospinal fluid (CSF) of patients harboring heterozygous variants in TREM2, which are known to lead to NHD in the homozygous state (Piccio, et al., 2016). Additionally, other rare variants in TREM2 that have been reported as risk factors for AD correlate with a lower abundance of TREM2 in the CSF. Our results for the p.D50_L51ins14 mutation align with those findings and reinforce the idea that a loss of TREM2/TYROBP function might contribute to the physiopathology of AD. Whether and how the other 5 observed TYROBP rare variants affect TYROBP biology remains to be studied. Two of the TYROBP variants (p.G2E and p.R23C) are located in the signal peptide segment of TYROBP and might affect TYROBP localization or processing. Moreover, variant p.V47A is located in close proximity to p.D50_L51ins14 and might similarly affect the TREM2-TYROBP interaction and TREM2 stabilization. Finally, we note that we also detected rare coding TYROBP variants in 3 EOAD patients already harboring pathogenic mutations in APP and PSEN1. It remains possible that these TYROBP variants contribute to the disease in these patients, especially in light of the oligogenic disease mechanism that is gaining attention in other neurodegenerative disorders such as amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (van Blitterswijk, et al., 2012).
In addition to TYROBP, we provide a list of 28 additional potential EOAD genes. Two notable candidates, which we have not yet followed-up genetically, are AMPH (MIM #600418) and CD300E (MIM #609801). Specifically, we identified in two unrelated individuals a non-sense variant in AMPH which encodes the amphiphysin-1 protein involved in synaptic endocytosis (Wu, et al., 2009). AMPH1 has been shown to be reduced in frontal and temporal cortex of AD patients as well as in the JNPL3 tauopathy mouse model (De Jesus-Cortes, et al., 2012), suggestive of a role in tau-mediated neurodegeneration. Interestingly, amphiphysin-2, also known as BIN1 (MIM #601248), resides at one of the most significant genome-wide association loci for LOAD risk (Lambert, et al., 2013). We also detected a non-sense variant in CD300E, which was present in two unrelated individuals. CD300E is a type I transmembrane protein and was shown to interact with TYROBP (Brckalo, et al., 2010). Whether AMPH, CD300E or any of the other candidate genes identified in our study are implicated in EOAD will require systematic mutation and association analyses in large EOAD and control cohorts. Similarly, in the near future, large scale on-going sequencing projects focused on LOAD may allow evaluating the impact on LOAD risk of the candidate genes identified in the current study.
5. Conclusions
In summary, we propose the use of extreme phenotypes such as very young EOAD patients or EOAD patients with a family history, as well as the use of multiple prioritization strategies as an efficient strategy to identify candidate genes for neurodegenerative disorders. We further present, for the first time, an association of rare coding variants in TYROBP with EOAD, which adds to the growing body of evidence suggesting that TYROBP is an important player in the physiopathology of AD.
Supplementary Material
Highlights.
Exome sequencing of highly selected EOAD patients nominates 29 candidate genes
Rare coding variants in TYROBP are genetically associated with EOAD
TYROBP p.D50_L51ins14 is a potentially pathogenic variant altering TREM2 levels
Acknowledgments
This work was funded by the US National Institutes of Health (P50 AG016574, R01 NS080882, P50 AG016574, R01-AG032990, R01-NS080820, U01 AG046139, R21 AG048101, and RF1 AG051504), and the Florida State Ed and Ethel Moore Alzheimer’s Disease Research Program. J.T.B. and R.G. are supported by Research Fellowships from Alzheimer’s Society. D.C, G.N. and A.-C.R. were supported by France Génomique National infrastructure, funded as part of “Investissement d’avenir” program managed by Agence Nationale pour la Recherche (contrat ANR-10-INBS-09), by the JPND “ Perades Program” and by the Labex GENMED ANR- 10-LABX-0013.
Abbreviations
- AD
Alzheimer’s diease
- EOAD
early-onset Alzheimer’s disease
- LOAD
late-onset Alzheimer’s disease
- ADRC
Alzheimer’s disease research center
- CNR-MAJ
centre national de référence pour les malades Alzheimer jeunes
- NINCDS-ADRDA
National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s Disease and Related Disorders Association
- UTR
untranslated region
- GATK
Genome Analysis Toolkit
- VQSR
Variant Quality Score Recalibration
- PCR
polymerase chain reaction
- CMC
Combined and Multivariate Collapsing
- MAF
minor allele frequency
- NHD
Nasu-Hakola disease
- HDLS
hereditary diffuse leukoencephalopathy with spheroids
- CSF
cerebrospinal fluid
- ALS
amyotrophic lateral sclerosis
- FTD
frontotemporal dementia
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement
The authors declare have no conflicts of interest to disclose. The ethics committees of all participating institutions have approved this study.
Appendix A. Supplementary data
References
- Benitez BA, Cooper B, Pastor P, Jin SC, Lorenzo E, Cervantes S, Cruchaga C. TREM2 is associated with the risk of Alzheimer’s disease in Spanish population. Neurobiology of aging. 2013;34:1711, e1715–e1717. doi: 10.1016/j.neurobiolaging.2012.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchon A, Hernandez-Munain C, Cella M, Colonna M. A DAP12-mediated pathway regulates expression of CC chemokine receptor 7 and maturation of human dendritic cells. J Exp Med. 2001;194:1111–1122. doi: 10.1084/jem.194.8.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brckalo T, Calzetti F, Perez-Cabezas B, Borras FE, Cassatella MA, Lopez-Botet M. Functional analysis of the CD300e receptor in human monocytes and myeloid dendritic cells. Eur J Immunol. 2010;40:722–732. doi: 10.1002/eji.200939468. [DOI] [PubMed] [Google Scholar]
- Cruchaga C, Karch CM, Jin SC, Benitez BA, Cai Y, Guerreiro R, Harari O, Norton J, Budde J, Bertelsen S, Jeng AT, Cooper B, Skorupa T, Carrell D, Levitch D, Hsu S, Choi J, Ryten M, Hardy J, Trabzuni D, Weale ME, Ramasamy A, Smith C, Sassi C, Bras J, Gibbs JR, Hernandez DG, Lupton MK, Powell J, Forabosco P, Ridge PG, Corcoran CD, Tschanz JT, Norton MC, Munger RG, Schmutz C, Leary M, Demirci FY, Bamne MN, Wang X, Lopez OL, Ganguli M, Medway C, Turton J, Lord J, Braae A, Barber I, Brown K, Passmore P, Craig D, Johnston J, McGuinness B, Todd S, Heun R, Kolsch H, Kehoe PG, Hooper NM, Vardy ER, Mann DM, Pickering-Brown S, Kalsheker N, Lowe J, Morgan K, David Smith A, Wilcock G, Warden D, Holmes C, Pastor P, Lorenzo-Betancor O, Brkanac Z, Scott E, Topol E, Rogaeva E, Singleton AB, Kamboh MI, St George-Hyslop P, Cairns N, Morris JC, Kauwe JS, Goate AM. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature. 2014;505:550–554. doi: 10.1038/nature12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jesus-Cortes HJ, Nogueras-Ortiz CJ, Gearing M, Arnold SE, Vega IE. Amphiphysin-1 protein level changes associated with tau-mediated neurodegeneration. Neuroreport. 2012;23:942–946. doi: 10.1097/WNR.0b013e32835982ce. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez MA, Lebrigio RF, Van Booven D, Ulloa RH, Powell E, Speziani F, Tekin M, Schule R, Zuchner S. GEnomes Management Application (GEM.app): a new software tool for large-scale collaborative genome analysis. Hum Mutat. 2013;34:842–846. doi: 10.1002/humu.22305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JS, Younkin S, Hazrati L, Collinge J, Pocock J, Lashley T, Williams J, Lambert JC, Amouyel P, Goate A, Rademakers R, Morgan K, Powell J, St George-Hyslop P, Singleton A, Hardy J. TREM2 variants in Alzheimer’s disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro RJ, Lohmann E, Kinsella E, Bras JM, Luu N, Gurunlian N, Dursun B, Bilgic B, Santana I, Hanagasi H, Gurvit H, Gibbs JR, Oliveira C, Emre M, Singleton A. Exome sequencing reveals an unexpected genetic cause of disease: NOTCH3 mutation in a Turkish family with Alzheimer’s disease. Neurobiol Aging. 2012;33:1008, e1017–e1023. doi: 10.1016/j.neurobiolaging.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman SE, Kingery ND, Ohsumi TK, Borowsky ML, Wang LC, Means TK, El Khoury J. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SC, Benitez BA, Karch CM, Cooper B, Skorupa T, Carrell D, Norton JB, Hsu S, Harari O, Cai Y, Bertelsen S, Goate AM, Cruchaga C. Coding variants in TREM2 increase risk for Alzheimer’s disease. Hum Mol Genet. 2014;23:5838–5846. doi: 10.1093/hmg/ddu277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, Rujescu D, Hampel H, Giegling I, Andreassen OA, Engedal K, Ulstein I, Djurovic S, Ibrahim-Verbaas C, Hofman A, Ikram MA, van Duijn CM, Thorsteinsdottir U, Kong A, Stefansson K. Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher JP, Quest DJ, Duffy P, Meiners MA, Moore RM, Rider D, Hossain A, Hart SN, Dinu V. The Biological Reference Repository (BioR): a rapid and flexible system for genomics annotation. Bioinformatics. 2014;30:1920–1922. doi: 10.1093/bioinformatics/btu137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli MA, Cukier HN, Hamilton-Nelson KL, Rolati S, Kunkle BW, Whitehead PL, Zuchner SL, Farrer LA, Martin ER, Beecham GW, Haines JL, Vance JM, Cuccaro ML, Gilbert JR, Schellenberg GD, Carney RM, Pericak-Vance MA. Segregation of a rare TTC3 variant in an extended family with late-onset Alzheimer disease. Neurol Genet. 2016;2:e41. doi: 10.1212/NXG.0000000000000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thorton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Moron FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fievet N, Huentelman MW, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuiness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossu P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Deniz Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannefelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH, Jr, Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltuenen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nothen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Jiang T, Tan L, Yu JT. TYROBP in Alzheimer’s disease. Mol Neurobiol. 2015;51:820–826. doi: 10.1007/s12035-014-8811-9. [DOI] [PubMed] [Google Scholar]
- Malik M, Parikh I, Vasquez JB, Smith C, Tai L, Bu G, LaDu MJ, Fardo DW, Rebeck GW, Estus S. Genetics ignite focus on microglial inflammation in Alzheimer’s disease. Mol Neurodegener. 2015;10:52. doi: 10.1186/s13024-015-0048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Jr, Kawas CH, Klunk WE, Koroshetz WJ, Manly JJ, Mayeux R, Mohs RC, Morris JC, Rossor MN, Scheltens P, Carrillo MC, Thies B, Weintraub S, Phelps CH. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s & dementia : the journal of the Alzheimer’s Association. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H, Daly MJ. Variant TREM2 as risk factor for Alzheimer’s disease. The New England journal of medicine. 2013;368:182–184. doi: 10.1056/NEJMe1213157. [DOI] [PubMed] [Google Scholar]
- Nicolas G, Wallon D, Charbonnier C, Quenez O, Rousseau S, Richard AC, Rovelet-Lecrux A, Coutant S, Le Guennec K, Bacq D, Garnier JG, Olaso R, Boland A, Meyer V, Deleuze JF, Munter HM, Bourque G, Auld D, Montpetit A, Lathrop M, Guyant-Marechal L, Martinaud O, Pariente J, Rollin-Sillaire A, Pasquier F, Le Ber I, Sarazin M, Croisile B, Boutoleau-Bretonniere C, Thomas-Anterion C, Paquet C, Sauvee M, Moreaud O, Gabelle A, Sellal F, Ceccaldi M, Chamard L, Blanc F, Frebourg T, Campion D, Hannequin D. Screening of dementia genes by whole-exome sequencing in early-onset Alzheimer disease: input and lessons. Eur J Hum Genet. 2015 doi: 10.1038/ejhg.2015.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paloneva J, Kestila M, Wu J, Salminen A, Bohling T, Ruotsalainen V, Hakola P, Bakker AB, Phillips JH, Pekkarinen P, Lanier LL, Timonen T, Peltonen L. Loss-of-function mutations in TYROBP (DAP12) result in a presenile dementia with bone cysts. Nat Genet. 2000;25:357–361. doi: 10.1038/77153. [DOI] [PubMed] [Google Scholar]
- Paradowska-Gorycka A, Jurkowska M. Structure, expression pattern and biological activity of molecular complex TREM-2/DAP12. Hum Immunol. 2013;74:730–737. doi: 10.1016/j.humimm.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Piccio L, Deming Y, Del-Aguila JL, Ghezzi L, Holtzman DM, Fagan AM, Fenoglio C, Galimberti D, Borroni B, Cruchaga C. Cerebrospinal fluid soluble TREM2 is higher in Alzheimer disease and associated with mutation status. Acta Neuropathol. 2016 doi: 10.1007/s00401-016-1533-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pottier C, Hannequin D, Coutant S, Rovelet-Lecrux A, Wallon D, Rousseau S, Legallic S, Paquet C, Bombois S, Pariente J, Thomas-Anterion C, Michon A, Croisile B, Etcharry-Bouyx F, Berr C, Dartigues JF, Amouyel P, Dauchel H, Boutoleau-Bretonniere C, Thauvin C, Frebourg T, Lambert JC, Campion D. High frequency of potentially pathogenic SORL1 mutations in autosomal dominant early-onset Alzheimer disease. Mol Psychiatry. 2012;17:875–879. doi: 10.1038/mp.2012.15. [DOI] [PubMed] [Google Scholar]
- Pottier C, Wallon D, Rousseau S, Rovelet-Lecrux A, Richard AC, Rollin-Sillaire A, Frebourg T, Campion D, Hannequin D. TREM2 R47H variant as a risk factor for early-onset Alzheimer’s disease. J Alzheimers Dis. 2013;35:45–49. doi: 10.3233/JAD-122311. [DOI] [PubMed] [Google Scholar]
- Rademakers R, Baker M, Nicholson AM, Rutherford NJ, Finch N, Soto-Ortolaza A, Lash J, Wider C, Wojtas A, DeJesus-Hernandez M, Adamson J, Kouri N, Sundal C, Shuster EA, Aasly J, MacKenzie J, Roeber S, Kretzschmar HA, Boeve BF, Knopman DS, Petersen RC, Cairns NJ, Ghetti B, Spina S, Garbern J, Tselis AC, Uitti R, Das P, Van Gerpen JA, Meschia JF, Levy S, Broderick DF, Graff-Radford N, Ross OA, Miller BB, Swerdlow RH, Dickson DW, Wszolek ZK. Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids. Nat Genet. 2012;44:200–205. doi: 10.1038/ng.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A, Dols-Icardo O, Bullido MJ, Pastor P, Rodriguez-Rodriguez E, Lopez de Munain A, de Pancorbo MM, Perez-Tur J, Alvarez V, Antonell A, Lopez-Arrieta J, Hernandez I, Tarraga L, Boada M, Lleo A, Blesa R, Frank-Garcia A, Sastre I, Razquin C, Ortega-Cubero S, Lorenzo E, Sanchez-Juan P, Combarros O, Moreno F, Gorostidi A, Elcoroaristizabal X, Baquero M, Coto E, Sanchez-Valle R, Clarimon J. Assessing the role of the TREM2 p.R47H variant as a risk factor for Alzheimer’s disease and frontotemporal dementia. Neurobiology of aging. 2014;35:444, e441–e444. doi: 10.1016/j.neurobiolaging.2013.08.011. [DOI] [PubMed] [Google Scholar]
- van Blitterswijk M, DeJesus-Hernandez M, Rademakers R. How do C9ORF72 repeat expansions cause amyotrophic lateral sclerosis and frontotemporal dementia: can we learn from other noncoding repeat expansion disorders? Curr Opin Neurol. 2012;25:689–700. doi: 10.1097/WCO.0b013e32835a3efb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtas A, Heggeli KA, Finch N, Baker M, Dejesus-Hernandez M, Younkin SG, Dickson DW, Graff-Radford NR, Rademakers R. C9ORF72 repeat expansions and other FTD gene mutations in a clinical AD patient series from Mayo Clinic. Am J Neurodegener Dis. 2012;1:107–118. [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Matsui H, Tomizawa K. Amphiphysin I and regulation of synaptic vesicle endocytosis. Acta Med Okayama. 2009;63:305–323. doi: 10.18926/AMO/31822. [DOI] [PubMed] [Google Scholar]
- Wunderlich P, Glebov K, Kemmerling N, Tien NT, Neumann H, Walter J. Sequential proteolytic processing of the triggering receptor expressed on myeloid cells-2 (TREM2) protein by ectodomain shedding and gamma-secretase-dependent intramembranous cleavage. J Biol Chem. 2013;288:33027–33036. doi: 10.1074/jbc.M113.517540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, Zhang C, Xie T, Tran L, Dobrin R, Fluder E, Clurman B, Melquist S, Narayanan M, Suver C, Shah H, Mahajan M, Gillis T, Mysore J, MacDonald ME, Lamb JR, Bennett DA, Molony C, Stone DJ, Gudnason V, Myers AJ, Schadt EE, Neumann H, Zhu J, Emilsson V. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Chen XF, Zhang ZL, Wang Z, Shi XZ, Xu K, Zhang YW, Xu H, Bu G. DAP12 Stabilizes the C-terminal Fragment of the Triggering Receptor Expressed on Myeloid Cells-2 (TREM2) and Protects against LPS-induced Pro-inflammatory Response. J Biol Chem. 2015;290:15866–15877. doi: 10.1074/jbc.M115.645986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.