Abstract
Ceramide-1-phosphate (C1P) is an important signaling sphingolipid and a metabolite of ceramide. C1P contains an anionic phosphomonoester head group and has been shown to regulate physiological and pathophysiological processes such as cell proliferation, inflammation, apoptosis, phagocytosis, and macrophage chemotaxis. Despite this mechanistic information on its role in intra- and intercellular communication, little information is available on the biophysical properties of C1P in biological membranes and how it interacts with effector proteins. Fluorescently labeled lipids have been a useful tool to understand the membrane behavior properties of lipids such as phosphatidylserine, cholesterol, and some phosphoinositides. However, to the best of our knowledge, fluorescently labeled C1P hasn’t been implemented to investigate its ability to serve as a mimetic of endogenous C1P in cells or untagged C1P in in vitro experiments. Cellular and in vitro assays demonstrate TopFluor-C1P harbors a fluorescent group that is fully buried in the hydrocarbon core and fluoresces across the spectrum of physiological pH values. Moreover, TopFluor-C1P didn’t affect cellular toxicity at concentrations employed, was as effective as unlabeled C1P in recruiting an established protein effector to intracellular membranes, and its subcellular localization recapitulated what is known for endogenous C1P. Notably, the diffusion coefficient of TopFluor-C1P was slower than that of TopFluor-phosphatidylserine or TopFluor-cholesterol in the plasma membrane and similar to that of other fluorescently labeled sphingolipids including ceramide and sphingomyelin. These studies demonstrate that TopFluor-C1P should be a reliable mimetic of C1P to study C1P membrane biophysical properties and C1P interactions with proteins.
Keywords: ceramide-1-phosphate, cytosolic phospholipase A2, fluorescence, membrane dynamics, TopFluor
1. Introduction
Sphingolipids play roles in cell signaling and membrane trafficking (Gault et al., 2010, Hannun and Obeid, 2011) with main players including sphingosine, sphingosine-1-phosphate (S1P), ceramide, and ceramide-1-phosphate (C1P). C1P has been implicated in several intra- and extracellular events including cell proliferation (Gangoiti et al., 2010, Gangoiti et al., 2008b, Gomez-Munoz et al., 2010), phagocytosis (Hinkovska-Galcheva et al., 2005), inflammation (Arana et al., 2010, Gomez-Munoz et al., 2013, Gomez-Munoz et al., 2010), tumor metastasis (Gangoiti et al., 2008a), and migration of macrophages (Ouro et al., 2014), pre-adipocytes, human monocytes (Arana et al., 2013), and human pancreatic cancer cells (Rivera et al., 2016). C1P is generated in mammalian cells at the trans Golgi network (TGN) by the enzyme ceramide kinase (Cerk) (Lamour et al., 2007), where it has been shown to activate the enzyme cytosolic phospholipase A2α (Lamour et al., 2009). C1P that is synthesized in the TGN is trafficked to the plasma membrane through a vesicular pathway (Boath et al., 2008) and via a putative C1P transport protein (Simanshu et al., 2013). Disruption of the C1P transport to the plasma membrane led to an increase in the TGN/endosome/nuclear fraction of C1P and a decrease in the C1P in the plasma membrane (Simanshu et al., 2013). The increased C1P in the TGN significantly impacted synthesis of eicosanoids through cPLA2α activation (Simanshu et al., 2013). Despite the identification of C1P binding proteins such as cPLA2α (Subramanian et al., 2005, Subramanian et al., 2007, Ward et al., 2013) and more recently Annexin A2 (Hankins et al., 2013), diacylglycerol kinase γ (Takeshita et al., 2014), Tumor Necrosis Factor-α converting enzyme (TACE) (Lamour et al., 2011) and prostaglandin D synthase (Bidlingmaier et al., 2016), there are still many gaps to fill in our understanding of C1P signaling and identification of binding partners.
The physical properties of C1P in membranes of mixed lipid compositions are also not well understood. C1P is an anionic phosphomonoester and will form a spontaneous monolayer on water despite its anionic charge (Kooijman et al., 2008). When C1P is fully hydrated it is able to form a bilayer with a crystalline state at low temperatures, a gel phase at ~45°C and a transition to fluid phase at ~65°C (Kooijman et al., 2008). In addition, calcium is able to bind to the C1P head group (Kooijman et al., 2009) somewhat akin to Ca2+-phosphatidylserine interactions (Verdaguer et al., 1999), which could be an important factor in recruitment of some binding proteins such as Annexin A2 (Hankins et al., 2013). C1P has two ionizable groups on its phosphate head group suggesting the local membrane environment and pH can influence its overall charge. The pKA2 of C1P is 7.39 in membrane containing phosphatidylcholine, while the introduction of phosphatidylethanolamine (PE) to the membranes significantly lowered the pKA2 to 6.64 (Kooijman et al., 2008). This has also been observed for the phosphate head group of phosphatidic acid when PE is found in membranes and is attributed to an electrostatic/hydrogen bond switch (Kooijman et al., 2007). Since PE is an abundant lipid in mammalian cells and found both in the Golgi and plasma membrane where C1P resides, a significant population of the cellular C1P may carry a −2 charge at cytoplasmic pH. A small mol% of C1P is sufficient to induce negative membrane curvature generation (Kooijman et al., 2008), which is similar to reported effects of phosphatidic acid (Kooijman et al., 2005), which is structurally very similar to C1P. The cellular role of C1P in membrane curvature changes is still an unexplored area of research, however, down regulation of C1P transport from the Golgi to the plasma membrane led to an increased fragmentation pattern of the Golgi when C1P levels increased (Simanshu et al., 2013). Thus, the effects of C1P on membrane structure or the interactions with their effector proteins may play a role in membrane curvature changes and/or membrane stability.
Fluorescent labeling of lipids has had significant impact on studying lipid distribution (Kay et al., 2012, Maekawa and Fairn, 2014), lipid diffusion rates (Golebiewska et al., 2011, Kay et al., 2012), and lipid dynamics via single particle tracking (Kay et al., 2012, Knight and Falke, 2009). However, labeling lipids comes with some difficulty, as they could behave differently from their parent lipid due to the inherent biophysical properties and location of the fluorophore. For example, several lipids have been labeled with NBD on the acyl chain, but due to its moderately polar nature it can alter their physical properties, which is most well characterized for phosphatidylserine (Kay et al., 2012). The more hydrophilic nature of the labelled acyl chain has been shown to change the position of the labeled acyl chain towards the membrane interface (Abrams and London, 1993, Chattopadhyay and London, 1987, Kay et al., 2012). Here we examined a new fluorescent derivative of C1P, TopFluor-C1P, and examined its use as a fluorescent lipid mimicking unlabeled C1P. We have investigated the in vitro and cellular behaviors of TopFluor labeled on the acyl chain tail of C1P for accessibility in the lipid bilayer, cellular localization, recruitment of a C1P effector protein, and membrane dynamics using fluorescence recovery after photobleaching (FRAP). These assays together suggest TopFluor-C1P is a sufficient analog of C1P for the study of biochemical and biophysical properties of C1P and its effector proteins in vitro and in cells. The studies also provide new knowledge on cellular C1P dynamics demonstrating the diffusion coefficient of C1P is less than other important signaling lipids such as PS and cholesterol but similar to that of other sphingolipids.
2. Materials and methods
2.1 Chemicals and Reagents
All lipids, including TopFluor-C1P, were obtained from Avanti Polar Lipids (Alabaster, AL). A549 cellular transfection reagents, PLUS reagent and Lipofectamine LTX, were from Life Technologies (Grand Island, NY). 7-Aminoactinomycin D was acquired from BD Biosciences (San Jose, CA). A23187, BAPTA AM, camptothecin, laurdan, and potassium iodide were acquired from Sigma-Aldrich (St. Louis, MO).
2.2 Liposome Assays
Liposomes were prepared by adding the required amount of stock lipid (in chloroform) into a glass vial and drying under a stream of N2 gas. The dried lipid was re-suspended in liposome buffer with different buffering agents to prepare appropriate pH values (25 mM HEPES, 25 mM sodium acetate, or 25 mM sodium phosphate containing 140 mM KCl, 0.5 mM MgCl2, 15 mM NaCl), followed by vortexing. Large unilamellar vesicles (LUVs) were prepared by passing lipids in liposome buffer 50 times through an Avanti® Mini-extruder (Avanti Polar Lipids, Alabaster, AL) containing a 100 nm filter pore size. Liposomes were prepared at 100 μM concentration with 1% TopFluor-labeled lipid and 99% POPC. LUVs were added to a Costar black with clear flat bottom 96-well plate (Corning, Kennebunk, ME) to a total volume of 200 μl. For the potassium iodide quenching assay, increasing concentrations of KI (0 to 0.5 M) were added to the appropriate wells and fluorescence was measured using a SpectraMax M5 microplate reader (Sunnyvale, CA). Fluorescence of TopFluor-labeled lipids was excited at 470 nm and emission was measured at 510 nm. Fluorescence was measured before (F0) and after (F) the addition of KI. Data was plotted F/F0 vs. KI (M). For the laurdan assay, multilamellar vesicles (MLVs) were prepared at 1 mM with indicated lipid composition. MLVs were added to a 96 well plate and excited at 340 nm, and emission was measured from 390 to 600 nm. General polarization (GP) was measured using the equation, GP = (I435 − I480)/(I435 + I480).
2.3 Surface Plasmon Resonance (SPR) Binding Assay
To gauge the ability of the C2 domain of cPLA2α to bind TopFluor-C1P in liposomes, we performed SPR analysis with liposomes containing either POPC:POPE:TopFluor-C1P (77:20:3) or POPC:POPE:C1P (77:20:3). Using a Biacore X instrument and a L1 sensor chip, liposomes containing the aforementioned compositions were coated on flow channel two at ~4000 RU. Flow channels one and two were then coated with 0.1 mg/mL BSA to greatly reduce nonspecific binding to the sensor chip surface. Binding experiments with the C2 domain were run in 10 mM HEPES, pH 7.4 containing 0.16 M KCl and 1 μM CaCl2. Increasing concentrations of the C2 domain were injected over both flow cell 1 and flow cell 2 for both lipid conditions to monitor the association and dissociation phase of liposome binding. Flow channel 1 was then subtracted out from flow channel 2 for each protein concentration to account for any nonspecific association with the sensor chip surface. The saturation response (RU) was determined following subtraction of the control flow cell. The saturation response (RU) was then plotted versus C2 domain concentration in order to determine the apparent Kd value by a nonlinear least-squares analysis of the binding isotherm using an equation, Req= Rmax/(1 + Kd/C).
2.4 Cell Culture, Transfection, and Cell Treatments
A549 lung adenocarcinoma cells were grown in 50:50 DMEM:RPMI supplemented with 10% FBS and 1% penicillin-streptomycin at 37°C with 5% CO2. For imaging experiments, cells were seeded in Nunc Lab-Tek™ II 8-well imaging plates (Fisher Scientific) in growth medium and grown to ~60–70% confluency. Cells were then transfected with 1 μg of DNA using PLUS reagent and lipofectamine LTX according to the manufactures’ protocol (Life Technologies, Grand Island, NY) and replaced with Opti-MEM growth medium. 18 hours post-transfection, cells were treated with lipid or vehicle and imaged 1 hour later. Lipid was prepared by adding the required amount of stock lipid into a glass vial and dried under a stream of N2. Lipid was re-suspended in a solution of ethanol:dodecane (98:2) (Wijesinghe et al., 2009) to a concentration of 100 μM, followed by sonication for ten cycles of 10 s on, 10 s off, followed by incubation in 37°C for 20 minutes. Lipids were added to cells in 8-well imaging plates to a final concentration of 1 μM. For vehicle experiments, cells were treated with an equivalent volume of ethanol:dodecane (98:2).
2.5 Confocal Microscopy – Colocalization and FRAP
Cells were imaged using confocal microscopy (Zeiss LSM 710) using an oil 63× 1.4 numerical aperture objective. For organelle colocalization experiments, mTurquoise2 constructs were excited with a 434 nm laser line and emission collected using cyan fluorescent protein (CFP) emission from 450–510 nm, and TopFluor-C1P excitation was at 488 nm with emission measured using green fluorescent protein (GFP) settings from 510 – 600 nm. Colocalization experiments were measured using the Zeiss LSM software for determination of the Mander’s overlap coefficient by outlining transfected cells of interest in the image. Values greater than 0.6 indicate colocalization between the two fluorophores. For % translocation assays, cells were counted in 3 separate experiments, where 20–70 cells were counted in each experiment. For cPLA2α experiments inducing translocation with C1P, A23187 dissolved in DMSO, was added to cells at a final concentration of 10 μM.
Fluorescence recovery after photo-bleaching (FRAP) experiments bleached a circular region of interest (ROI) of 2.2 μm diameter. After 3 pre-bleach scans, the ROI was bleached for 10 iterations at 488 nm with 100% laser power. The post-bleach profile was scanned for 30 seconds. Fluorescence data was extrapolated using ImageJ software to measure fluorescence in the bleached area over time. The mobile fraction was measured using the following equation where F1 is normalized to 1 for the pre-bleach scans. The diffusion coefficient was measured using a simplified equation described by Kang, M, et al. (Kang et al., 2010) where the nominal bleach region, rn, is kept constant among all samples, and the re is determined from accurate measurements of t1/2, and the half width at 14% of bleaching depth from the top. These measurements were determined from ImageJ analysis of the bleach depth profile in fluorescence intensity of the ROI immediately following the pre-bleach scans.
2.6 Toxicity Assay
Cells were seeded in growth medium at 5 × 104 cells per 15 × 60 mm dish. 24 hours prior to experiment, cells were washed with PBS and replaced with 50:50 DMEM:RPMI and appropriate dishes were treated with 35 μM camptothecin. Two hours prior to staining, cells were treated with 1 μM lipid. Cells were washed with PBS, treated with 0.5 mL Trypsin for 3–5 minutes at 37°C and collected via centrifugation. Cells were washed twice with PBS and re-suspended in 300 μl 1X Binding Buffer. Appropriate dishes were treated with 7-Aminoactinomycin D (7-AAD) according to manufacture’s protocol. Cells were collected using the FC500 flow cytometer (Beckman) and analyzed using FlowJo software. 7-AAD was excited at 488 nm and emission collected between 660–690 nm.
2.7 Statistical Analysis
Student’s t-test was used for statistical analysis, and p < 0.05 was used as the significance threshold.
3. Results
3.1 Biophysical properties of TopFluor-C1P as a fluorescent analog of C1P
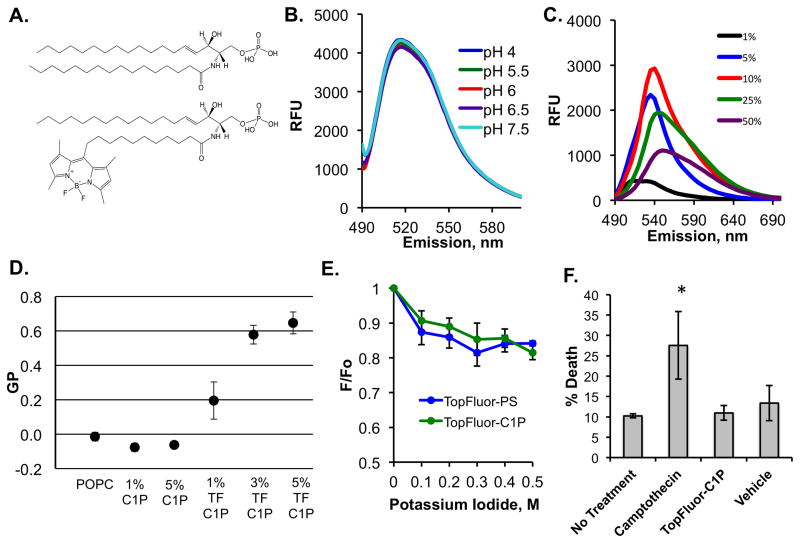
TopFluor is a fluorescent molecule characterized by its dipyrromethene boron difluoride moiety, which has been covalently attached to different lipid molecules, including the C11 acyl chain tail of C1P as shown in figure 1A. To first assess the fluorescence of TopFluor C1P, we monitored the fluorescence emission profile of 1 mol% TopFluor C1P in liposomes containing 99 mol% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) at pH 7.5 (Figure 1B). TopFluor-C1P exhibited an emission maxima at ~510 nm when exited at 470 nm. In order to determine if the fluorescence properties of TopFlour-C1P differed when the pH of the solution changed, we examined the fluorescence emission profile of 1 mol% TopFluor-C1P in buffers with pH values of 4 to 7.5 (Figures 1B and S1A). This is an important assessment and control for potential cellular studies with TopFluor-C1P, as when TopFluor-C1P is added to cell culture, it may encounter lower pH environments if it’s incorporated into the endosomal/lysosomal pathway. As expected, the TopFluor-C1P emission spectra behaved similarly from pH 4 to pH 7.5 suggesting the TopFluor fluorescence won’t be suppressed in the more acidic physiological environment. The TopFluor fluorescence with respect to pH dependency was similar in POPC or DOPC vesicles (Figures 1B and S1A).
Figure 1. Biophysical Properties of TopFluor-C1P.
A) The structures of C1P (d18:0/16:0) (top) and TopFluor-C1P (bottom), which harbors a dipyrromethene boron difluoride (TopFluor) moiety at the tail end of the amine-linked acyl chain, are shown in their fully protonated state. B) The emission spectra (excitation at 470 nm and emission 490–600 nm) of 100 nm liposomes containing 1% TopFluor-C1P and 99% POPC at varying physiological pH values are shown. RFU = relative fluorescence unit. C) Increasing mol% of TopFluor-C1P was incorporated into POPC vesicles and the emission spectra (490–600 nm) was collected following excitation at 470 nm. D) The general polarization (GP) values from a laurdan assay containing MLVs with the indicated lipid concentrations in POPC. GP was measured using the equation, GP = (I435 − I480)/(I435 + I480). MLVs were excited at 340 nm and the emission was measured from 390 to 600 nm. E) A potassium iodide quenching assay was performed for liposomes containing either 1 mol% TopFluor-C1P or 1 mol% TopFluor-PS in DOPC vesicles. The emission (510 nm) was collected following excitation at 470 nm and the data were plotted as a ratio of (F/F0) versus the concentration of potassium iodide. F0 = fluorescence before KI addition, F = fluorescence after KI addition. F) Flow cytometry using the dye 7-AAD was employed to assess cell death for A549 cells treated with nothing, camptothecin, TopFluor-C1P or vehicle control. * p < 0.05. TF = TopFluor.
To determine if TopFluor-C1P could recapitulate C1P in liposomes to support binding of a known C1P effector protein, we employed SPR analysis for the C2 domain of cPLA2α (Stahelin et al., 2007, Subramanian et al., 2005, Ward et al., 2013). Liposomes containing either 3 mol% TopFluor-C1P or 3 mol% C1P were used to test C2 domain binding at varying concentrations of protein. SPR sensorgrams for C2 domain association with each liposome following subtraction of a control flow cell are shown in the supplemental data (Figure S2). Determination of the apparent Kd value from a non-linear least squares analysis of the saturation response (RU) versus C2 domain concentrations yielded an apparent Kd for of 730 ± 140 nM for TopFluor-C1P containing liposomes and 230 ± 50 nM for C1P containing liposomes (Figure S2). TopFluor-C1P supports binding of the C2 domain to a similar magnitude to that of unlabeled C1P.
We also examined the TopFluor-C1P emission spectra at increasing concentrations of TopFluor-C1P (1–50 mol%) in the liposomes (Figure 1C). The fluorescence emission signal increased at ~510 nm from 1–10 mol% TopFluor-C1P in the liposomes, however, the emission spectra shifted and fluorescence emission decreased at 25 and 50 mol% TopFluor-C1P (Figures 1C and S1B). The decrease in emission spectra above 10 mol% TopFluor-C1P in liposomes is most likely due to self-quenching of the fluorophore. Notably a similar trend was detected for TopFluor-C1P in either POPC or DOPC vesicles (Figures 1C and S1B).
To investigate if the fluidity of membranes with TopFluor-C1P and native C1P were similar, a laurdan assay was employed. The general polarization (GP) was analyzed using the intensity values at 435 nm and 480 nm, rather than the traditional 440 nm and 490 nm, to avoid emission shift overlap from the fluorescent TopFluor-C1P. GP values range from −1 to +1, where a more positive value indicates a very rigid membrane and a more negative value a more fluid membrane. POPC and C1P MLVs displayed a GP value between 0 and −0.1. These data are consistent with previously published data showing a GP value of ~ −0.5 for 5 mol% C1P (Stock et al., 2012). However, MLVs containing TopFluor-C1P showed significantly increased GP values: 0.2, 0.58, and 0.65, for 1, 3, and 5 mol% respectively, indicating that TopFluor-C1P creates a more rigid and ordered membrane structure.
To further assess the physical properties of the TopFluor moiety on C1P, we performed a potassium iodide quenching assay to determine if the TopFluor moiety was near the membrane-water interface. For these experiments, we compared TopFluor-C1P to TopFluor-PS as Kay and colleagues elegantly compared TopFluor-PS and NBD-PS to show the TopFluor probe was more hydrophobic, sat at the interior of the membrane, and was not quenched by potassium iodide (Kay et al., 2012). In the iodide assay, the fluorescent probe will be quenched when exposed to aqueous medium at increasing concentration of iodide. In contrast, if the probe is not quenched, it sits tightly within the hydrophobic core of the lipid bilayer. As shown in figure 1E, as increasing concentrations of potassium iodide were added we observed little change in the fluorescence emission of TopFluor-PS or TopFluor-C1P (Figure 1E). Fluorescence was measured before (F0) and after (F) the addition of KI to yield the data plotted as F/F0 vs. KI concentration in Figure 1E.
3.2 TopFluor-C1P cellular toxicity
To assess if exogenously added TopFluor-C1P was toxic to cells, we performed assessment of cellular death using flow cytometry. 7-AAD, which stains dead cells, was used to measure the percentage of cells that were dead or dying after no treatment, treatment with camptothecin, treatment with vehicle (ethanol:dodecane 98:2), or treatment with TopFluor-C1P. 7-AAD stained cells were gated based on their staining of 7-AAD and quantified based on percentage of cell death. Camptothecin treated cells were used as a positive control to ensure cell death was possible within our experimental procedures (Figure 1F) and induced nearly 30% cell death under these conditions. We didn’t observe a significant increase in cell death when cells were exogenously treated with TopFluor-C1P versus 18:1/16:0 C1P compared to untreated cells (Figure 1F). Additionally, the vehicle (added at less than 1% of total volume) control seems to play no significant role in cell viability with these experiments (Figure 1F).
3.3 TopFluor-C1P cellular localization and activation of cPLA2α
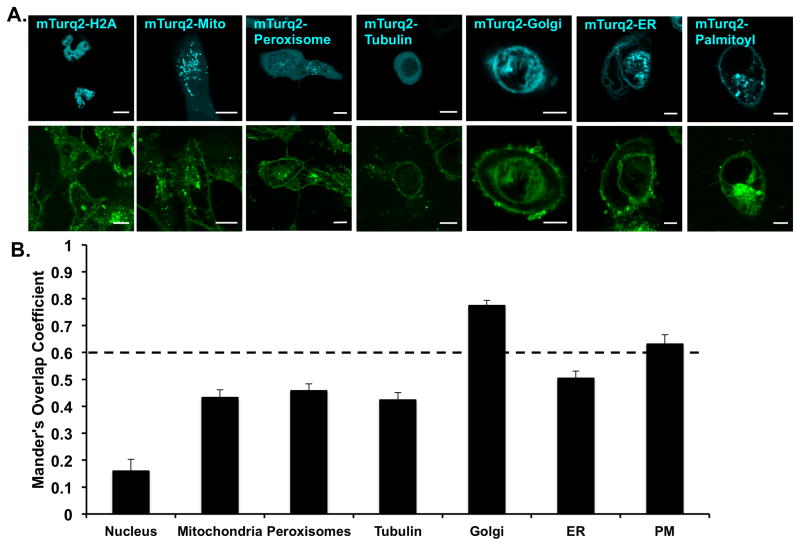
C1P was shown to be synthesized in the TGN and then transported to the plasma membrane but may also be found at low levels in endosomes and nuclear membranes (Simanshu et al., 2013). In order to determine the cellular distribution of TopFluor-C1P, we treated A549 cells with TopFluor-C1P or vehicle control and imaged cells expressing various organelle and membrane markers one hour after lipid or vehicle treatment (Figure 2). Using Mander’s correlation coefficient at a cutoff of above 0.6 indicating colocalization (Zinchuk et al., 2007), we determined that TopFluor-C1P co-localized with the Golgi and plasma membrane, but not significantly with the mitochondria, peroxisome, tubulin, endoplasmic reticulum (ER), or nucleus. The distribution of TopFluor-C1P was similar to that known for endogenous C1P in mammalian cells (Simanshu et al., 2013). Similar cellular distribution was observed with either ultra pure H2O or ethanol:dodecane (98:2) as the TopFluor-C1P delivery vehicle (Figure S3).
Figure 2. Cellular distribution of TopFluor-C1P in A549 cells.
A) A549 cells were transfected with mTurquiose2 organelle specific markers (cyan) to investigate the cellular distribution of TopFluor-C1P (green). 1 μM TopFluor-C1P was added to cells 1 hour prior to imaging. Representative images for the mTurquiose2 and TopFluor channels are shown for each cellular marker. Scale bar = 10 μm. B) Cellular images were analyzed for colocalization using Mander’s overlap coefficient. Significant colocalization is above 0.6 and data shown is ± standard error of the mean (SEM).
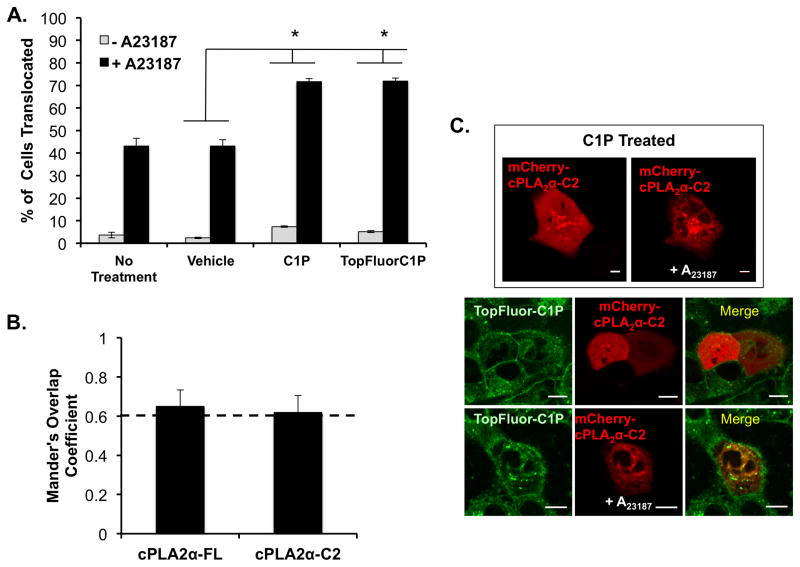
C1P has previously been shown to activate cytosolic phospholipase A2α (cPLA2α) in a calcium dependent manner (Subramanian et al., 2005). In addition, cPLA2α associates with C1P using a cluster of cationic residues (Stahelin et al., 2007, Ward et al., 2013) that are important for the C1P-dependent translocation of cPLA2α (Lamour et al., 2009, Ward et al., 2013). Thus, we employed cellular translocation assays of cPLA2α to assess the ability of TopFluor-C1P to recruit a C1P protein effector to the membrane when compared to C1P without a fluorescent tag. As shown in figure 3, we first assessed the number of cells displaying detectable translocation by performing 3 independent experiments with 20–70 cells assessed per experiment. Four different treatment regimens (with or without the calcium ionophore A23187) were used: no treatment (with or without A23187), vehicle (with or without A23187), C1P (with or without A23187), or TopFluor-C1P (with or without A23187). As previously reported (Lamour et al., 2009), A23187 was sufficient to induce cPLA2α translocation to internal membranes in >40% of cells. No significant translocation was detectable with vehicle in the absence of A23187. C1P in combination with A23187 induced a statistically significant increase in the translocation efficiency (>70% of cells) of cPLA2α as previously reported (Ward et al., 2013). Notably, the TopFluor-C1P was able to induce significant cPLA2α translocation in the presence of A23187 indicating the TopFluor-C1P was able to mimic C1P in the cPLA2α cellular membrane binding mechanism.
Figure 3. Translocation and colocalization of cPLA2α with C1P and TopFluor-C1P.
A) A549 cells expressing cPLA2α were assessed for translocation to intracellular membranes following treatment with or without the calcium ionophore A23187. In addition to A23187, cells were either treated with vehicle control, C1P, or TopFluor-C1P. The percentage of cells exhibiting cPLA2α translocation from the cytosol to the Golgi were counted. In three separate experiments, 20–70 cells were counted for each condition to display the average ± SEM. B) Colocalization analysis was performed for the entire cell image using TopFluor-C1P with both mCherry fusion constructs of full length and the isolated C2 domain of cPLA2α. C) Several representative images are displayed with the addition of 10 μM A23187 between C1P treated cells and TopFluor-C1P treated cells. The merged images display colocalization with TopFluor-C1P when A23187 is added to cells to promote binding and translocation of cPLA2α. Scale bar = 10 μm. * Indicates p <0.05 statistical significance.
To further validate the localization of TopFluor-C1P with respect to cPLA2α, we performed colocalization analysis of either cPLA2α or the isolated C2 domain with TopFluor-C1P. Cells were treated with A23187 and TopFluor-C1P to monitor the translocation of mCherry-cPLA2α or mCherry-cPLA2α-C2. The Mander’s overlap coefficient analysis indicated TopFluor-C1P co-localized with cPLA2α or the isolated C2 domain above the 0.6 coefficient threshold (Figure 3B and C). Note the lower but statistically significant Mander’s coefficient for this analysis isn’t unexpected as the whole cell was analyzed and TopFluor-C1P has a significant localization to the plasma membrane, whereas cPLA2α and C2 domain membrane translocation is restricted to internal membranes.
3.4 Membrane Dynamics of TopFluor-C1P
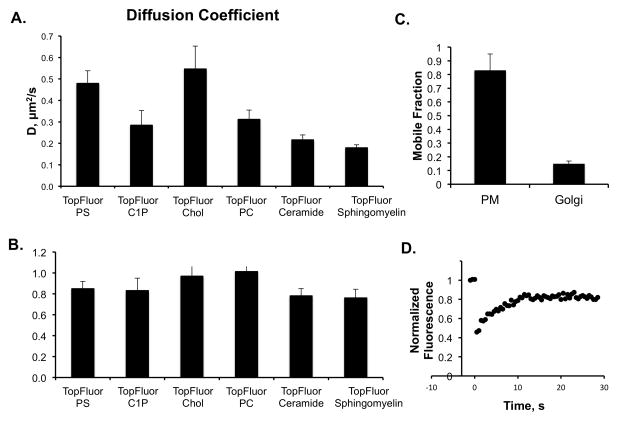
Little is known regarding the membrane dynamics of C1P in cellular membranes and how its lipid dynamics compare to those of other more well characterized lipids. We employed FRAP to determine the diffusion coefficient and mobile fraction of TopFluor-C1P in the plasma membrane and the Golgi of A549 cells. We also determined the diffusion coefficient for TopFluor-PS, TopFluor-cholesterol, TopFluor-phosphatidylcholine (PC), TopFluor-Ceramide, and TopFluor-Sphingomyelin as cellular diffusion of some of these lipids has been investigated previously (Kay et al., 2012, Maekawa and Fairn, 2014, Ries et al., 2009).
We performed FRAP using a ROI of 2.2 μm diameter in the plasma membrane or the Golgi (TopFluor-C1P only) where after 3 pre-bleach scans, the ROI was bleached for 10 iterations at 488 nm with 100% laser power. The post-bleach profile was then scanned for 30 seconds for each lipid treatment. This allowed us to determine the diffusion coefficient and mobile fraction (see methods) for each fluorescently labeled lipid. As previously reported, TopFluor-PS (0.48 ± 0.06 μm2/s) and TopFluor-cholesterol (0.55 ± 0.11 μm2/s) (See table 1) had a similar diffusion coefficient in the plasma membrane (Kay et al., 2012, Maekawa and Fairn, 2014, Ries et al., 2009), 0.49 μm2/s and 0.33 μm2/s, respectively. Some differences may be expected due to different cell lines employed and the use of Bodipy-cholesterol in the prior work (Ries et al., 2009). In contrast to TopFluor-PS and TopFluor-cholesterol, TopFluor-C1P exhibited an even slower diffusion coefficient of 0.28 ± 0.07 μm2/s TopFluor-PC also displayed a slower dissociation rate in the plasma membrane (0.31 ± 0.04 μm2/s) similar to that of TopFluor-C1P. TopFluor-ceramide and TopFluor-sphingomyelin, which display more structure similarity to TopFluor-C1P displayed even slower diffusion coefficients, 0.22 ± 0.02 μm2/s and 0.18 ± 0.01 μm2/s, respectively.
Table 1.
Diffusion coefficient of different TopFluor lipids in the plasma membrane of A549 cells.
| Diffusion Coeff. (μm2/s) | Average | SEM |
|---|---|---|
| TopFluor-PS | 0.48 | 0.06 |
| TopFluor-C1P | 0.28 | 0.07 |
| TopFluor-Chol | 0.55 | 0.11 |
| TopFluor-PC | 0.31 | 0.04 |
| TopFluor-Ceramide | 0.22 | 0.02 |
| TopFluor-Sphingomyelin | 0.18 | 0.01 |
We also determined the mobile fraction of each lipid in the FRAP experiments (See table 2). All four fluorescently labeled lipids had a mobile fraction of greater than 75% in the plasma membrane. TopFluor-cholesterol and TopFluor-PC were nearly fully mobile with a mobile fraction near 1 while TopFluor-PS and TopFluor-C1P had mobile fractions of 0.85 ± 0.07 and 0.83 ± 0.12, respectively. Similarly, TopFluor-ceramide and TopFluor-sphingomyelin had mobile fractions of 0.78 ± 0.07 and 0.76 ± 0.09, respectively. We also attempted to measure the diffusion coefficient of TopFluor-C1P in the Golgi membrane, however, this was difficult as only ~15% of TopFluor-C1P was mobile in the Golgi.
Table 2.
Mobile fraction of different TopFluor lipids in the plasma membrane of A549 cells.
| Mobile Fraction | Average | SEM |
|---|---|---|
| TopFluor-PS | 0.85 | 0.07 |
| TopFluor-C1P | 0.83 | 0.12 |
| TopFluor-Chol | 0.97 | 0.13 |
| TopFluor-PC | 1.01 | 0.09 |
| TopFluor-Ceramide | 0.78 | 0.07 |
| TopFluor-Sphingomyelin | 0.76 | 0.09 |
4. Discussion
4.1 Biophysical properties of TopFluor-C1P
Fluorescent probes have been valuable tools to study lipid biophysics and lipid dynamics in biological systems (Maekawa and Fairn, 2014). Fluorescently labeling lipids is a challenging task as head group labeling may affect binding and specificity of peripheral proteins while acyl chain labeling may alter lipid packing in the hydrophobic environment (Abrams and London, 1993, Chattopadhyay and London, 1987). The goal has been to find a probe that is sensitive for desired fluorescence experiments but does not significantly perturb the membrane. Recently, the TopFluor fluorescent label has become available on several glycerophospholipids, sterols, and sphingolipids and has been shown to be superior to the NBD label for studying PS in vitro and in cells (Kay et al., 2012). Here, using an array of in vitro and cellular imaging techniques, we have found that TopFluor-C1P successfully mimics unlabeled C1P in lipid-protein interaction, subcellular distribution, and further is not toxic to a human cancer cell line. We also found that TopFluor-C1P diffusion in the plasma membrane of A549 cells is similar to other sphingolipids.
Fluorescence experiments at different pH values, meant to represent the pH values found from the cellular cytoplasm down to the acidic pH of the lysosomes, demonstrated TopFluor-C1P robustly fluoresces over a wide physiological pH range. This was also investigated because Kooijman and colleagues hypothesized that C1P binds to proteins in both a pH and calcium dependent manner (Kooijman et al., 2008, Kooijman et al., 2009), which affects the microenvironment and could affect fluorescence, but our data indicate TopFluor-C1P should be stable under these different conditions.
The fluorescent label on TopFluor-C1P, which is not readily accessible at the membrane-water interface, seems to be buried in the hydrocarbon core similar to the TopFluor-PS shown in figure 1 and in previous work (Kay et al., 2012). TopFluor-C1P also displays typical fluorescence spectra patterns when varying the C1P mol% in liposomes. At TopFluor-C1P concentrations above 10% we observed a self-quenching event, which is typical of Top-Fluor labeled lipids (Kay et al., 2012). This might be explained by the observation that at increasing concentrations of C1P, there is a phase change and formation of non-lamellar structure (Kooijman et al., 2008). Additionally, quenching can result from transient excited state interactions, which commonly occurs in high loading concentrations of a fluorophore (Ogawa et al., 2009). Since C1P is thought to be a minor component of lipid membranes, and a small mol% of C1P (1–3 mol%) is sufficient to recruit protein effectors to liposomes (Hankins et al., 2013, Lamour et al., 2011, Ward et al., 2013), it’s recommended to study TopFluor-C1P under 5 mol% in liposomes. TopFluor-C1P also seems to be a valuable probe because of its hydrophobic character and ability to tightly fit within membranes with little disruption. This was shown with iodide quenching assays, where at increasing concentration of iodide (Figure 1E), the fluorophore was not significantly quenched. This means the fluorophore is able to sit tightly within he hydrophobic core of the membrane without forcing the acyl chain to migrate towards the membrane-water interface. This also suggests that TopFluor-C1P does not affect the head group orientation of C1P in membranes, allowing head group recognition and binding of proteins to remain unaltered (Figures 1C and 3).
4.2 TopFluor-C1P cellular and trafficking data
In mammalian cells, ceramide is trafficked from the ER to the TGN (Hanada, 2006), where it is converted to C1P by ceramide kinase (Lamour et al., 2007). From the TGN, C1P is trafficked to the plasma membrane by the recently discovered CPTP protein (Simanshu et al., 2013). We performed organelle co-localization analysis to determine where TopFluor-C1P localized within the A549 cells. Our findings indicate TopFluor-C1P was found predominately at the Golgi and the plasma membrane, consistent with previous studies on C1P localization (Simanshu et al., 2013). How TopFluor-C1P is trafficked from the plasma membrane to the Golgi of a mammalian cell from a microemulsion/organic solvent solution addition is still unknown. We speculate that high concentrations of C1P in the plasma membrane may trigger a retrograde transport of C1P through CPTP from the plasma membrane to the TGN. Silencing of CPTP previously lead to accumulation of C1P in the TGN (Simanshu et al., 2013) but it is unknown how cellular membranes and trafficking mechanisms respond when the C1P levels in the plasma membrane rise about those typically found in healthy cells. There also could be alternative lipid transport proteins of vesicular trafficking mechanisms for C1P that have yet to be elucidated.
Extensive work has previously described that C1P binds and activates cPLA2α in a calcium-dependent manner (Subramanian et al., 2005). cPLA2α activity liberates arachidonic acid and activates the COX2 pathway involved inflammation. Eicosanoids are further metabolized by downstream enzymes such as prostaglandin synthase D, which may also interact with C1P in the Golgi (Bidlingmaier et al., 2016). Here we show that TopFluor-C1P can recruit cPLA2α or its isolated N-terminal C2 domain, which harbors the primary C1P interaction site (Lamour et al., 2009, Stahelin et al., 2007, Ward et al., 2013). SPR experiments for the C2 domain displayed binding for TopFluor-C1P or C1P in POPC vesicles to be similar (~2.5-fold weaker for TopFluor-C1P). The modest difference in affinity for the two C1P molecules embedded in liposomes is still unknown, but TopFluor-C1P did cause membranes to be more rigid compared to native C1P as evidenced from the laurdan assay. This difference in membrane rigidity between labeled and unlabeled C1P may help explain the ~2.5-fold difference in the apparent Kd for the cPLA2α C2 domain. Since TopFluor-C1P mimicked unlabeled C1P in A549 cells, it strongly suggests TopFluor-C1P can recapitulate C1P in recruitment of some C1P effector proteins. The C2 domain of C1P is thought to bind to cPLA2α through head group recognition at the conserved RxRH sequence in the C2 domain (Ward et al., 2013), but the mechanisms of how other C1P binding proteins such as Annexin A2 (Hankins et al., 2013) or TACE (Lamour et al., 2011) bind C1P is unknown. This suggests further studies are needed to decipher if C1P can interact with binding effectors that harborg different modes of C1P recognition.
4.3 TopFluor diffusion coefficients and interactions at the plasma membrane
FRAP experiments were employed to investigate the diffusion coefficient of Topfluor-C1P in the plasma membrane and Golgi membranes. TopFluor-C1P is much more dynamic at the plasma membrane than at the Golgi, with an almost 2-fold increase in the mobile fraction. By taking a further look into TopFluor-C1P at the plasma membrane, we calculated the diffusion coefficient of TopFluor-C1P, TopFluor-cholesterol, TopFluor-PC, TopFluor-PS, TopFluor-ceramide, and TopFluor-sphingomyelin. TopFluor-C1P had a diffusion coefficient slower than that of PS or cholesterol, and notably TopFluor-PS and TopFluor-cholesterol had similar diffusion coefficients as reported previously (Kay et al., 2012). Interestingly, TopFluor-PC also had a slower diffusion coefficient than PS or cholesterol in the plasma membrane but to the best of our knowledge, fluorescent PC dynamics in the plasma membrane aren’t well studied. What is clear is that TopFluor-C1P has as slower diffusion coefficient in the plasma membrane than other anionic lipids such as PS (Kay et al., 2012, Maekawa and Fairn, 2014) and PI(4,5)P2 (Golebiewska et al., 2011) which are known to have hundreds of effectors proteins (Moravcevic et al., 2012, Stahelin et al., 2014). TopFluor-C1P had more comparable diffusion coefficient values with Topfluor-ceramide and TopFluor-sphingomyelin, which are known to have spontaneous movement into micrometric domains (Tyteca et al., 2010). This slower diffusion coefficient of TopFluor-C1P compared to glycerophopholipids is consistent with the sphingosine backbone on C1P, as ceramide and sphingomyelin have been implicated in membrane raft (Castro et al., 2014, Rog and Vattulainen, 2014) and gel-like domain structures (Taniguchi et al., 2006). Our data also coincide with previous studies showing higher mobility for NBD-cholesterol than NBD-ceramide (Oghalai et al., 1999). Importantly, TopFluor-C1P was shown to increase the rigidity of membrane in vitro compared to unlabeled C1P so its important to consider that TopFlour-C1P may be more readily incorporated into raft-like regions of the plasma membrane compared to native C1P.
Since TopFluor-C1P recruited cPLA2α to the Golgi, we also hypothesize that TopFluor-C1P should be successfully transported by CPTP (Simanshu et al., 2013) from the TGN to the plasma membrane in cells. It was previously shown by Zhai et al. that the human glycolipid transfer protein, with structural similarity to CPTP, was successful in transfer of BODIPY-GalCer (Zhai et al., 2013) in a similar manner to GalCer. If CPTP is able to transport TopFluor-C1P from the TGN to the plasma membrane, this may contribute to the restoration of some of the TopFluor-C1P in the plasma membrane following photobleaching.
5. Conclusions
In closing, TopFluor-C1P is a valuable lipid mimetic that can be further utilized to study many important biophysical and cellular questions regarding C1P properties in membranes and its role in lipid-lipid and lipid-protein interactions. The bilayer leaflet distribution of TopFluor-C1P is unknown in the plasma membrane and TopFluor-C1P may be a favorable tool to study C1P dynamics across the plasma membrane interface, somewhat akin to what has been done to study PS movement and asymmetry (Daleke, 2007).
Supplementary Material
Figure 4. Fluorescence Recovery After Photo-bleaching (FRAP) with TopFluor lipids.
A) The diffusion coefficient was determined from three independent experiments for each TopFluor lipid shown in the plasma membrane of A549 cells. B) The mobile fraction of the four different TopFluor lipids was calculated from photo bleaching of a 1.1 μm radius circle at the plasma membrane followed by analysis in ImageJ and Kaleidagraph. C) Mobile fraction of TopFluor-C1P after FRAP at the plasma membrane or Golgi of A549 cells. D) A normalized fluorescence versus time plot is shown for the FRAP of TopFluor-C1P in the plasma membrane of A549 cells.
Acknowledgments
This work has been supported by a grant from the American Heart Association (GRNT12080254) and Walther Cancer Foundation to R.V.S. The imaging studies have been supported by the Indiana University School of Medicine-South Bend Imaging and Flow Cytometry Core. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE-1313583 (C.M.S.). Any opinion, findings, and conclusions or recommendations expressed in this material are those of the authors(s) and do not necessarily reflect the views of the National Science Foundation. Additionally, a Notre Dame HCRI Research Like a Champion award supported the work (C.M.S.) K.E.W was supported by an American Heart Association predoctoral fellowship (11PRE7640028) and a NIH CBBI training fellowship (T32GM075762).
Footnotes
Author Contributions
C.M.S. and R.V.S. conceived of and designed the studies. C.M.S. performed all experiments except SPR. K.E.W. performed SPR experiments. C.M.S. analyzed the data. C.M.S. and R.V.S. interpreted the data. C.M.S. and R.V.S. wrote the manuscript.
Conflict of interest
The authors report no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrams FS, London E. Extension of the parallax analysis of membrane penetration depth to the polar region of model membranes: use of fluorescence quenching by a spin-label attached to the phospholipid polar headgroup. Biochemistry. 1993;32:10826–10831. doi: 10.1021/bi00091a038. [DOI] [PubMed] [Google Scholar]
- Arana L, Gangoiti P, Ouro A, Trueba M, Gomez-Munoz A. Ceramide and ceramide 1-phosphate in health and disease. Lipids in health and disease. 2010;9:15. doi: 10.1186/1476-511X-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana L, Ordonez M, Ouro A, Rivera IG, Gangoiti P, Trueba M, Gomez-Munoz A. Ceramide 1-phosphate induces macrophage chemoattractant protein-1 release: involvement in ceramide 1-phosphate-stimulated cell migration. American journal of physiology. Endocrinology and metabolism. 2013;304:E1213–1226. doi: 10.1152/ajpendo.00480.2012. [DOI] [PubMed] [Google Scholar]
- Bidlingmaier S, Ha K, Lee NK, Su Y, Liu B. Proteome-wide Identification of Novel Ceramide-binding Proteins by Yeast Surface cDNA Display and Deep Sequencing. Molecular & cellular proteomics: MCP. 2016;15:1232–1245. doi: 10.1074/mcp.M115.055954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boath A, Graf C, Lidome E, Ullrich T, Nussbaumer P, Bornancin F. Regulation and traffic of ceramide 1-phosphate produced by ceramide kinase: comparative analysis to glucosylceramide and sphingomyelin. The Journal of biological chemistry. 2008;283:8517–8526. doi: 10.1074/jbc.M707107200. [DOI] [PubMed] [Google Scholar]
- Castro BM, Prieto M, Silva LC. Ceramide: a simple sphingolipid with unique biophysical properties. Progress in lipid research. 2014;54:53–67. doi: 10.1016/j.plipres.2014.01.004. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay A, London E. Parallax method for direct measurement of membrane penetration depth utilizing fluorescence quenching by spin-labeled phospholipids. Biochemistry. 1987;26:39–45. doi: 10.1021/bi00375a006. [DOI] [PubMed] [Google Scholar]
- Daleke DL. Phospholipid flippases. The Journal of biological chemistry. 2007;282:821–825. doi: 10.1074/jbc.R600035200. [DOI] [PubMed] [Google Scholar]
- Gangoiti P, Granado MH, Alonso A, Goni FM, Gomez-Munoz A. Implication of ceramide, ceramide 1-phosphate and sphingosine 1-phosphate in tumorigenesis. Translational oncogenomics. 2008a;3:81–98. [PMC free article] [PubMed] [Google Scholar]
- Gangoiti P, Granado MH, Arana L, Ouro A, Gomez-Munoz A. Activation of protein kinase C-alpha is essential for stimulation of cell proliferation by ceramide 1-phosphate. FEBS letters. 2010;584:517–524. doi: 10.1016/j.febslet.2009.11.086. [DOI] [PubMed] [Google Scholar]
- Gangoiti P, Granado MH, Wang SW, Kong JY, Steinbrecher UP, Gomez-Munoz A. Ceramide 1-phosphate stimulates macrophage proliferation through activation of the PI3-kinase/PKB, JNK and ERK1/2 pathways. Cellular signalling. 2008b;20:726–736. doi: 10.1016/j.cellsig.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Gault CR, Obeid LM, Hannun YA. An overview of sphingolipid metabolism: from synthesis to breakdown. Advances in experimental medicine and biology. 2010;688:1–23. doi: 10.1007/978-1-4419-6741-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golebiewska U, Kay JG, Masters T, Grinstein S, Im W, Pastor RW, Scarlata S, McLaughlin S. Evidence for a fence that impedes the diffusion of phosphatidylinositol 4,5-bisphosphate out of the forming phagosomes of macrophages. Molecular biology of the cell. 2011;22:3498–3507. doi: 10.1091/mbc.E11-02-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Munoz A, Gangoiti P, Arana L, Ouro A, Rivera IG, Ordonez M, Trueba M. New insights on the role of ceramide 1-phosphate in inflammation. Biochimica et biophysica acta. 2013;1831:1060–1066. doi: 10.1016/j.bbalip.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Gomez-Munoz A, Gangoiti P, Granado MH, Arana L, Ouro A. Ceramide-1- phosphate in cell survival and inflammatory signaling. Advances in experimental medicine and biology. 2010;688:118–130. doi: 10.1007/978-1-4419-6741-1_8. [DOI] [PubMed] [Google Scholar]
- Hanada K. Discovery of the molecular machinery CERT for endoplasmic reticulum- to-Golgi trafficking of ceramide. Molecular and cellular biochemistry. 2006;286:23–31. doi: 10.1007/s11010-005-9044-z. [DOI] [PubMed] [Google Scholar]
- Hankins JL, Ward KE, Linton SS, Barth BM, Stahelin RV, Fox TE, Kester M. Ceramide 1-phosphate mediates endothelial cell invasion via the annexin a2-p11 heterotetrameric protein complex. The Journal of biological chemistry. 2013;288:19726–19738. doi: 10.1074/jbc.M113.481622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannun YA, Obeid LM. Many ceramides. The Journal of biological chemistry. 2011;286:27855–27862. doi: 10.1074/jbc.R111.254359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinkovska-Galcheva V, Boxer LA, Kindzelskii A, Hiraoka M, Abe A, Goparju S, Spiegel S, Petty HR, Shayman JA. Ceramide 1-phosphate, a mediator of phagocytosis. The Journal of biological chemistry. 2005;280:26612–26621. doi: 10.1074/jbc.M501359200. [DOI] [PubMed] [Google Scholar]
- Kang M, Day CA, DiBenedetto E, Kenworthy AK. A quantitative approach to analyze binding diffusion kinetics by confocal FRAP. Biophysical journal. 2010;99:2737–2747. doi: 10.1016/j.bpj.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JG, Koivusalo M, Ma X, Wohland T, Grinstein S. Phosphatidylserine dynamics in cellular membranes. Molecular biology of the cell. 2012;23:2198–2212. doi: 10.1091/mbc.E11-11-0936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JD, Falke JJ. Single-molecule fluorescence studies of a PH domain: new insights into the membrane docking reaction. Biophysical journal. 2009;96:566–582. doi: 10.1016/j.bpj.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooijman EE, Chupin V, Fuller NL, Kozlov MM, de Kruijff B, Burger KN, Rand PR. Spontaneous curvature of phosphatidic acid and lysophosphatidic acid. Biochemistry. 2005;44:2097–2102. doi: 10.1021/bi0478502. [DOI] [PubMed] [Google Scholar]
- Kooijman EE, Sot J, Montes LR, Alonso A, Gericke A, de Kruijff B, Kumar S, Goni FM. Membrane organization and ionization behavior of the minor but crucial lipid ceramide-1-phosphate. Biophysical journal. 2008;94:4320–4330. doi: 10.1529/biophysj.107.121046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooijman EE, Tieleman DP, Testerink C, Munnik T, Rijkers DT, Burger KN, de Kruijff B. An electrostatic/hydrogen bond switch as the basis for the specific interaction of phosphatidic acid with proteins. The Journal of biological chemistry. 2007;282:11356–11364. doi: 10.1074/jbc.M609737200. [DOI] [PubMed] [Google Scholar]
- Kooijman EE, Vaknin D, Bu W, Joshi L, Kang SW, Gericke A, Mann EK, Kumar S. Structure of ceramide-1-phosphate at the air-water solution interface in the absence and presence of Ca2+ Biophysical journal. 2009;96:2204–2215. doi: 10.1016/j.bpj.2008.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamour NF, Stahelin RV, Wijesinghe DS, Maceyka M, Wang E, Allegood JC, Merrill AH, Jr, Cho W, Chalfant CE. Ceramide kinase uses ceramide provided by ceramide transport protein: localization to organelles of eicosanoid synthesis. Journal of lipid research. 2007;48:1293–1304. doi: 10.1194/jlr.M700083-JLR200. [DOI] [PubMed] [Google Scholar]
- Lamour NF, Subramanian P, Wijesinghe DS, Stahelin RV, Bonventre JV, Chalfant CE. Ceramide 1-phosphate is required for the translocation of group IVA cytosolic phospholipase A2 and prostaglandin synthesis. The Journal of biological chemistry. 2009;284:26897–26907. doi: 10.1074/jbc.M109.001677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamour NF, Wijesinghe DS, Mietla JA, Ward KE, Stahelin RV, Chalfant CE. Ceramide kinase regulates the production of tumor necrosis factor alpha (TNFalpha) via inhibition of TNFalpha-converting enzyme. The Journal of biological chemistry. 2011;286:42808–42817. doi: 10.1074/jbc.M111.310169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa M, Fairn GD. Molecular probes to visualize the location, organization and dynamics of lipids. Journal of cell science. 2014;127:4801–4812. doi: 10.1242/jcs.150524. [DOI] [PubMed] [Google Scholar]
- Moravcevic K, Oxley CL, Lemmon MA. Conditional peripheral membrane proteins: facing up to limited specificity. Structure. 2012;20:15–27. doi: 10.1016/j.str.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Kosaka N, Choyke PL, Kobayashi H. H-type dimer formation of fluorophores: a mechanism for activatable, in vivo optical molecular imaging. ACS chemical biology. 2009;4:535–546. doi: 10.1021/cb900089j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oghalai JS, Tran TD, Raphael RM, Nakagawa T, Brownell WE. Transverse and lateral mobility in outer hair cell lateral wall membranes. Hearing research. 1999;135:19–28. doi: 10.1016/s0378-5955(99)00077-5. [DOI] [PubMed] [Google Scholar]
- Ouro A, Arana L, Rivera IG, Ordonez M, Gomez-Larrauri A, Presa N, Simon J, Trueba M, Gangoiti P, Bittman R, Gomez-Munoz A. Phosphatidic acid inhibits ceramide 1-phosphate-stimulated macrophage migration. Biochemical pharmacology. 2014;92:642–650. doi: 10.1016/j.bcp.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Ries J, Chiantia S, Schwille P. Accurate determination of membrane dynamics with line-scan FCS. Biophysical journal. 2009;96:1999–2008. doi: 10.1016/j.bpj.2008.12.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera IG, Ordonez M, Presa N, Gangoiti P, Gomez-Larrauri A, Trueba M, Fox T, Kester M, Gomez-Munoz A. Ceramide 1-phosphate regulates cell migration and invasion of human pancreatic cancer cells. Biochemical pharmacology. 2016;102:107–119. doi: 10.1016/j.bcp.2015.12.009. [DOI] [PubMed] [Google Scholar]
- Rog T, Vattulainen I. Cholesterol, sphingolipids, and glycolipids: what do we know about their role in raft-like membranes? Chemistry and physics of lipids. 2014;184:82–104. doi: 10.1016/j.chemphyslip.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Simanshu DK, Kamlekar RK, Wijesinghe DS, Zou X, Zhai X, Mishra SK, Molotkovsky JG, Malinina L, Hinchcliffe EH, Chalfant CE, Brown RE, Patel DJ. Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids. Nature. 2013;500:463–467. doi: 10.1038/nature12332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahelin RV, Scott JL, Frick CT. Cellular and molecular interactions of phosphoinositides and peripheral proteins. Chemistry and physics of lipids. 2014;182:3–18. doi: 10.1016/j.chemphyslip.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahelin RV, Subramanian P, Vora M, Cho W, Chalfant CE. Ceramide-1- phosphate binds group IVA cytosolic phospholipase a2 via a novel site in the C2 domain. The Journal of biological chemistry. 2007;282:20467–20474. doi: 10.1074/jbc.M701396200. [DOI] [PubMed] [Google Scholar]
- Stock RP, Brewer J, Wagner K, Ramos-Cerrillo B, Duelund L, Jernshoj KD, Olsen LF, Bagatolli LA. Sphingomyelinase D activity in model membranes: structural effects of in situ generation of ceramide-1-phosphate. PloS one. 2012;7:e36003. doi: 10.1371/journal.pone.0036003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian P, Stahelin RV, Szulc Z, Bielawska A, Cho W, Chalfant CE. Ceramide 1-phosphate acts as a positive allosteric activator of group IVA cytosolic phospholipase A2 alpha and enhances the interaction of the enzyme with phosphatidylcholine. The Journal of biological chemistry. 2005;280:17601–17607. doi: 10.1074/jbc.M414173200. [DOI] [PubMed] [Google Scholar]
- Subramanian P, Vora M, Gentile LB, Stahelin RV, Chalfant CE. Anionic lipids activate group IVA cytosolic phospholipase A2 via distinct and separate mechanisms. Journal of lipid research. 2007;48:2701–2708. doi: 10.1194/jlr.M700356-JLR200. [DOI] [PubMed] [Google Scholar]
- Takeshita E, Kume A, Maeda Y, Sakai H, Sakane F. Diacylglycerol kinase gamma is a novel anionic phospholipid binding protein with a selective binding preference. Biochemical and biophysical research communications. 2014;444:617–621. doi: 10.1016/j.bbrc.2014.01.116. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Ohba T, Miyata H, Ohki K. Rapid phase change of lipid microdomains in giant vesicles induced by conversion of sphingomyelin to ceramide. Biochimica et biophysica acta. 2006;1758:145–153. doi: 10.1016/j.bbamem.2006.02.026. [DOI] [PubMed] [Google Scholar]
- Tyteca D, D’Auria L, Der Smissen PV, Medts T, Carpentier S, Monbaliu JC, de Diesbach P, Courtoy PJ. Three unrelated sphingomyelin analogs spontaneously cluster into plasma membrane micrometric domains. Biochimica et biophysica acta. 2010;1798:909–927. doi: 10.1016/j.bbamem.2010.01.021. [DOI] [PubMed] [Google Scholar]
- Verdaguer N, Corbalan-Garcia S, Ochoa WF, Fita I, Gomez-Fernandez JC. Ca(2+) bridges the C2 membrane-binding domain of protein kinase Calpha directly to phosphatidylserine. The EMBO journal. 1999;18:6329–6338. doi: 10.1093/emboj/18.22.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward KE, Bhardwaj N, Vora M, Chalfant CE, Lu H, Stahelin RV. The molecular basis of ceramide-1-phosphate recognition by C2 domains. Journal of lipid research. 2013;54:636–648. doi: 10.1194/jlr.M031088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijesinghe DS, Subramanian P, Lamour NF, Gentile LB, Granado MH, Bielawska A, Szulc Z, Gomez-Munoz A, Chalfant CE. Chain length specificity for activation of cPLA2alpha by C1P: use of the dodecane delivery system to determine lipid-specific effects. Journal of lipid research. 2009;50:1986–1995. doi: 10.1194/jlr.M800367-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai X, Momsen WE, Malakhov DA, Boldyrev IA, Momsen MM, Molotkovsky JG, Brockman HL, Brown RE. GLTP-fold interaction with planar phosphatidylcholine surfaces is synergistically stimulated by phosphatidic acid and phosphatidylethanolamine. Journal of lipid research. 2013;54:1103–1113. doi: 10.1194/jlr.M034744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinchuk V, Zinchuk O, Okada T. Quantitative colocalization analysis of multicolor confocal immunofluorescence microscopy images: pushing pixels to explore biological phenomena. Acta histochemica et cytochemica. 2007;40:101–111. doi: 10.1267/ahc.07002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.