Abstract
Hepatoblastoma (HB) is associated with aberrant activation of the β-catenin and Hippo/YAP signaling pathways. Overexpression of mutant β-catenin and YAP in mice induces HBs that express high levels of c-Myc (Myc). In light of recent observations that Myc is unnecessary for long-term hepatocyte proliferation, we have now examined its role in HB pathogenesis using the above model. Although Myc was found to be dispensable for in vivo HB initiation, it was necessary to sustain rapid tumor growth. Gene expression profiling identified key molecular differences between myc+/+ (WT) and myc−/− (KO) hepatocytes and HBs that explain these behaviors. In HBs, these included both Myc-dependent and Myc-independent increases in families of transcripts encoding ribosomal proteins, non-structural factors affecting ribosome assembly and function, and enzymes catalyzing glycolysis and lipid bio-synthesis. In contrast, transcripts encoding enzymes involved in fatty acid β-oxidation were mostly down-regulated. Myc-independent metabolic changes associated with HBs included dramatic reductions in mitochondrial mass and oxidative function, increases in ATP content and pyruvate dehydrogenase activity, and marked inhibition of fatty acid β-oxidation (FAO). Myc-dependent metabolic changes included higher levels of neutral lipid and acetyl-CoA in WT tumors. The latter correlated with higher histone H3 acetylation. Collectively, our results indicate that the role of Myc in HB pathogenesis is to impose mutually dependent changes in gene expression and metabolic reprogramming that are unattainable in non-transformed cells and that cooperate to maximize tumor growth.
Keywords: beta-catenin (B-catenin), hepatocellular carcinoma, metabolism, Myc (c-Myc), yes-associated protein (YAP), oxidative phosphorylation
Introduction
Hepatoblastoma (HB)2 is the most common pediatric liver tumor (1). Although quite rare, its incidence is increased ∼15-fold in very low birth weight infants (2). An even higher incidence (up to 5000-fold relative risk) occurs in individuals with genetic predispositions such as familial adenomatous polyposis (FAP) who also have high rates of colorectal cancer (CRC) later in life (3, 4). FAP is often associated with inactivating mutations in the adenomatous polyposis coli (APC) tumor suppressor gene, which encodes a critical component of the Wnt/β-catenin signaling pathway (5). In these cases, mutant APC fails to interact with β-catenin, which is instead stabilized and translocated to the nucleus, thus mimicking constitutive Wnt signaling. Intranuclear β-catenin associates with the transcription factor LEF/Tcf4 (6) and activates numerous target genes including c-Myc (Myc), which collectively promote transformation (7).
The molecular underpinnings of FAP and its HB and CRC predisposition (8) have guided our understanding of the more common sporadic form of HB. In contrast to the germ line APC mutations associated with familial HB and CRC, ∼85% of sporadic HBs harbor acquired β-catenin mutations, mostly in exon 3-encoded amino acids, that also relieve β-catenin of its APC-dependent regulation and allow for its constitutive nuclear localization (9, 10).
Most sporadic HBs also demonstrate nuclear localization of Yes-associated protein (YAP), a component of the Hippo tumor suppressor pathway that communicates with the Wnt/β-catenin pathway (10, 11). Hippo regulates the size, proliferation, and survival of normal organs and tissues and affects tumor growth (12). Hippo activation promotes the cytoplasmic sequestration of YAP, whereas Hippo inactivation causes nuclear translocation of YAP, association with the transcriptional enhancer associate domain (TEAD) factor, and activation of genes such as cyclin E and survivin (13, 14). YAP nuclear localization does occur in other liver cancers, but it is only in HB that it interacts with β-catenin (10). Overexpression of mutant forms of β-catenin and YAP in murine livers closely recapitulates pediatric HB (10, 11). Myc is a key target of β-catenin/Tcf4 and is deregulated in these experimental HBs and human HBs, and it could be a significant determinant of HB tumorigenesis (15, 16).
Myc is frequently overexpressed in other tumors where it has been proposed to promote the synthesis of macromolecular precursors and other anabolic changes necessary to support biomass accumulation and growth (17, 18). Consistent with this, genes encoding ribosomal proteins and rRNAs are primary targets of Myc and are consistently up-regulated in response to enforced Myc overexpression, as are genes involved in glycolysis and glutaminolysis (18, 19). Mitochondrial biogenesis and oxidative phosphorylation (Oxphos) are also Myc-dependent, particularly in vitro (20–22). However, much less is known about the role of Myc in tumors such as HB where it is not necessarily the inciting oncogene but is nonetheless deregulated (10, 15, 16).
The function of Myc in normal cells appears to be more context- and tissue-dependent than in transformed cells. For example, myc haplo-insufficient mice experience longer survival and accumulate less severe age-related degenerative changes than their wild-type counterparts (23). In contrast, global myc loss is embryonic lethal (24), whereas chronic Myc inhibition in adult mice is associated with long-term survival and only mild and reversible toxicities (25). Consistent with this latter finding, we have shown Myc to be dispensable for the ability of hepatocytes to replicate 50–100-fold and repopulate the diseased liver in a murine model of hereditary tyrosinemia (26). In this latter context, myc−/− (KO) hepatocytes showed modestly lower expression of ∼15% of transcripts encoding 40S and 60S ribosomal subunit proteins, a tendency to be more reliant on fatty acid oxidation as an energy source following fasting and a propensity to accumulate intracellular neutral lipid. These features were more pronounced following transplantation.
We now demonstrate that, in contrast to normal hepatic regeneration, Myc is an important determinant of HB pathogenesis in the context of mutant β-catenin and YAP overexpression. Rather than being required for tumor initiation, however, endogenous Myc maintains high rates of tumor growth. This role involves coordinating and maximizing ribosomal biogenesis, glycolysis, and lipid metabolism to keep space with increased biosynthetic needs.
Results
Tumor Growth but Not Initiation Is Myc-dependent
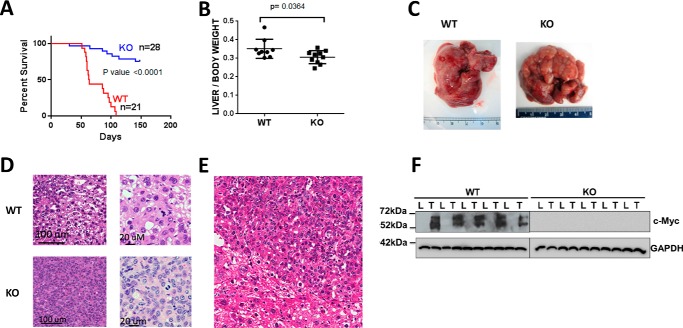
Sleeping Beauty (SB) vectors encoding oncogenic forms of β-catenin and YAP (10, 27) were delivered to the liver by hydrodynamic tail vein injection (HDTVI) to KO mice and age-matched mycflox/flox (WT) littermates (26). All WT mice succumbed to aggressive, multi-focal HBs within ∼16 weeks (WT HBs), whereas mean survival of KO mice exceeded 22 weeks, at which time the study was terminated (Fig. 1, A–C). The smaller size of tumors in this latter group (KO HBs) reflected this slower growth rate (Fig. 1, A and B).
FIGURE 1.
Properties of WT and KO tumors. A, survival curves of WT or KO mice inoculated with mutant β-catenin + YAP SB vectors by HDTVI. The study was terminated at week 22. B, liver weights from the survival curves shown in A. The results include all surviving KO mice. C, gross appearance of typical tumors arising in WT and KO livers. Note that although the depicted WT and KO tumor are of comparable size and appearance, they were obtained at different times due to the slower growth of the latter. D, histologic appearance of WT and KO HBs. Sections were stained with H&E. WT tumors showed small indistinct nodules with variation in nuclear size and frequent mitoses resembling pleomorphic fetal or HCC-like morphology. KO tumors were more distinct with a predominance of small uniform cells with abundant, eosinophilic cytoplasm and uniform small nuclei with inconspicuous to absent nucleoli and many mitoses. E, lower power view of H&E-stained section of normal liver-tumor border emphasizing the size differences between tumor cells (center and upper right) and normal hepatocytes (lower left). The crowded fetal morphology is more apparent. There was also significant nuclear unrest and variation in size in the liver surrounding the nodules. F, immunoblots for Myc protein in livers (L) and HBs (T) from WT and KO mice. GAPDH was included as a loading control. Error bars indicate ± S.E.
In WT HBs, the normal lobular architecture was replaced by small nodules that were indistinct from the surrounding parenchyma and were identifiable only by their appearance. Some areas resembled fetal HB with mitoses, whereas others resembled pleomorphic fetal HB or even hepatocellular carcinoma (HCC) (Fig. 1D). Individual cells varied in size from small with scant cytoplasm and uniform nuclei without nucleoli to larger cells with moderate nuclear pleomorphism and mitoses. Marked nuclear unrest and size variation were a common finding in the surrounding liver parenchyma.
KO HBs were more distinct and readily distinguishable from surrounding liver and also possessed a more pronounced nodular pattern. Microscopically, tumors were composed of uniform cells with eosinophilic cytoplasm, small round to oval nuclei, and indistinct nucleoli. Scattered mitoses were evident with virtually no atypical forms. Overall, the HBs resembled the fetal subtype with mitoses (crowded fetal pattern). Other KO HBs showed a more trabecular arrangement of tumor cells with moderate to marked nuclear pleomorphism and prominent nucleoli and increased nuclear-cytoplasmic ratio. This pattern had features resembling pleomorphic fetal HB with a close resemblance to HCC in some areas. Individual cells in both WT and KO HBs were also notably smaller than normal hepatocytes (Fig. 1E).
Myc protein was highly expressed by WT HBs but not by KO HBs (Fig. 1F), indicating that the latter did not originate from a minority population of hepatocytes that failed to excise myc. Transcriptional profiling showed some changes in N-Myc and L-Myc transcripts in KO tumors (supplemental Fig. S1A) and 2–3-fold increases in L-Myc protein in HBs (supplemental Fig. S1B). No expression differences were detected in other genes previously shown to complement the growth defect of myc−/− fibroblasts (28, 29) (supplemental Fig. S1B).
Reduced Mitochondrial Function and Mass in WT and KO HBs
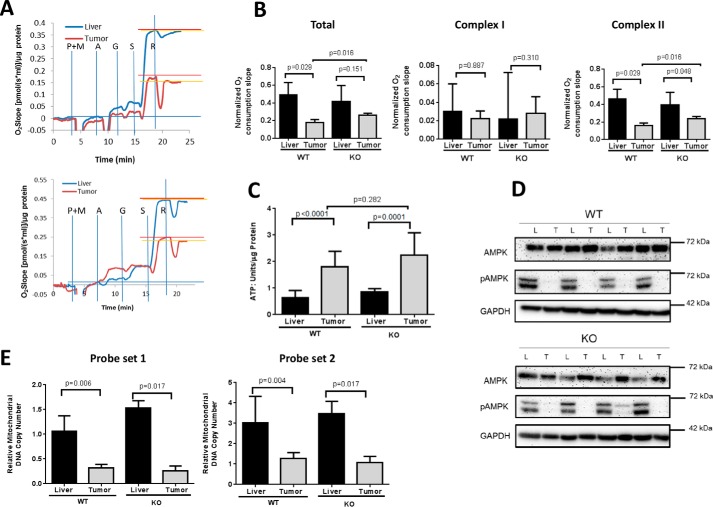
Myc supports mitochondrial energy metabolism, which can affect tumor growth (17, 18, 21, 22, 30). However, oxygen consumption rates (OCRs) in both WT and KO HBs were reduced relative to their corresponding livers but somewhat more so in the former group (Fig. 2, A and B). Virtually all the difference was attributable to the attenuated activity of Complex II. Non-denaturing, blue native gel electrophoresis and enzymatic activity profiling of isolated mitochondria showed no differences among electron transport chain Complexes I-IV. However, Complex V activity was slightly but significantly higher in WT HBs (supplemental Fig. S2). These results were consistent with the Warburg effect (31) and perhaps reflected an increased need for ATP production by the more rapidly growing WT HBs.
FIGURE 2.
Mitochondrial function in WT and KO livers and HBs. A, typical Oroboros Oxygraph 2k respirometer tracings in paired sets of WT (top) and KO (bottom) livers and tumors. Vertical blue lines indicate the points of addition of the Complex I substrates pyruvate (P), malate (M), and glutamine (G) and the Complex II substrate succinate (S). A = ADP; R = the Complex I inhibitor rotenone, the concentration of which had been previously titrated to provide maximal Complex I inhibition. The results depicted here were adjusted for differences in total protein levels. B, quantification of the results depicted in A. Each point represents total rates of oxygen consumption or the individual activities of Complex I or Complex II in response to their respective substrates. Total activity was calculated based on the spike in oxygen consumption following the addition of succinate. Complex II activity was derived by calculating the residual O2 consumption remaining after rotenone addition (horizontal orange lines in A). C, ATP levels. n = 4–5 samples/group. D, AMPK and phosphorylated AMPK (pAMPK) levels in tumors (T) and livers (L). Liver and tumor lysates, like those shown in in Fig. 1F, were assessed for total AMPK or its active (phospho-Thr172) form. GAPDH served as a control for protein loading. E, mitochondrial mass in livers and HBs. Two different probe sets (probe set 1 and probe set 2) were used to amplify mtDNA from two unique genomic regions using a TaqMan-based approach. mtDNA content was normalized to a nuclear DNA target that was amplified by a similar approach. n = 4–6 samples/group. Error bars indicate ± S.E.
Similarly elevated levels of ATP in WT and KO HBs were also consistent with the Warburg effect and suggested that HBs derive more ATP from glycolysis than normal livers (Fig. 2C). This implied that the slow growth rate of KO HBs was not due to a chronic lack of ATP as reported for Myc-KO fibroblasts (22). Further supporting this idea was the finding that levels of the phosphorylated, activated form of AMP-dependent protein kinase, which helps to maintain energy homeostasis by down-regulating energy-consuming biosynthetic processes and enabling energy-consuming processes such as proliferation (21, 31), were markedly and equally decreased in both WT and KO HBs. (Fig. 2D).
The similarity of electron transport chain function in livers and HBs (supplemental Fig. S2), suggested that the attenuated Oxphos of HBs might not only be a result of reduced AMPK activity but also of fewer mitochondria per cell. Indeed, mitochondrial DNA content was reduced by ∼65–80% in all tumors regardless of Myc status (Fig. 2E). This was confirmed by transmission electron microscopy, which also showed similarly sized mitochondria in all cases examined (supplemental Fig. S3).
Potential Rate-limiting Pathways for Tumorigenesis
To identify gene expression differences between WT and KO HBs that might explain their disparate growth rates, we performed RNA sequencing on representative tumors. Rather than the 102 differentially expressed transcripts that distinguish WT and KO hepatocytes (26), WT and KO HBs differed by 685 transcripts (q < 0.05, where q represents adjusted p values) (supplemental Fig. S4). Thus, either directly or indirectly, Myc regulates the expression of a much larger subset of genes in rapidly dividing HBs than in normal hepatocytes.
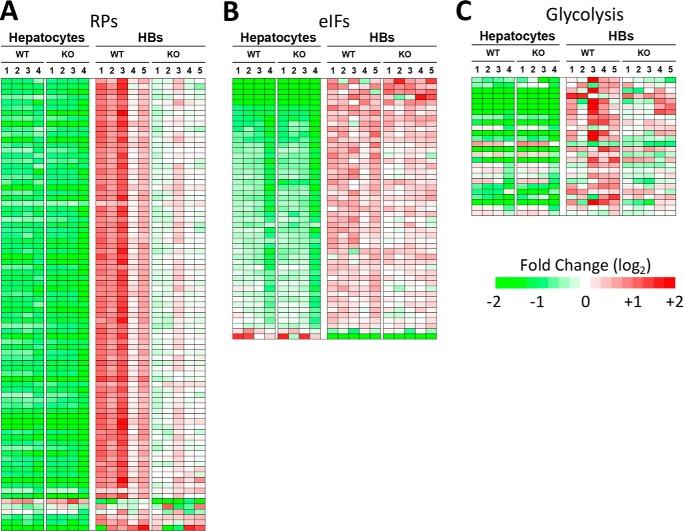
Ingenuity Pathway Analysis (IPA) was used to group these and additional relevant transcripts (p < 0.05 but q > 0.05) into functional categories. Of >600 queried pathways, the top three (p = 10−13–10−37) involved signaling by eukaryotic initiation factors (eIFs), p70S6K, and the mammalian target of rapamycin (mTOR) (32, 33). These included transcripts encoding 74 of 86 ribosomal proteins (RPs) comprising the 40S and 60S ribosomal subunits, nearly all of which were up-regulated in HBs (Fig. 3A and supplemental Fig. S5). As a group, these were increased more in WT HBs than in KO HBs relative to their corresponding hepatocytes (5.2-fold increase versus 3.6-fold increase, respectively; p < 0.0001 as determined by ratio t test).
FIGURE 3.
Transcriptional pathways that distinguish WT and KO HBs include those involved in ribosomal biogenesis, translational control and glycolysis. A, transcripts encoding RPs. Relative to WT hepatocytes, WT tumors up-regulated RP transcripts by an average of 5.2-fold versus 3.6-fold for KO HB (p < 0.0001). Red = up-regulated; green = down-regulated. B, transcripts identified by IPA as belonging to pathways involved in signaling by eIF2, eIF4, p70S6KJ, and mTOR. Each of these pathways also contained numerous RP transcripts, which are now included in A. As a group, these transcripts were similarly up-regulated in WT and KO HBs. C, transcripts encoding glycolytic enzyme. Relative to their corresponding livers, WT HB transcripts were up-regulated by an average of 11.2-fold, whereas KO HB transcripts were up-regulated by an average of 8.9-fold (p = 0.0004). Panels A–C also contain additional transcripts that did not meet the false discovery rate threshold but were nevertheless differentially expressed at a level of p < 0.05 (see supplemental Figs. S5–S7 for the identities of each set of transcripts).
Transcripts encoding non-structural ribosomal factors that positively affect translation were also up-regulated in tumors but were only modestly affected by Myc levels (p = 0.049) (Fig. 3B). Importantly, eIF4EBP3, a negative regulator of eIF4E, which represses translation by inhibiting the assembly of the eIF4F initiation complex (32, 33), was the only down-regulated transcript in this category (supplemental Fig. S6). Thus, WT and KO HBs differ significantly in the degree of RP transcript up-regulation but hardly at all with respect to transcripts encoding non-structural proteins involved in preinitiation complex formation and polysome assembly and stability.
Transcripts encoding glucose transporters and glycolytic enzymes were also more highly overexpressed in WT HBs (11.2-fold versus 8.9-fold in KO HBs: p = 0.0004: Fig. 3C and supplemental Fig. S7, A and B). Thus, as was true for RP transcripts, those involved in glycolysis were regulated by both Myc-dependent and Myc-independent processes.
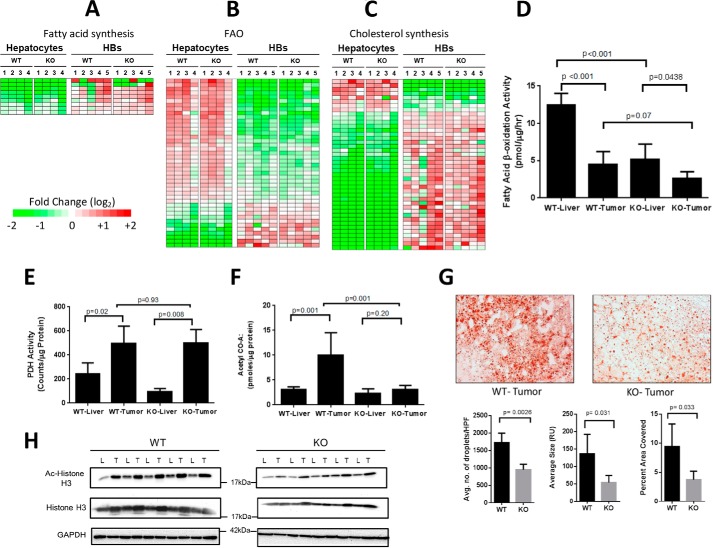
Aside from several pathways deemed unlikely to be hepatocyte-specific, such as those involving leukocyte activation (26), 8 of the top 14 remaining pathways distinguishing WT and KO HBs from hepatocytes involved fatty acid metabolism or cholesterol synthesis (supplemental Fig. S8) (p < 10−3). The majority of transcripts involved in fatty acid biosynthesis were up-regulated in both WT and KO tumors and encoded such critical proximal enzymes as ATP citrate lyase (ACLY), acetyl-Co-A carboxylase (ACAC), and fatty acid synthase (FASN) (Fig. 4A and supplemental Fig. S9). Concomitantly, multiple transcripts in the reciprocal fatty acid β-oxidation (FAO) pathway were down-regulated in tumors including carnitine palmitoyltransferase-2 (CPT2), very long-chain acyl-CoA dehydrogenase (VLCAD), and trifunctional protein (HADHA/HADHB) (Fig. 4B and supplemental Fig. S10).
FIGURE 4.
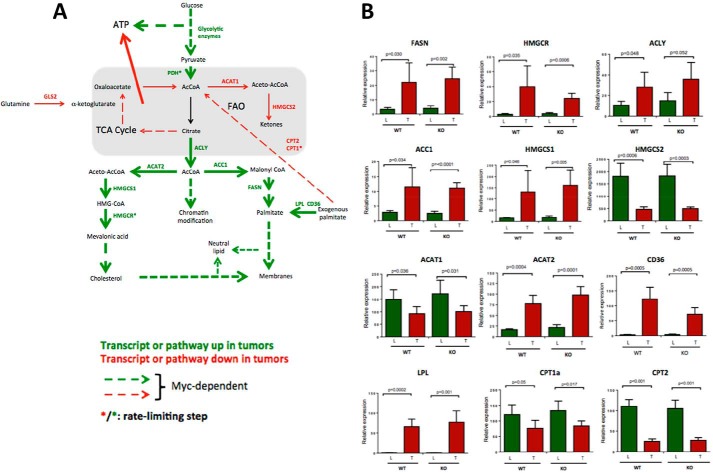
Deregulation of pathways involving lipid and acetyl-CoA metabolism in WT and KO HBs. A–C, differential expression of transcripts related to fatty acid synthesis, FAO, and cholesterol synthesis in WT and KO hepatocytes and HBs. Non-overlapping sets of transcripts from the relevant IPAs depicted in supplemental Fig. S8 are shown as are additional members of these pathways (q > 0.05> p: see supplemental Figs. S9–S11 for absolute expression differences among the transcripts depicted here). D, β-oxidation of [3H]palmitate in WT and KO livers and tumors. E, PDH assays were performed in triplicate on four samples from each of the indicated groups. F, acetyl Co-A levels in WT and KO livers and tumors. The results represent the mean of at least five samples/group each performed in triplicate. G, Oil Red O staining showing typical examples of neutral lipid staining in WT and KO HBs. Histograms beneath the micrographs show quantification for lipid droplet number, size, and intracellular area composed of lipid in a series of sections compiled from five representative tumors from each group. H, immunoblots of representative WT and KO livers (L) and tumors (T) probed with anti-acetyl histone H3 (Lys-9/Lys-14) or anti-histone H3 antibodies. Calculations of the Ac-H3:Total H3 ratios in each sample showed them to be higher in WT tumors than in KO tumors after adjusting to total histone H3 (p = 0.038). Error bars indicate ± S.E.
FAO also occurs in peroxisomes, which, although incapable of producing energy, can still pass chain-shortened medium-chain fatty acids and acetyl-CoA to mitochondria for further oxidation. Peroxisomal FAO-related transcripts were also down-regulated in tumors including those for the fatty acid transporter ATP-binding cassette-D3 (ABCD3), acyl-CoA oxidase-1 (ACOX1), and peroxisomal bifunctional protein (EHHADH) (Fig. 4B and supplemental Fig. S10).
Mostly up-regulated in tumors were transcripts encoding enzymes participating in cholesterol biosynthesis including 3-hydroxy-3-methylglutaryl-CoA synthase 1 (HMGCS1) and HMG coenzyme A reductase (HMGCR), the rate-limiting enzyme of the mevalonate pathway (Fig. 4C and supplemental Fig. S11). Transcripts for Cyp7b1 and Cyp46a1, needed for the production of bile acids from cholesterol (34), were the most down-regulated in tumors, implying that HBs are more likely to utilize free cholesterol for anabolic purposes than for excreted products such as bile acids. The extent to which fatty acid synthesis, FAO, and cholesterol synthesis transcripts were regulated was similar in WT and KO tumors (p ≥ 0.05).
Our transcriptional profiling indicated that HBs undergo both Myc-dependent and Myc-independent up-regulation of glucose utilization. The resulting carbon is then channeled into lipid biosynthetic pathways, which are up-regulated in a largely Myc-independent manner. Newly synthesized lipids are protected from metabolic consumption by the Myc-independent down-regulation of FAO. In support of this, the rate of [3H]palmitate β-oxidation was lower in HBs, particularly in KO HBs (Fig. 4D).
HBs also demonstrated significant Myc-independent up-regulation of pyruvate dehydrogenase (PDH)-mediated oxidation of [14C]pyruvate (Fig. 4E). This suggests that HB metabolic reprogramming drives glucose to acetyl-CoA, which is then converted to citrate, shunted to the cytoplasm, and reconverted to acetyl-CoA by ACLY for use in fatty acid synthesis. However, acetyl-CoA levels were significantly lower in KO HBs than WT HBs (Fig. 4F). Downstream products of acetyl-CoA metabolism were also lower in KO HBs, namely intracellular lipid stores (Fig. 4G) and acetylated nuclear histones (Fig. 4H). The latter utilize cytoplasmic acetyl-CoA as the source of their acetyl groups (35). These findings suggest a paucity of glucose-derived acetyl-CoA as being a critical factor that limits tumor growth in Myc KO HBs.
Glutamine uptake and conversion to α-ketoglutarate are frequently elevated in tumors and serve as a key source of citrate-derived acetyl-CoA and TCA cycle intermediates, which cannot otherwise be adequately provided by the diminished supply of glucose-derived pyruvate (36, 37). We therefore examined the expression of six transcripts involved in glutamine transport and glutaminolysis (supplemental Fig. S12A). Only those encoding the liver-specific glutamine transporter Slc1A5 and glutamine dehydrogenase (Glud1) were elevated in HBs. Most notably, transcripts encoding the rate-limiting enzyme glutaminase 2 (Gls2) were markedly reduced in HBs (supplemental Fig. S12B).
Next, we assessed by immunoblotting the expression of Gls2 and Glud1 and showed that both enzymes, but particularly Gls2, were reduced in both WT and KO HBs (supplemental Fig. S12C). Finally, we quantified the combined activities of these enzymes, which sequentially release two ammonium ions during the glutamine-to-α-ketoglutarate conversion process. Consistent with the foregoing results, overall conversion was significantly reduced in both WT and KO HBs (supplemental Fig. S12D). Taken together, these findings indicate that glutaminolysis does not appear to play a significant role in supplying alternate TCA substrates in HBs. More likely, any increase in glutamine uptake likely reflects increased protein and/or nucleic acid biosynthetic demands.
Discussion
β-Catenin and YAP deregulation is a hallmark of HBs, which commonly overexpress Myc (10, 15). A direct causal role for β-catenin and YAP in HB pathogenesis has recently been demonstrated (10), and we have confirmed that Myc is highly expressed in WT HBs (Fig. 1F and supplemental Fig. S1A).
Relative to control livers, most WT and KO HBs expressed higher levels of L-Myc (supplemental Fig. S1B), which can rescue myc−/− murine fibroblasts (38). However, the transcriptional activation domain of L-Myc is only 5–10% as active as that of Myc, and L-Myc is poorly transforming (39). It seems possible that the slower growth rates of KO tumors and their overall lower level of expression of Myc targets may reflect the inferiority of L-Myc as a transcriptional activator. We were unable to document differential expression of any other transcripts that encode negative regulators of Myc proteins or that complement myc−/− fibroblasts (28, 29). Thus, like myc−/− fibroblasts, KO tumors grow at a low basal rate, suggesting that Myc provides an additional growth stimulus beyond that needed to support basal, non-neoplastic functions. Ribosomal biogenesis and protein translation, both of which are highly energy-consuming processes, increase as a consequence of Myc overexpression and are rate-limiting determinants of many tumor types (40, 41). Unsurprisingly, therefore, transcripts encoding nearly 90% of all structural components of the 40S and 60S ribosomal subunits were elevated in both WT and KO HBs, although as a group, they were more highly expressed in the former (Fig. 3A and supplemental Fig. S5). This indicated that both Myc-dependent and Myc-independent pathways play distinct roles in this regulation. In contrast, transcripts encoding factors involved in translation-related processes such as cap-dependent initiation, mRNA start codon selection, and preinitiation complex assembly and stabilization (32, 33) were up-regulated in HBs regardless of Myc status, whereas a prominent negative regulator, eIF4EBP3, was down-regulated (Fig. 3B and supplemental Fig. S6). Thus, the control of HB translation involves the coordination of ribosomal structural components and associated factors that are necessary for the assembly of functional polysomes. That the former group is subject to significant Myc-dependent regulation, whereas the latter group is not, may reflect the participation of many of its members in processes other than translation (33, 42) Both the inability to fully maximize ribosomal biogenesis and the disparate stoichiometries of the structural and functional components of the translational machinery may contribute to the slower growth rate of KO HBs and perhaps to inefficiencies or loss of fidelity in global protein synthetic rates.
Our studies on mitochondrial function, substrate flux, acetyl-CoA levels, and metabolic pathway transcriptional profiling also point to a growth-limiting role for glucose-derived acetyl-CoA in Myc KO HBs. The Warburg effect involves a shift of ATP synthesis from the mitochondria to the cytosol, which was clearly manifested by changes in Oxphos, particularly in more rapidly growing WT HBs (Fig. 2B). An often under-appreciated aspect of the Warburg effect is that, even in the face of oxidative glycolysis, the TCA cycle continues to produce essential substrates such as citrate for biosynthetic purposes. Cytosolic acetyl-CoA-derived citrate is used to synthesize lipids needed by rapidly proliferating cells. The primary source of mitochondrial acetyl-CoA is pyruvate, produced by glycolysis and provided to the TCA cycle by PDH. PDH generates NADH, which must be reoxidized to NAD+ by Complex I. We found that HBs up-regulated PDH activity in a Myc-independent manner (Fig. 4E) and maintained Complex I-driven respiration but displayed reduced Complex II-driven respiration (Fig. 2, A and B). Because Complex II also fulfills the role of succinate dehydrogenase, HBs simultaneously down-regulate the distal TCA cycle while continuing to drive pyruvate through the proximal TCA cycle, thereby allowing the shunting of citrate to the cytosol for conversion to acetyl-CoA and, ultimately, lipids. In turn, these are protected from degradation by down-regulating both mitochondrial and peroxisomal FAO. Although Myc is not required for significant reprogramming of either the mitochondrial machinery that produces the acetyl-CoA or the cytosolic machinery that utilizes it, it is clearly required for reprogramming glycolysis, the pathway that produces the starting substrate (pyruvate). We thus propose that KO HBs have a pyruvate supply chain deficit that limits the production of acetyl-CoA, which in turn compromises lipid synthesis, a process that is ATP-dependent. Interestingly, KO HBs maintained steady-state ATP levels that were equivalent to WT HBs (Fig. 2C). The somewhat higher Complex II activity in KO versus WT HBs (Fig. 2B) may indicate that the former balance their lower glycolytic capacity in a Myc-independent manner by up-regulating Oxphos to maintain ATP levels in the face of reduced glycolysis.
Also Myc independent was the ∼65–80% reduction of mitochondrial mass seen in all HBs (Fig. 2E and supplemental Fig. S3). Although perhaps reflecting the overall smaller size of HB cells (Fig. 1E), recent findings conducted on numerous human cancers of various types (including HCCs) compiled from The Cancer Genome Atlas indicate that this may in fact be a more widespread phenomenon (43). The reduced mitochondrial content of HBs may therefore represent a general strategy for altering metabolism, particularly with regard to adopting the Warburg effect and its coincident down-regulation of Oxphos. Another possibility is that, relative to HBs, hepatocytes may possess inherent excess mitochondrial capacity as a way of rapidly adapting to the metabolic fluctuations that the liver typically experiences in response to fasting, feeding, and xenobiotic-mediated tissue damage. Such plasticity could well be subverted by tumor cells to provide a highly responsive and energetically advantageous way to adapt rapidly to constant fluctuations in environmental and metabolic cues (21, 22).
In summary, our work shows murine HBs to be highly Myc-dependent for growth but not initiation. Metabolic studies and transcriptional profiling revealed three categories of alterations that distinguish tumors and livers in general and WT and KO tumors specifically. The first includes Myc-independent alterations involving ATP levels, mitochondrial mass, PDH activity, and certain gene expression profiles. Because these are quite similar in WT and KO HBs, they do not appear to explain their differential growth rates. The second category of changes is Myc-dependent and affects lipid and acetyl Co-A content. Acetyl-Co-A is a key substrate that connects catabolic pathways (primarily glycolysis and FAO) to those providing energy (the TCA cycle) and anabolic substrates. Acetyl Co-A also coordinates the expression of a large suite of genes involved in cell cycle control, ribosomal biogenesis, and metabolism via epigenetic regulation at the level of histone acetylation (44). The acetyl Co-A disparity between WT and KO tumors (Fig. 4F) could potentially account for many of the observed differences that would be reinforced by Myc and its acetylation-dependent activation of gene expression (Fig. 4H). Finally, the third category of changes is clearly Myc-dependent and includes changes in Complex II activity and transcripts encoding RPs and glycolytic enzymes (Figs. 2B and 3, A and C). Fig. 5A summarizes some of these ideas, and Fig. 5B depicts the actual expression levels obtained for several of the key transcripts.
FIGURE 5.
Myc-dependent and Myc-independent pathways in HB tumorigenesis. A, summary of relevant tumor pathways and targets. ATP generated by the up-regulation of glycolysis in tumors is sufficient to offset the overall loss of ATP generated by reduced Oxphos. B, relevant levels of expression in hepatocytes and HBs of the transcripts encoding the enzymes depicted in A. Data were taken from the results shown in Fig. 4 and supplemental Fig. S9. Error bars indicate ± S.E. The abbreviations used are: FASN, fatty acid synthase; HMGCR, HMG-coenzyme A reductase; ACLY, ATP citrate lyase; ACC1, acetyl-CoA carboxylase; HMGCS1 & 2, HMG-coenzyme S synthase, cytoplasmic and mitochondrial, respectively; ACAT1&2, acetyl-CoA acetyltransferase: mitochondrial and cytoplasmic, respectively; CD36, receptor for thrombospondin, oxidized low density lipoprotein, oxidized phospholipids, long-chain fatty acids, and native lipoproteins; LPL, lipoprotein lipase; CPT1a, carnitine palmitoyltransferase 1A; CPT2, carnitine palmitoyltransferase; PDH, pyruvate dehydrogenase.
Notably, many of the differences between WT and KO HBs represent an extension or exaggeration of those seen in non-transformed hepatocytes (26). For example, WT and KO hepatocytes differentially express only 12 RP transcripts, and the latter accumulate neutral lipids only under fasting conditions. These results argue that many Myc targets are subject to a basal level of Myc-independent regulation that suffices to address the proliferative and metabolic demands imposed by normal stresses such as regeneration. In contrast, the unremitting proliferative and anabolic demands imposed by transformation may stress these capabilities beyond typical physiologic limits. These changes require high levels of Myc expression and potentially explain the phenomenon that has been termed “oncogene addiction” (45).
Experimental Procedures
Animal Studies
Animal studies were performed in accordance with the Public Health Service Policy on Humane Care and Use of Laboratory Animals Institute for Laboratory Animal Research (ILAR) Guide for Care and Use of Laboratory Animals. All experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Pittsburgh.
C57BL6 mycfl/fl (WT) mice (46) and hepatocyte-specific KO mice were housed and maintained under standard conditions (26). Genotyping was confirmed on DNA isolated from livers at the time of sacrifice (26).
SB vectors encoding mutant forms of human β-catenin (Δ90) and YAP (S27A) were delivered to the livers of 6–8-week-old WT and KO mice via HDTVI (10, 27). Animals were euthanized at the recommendation of a veterinarian when tumor burdens caused obvious distress. Otherwise healthy-appearing animals were euthanized at the study's conclusion (22 weeks).
Respirometry Studies
∼50 mg of tumor or liver was disrupted in ice-cold MiR05 buffer (110 mm sucrose; 0.5 mm EGTA; 3 mm MgCl2; 60 mm potassium lactobionate; 20 mm taurine; 10 mm KH2PO4; 20 mm HEPES, pH 7.2; and 1 mg/ml fatty acid free BSA) (20). OCR was quantified in 2 ml of Mir05 buffer containing ∼40 μg of tissue lysate and 10 μm cytochrome c using an Oroboros Oxygraph 2k instrument (Oroboros Instruments, Inc., Innsbruck, Austria). Malate (5 mm), pyruvate (5 mm), ADP (5 mm), and glutamate (10 mm) were sequentially added to initiate electron flow. Upon reaching a plateau OCR, 10 mm succinate was added to assess the combined activity of Complex I + II. Finally, rotenone (0.5 μm) was added to inhibit Complex I and to obtain a pure assessment of Complex II activity. Final activities were normalized to total protein (47).
RNA Sequencing and Analyses
RNA purification, processing, and sequencing were performed as described previously using Qiagen RNeasy columns (Qiagen, Inc., Valencia, CA) (26). Paired-end single-indexed sequencing (∼75 bp) was performed on an Illumina NextSeq 500 sequencer (San Diego, CA). Read counts were normalized across all samples, and differentially expressed genes were determined by adjusted p values (q values) with a threshold of 0.05. Raw and processed original data have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (48) and accessible through GEO (https://www.ncbi.nlm.nih.gov/) at accession number GSE87578.
PDH, Acetyl Coenzyme A, FAO, and Glutaminase Assays
PDH complex activity was measured in fresh liver and tumors. 50–80 mg of tissue was minced in 4 ml of ice-cold DMEM lacking glucose, glutamine, and pyruvate and passed several times through an 18-gauge needle. 0.5-ml aliquots were placed into 0.5 ml of 2× assay buffer (final concentration: 1.1 mm EDTA, 1.2 mm DTT, 1.6 mm NAD, 0.0075% sodium pyrophosphate, 0.6 mm pyruvate, and 0.15 μCi of [1-14C]pyruvate/ml in DMEM). Tubes were sealed with rubber stoppers fitted with a central hanging basket (Kimble-Chase Life Science and Research Products, Rockwood, TN) containing a 25-mm glass microfiber filter (Whatman/GE Healthcare) soaked in 0.5 m KOH. Reactions were incubated in a rocking 37 °C water bath and stopped by injecting 0.5 ml of 4.0 m perchloric acid through the stopper. 14CO2 was collected on the filters for 40 min at 37 °C and quantified by scintillation counting.
Acetyl-Co-A was measured using a fluorometry-based assay as recommended (Sigma-Aldrich, MAK039). 200–600 μg of total liver and HB lysates was incubated with the assay mix containing acetyl-Co-A substrates, conversion enzyme mix, and fluorescent probe detector. Reactions were performed in triplicate in 96-well plates at 37 °C for 15 min, and fluorescence intensities were measured at λex = 535/λem = 587 nm. The readings were normalized to input protein content.
FAO was measured as previously described (26). ∼50 mg of tissue was minced in SET buffer (250 mm sucrose, 1 mm EDTA, 10 μm Tris-HCl, pH 7.4) and subjected to five passes in a Potter-Elvehjem homogenizer on ice. Reactions contained 5 μl of homogenate in 195 μl of reaction buffer (100 mm sucrose, 10 mm Tris-HCl, 5 mm KH2PO4, 0.2 mm EDTA, 0.3% fatty acid-free BSA, 80 mm KCl, 1 mm MgCl2, 2 mm l-carnitine, 0.1 mm malate, 0.05 mm coenzyme A, 2 mm ATP, 1 mm DTT, pH 8.0, 125 μm palmitate, and 1 μCi of 3H-labeled BSA-conjugated palmitate (Perkin Elmer)). Reactions were incubated at 37 °C, and then stopped with 40 μl of 1 m KOH. Following incubation at 60 °C for 1 h to hydrolyze newly synthesized acyl-carnitine esters, 40 μl of 4 mm perchloric acid was added for an additional hour on ice. Following organic extraction(49), water-soluble FAO products, including 3H2O and [3H]acetyl-CoA, were measured by scintillation counting. Glutaminase assays were performed with a Glutaminase Microplate Assay Kit (Cohesion Biosciences, Inc., London, UK) using the directions provided by the supplier.
Immunoblotting and Immunofluorescence Staining
Immunoblotting was performed as previously described (26). All antibody information is shown in supplemental Table S1. Immunoblots were developed using a SuperSignalTM West Pico Chemiluminescent Substrate kit (Thermo Fisher).
Quantification of mtDNA
10 ng of total DNA was used in a TaqMan-based assay. Two PCR primer sets were used to amplify mtDNAs. Probe set 1 amplified 101 bp of the cytochrome c oxidase I gene encoded by the 16,295-bp murine mitochondrial reference genome (GenBankTM accession no.: NC_005089) (forward primer, 5′-CCAGATATAGCATTCCCACGAATA-3′; reverse primer, 5′-CCTGCTCCTGCTTCTACTATT-3′). The product was detected with the probe: 5′-/56-FAM/TCCTACCAC/ZEN-CATCATTTCTCCTTCTCCT-3IABkFQ/-3′). Probe set 2 amplified 90 bp of themitochondrial D-loop region (forward primer, 5′-AATCTACCATCCTCCGTGAAACC-3′; reverse primer, 5′-TCAGTTTAGCTACCCCCAAGTTTAA-3′) (50) and was detected with the probe: 5′-/56-FAM-CGCCCACCA/ZEN/ATGCCCCTCTTC-3IABkFQ/-3′. Reactions were normalized to a 73-bp PCR product of the nuclear apolipoprotein B gene using the following primers: forward, 5′-CACGTGGGCTCCAGCATT-3′; and reverse, 5′-TCACCAGTCATTTCTGCCTTTG-3′ whose product was detected with the TaqMan probe: 5′-/Cy5-CCAATGGTCGGGCACTGCTCAA-Black Hole Quencher 2-3′. All primers and probes were synthesized by IDT, Inc. (Coralville, IA). PCR reactions were performed on a CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad, Inc.) using the following conditions: 95 °C for 10 s; 40 cycles at 95 °C for 15 s; and 60 °C for 60 s.
Author Contributions
H. W., J. L., L. R. E., S. K., J. D., J. T., L. J., M. F., R. U., and S. B. carried out the experiments. H. W., S. P. M., E. S. G., and E. V. P. analyzed the data. S. R. and D. B.-S. carried out histopathological and electron microscopic examination of tumors. E. V. P., E. S. G., and H. W. devised experiments and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Note Added in Proof
The heat maps in the version of this article that was published as a Paper in Press on October 13, 2016 have been revised to conform to the more standard format of displaying gene expression changes.
This work was supported by National Institutes of Health Grants 5RO1 CA174713 (to E. V. P.) and 1R01 CA204586 and 1R01 DK100287 (to S. P. M.). This work was also supported by a predoctoral research award from The Children's Hospital of Pittsburgh of UPMC Research Advisory Committee (to L. R. E.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as a Paper of the Week.

This article contains supplemental Figs. S1–S12 and supplemental Table S1.
The raw and processed original data reported in this paper have been deposited in the NCBI Gene Expression Omnibus under GEO Series accession number GSE87578.
- HB
- hepatoblastoma
- HCC
- hepatocellular carcinoma
- CRC
- colorectal cancer
- FAP
- familial adenomatous polyposis
- YAP
- Yes-associated protein
- Oxphos
- oxidative phosphorylation
- HDTVI
- hydrodynamic tail vein injection
- SB
- Sleeping Beauty
- OCR
- O2 consumption rate
- PDH
- pyruvate dehydrogenase
- FAO
- fatty acid (palmitate) β-oxidation
- RP
- ribosomal protein
- AMPK
- AMP-activated protein kinase
- ACLY
- ATP citrate lyase
- FAM
- 6-carboxyfluorescein.
References
- 1.De Ioris M., Brugieres L., Zimmermann A., Keeling J., Brock P., Maibach R., Pritchard J., Shafford L., Zsiros J., Czaudzerna P., and Perilongo G. (2008) Hepatoblastoma with a low serum α-fetoprotein level at diagnosis: the SIOPEL group experience. Eur. J. Cancer 44, 545–550 [DOI] [PubMed] [Google Scholar]
- 2.Heck J. E., Meyers T. J., Lombardi C., Park A. S., Cockburn M., Reynolds P., and Ritz B. (2013) Case-control study of birth characteristics and the risk of hepatoblastoma. Cancer Epidemiol. 37, 390–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamada Y., Takada K., Fukunaga S., and Hioki K. (2003) Hepatoblastoma associated with Beckwith-Wiedemann syndrome and hemihypertrophy. Pediatr. Surg. Int. 19, 112–114 [DOI] [PubMed] [Google Scholar]
- 4.Spector L. G., and Birch J. (2012) The epidemiology of hepatoblastoma. Pediatr. Blood Cancer 59, 776–779 [DOI] [PubMed] [Google Scholar]
- 5.Karim R., Tse G., Putti T., Scolyer R., and Lee S. (2004) The significance of the Wnt pathway in the pathology of human cancers. Pathology 36, 120–128 [DOI] [PubMed] [Google Scholar]
- 6.Korinek V., Barker N., Morin P. J., van Wichen D., de Weger R., Kinzler K. W., Vogelstein B., and Clevers H. (1997) Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science 275, 1784–1787 [DOI] [PubMed] [Google Scholar]
- 7.Monga S. P. (2015) β-Catenin signaling and roles in liver homeostasis, injury, and tumorigenesis. Gastroenterology 148, 1294–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White B. D., Chien A. J., and Dawson D. W. (2012) Dysregulation of Wnt/β-catenin signaling in gastrointestinal cancers. Gastroenterology 142, 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Armengol C., Cairo S., Fabre M., and Buendia M. A. (2011) Wnt signaling and hepatocarcinogenesis: the hepatoblastoma model. Int. J. Biochem. Cell Biol. 43, 265–270 [DOI] [PubMed] [Google Scholar]
- 10.Tao J., Calvisi D. F., Ranganathan S., Cigliano A., Zhou L., Singh S., Jiang L., Fan B., Terracciano L., Armeanu-Ebinger S., Ribback S., Dombrowski F., Evert M., Chen X., and Monga S. P. (2014) Activation of β-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology 147, 690–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sylvester K. G., and Colnot S. (2014) Hippo/YAP, β-catenin, and the cancer cell: a “ménage à trois” in hepatoblastoma. Gastroenterology 147, 562–565 [DOI] [PubMed] [Google Scholar]
- 12.Kango-Singh M., and Singh A. (2009) Regulation of organ size: insights from the Drosophila Hippo signaling pathway. Dev. Dyn. 238, 1627–1637 [DOI] [PubMed] [Google Scholar]
- 13.Huang J., Wu S., Barrera J., Matthews K., and Pan D. (2005) The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122, 421–434 [DOI] [PubMed] [Google Scholar]
- 14.Pan D. (2010) The hippo signaling pathway in development and cancer. Dev. Cell 19, 491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cairo S., Armengol C., and Buendia M. A. (2012) Activation of Wnt and Myc signaling in hepatoblastoma. Front. Biosci. (Elite Ed.) 4, 480–486 [DOI] [PubMed] [Google Scholar]
- 16.Ranganathan S., Tan X., and Monga S. P. (2005) β-Catenin and Met deregulation in childhood hepatoblastomas. Pediatr. Dev. Pathol. 8, 435–447 [DOI] [PubMed] [Google Scholar]
- 17.Dang C. V. (2010) Rethinking the Warburg effect with Myc micromanaging glutamine metabolism. Cancer Res. 70, 859–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dang C. V. (2011) Therapeutic targeting of Myc-reprogrammed cancer cell metabolism. Cold Spring Harb. Symp. Quant. Biol. 76, 369–374 [DOI] [PubMed] [Google Scholar]
- 19.Campbell K. J., and White R. J. (2014) MYC regulation of cell growth through control of transcription by RNA polymerases I and III. Cold Spring Harb. Perspect. Med. 4, a018408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edmunds L. R., Sharma L., Kang A., Lu J., Vockley J., Basu S., Uppala R., Goetzman E. S., Beck M. E., Scott D., and Prochownik E. V. (2014) c-Myc programs fatty acid metabolism and dictates acetyl-CoA abundance and fate. J. Biol. Chem. 289, 25382–25392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edmunds L. R., Sharma L., Wang H., Kang A., d'Souza S., Lu J., McLaughlin M., Dolezal J. M., Gao X., Weintraub S. T., Ding Y., Zeng X., Yates N., and Prochownik E. V. (2015) c-Myc and AMPK control cellular energy levels by cooperatively regulating mitochondrial structure and function. PLoS ONE 10, e0134049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graves J. A., Wang Y., Sims-Lucas S., Cherok E., Rothermund K., Branca M. F., Elster J., Beer-Stolz D., Van Houten B., Vockley J., and Prochownik E. V. (2012) Mitochondrial structure, function and dynamics are temporally controlled by c-Myc. PLoS ONE 7, e37699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofmann J. W., Zhao X., De Cecco M., Peterson A. L., Pagliaroli L., Manivannan J., Hubbard G. B., Ikeno Y., Zhang Y., Feng B., Li X., Serre T., Qi W., Van Remmen H., Miller R. A., et al. (2015) Reduced expression of MYC increases longevity and enhances healthspan. Cell 160, 477–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis A. C., Wims M., Spotts G. D., Hann S. R., and Bradley A. (1993) A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 7, 671–682 [DOI] [PubMed] [Google Scholar]
- 25.Soucek L., Whitfield J., Martins C. P., Finch A. J., Murphy D. J., Sodir N. M., Karnezis A. N., Swigart L. B., Nasi S., and Evan G. I. (2008) Modelling Myc inhibition as a cancer therapy. Nature 455, 679–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edmunds L. R., Otero P. A., Sharma L., D'Souza S., Dolezal J. M., David S., Lu J., Lamm L., Basantani M., Zhang P., Sipula I. J., Li L., Zeng X., Ding Y., Ding F., et al. (2016) Abnormal lipid processing but normal long-term repopulation potential of myc−/− hepatocytes. Oncotarget 7, 30379–30395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X., and Calvisi D. F. (2014) Hydrodynamic transfection for generation of novel mouse models for liver cancer research. Am. J. Pathol. 184, 912–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikiforov M. A., Chandriani S., O'Connell B., Petrenko O., Kotenko I., Beavis A., Sedivy J. M., and Cole M. D. (2002) A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol. Cell Biol. 22, 5793–5800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothermund K., Rogulski K., Fernandes E., Whiting A., Sedivy J., Pu L., and Prochownik E. V. (2005) C-Myc-independent restoration of multiple phenotypes by two C-Myc target genes with overlapping functions. Cancer Res. 65, 2097–2107 [DOI] [PubMed] [Google Scholar]
- 30.Li F., Wang Y., Zeller K. I., Potter J. J., Wonsey D. R., O'Donnell K. A., Kim J. W., Yustein J. T., Lee L. A., and Dang C. V. (2005) Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol. Cell Biol. 25, 6225–6234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ward P. S., and Thompson C. B. (2012) Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell 21, 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klann E., and Dever T. E. (2004) Biochemical mechanisms for translational regulation in synaptic plasticity. Nat. Rev. Neurosci. 5, 931–942 [DOI] [PubMed] [Google Scholar]
- 33.Livingstone M., Atas E., Meller A., and Sonenberg N. (2010) Mechanisms governing the control of mRNA translation. Phys. Biol. 7, 021001 [DOI] [PubMed] [Google Scholar]
- 34.Lorbek G., Lewinska M., and Rozman D. (2012) Cytochrome P450s in the synthesis of cholesterol and bile acids: from mouse models to human diseases. FEBS J. 279, 1516–1533 [DOI] [PubMed] [Google Scholar]
- 35.Workman J. L., and Kingston R. E. (1998) Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67, 545–579 [DOI] [PubMed] [Google Scholar]
- 36.Wise D. R., and Thompson C. B. (2010) Glutamine addiction: a new therapeutic target in cancer. Trends Biochem. Sci. 35, 427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stine Z. E., Walton Z. E., Altman B. J., Hsieh A. L., and Dang C. V. (2015) MYC, metabolism, and cancer. Cancer Discov. 5, 1024–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landay M., Oster S. K., Khosravi F., Grove L. E., Yin X., Sedivy J., Penn L. Z., and Prochownik E. V. (2000) Promotion of growth and apoptosis in c-myc nullizygous fibroblasts by other members of the myc oncoprotein family. Cell Death Differ. 7, 697–705 [DOI] [PubMed] [Google Scholar]
- 39.Barrett J., Birrer M. J., Kato G. J., Dosaka-Akita H., and Dang C. V. (1992) Activation domains of L-Myc and c-Myc determine their transforming potencies in rat embryo cells. Mol. Cell Biol. 12, 3130–3137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhat M., Robichaud N., Hulea L., Sonenberg N., Pelletier J., and Topisirovic I. (2015) Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 14, 261–278 [DOI] [PubMed] [Google Scholar]
- 41.van Riggelen J., Yetil A., and Felsher D. W. (2010) MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer 10, 301–309 [DOI] [PubMed] [Google Scholar]
- 42.Leprivier G., Rotblat B., Khan D., Jan E., and Sorensen P. H. (2015) Stress-mediated translational control in cancer cells. Biochim. Biophys. Acta 1849, 845–860 [DOI] [PubMed] [Google Scholar]
- 43.Reznik E., Miller M. L., enbabaoğlu Y., Riaz N., Sarungbam J., Tickoo S. K., Al-Ahmadie H. A., Lee W., Seshan V. E., Hakimi A. A., and Sander C. (2016) Mitochondrial DNA copy number variation across human cancers. Elife 5, e10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cai L., Sutter B. M., Li B., and Tu B. P. (2011) Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell 42, 426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vivanco I. (2014) Targeting molecular addictions in cancer. Br. J. Cancer 111, 2033–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Alboran I. M., O'Hagan R. C., Gärtner F., Malynn B., Davidson L., Rickert R., Rajewsky K., DePinho R. A., and Alt F. W. (2001) Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity 14, 45–55 [DOI] [PubMed] [Google Scholar]
- 47.Eigentler A., Draxl A., Wiethüchter A., Kuznetsov A. V., Lassing B., and Gnaiger E. (2015) Laboratory protocol: Citrate synthase: A mitochondrial marker enzyme. Mitochondrial Physiology Network 17.04, 1–11 [Google Scholar]
- 48.Edgar R., Domrachev M., and Lash A. E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bligh E. G., and Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 50.Trinei M., Berniakovich I., Pelicci P. G., and Giorgio M. (2006) Mitochondrial DNA copy number is regulated by cellular proliferation: a role for Ras and p66Shc. Biochim. Biophys. Acta 1757, 624–630 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.