Abstract
SAMHD1 (sterile α motif and HD domain-containing protein 1) is a mammalian protein that regulates intracellular dNTP levels through its hydrolysis of dNTPs. SAMHD1 functions as an important retroviral restriction factor through a mechanism relying on its dNTPase activity. We and others have reported that human SAMHD1 interacts with the cell cycle regulatory proteins cyclin A, CDK1, and CDK2, which mediates phosphorylation of SAMHD1 at threonine 592, a post-translational modification that has been implicated in abrogating SAMHD1 restriction function and ability to form stable tetramers. Utilizing co-immunoprecipitation and co-localization approaches, we show that endogenous SAMHD1 is able to interact with the cyclin A-CDK1-CDK2 complexin monocytic THP-1 cells and primary monocyte-derived macrophages. Sequence analysis of SAMHD1 identifies a putative cyclin-binding motif found in many cyclin-CDK complex substrates. Using a mutagenesis-based approach, we demonstrate that the conserved residues in the putative cyclin-binding motif are important for protein expression, protein half-life, and optimal phosphorylation of SAMHD1 at Thr592. Furthermore, we observed that SAMHD1 mutants of the cyclin-binding motif mislocalized to a nuclear compartment and had reduced ability to interact with cyclin A-CDK complexes and to form the tetramer. These findings help define the mechanisms by which SAMHD1 is phosphorylated and suggest the contribution of cyclin binding to SAMHD1 expression and stability in dividing cells.
Keywords: cyclin, cyclin-dependent kinase (CDK), phosphorylation, protein stability, SAM domain and HD domain-containing protein 1 (SAMHD1), CDK1, CDK2, cyclin-binding
Introduction
SAMHD1 (sterile α motif and HD domain containing protein 1) is the only mammalian protein characterized as a dNTP triphosphohydrolase (dNTPase)2 enzyme (1–4) and has a clear function in the hydrolysis of excessive dNTPs and therefore is important for the regulation of dNTP homeostasis in cells (5). SAMHD1 is ubiquitously expressed across cell types but is most highly expressed in quiescent cells such as macrophages, dendritic cells, and resting CD4+ T cells (2, 3, 6). Mutations in SAMHD1 have been associated with Aicardi-Goutières syndrome, an autoimmune disorder in which the loss of SAMHD1 function is correlated with triggering aberrant type I interferon responses (7, 8). SAMHD1 is also thought to have a role in maintenance of genome stability (5, 9), and its down-regulation is associated with progression of some cancers (reviewed in Ref. 10).
SAMHD1 has been characterized as a viral restriction factor that blocks the replication of several retroviruses, including HIV-1, and DNA viruses in quiescent cells such as resting T cells, macrophages, and dendritic cells (2, 3, 11, 12). This mechanism is thought to occur as a result of its dNTPase function and depletion of the intracellular dNTP pool required for reverse transcription or synthesis of viral genomes (1, 12–17). Some studies have reported that SAMHD1 binds nucleic acids (18–20). SAMHD1 has also been described to function as a ribonuclease (21), and it has been suggested that degradation of incoming viral genomic RNA is the primary mode of restriction against HIV-1 and other retroviruses (21, 22). However, this finding remains controversial, and other studies have failed to identify RNase activity (23–25).
It is unclear how SAMHD1 function is regulated in cells, although it is known to be ineffective as a lentiviral restriction factor in dividing cells because of SAMHD1 phosphorylation (26–29). The HD domain of SAMHD1 contains the catalytic site of the dNTPase enzyme (1, 30). Tetramer formation is required for dNTPase activity and HIV-1 restriction (31, 32), although it has also been reported that oligomerization-deficient SAMHD1 mutants are capable of restricting HIV-1 (33). Phosphorylation of SAMHD1 at Thr592 is involved in negative regulation of SAMHD1-mediated HIV-1 restriction; however, there are conflicting data on whether dNTPase function is impaired by Thr592 phosphorylation (26–28, 34, 35).
Knock-out of SAMHD1 in THP-1 cells correlates with changes in cell cycle distribution (36), suggesting that SAMHD1 expression contributes to cell cycle regulation. SAMHD1 expression has been reported to decrease as cells progress through the S phase (5). However, another study reported that SAMHD1 expression did not change during the cell cycle and that expression level was not regulated by phosphorylation in THP-1, U937 cell lines, and primary CD4+ T cells (37).
We and others previously identified SAMHD1 interaction with cyclin A2, CDK1, and CDK2 (26, 27, 29) and demonstrated the cyclin A-CDK proteins mediate the phosphorylation of SAMHD1 at Thr592 (38), but the amino acid determinants responsible for the interaction of SAMHD1 with cyclin A remain unclear. Several cyclin-CDK substrates possess cyclin-binding or RXL motifs, which allow association of target proteins with cyclin-CDK complexes. Examples can be found in Myt1 (39, 40), CDC6 (41), and human papillomavirus E1 protein (42). Sequence analysis of conserved residues indicates that the RXL motif consists of conserved arginine and leucine residues separated by a single amino acid (43, 44).
The mechanisms of SAMHD1 regulation in dividing cells or how Thr592 phosphorylation is modulated remain not fully understood. We aimed to better understand the molecular mechanism of SAMHD1 phosphorylation in dividing cells. In this study, we identified a cyclin-binding motif in SAMHD1 and further analyzed the structural basis underlying the mechanism of the cyclin A2-CDK complex interaction with SAMHD1. Our data on SAMHD1 RXL motif and its interaction with cyclin A suggest effects on SAMHD1 protein expression, phosphorylation, localization, protein stability, and tetramerization of SAMHD1.
Results
Endogenous SAMHD1 Interacts and Co-localizes with Cyclin A, CDK1, and CDK2 in THP-1 Cells and Primary Macrophages
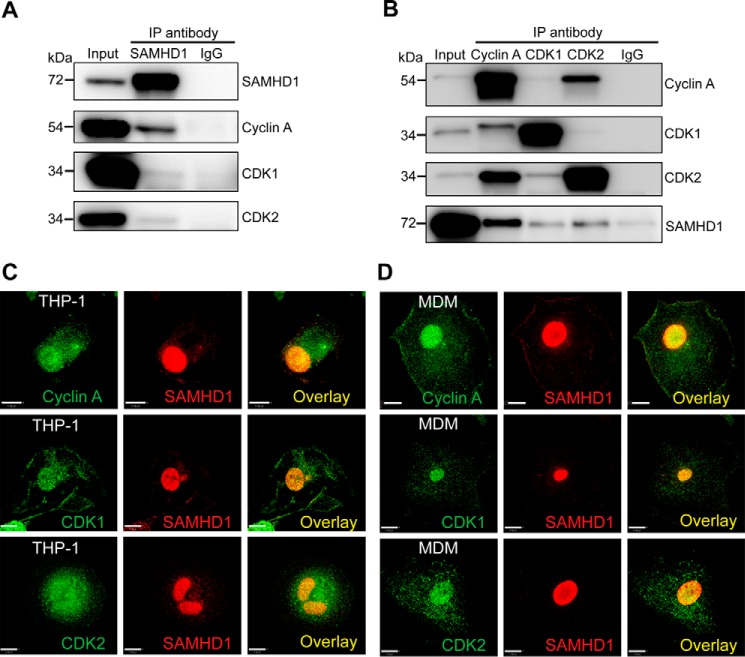
We have previously shown that overexpressed SAMHD1 interacts with cyclin A1, CDK1, and CDK2 in HEK293T cells (29). However, the validity of these interactions at physiological expression levels in cells is not known. To determine whether endogenous SAMHD1 interacts with cyclin A, CDK1, and CDK2, we utilized a THP-1 cell line that expresses endogenous SAMHD1 at a level comparable with primary monocyte-derived macrophages (MDM). Using a co-immunoprecipitation (co-IP) approach, we first immunoprecipitated SAMHD1 and confirmed enrichment of SAMHD1 in the IP products (Fig. 1A). Next, we probed for interacting proteins using antibodies to cyclin A, CDK1, and CDK2 and found that cyclin A interacted clearly with SAMHD1, whereas CDK1 and CDK2 showed a weaker interaction. A mouse IgG negative control indicated low levels of nonspecific binding (Fig. 1A). To further validate these interactions, we performed reverse IP for cyclin A, CDK1, and CDK2 and confirmed a strong interaction between cyclin A and SAMHD1. Consistent with Fig. 1A, CDK1 and CDK2 showed a weak interaction with SAMHD1 (Fig. 1B).
FIGURE 1.
Endogenous SAMHD1 interacts and co-localizes with cyclin A, CDK1, and CDK2 in THP-1 cells and MDMs. A, cell lysates were harvested and co-immunoprecipitated using anti-SAMHD1 antibody or mouse IgG coupled to protein G Dynabeads. Interacting proteins were eluted by boiling in SDS sample buffer, and samples were analyzed by SDS-PAGE and immunoblotting for SAMHD1, cyclin A, CDK1, and CDK2. B, reverse co-IP in THP-1 cells was performed as described for A except antibodies for cyclin A, CDK1, CDK2, or a rabbit IgG were bound to protein G Dynabeads, and eluted lysates were analyzed via immunoblotting for SAMHD1, cyclin A, CDK1, and CDK2. All experiments were performed a minimum of three times. The data shown are from one representative experiment. C, co-localization of endogenous SAMHD1 with cyclin A, CDK1, and CDK2 in differentiated THP-1 cells. D, co-localization of endogenous SAMHD1 with cyclin A, CDK1, and CDK2 in primary MDMs. Immunofluorescence experiments were performed with specific antibodies at least three times. The data shown are from one representative experiment. Scale bars, 11 μm.
To visualize the co-localization of SAMHD1 with cyclin and CDK1/2, we performed immunofluorescence analysis in THP-1 cells and MDM. We observed nuclear localization of SAMHD1, with some cytoplasmic puncta visible in THP-1 cells (Fig. 1C) and MDM (Fig. 1D). As expected, cyclin A showed predominantly nuclear staining with some diffuse staining in the cytoplasm. Analysis of the SAMHD1 and cyclin A signals confirmed co-localization of the two proteins within the nucleus of THP-1 cells and MDMs (Fig. 1, C and D). Quantitation of co-localization demonstrated strong co-localization of SAMHD1 with cyclin A in both THP-1 cells and MDMs (Mander's coefficient of 0.88 ± 0.02 and 0.87 ± 0.06, respectively). Similarly, we observed clear co-localization of endogenous SAMHD1 with CDK1/CDK2 within the nucleus of THP-1 cells and MDM (Fig. 1, C and D). Similar values of SAMHD1/CDK2 signal were noted in MDMs (Mander's coefficient of 0.93 ± 0.02, whereas co-localization of SAMHD1/CDK1 was somewhat less (Mander's coefficient of 0.81 ± 0.04, MDM). Together, the co-IP and localization data suggest that endogenous SAMHD1 interacts with cyclin A, CDK1, and CDK2 in cells.
A Cyclin-binding Motif in SAMHD1 Affects Protein Expression and Phosphorylation
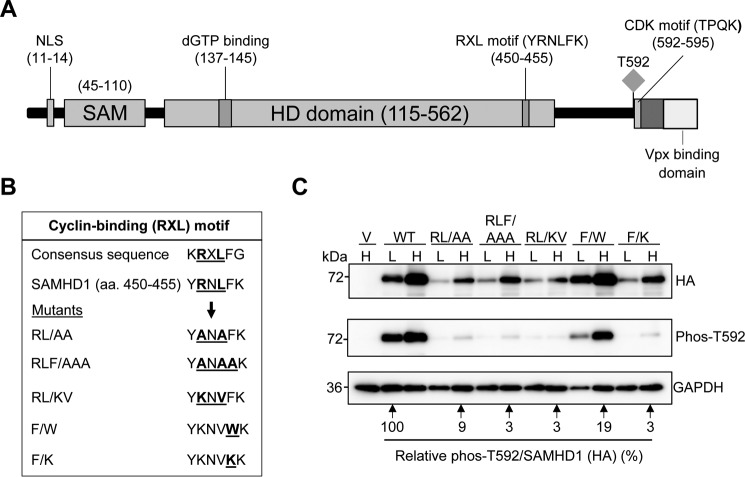
Previous publications have demonstrated that SAMHD1 is a substrate of CDK1/2, mediated through a CDK motif located in the C-terminal region of SAMHD1 at residues 592–595 (16) (Fig. 2A). We hypothesized that SAMHD1 phosphorylation occurs through interaction with cyclin A via a putative cyclin-binding motif. Sequence alignment of SAMHD1 to proteins that contain a consensus cyclin-binding motif (RXL) highlighted a sequence of conserved residues in SAMHD1 that are similar to proteins containing cyclin-CDK substrate recognition motifs (Fig. 2B) (43, 45). To understand the structural basis of SAMHD1 binding to cyclin A-CDK complexes, we generated a panel of mutants targeting the conserved Arg451, Leu453, and Phe454 residues (Fig. 2B), which we predicted would affect cyclin A binding to SAMHD1, prevent recruitment of the CDK1/2 to the C-terminal CDK motif, and thus negatively affect SAMHD1 phosphorylation at Thr592.
FIGURE 2.
A cyclin-binding (RXL) motif in SAMHD1 decreases SAMHD1 protein expression and Thr592 phosphorylation. A, schematic of the SAMHD1 protein highlighting key domains and residues. The RXL motif (YRNLFK) is in the C-terminal region of the HD domain and positions 450–455. The Thr592 phosphorylation site, the CDK consensus motif (TPQK), and the VPx-binding domain are indicated. B, sequence alignment of the WT SAMHD1 RXL motif with a consensus cyclin-binding motif is shown. Mutations of the conserved residues are also shown. C, immunoblot analysis of HEK293T cells overexpressing HA-tagged WT or SAMHD1 mutants. The cells were transfected with plasmid DNA, and the lysates were harvested 24 h post-transfection. Plasmid DNA input was low (L) at 0.25 μg and high (H) at 2.5 μg. The empty vector was used as a negative control. All transfections were normalized to the same total amount of DNA using empty vector. Membranes were probed for HA and Thr(P)592 SAMHD1. GAPDH was used as a loading control. Densitometry was performed using ImageJ software. Overexpressed HA-tagged SAMHD1 and phospho-SAMHD1 probed membranes were normalized to GAPDH, and the relative amount of phospho-SAMHD1 per total SAMHD1 (HA) was calculated. WT low (L) was set as 100%, and everything was calculated relative to this. Lysates from three independent transfections were used. The data shown are from one representative experiment.
We overexpressed mutant and WT SAMHD1 in HEK293T cells and immunoblotted HA-tagged exogenous SAMHD1 and Thr(P)592 SAMHD1. Interestingly, we found that theRL → AA, RL → KV, and RLF → AAA mutants expressed at lower levels compared with the WT protein (Fig. 2C), suggesting either reduced protein stability or increased degradation. When the plasmid DNA input of mutants was increased 10-fold to compensate for low expression levels, we were able to express the mutant proteins to levels near that of the WT protein. However, the phosphorylation status of the mutants remained low compared with the WT SAMHD1 (Fig. 2C). These results suggest that the RXL motif is likely important for Thr592 phosphorylation of SAMHD1. Previous data has shown that a R451E mutant affected SAMHD1 tetramerization and in vitro dNTPase activity (46) and that this residue could be involved in binding nucleotides in the allosteric site (32).
Although the RXL residues are predicted to be the principal conserved residues involved in cyclin binding, it is possible that adjoining residues could have roles in mediating cyclin binding or specificity (45). To this end, we mutated another conserved residue contained within the cyclin-binding motif (Fig. 2B) that was predicted, based on SAMHD1 structure (30), to have no effect on SAMHD1 tetramer formation or stability but could affect cyclin A interaction. F454W is a conservative mutation maintaining the hydrophobic aromatic side chain, whereas F454K is a less conservative mutation substituting the aromatic ring with a basic charge. Interestingly, the expression level of F → W was comparable with that of WT (Fig. 2C); however, its phosphorylation was still reduced, although not as severely as the other mutants. The F → K mutant displayed a similar phenotype to the other mutants, expressing at low levels and with dramatically reduced Thr592 phosphorylation (Fig. 2C).
Expression of SAMHD1 RXL Mutants Is Not Affected at the Transcriptional Level
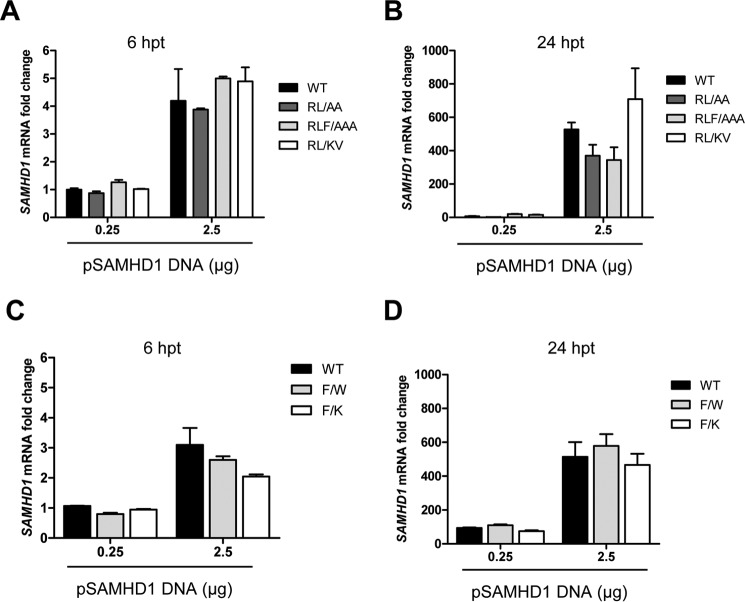
To eliminate the possibility that the SAMHD1 mutants inefficiently transcribed in cells, we transfected HEK293T cells and measured SAMHD1 mRNA levels by quantitative PCR. We found that at either 6 or 24 h post-transfection, the levels of mRNA of mutant SAMHD1 (RL → AA, RL → KV, RLF → AAA, F → W, and F → K) were comparable with that of the WT SAMHD1 (Fig. 3, A and B). Together, these data suggest that reduced SAMHD1 protein expression phenotype of the mutants occurs at a post-transcriptional step and could involve translational regulation or a protein instability mechanism.
FIGURE 3.
Mutation of the RXL motif in SAMHD1 does not affect the levels of SAMHD1 mRNA transcripts. HEK293T cells were transfected with either 0.25 or 2.5 μg of plasmid DNA expressing WT or mutant SAMHD1, and total cellular RNA was extracted at 6 or 24 h post-transfection (hpt). RNA input was normalized for cDNA synthesis, and these products were utilized in quantitative PCR using SAMHD1 specific primers. A and B, Relative SAMHD1 mRNA levels for WT, RL → AA, RL → KV, and RLF → AA mutants are shown at 6 hpt (A) or 24 hpt (B), respectively. C and D, relative SAMHD1 mRNA levels for WT, F → W, and F → K mutants are shown at 6 hpt (C) or 24 hpt (D), respectively. GAPDH was used to normalize quantification, and Renilla luciferase activity was used as an internal control to normalize transfection efficiency. The data shown are n = 2 for a single experiment and are representative of three independent experiments.
SAMHD1 RXL Mutants Have Decreased Interaction with Cyclin A, CDK1, and CDK2
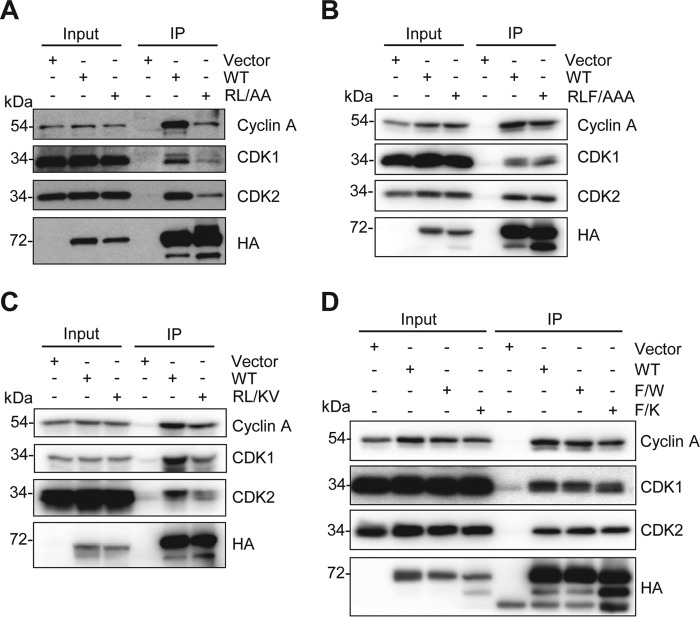
To determine the importance of the RXL motif in mediating binding of SAMHD1 to cyclin A-CDK complexes, WT and mutant SAMHD1 were overexpressed in HEK293T cells, and SAMHD1 co-IP experiments were performed. For these experiments, the expression level of each mutant was increased by increasing plasmid DNA amounts, so that it was comparable with WT to ensure equal IP efficiency. The RL → AA mutant demonstrated reduced interaction with cyclin A, CDK1, and CDK2, although the interaction was not completely abrogated (Fig. 4A). The RLF → AAA and RL → KV mutants (Fig. 4, B and C) also showed a slight reduction in cyclin A binding. Neither of the Phe454 mutants, F → W or F → K, had an impact on interaction with CDK2, although cyclin A and CDK1 were slightly reduced for the F → K mutant (Fig. 4D). Interestingly, for the RXL mutants, a more abundant lower molecular weight species was evident, which was not as abundant in WT or F → W SAMHD1 (Fig. 4, A–D), suggesting that the mutants could be undergoing enhanced or accelerated degradation.
FIGURE 4.
The RXL mutants of SAMHD1 have decreased interaction with cyclin A, CDK1, and CDK2. HEK293T cells were transfected with empty vector or constructs expressing HA-tagged WT or mutant SAMHD1 and 24 h post-transfection, co-IP performed using HA-conjugated agarose. Bound proteins were eluted by boiling in SDS sample buffer, analyzed by SDS-PAGE, and immunoblotted for HA, cyclin A, CDK1, and CDK2. A, vector, WT, and RL → AA mutant. B, vector, WT, and RLF → AAA mutant. C, vector, WT, and RL → KV mutant. D, vector, WT, F → W, and F → K mutants. To compensate for lower expression levels of the mutants compared with WT, 10-fold higher mutant SAMHD1 or empty vector plasmids were transfected compared with the WT SAMHD1 transfection. Co-IP experiments were repeated three times, and one representative experiment is shown.
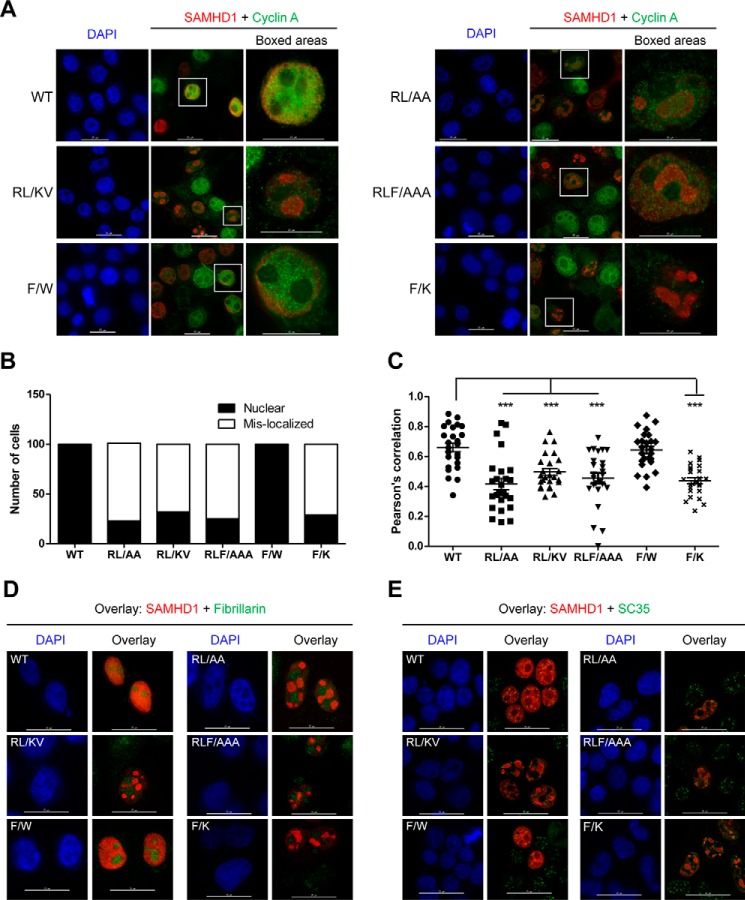
SAMHD1 RXL Mutants Are Mislocalized within the Nucleus in HEK293T Cells
To further validate the co-IP data, we determined the subcellular localization of each mutant compared with WT SAMHD1. The mutations are located in the C-terminal region of the HD domain, leaving the N-terminal nuclear localization signal of SAMHD1 intact (47). In transfected HEK293T cells, WT SAMHD1 signal overlapped with DAPI, indicating nuclear localization, and showed strong co-localization with cyclin A (Fig. 5A). Surprisingly, the RL → AA, RLF → AAA, RL → KV, and F → K mutants displayed a mislocalized phenotype, with SAMHD1 accumulated within an intranuclear region, whereas the cyclin A signal remained throughout the nucleus (Fig. 5A). We then counted and scored 100 cells of each WT or mutant SAMHD1 according to their localization pattern. WT SAMHD1 was exclusively nuclear (Fig. 5B). Mutant SAMHD1 was predominantly mislocalized (71–78% of cells expressing RL → AA, RL → KV, RLF → AAA, and F → K). The remainder of the cells (≤32%) had nuclear localization (Fig. 5B). The F → W mutant exhibited the same nuclear localization phenotype as WT SAMHD1 (Fig. 5, A and B).
FIGURE 5.
The RXL mutants of SAMHD1 are mislocalized to a nuclear compartment. HEK293T cells were transfected with empty vector or constructs expressing HA-tagged WT or mutant SAMHD1. At 24 h post-transfection, immunofluorescence staining was performed using HA-specific antibodies for SAMHD1 detection, and endogenous cyclin A was detected using a specific antibody. DAPI was used to stain nuclei. A, WT and mutant SAMHD1-expressing cells were co-stained for HA (SAMHD1) and cyclin A. Red, SAMHD1; green, cyclin A; blue, DAPI. Scale bars in left and middle panels of WT and mutants, 25 μm; scale bars in boxed areas, 15 μm. B, 100 SAMHD1-expressing cells were examined, counted for WT and each mutant, and scored according to the visual localization. Nuclear, exclusively nuclear localized cells; Mis-localized, cells with mislocalized SAMHD1 signal. C, graph representing Pearson's correlation co-efficient values for SAMHD1 and cyclin A localization. 25 cells expressing WT or mutant SAMHD1 were counted and analyzed for co-localization. The images were captured on a DeltaVision Elite microscope. Deconvoluted images were analyzed using the SoftWoRx software. Statistical analysis was performed using a one-way analysis of variance with Dunnet multiple comparison test. ***, p < 0.0001. D, WT and mutant SAMHD1-expressing cells were co-stained for either HA (SAMHD1) or fibrillarin. Red, SAMHD1; green, fibrillarin; blue, DAPI. Scale bars, 25 μm. E, WT and mutant SAMHD1-expressing cells were co-stained for either HA (SAMHD1) or SC-35. Red, SAMHD1; green, SC-35; blue, DAPI. Scale bars, 25 μm.
Co-localization analysis of cyclin A and SAMHD1 showed that WT SAMHD1, and the F → W mutant, co-localized with cyclin A, as expected, with a mean Pearson's correlation = 0.7 and 0.6, respectively (n = 25) (Fig. 5C). Analysis of cells expressing the mutants showed significantly reduced co-localization with cyclin A, compared with WT SAMHD1; mean Pearson's correlation ranging from 0.4 to 0.5 (n = 25) (Fig. 5C). Together, these data reveal that SAMHD1 RXL mutants predominantly mislocalize within the nucleus and that in these cells, co-localization with cyclin A is reduced. To delineate the subcellular compartment in which the mutants accumulated, we co-stained transfected cells for HA-tagged SAMHD1 and fibrillarin. Fibrillarin is located in fibrillar components and Cajal bodies contained within the nucleolus. We observed that the SAMHD1 mutants did not co-localize with fibrillarin in the nucleolus. Importantly, the localization of fibrillarin did not alter upon expression with WT or mutant SAMHD1 (Fig. 5D). Further analysis of the nuclear speckle marker SC-35 indicated that SC-35 partially localized with SAMHD1 (WT or mutant) in small puncta in the nucleus; however, SAMHD1 RXL mutants were not relocalized to regions of abundant nuclear speckles (Fig. 5E).
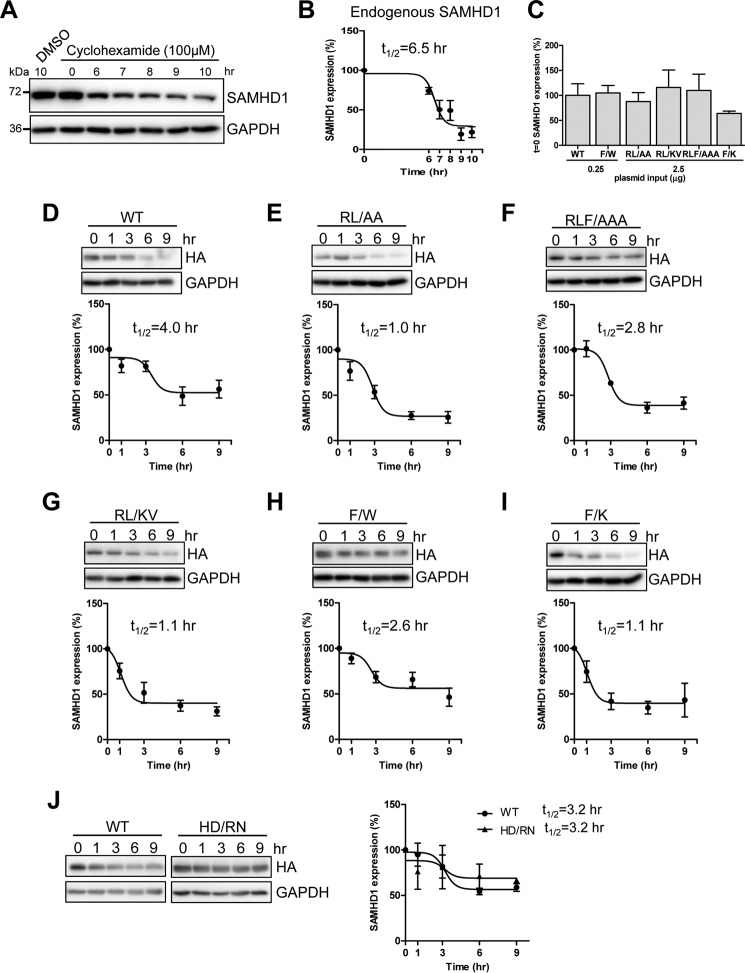
SAMHD1 RXL Mutants Have Decreased Protein Half-life Compared with WT SAMHD1
To further investigate the effects of mislocalization and the low protein expression of the RXL mutants compared with WT SAMHD1, we determined the half-life of either endogenous SAMHD1 in THP-1 cells or overexpressed WT and mutant SAMHD1 in HEK293T cells using a cyclohexamide treatment assay (48). Endogenous SAMHD1 in THP-1 cells was found to decrease steadily from 6 to 8 h post-treatment, after which protein levels remained constant, whereas the DMSO treated cells maintained constant SAMHD1 expression (Fig. 6A). Densitometry analysis from four independent experiments was plotted, and the t½ of endogenous WT SAMHD1 was determined to be 6.5 h (Fig. 6B).
FIGURE 6.
SAMHD1 mutants have decreased half-life compared with WT SAMHD1. Shown are THP-1 or transfected HEK293T cells treated with cyclohexamide (100 μm) and lysates harvested at the indicated times. Immunoblot analysis was used to determine levels of endogenous SAMHD1 protein. A, analysis of endogenous SAMHD1 expression in cyclohexamide treated THP-1 cells at 0, 6, 7, 8, 9, and 10 h post-treatment. The lysates were immunoblotted for SAMHD1 and GAPDH. B, non-linear curve of densitometry from four independent experiments in THP-1 cells. Endogenous SAMHD1 protein half-life (t½) = 6.5 h is indicated. C, comparison of expression levels of WT and mutant SAMHD1 at t = 0 from four experiments used to calculate half-life in D–I. SAMHD1 levels at t = 0 were normalized to GAPDH, and each mutant was calculated as a percentage of WT SAMHD1, which was set as 100%. D–I, HEK293T cells transfected with constructs expressing HA-tagged WT or mutant SAMHD1 to normalize protein expression to equal levels. At 48 h post-transfection, cyclohexamide treatment was started, and lysates were collected at the indicated time points post-transfection. Lysates were immunoblotted for HA and GAPDH. D–I, non-linear curves, t½ calculated from immunoblotting densitometry, and one representative immunoblot. Densitometry data and t½ was calculated from a minimum of four independent experiments. J, comparison of WT and the dNTPase defective HD/RN mutant (H206R and D207N) showing immunoblotting analysis (left panel) and non-linear curves with calculated t½ (right panel). The data represent two independent experiments.
To analyze the half-life of WT and mutant SAMHD1, HEK293T cells were transfected with WT or mutant SAMHD1-expressing vectors to normalize for equal protein expression, and cells were treated with cyclohexamide 48 h post-transfection. Comparison of densitometry levels of WT and mutants confirmed comparable expression at t = 0, for the SAMHD1 half-life experiments (Fig. 6C). WT SAMHD1 began to decrease steadily from 3 h post-treatment, and the t½ was calculated to be 4.0 h (Fig. 6D). The half-life of WT SAMHD1 in HEK293T cells was lower compared with THP-1 cells, which could be attributed to different cell types. Interestingly, SAMHD1 RXL mutant protein levels decreased more than WT protein overall and also decreased more quickly to lower levels than WT protein between 1 and 3 h post-treatment (Fig. 6, E–I). The t½ of mutants RL → AA, RLF → AAA, RL → KV, F → W, and F → K was 1.0, 2.8, 1.1, 2.6, and 1.1 h, respectively (Fig. 6, E–I). Comparison of WT SAMHD1 with the dNTPase-defective HD/RN mutant (H206R and D207N), which is unrelated to the RXL motif, demonstrated a comparable expression level and half-life (Fig. 6J). Both proteins also decreased to similar levels and showed the same half-life. These data suggest that the reduced half-life of the RXL mutants was specific to mutations in the RXL motif. Of note, in the WT and HD/RN experiments, the half-life of WT SAMHD1 was slightly decreased (from 4.0 to 3.2 h) compared with the previous data set (Fig. 6, D and J), which could be attributed to experimental variation in protein expression and using a semiquantitative densitometry method. Together, these results suggest that mutation of the RXL motif in SAMHD1 has detrimental effects on SAMHD1 stability and expression within the cell, irrespective of the ability to interact with cyclin A-CDK complexes.
SAMHD1 RXL Mutants Have Reduced Ability to Form Tetramers Compared with WT SAMHD1 in Cells
To determine the potential effects of the RXL mutation on SAMHD1 structure that may contribute to the reduced protein expression and half-life of the mutants, we analyzed the capability of the proteins to tetramerize in cells. We used a cross-linking experiment to determine monomeric and oligomeric SAMHD1 proteins expressed in cells (31). Interestingly, our results showed that both WT and F → W SAMHD1 retained the ability to form tetramers in HEK293T cells (Fig. 7A). In contrast, the remaining RXL mutants showed no indication of tetramer formation, when expressed to comparable levels as WT or F → W (Fig. 7A). A non-cross-linked control confirmed the presence of monomeric SAMHD1 in the transfected (Fig. 7B). Of note, the reduced half-life of SAMHD1 correlates with the inability to form stable tetrameric structures within cells (Figs. 6 and 7). Together, these data suggest that mutation of residues within the RXL motif can also affect the ability of SAMHD1 to form stable tetramer, thereby altering protein trafficking and stability.
FIGURE 7.
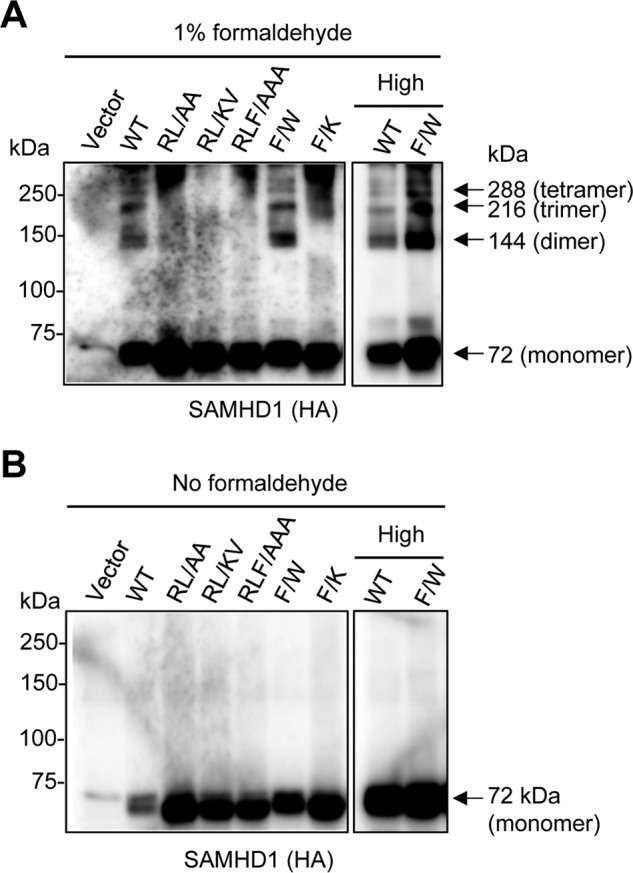
SAMHD1 RXL mutants have decreased ability to form tetramers in cells. HEK293T cells were transfected with empty vector, HA-tagged SAMHD1 WT- (1 μg), or mutant-expressing constructs (10 μg), or with high levels of WT or F → W mutant-expressing plasmid DNA (10 μg) and then were treated with 1% formaldehyde for cross-linking (A) or not treated (B) the day following transfection. After a 10-min incubation at 37 °C, the cross-linking was stopped by the addition of glycine. The cell lysates were harvested, and monomeric and oligomeric SAMHD1 proteins were assessed by immunoblotting using anti-HA. Arrows denote monomer, dimer, trimer, and tetramer forms of SAMHD1 at the respective predicted molecular weights. The experiments were repeated three times, and one representative experiment is shown.
Discussion
In the current study, we demonstrate that endogenous SAMHD1 interacts and co-localizes with cyclin A, CDK1, and CDK2 in THP-1 cells and primary macrophages, consistent with data showing that macrophages express detectable Thr(P)592 SAMHD1, compared with other non-dividing cell types (26). Our reverse IP data suggest that the proteins bind SAMHD1 as a complex. Although cyclin A-CDK2 complexes can shuttle between the nucleus and cytoplasm (49), we consistently observed that co-localization of SAMHD1 with cyclin A-CDK occurs predominantly in the nucleus. SAMHD1 has also been shown to interact with and co-localize with cyclin L2 in HeLa cells and phorbol 12-myristate 13-acetate-differentiated THP-1 (50); however, we were unable to confirm this interaction in dividing THP-1 cells (data not shown). Although no direct interaction has been demonstrated, CDK6 and cyclin D3 have been shown to be involved in regulating SAMHD1 phosphorylation and HIV-1 replication in macrophages (38, 51), and cyclin D2/CDK4 has been shown to affect HIV-1 replication through modulation of SAMHD1 activation (52).
Our previous study demonstrated that knock-out of SAMHD1in dividing THP-1 cells perturbed the cell cycle distribution (36), implicating a role for SAMHD1 in cell cycle regulation. Interestingly, a number of cell cycle regulatory proteins form stable complexes with cyclin-CDK partners, for example, E2F1 (53). Targeting sequences on CDK substrates can also target cyclin-CDK activity to specific periods of activity. We identified a similar RXL motif in SAMHD1 and found that mutation of the conserved Arg451 and Leu453 residues contained within the RXL motif affects SAMHD1 interaction with cyclin A, thereby contributing to reduced SAMHD1 Thr592 phosphorylation.
Cyclin A-CDK2 is predominantly active during the S phase (44), coinciding with the peak of SAMHD1 Thr592 phosphorylation (37). Cyclin A-CDK could function to phosphorylate SAMHD1 as the cell progresses into the S phase, serving to either restrict SAMHD1 function or down-regulate its expression through targeted degradation. Specific targeting of SAMHD1 for degradation has been observed with cyclin L2 in macrophages (50). RXL motifs are also found in CDK inhibitors (54), and it would be interesting to investigate whether a specific cellular CDK inhibitor controls SAMHD1 dephosphorylation at critical points during the cell cycle.
Surprisingly, the RXL mutants did not abrogate cyclin A-CDK association with SAMHD1, either with co-IP or co-localization studies. The cyclin-CDK interaction may not be exclusively dependent on the presence of an intact RXL motif. Alternatively, SAMHD1 may contain a second non-canonical cyclin-binding motif in the C terminus of SAMHD1, at residues Leu620 and Phe621 (37), allowing for some redundancy with cyclin binding. Other cyclin binding proteins such as cyclin-dependent kinase inhibitor 1 have been found to contain multiple cyclin-binding motifs (44). It is plausible that the mutants bind cyclin A but induce an allosteric change in the cyclin-CDK complex, reducing the efficiency of kinase activity, which could explain the strongly reduced phosphorylation in the presence of weaker cyclin A-CDK interactions.
Interestingly, co-localization analysis in asynchronous dividing HEK293T cells revealed a mixed population of cells with variable cyclin A expression, and atypical distribution of mutant SAMHD1, which had reduced Pearson's correlation coefficient with cyclin A. These data clarify the co-IP data, which suggested a reduced but variable interaction with cyclin A in total cell lysates. The presence of a more abundant lower molecular weight species of SAMHD1 expressed in mutant transfected cells coupled with the reduced half-life of the mutants suggests that these mutants might be targeted for rapid degradation. Although the mutants have decreased phosphorylation, we do not think this is causative of the decreased half-life. Cyclohexamide experiments analyzing the Thr(P)592 of SAMHD1 were found to exhibit degradation comparable with total protein (data not shown). Furthermore, phosphoablative mutants of SAMHD1 have been extensively studied, and there is no evidence these mutants express poorly or are rapidly degraded in cells. Experiments intended to elucidate a potential degradation pathway have proved inconclusive because neither proteasomal nor lysosomal degradation could be attributed to low expression of the SAMHD1 mutants (data not shown).
It has been shown that mutation of arginine 451 to glutamic acid of SAMHD1 (R451E) can affect its tetramerization (30, 46). Our cross-linking experiment confirmed this observation in cells, because the RXL mutants could not tetramerize, whereas the WT and F → W protein did. These data could suggest that SAMHD1 variants that are unable to form stable and functional tetramers in cells are not stably expressed in cells. Interestingly, the F → W mutant, which retained similarities to the WT SAMHD1 was found to maintain an HIV-1 restrictive phenotype in U937 cells (data not shown).
Overall, we have identified residues that are important for SAMHD1 interaction with cyclin A and phosphorylation, which could be important for regulation of SAMHD1. Our structure-based modeling suggests that SAMHD1 binds cyclin A-CDK in a monomeric form, which would phosphorylate Thr592, thus destabilizing SAMHD1 tetramerization (32, 34) and thus serving as a mechanism to regulate SAMHD1 structural based function in cells.
Experimental Procedures
Plasmids
The pLenti vectors expressing HA-tagged human SAMHD1 and the empty vector control were kind gifts from Nathaniel Landau (New York University). The pLenti vectors expressing HA-tagged mutant SAMHD1 were generated using a QuikChange mutagenesis kit (Agilent Technologies), based on the pLenti vector expressing WT SAMHD1 (29).
Cell Culture and Treatments
THP-1 cells were cultured in RPMI 1640 medium modified to contain 2 mm l-glutamine, 10 mm HEPES, 1 mm sodium pyruvate, 4500 mg/liter glucose, and 1500 mg/liter sodium bicarbonate, 10% fetal bovine serum, and 1% penicillin/streptomycin. HEK293T cells were maintained in complete DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. All the cell lines utilized for the presented studies were maintained at 37 °C, 5% CO2 and tested negative for mycoplasma contamination using a universal mycoplasma detection kit (ATCC, catalog no. 30-101-2K). MDMs were derived from normal healthy volunteers by isolation with the Monocyte Isolation Kit II (Miltenyi Biotec Inc.) and allowed to adhere to 35-mm2 MatTek dishes (MatTek Corporation) coated with collagen. MDMs were matured in RPMI 1640 supplemented with 10% FBS, 1% penicillin/streptomycin, 2 mm glutamine, and 5 ng/ml GM-CSF (R & D Systems). Staining was performed following 7 days of maturation.
Transfections and Immunoblotting
HEK293T cells were transfected using a calcium phosphate method to overexpress human WT SAMHD1, mutant SAMHD1, or vector controls. The cells were processed for downstream applications 24 h post-transfection, unless otherwise specified. The cells were harvested as indicated and lysed in cell lysis buffer (Cell Signaling, catalog no. 9803) supplemented with protease inhibitor mixture (Sigma-Aldrich). Cell lysates were quantified via BCA and subjected to immunoblotting as described. Antibodies specific to SAMHD1 (Abcam, catalog no. ab67820), HA (Covance, catalog no. 901501) and GAPDH (Bio-Rad, catalog no. AHP1628) have been described previously (29). Antibodies to Thr(P)592 SAMHD1 (1:1000, ProSci, catalog no. 8005), cyclin A (1:1000, Santa Cruz, catalog no. sc-751), CDK1 (1:1000, Santa Cruz, catalog no. sc-954), and CDK2 (1:1000, Santa Cruz, catalog no. sc-163). SuperSignal West Pico Chemiluminescence substrate (Pierce) was used to detect horseradish peroxidase-conjugated secondary antibodies. Immunoblotting images were captured and analyzed using the Amersham Biosciences imager 600 (GE Healthcare). Densitometry analysis of protein bands was performed using ImageJ.
Co-immunoprecipitation
HEK293T cells were transfected with an empty vector or constructs expressing HA-tagged WT SAMHD1 or mutants At 24 h post-transfection, the cells were collected and lysed for co-IP and analyzed by immunoblotting as described previously (29). For co-IP of SAMHD1 and interacting proteins from THP-1 cells, cell lysates were generated as described earlier. Protein G Dynabeads (Life Technologies) were incubated with 5 μg of IP antibody (SAMHD1, cyclin A, CDK1, or CDK2), or a rabbit or mouse IgG control, according to the manufactures directions. After binding antibody to the beads, cell lysates were incubated with the Dynabeads at 4 °C for 2 h. The beads were then washed three times with PBS + 0.1% Tween. Bound proteins were eluted from beads by boiling in 1- SDS sample buffer, and the supernatants were used for SDS-PAGE.
Immunofluorescence
Immunofluorescent detection of endogenous SAMHD1 and cyclin A, CDK1, or CDK2 was performed in THP-1 cells following differentiation with phorbol 12-myristate 13-acetate, and in MDMs. MDMs were grown on collagen-coated MatTek dishes as described above, stimulated by 10 ng/ml GM-CSF for 7 days. The cells were then fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100 for 10 min, and blocked with Dako blocking buffer (Dako) for 20 min. Primary antibodies used were as follows: SAMHD1 antibody from NOVUS (NBP1–31432); CDK1 from Cell Signaling (catalog no. 9111); CDK2 from Santa Cruz Biotechnology (sc-163); and cyclin A antibody also from Santa Cruz Biotechnology (sc-751). To determine the subcellular localization of SAMHD1 versus SAMHD1 mutants, HEK293T cells were transfected with WT or mutant SAMHD1. 24 h post-transfection, the cells were fixed in 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked in PBS containing 5% BSA. Antibodies for HA, cyclin A2, fibrillarin (Cell Signaling Technology catalog no. 14417), and SC-35 (Abcam, catalog no. 11826) were used as directed by the manufacturers, and the appropriate Alexa Fluor 568 or 488 secondary antibodies were used. Deconvolution analysis was performed using a DeltaVision Elite (GE Healthcare Life Sciences) imaging system. Fluorescent imaging was analyzed and processed using SoftWoRx software.
Quantitative PCR
To measure SAMHD1 mRNA in transfected HEK293T cells, WT or mutant SAMHD1 were co-transfected at either low or high DNA concentrations (0.25 or 2.5 μg), and a Renilla-TK plasmid (25 ng) and all samples were made up to a total of 2.525 μg of DNA with the empty pLenti plasmid. The cells were harvested either 6 or 24 h post-transfection, and total cellular RNA was extracted using the RNeasy minikit according to the manufacturer's guidelines (Qiagen). Total RNA from each sample used as a template for first strand cDNA synthesis using Superscript III first strand synthesis system and oligo(dT) primers (Thermo Fisher Scientific). SYBER Green-based quantitative real time PCR analysis was performed using SAMHD1 cDNA-specific primers. Quantification of spliced GAPDH mRNA was used for normalization as described previously (55). Calculation of relative gene expression was performed using the 2−ΔΔCT method as described previously (56). All values were also normalized to Renilla mRNA to control for transfection efficiency.
SAMHD1 Half-life Experiments
The half-life of endogenous SAMHD1 in THP-1 cells or overexpressed WT or mutant SAMHD1 was determined using cyclohexamide. THP-1 cells were plated in 12-well plates and incubated overnight. The following morning, 100 μm cyclohexamide or an equal volume of DMSO was added to cells, and lysates were harvested at the following time points: 0, 6, 7, 8, 9, and 10 h post-cyclohexamide or DMSO treatment. The lysates were quantified and analyzed by SDS-PAGE and immunoblotted for SAMHD1 and GAPDH, as described earlier. For HEK293T cells, multiple wells were transfected with WT (0.25 μg) or mutant SAMHD1 (2.5 μg), and the cells were incubated for 48 h. At this time, 100 μm cyclohexamide or an equal volume of DMSO was added to cells, and lysates were collected at the following time points: 0, 1, 3, 6, and 9 h post-cyclohexamide or DMSO treatment. The lysates were quantified and analyzed by SDS-PAGE, and immunoblotting for HA and GAPDH was performed as described earlier. Densitometry analysis was performed and the t½ calculated using Prism software.
Formaldehyde Cross-linking in HEK293T cells
HEK293T cells were transfected to express comparable levels of WT or mutant SAMHD1. The following day, the cells were treated with 1% formaldehyde for 10 min at 37 °C or not treated with formaldehyde as described (31). The reaction was stopped by the addition of glycine. The cell lysates were harvest and quantified by BCA. Lysates were analyzed by SDS-PAGE and immunoblotted for HA-tagged SAMHD1.
Author Contributions
L. W. conceived the study and designed experiments with C. S. G. and J. S. Y. C. S. G., L. D., and S. H. K. performed experiments and analyzed data with L. W. D. I. and P. S. contributed to experimental design and data analysis. C. S. and L. W. wrote the manuscript. All the authors read, revised, and approved the manuscript.
Acknowledgments
We thank Dr. Nathaniel Landau for reagents and the Wu lab members for valuable discussions.
This work was supported by National Institutes of Health Grants GM111027 (to P. S.) and AI104483, CA181997, AI120209, and AI127667 (to L. W.) and in part by the Public Health Preparedness for Infectious Diseases Program of the Ohio State University (to L. W.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- dNTPase
- dNTP triphosphohydrolase
- co-IP
- co-immunoprecipitation
- MDM
- monocyte-derived macrophages
- hpt
- h post-transfection.
References
- 1.Goldstone D. C., Ennis-Adeniran V., Hedden J. J., Groom H. C., Rice G. I., Christodoulou E., Walker P. A., Kelly G., Haire L. F., Yap M. W., de Carvalho L. P., Stoye J. P., Crow Y. J., Taylor I. A., and Webb M. (2011) HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 [DOI] [PubMed] [Google Scholar]
- 2.Hrecka K., Hao C., Gierszewska M., Swanson S. K., Kesik-Brodacka M., Srivastava S., Florens L., Washburn M. P., and Skowronski J. (2011) Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Ségéral E., Yatim A., Emiliani S., Schwartz O., and Benkirane M. (2011) SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Powell R. D., Holland P. J., Hollis T., and Perrino F. W. (2011) Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem. 286, 43596–43600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franzolin E., Pontarin G., Rampazzo C., Miazzi C., Ferraro P., Palumbo E., Reichard P., and Bianchi V. (2013) The deoxynucleotide triphosphohydrolase SAMHD1 is a major regulator of DNA precursor pools in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 110, 14272–14277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li N., Zhang W., and Cao X. (2000) Identification of human homologue of mouse IFN-γ induced protein from human dendritic cells. Immunol. Lett. 74, 221–224 [DOI] [PubMed] [Google Scholar]
- 7.Goutières F. (2005) Aicardi-Goutieres syndrome. Brain Dev 27, 201–206 [DOI] [PubMed] [Google Scholar]
- 8.Rice G. I., Bond J., Asipu A., Brunette R. L., Manfield I. W., Carr I. M., Fuller J. C., Jackson R. M., Lamb T., Briggs T. A., Ali M., Gornall H., Couthard L. R., Aeby A., Attard-Montalto S. P., et al. (2009) Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 41, 829–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kretschmer S., Wolf C., König N., Staroske W., Guck J., Häusler M., Luksch H., Nguyen L. A., Kim B., Alexopoulou D., Dahl A., Rapp A., Cardoso M. C., Shevchenko A., and Lee-Kirsch M. A. (2015) SAMHD1 prevents autoimmunity by maintaining genome stability. Ann. Rheum. Dis. 74, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohnken R., Kodigepalli K. M., and Wu L. (2015) Regulation of deoxynucleotide metabolism in cancer: novel mechanisms and therapeutic implications. Mol. Cancer 14, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Descours B., Cribier A., Chable-Bessia C., Ayinde D., Rice G., Crow Y., Yatim A., Schwartz O., Laguette N., and Benkirane M. (2012) SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4+) T-cells. Retrovirology 9, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baldauf H. M., Pan X., Erikson E., Schmidt S., Daddacha W., Burggraf M., Schenkova K., Ambiel I., Wabnitz G., Gramberg T., Panitz S., Flory E., Landau N. R., Sertel S., Rutsch F., et al. (2012) SAMHD1 restricts HIV-1 infection in resting CD4+ T cells. Nat. Med. 18, 1682–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lahouassa H., Daddacha W., Hofmann H., Ayinde D., Logue E. C., Dragin L., Bloch N., Maudet C., Bertrand M., Gramberg T., Pancino G., Priet S., Canard B., Laguette N., Benkirane M., et al. (2012) SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13, 223–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.St. Gelais C., de Silva S., Amie S. M., Coleman C. M., Hoy H., Hollenbaugh J. A., Kim B., and Wu L. (2012) SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology 9, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim E. T., White T. E., Brandariz-Núñez A., Diaz-Griffero F., and Weitzman M. D. (2013) SAMHD1 restricts herpes simplex virus 1 in macrophages by limiting DNA replication. J. Virol. 87, 12949–12956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White T. E., Brandariz-Nuñez A., Valle-Casuso J. C., Amie S., Nguyen L., Kim B., Brojatsch J., and Diaz-Griffero F. (2013) Contribution of SAM and HD domains to retroviral restriction mediated by human SAMHD1. Virology 436, 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gramberg T., Kahle T., Bloch N., Wittmann S., Müllers E., Daddacha W., Hofmann H., Kim B., Lindemann D., and Landau N. R. (2013) Restriction of diverse retroviruses by SAMHD1. Retrovirology 10, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beloglazova N., Flick R., Tchigvintsev A., Brown G., Popovic A., Nocek B., and Yakunin A. F. (2013) Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. J. Biol. Chem. 288, 8101–8110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tüngler V., Staroske W., Kind B., Dobrick M., Kretschmer S., Schmidt F., Krug C., Lorenz M., Chara O., Schwille P., and Lee-Kirsch M. A. (2013) Single-stranded nucleic acids promote SAMHD1 complex formation. J. Mol. Med. 91, 759–770 [DOI] [PubMed] [Google Scholar]
- 20.Goncalves A., Karayel E., Rice G. I., Bennett K. L., Crow Y. J., Superti-Furga G., and Bürckstümmer T. (2012) SAMHD1 is a nucleic-acid binding protein that is mislocalized due to Aicardi-Goutieres syndrome-associated mutations. Hum. Mutat. 33, 1116–1122 [DOI] [PubMed] [Google Scholar]
- 21.Ryoo J., Choi J., Oh C., Kim S., Seo M., Kim S. Y., Seo D., Kim J., White T. E., Brandariz-Nuñez A., Diaz-Griffero F., Yun C. H., Hollenbaugh J. A., Kim B., Baek D., et al. (2014) The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nat. Med. 20, 936–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi J., Ryoo J., Oh C., Hwang S., and Ahn K. (2015) SAMHD1 specifically restricts retroviruses through its RNase activity. Retrovirology 12, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wittmann S., Behrendt R., Eissmann K., Volkmann B., Thomas D., Ebert T., Cribier A., Benkirane M., Hornung V., Bouzas N. F., and Gramberg T. (2015) Phosphorylation of murine SAMHD1 regulates its antiretroviral activity. Retrovirology 12, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seamon K. J., Sun Z., Shlyakhtenko L. S., Lyubchenko Y. L., and Stivers J. T. (2015) SAMHD1 is a single-stranded nucleic acid binding protein with no active site-associated nuclease activity. Nucleic Acids Res. 43, 6486–6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antonucci J. M., St. Gelais C., de Silva S., Yount J. S., Tang C., Ji X., Shepard C., Xiong Y., Kim B., and Wu L. (2016) SAMHD1-mediated HIV-1 restriction in cells does not involve ribonuclease activity. Nat. Med. 22, 1072–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cribier A., Descours B., Valadão A. L., Laguette N., and Benkirane M. (2013) Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep 3, 1036–1043 [DOI] [PubMed] [Google Scholar]
- 27.White T. E., Brandariz-Nuñez A., Valle-Casuso J. C., Amie S., Nguyen L. A., Kim B., Tuzova M., and Diaz-Griffero F. (2013) The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 13, 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welbourn S., Dutta S. M., Semmes O. J., and Strebel K. (2013) Restriction of virus infection but not catalytic dNTPase activity is regulated by phosphorylation of SAMHD1. J. Virol. 87, 11516–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.St. Gelais C., de Silva S., Hach J. C., White T. E., Diaz-Griffero F., Yount J. S., and Wu L. (2014) Identification of cellular proteins interacting with the retroviral restriction factor SAMHD1. J. Virol. 88, 5834–5844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji X., Wu Y., Yan J., Mehrens J., Yang H., DeLucia M., Hao C., Gronenborn A. M., Skowronski J., Ahn J., and Xiong Y. (2013) Mechanism of allosteric activation of SAMHD1 by dGTP. Nat. Struct. Mol. Biol. 20, 1304–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan J., Kaur S., DeLucia M., Hao C., Mehrens J., Wang C., Golczak M., Palczewski K., Gronenborn A. M., Ahn J., and Skowronski J. (2013) Tetramerization of SAMHD1 is required for biological activity and inhibition of HIV infection. J. Biol. Chem. 288, 10406–10417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnold L. H., Groom H. C., Kunzelmann S., Schwefel D., Caswell S. J., Ordonez P., Mann M. C., Rueschenbaum S., Goldstone D. C., Pennell S., Howell S. A., Stoye J. P., Webb M., Taylor I. A., and Bishop K. N. (2015) Phospho-dependent regulation of SAMHD1 oligomerisation couples catalysis and restriction. PLoS Pathog. 11, e1005194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brandariz-Nuñez A., Valle-Casuso J. C., White T. E., Nguyen L., Bhattacharya A., Wang Z., Demeler B., Amie S., Knowlton C., Kim B., Ivanov D. N., and Diaz-Griffero F. (2013) Contribution of oligomerization to the anti-HIV-1 properties of SAMHD1. Retrovirology 10, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang C., Ji X., Wu L., and Xiong Y. (2015) Impaired dNTPase activity of SAMHD1 by phosphomimetic mutation of Thr-592. J. Biol. Chem. 290, 26352–26359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnold L. H., Kunzelmann S., Webb M. R., and Taylor I. A. (2015) A continuous enzyme-coupled assay for triphosphohydrolase activity of HIV-1 restriction factor SAMHD1. Antimicrob. Agents Chemother. 59, 186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonifati S., Daly M. B., St. Gelais C., Kim S. H., Hollenbaugh J. A., Shepard C., Kennedy E. M., Kim D. H., Schinazi R. F., Kim B., and Wu L. (2016) SAMHD1 controls cell cycle status, apoptosis and HIV-1 infection in monocytic THP-1 cells. Virology 495, 92–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan J., Hao C., DeLucia M., Swanson S., Florens L., Washburn M. P., Ahn J., and Skowronski J. (2015) CyclinA2-cyclin-dependent kinase regulates SAMHD1 protein phosphohydrolase domain. J. Biol. Chem. 290, 13279–13292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pauls E., Ruiz A., Badia R., Permanyer M., Gubern A., Riveira-Muñoz E., Torres-Torronteras J., Alvarez M., Mothe B., Brander C., Crespo M., Menéndez-Arias L., Clotet B., Keppler O. T., Martí R., et al. (2014) Cell cycle control and HIV-1 susceptibility are linked by CDK6-dependent CDK2 phosphorylation of SAMHD1 in myeloid and lymphoid cells. J. Immunol. 193, 1988–1997 [DOI] [PubMed] [Google Scholar]
- 39.Liu F., Rothblum-Oviatt C., Ryan C. E., and Piwnica-Worms H. (1999) Overproduction of human Myt1 kinase induces a G2 cell cycle delay by interfering with the intracellular trafficking of Cdc2-cyclin B1 complexes. Mol. Cell. Biol. 19, 5113–5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang W., Wells N. J., and Hunter T. (1999) Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc. Natl. Acad. Sci. U.S.A. 96, 6193–6198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Z., Zhu M., Pan X., Zhu Y., Yan H., Jiang T., Shen Y., Dong X., Zheng N., Lu J., Ying S., and Shen Y. (2014) Inhibition of hepatitis B virus replication by SAMHD1. Biochem. Biophys. Res. Commun. 450, 1462–1468 [DOI] [PubMed] [Google Scholar]
- 42.Ma T., Zou N., Lin B. Y., Chow L. T., and Harper J. W. (1999) Interaction between cyclin-dependent kinases and human papillomavirus replication-initiation protein E1 is required for efficient viral replication. Proc. Natl. Acad. Sci. U.S.A. 96, 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wohlschlegel J. A., Dwyer B. T., Takeda D. Y., and Dutta A. (2001) Mutational analysis of the Cy motif from p21 reveals sequence degeneracy and specificity for different cyclin-dependent kinases. Mol. Cell. Biol. 21, 4868–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pagano M., Pepperkok R., Verde F., Ansorge W., and Draetta G. (1992) Cyclin A is required at two points in the human cell cycle. EMBO J. 11, 961–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams P. D., Sellers W. R., Sharma S. K., Wu A. D., Nalin C. M., and Kaelin W. G. Jr. (1996) Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol. Cell. Biol. 16, 6623–6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu C., Gao W., Zhao K., Qin X., Zhang Y., Peng X., Zhang L., Dong Y., Zhang W., Li P., Wei W., Gong Y., and Yu X. F. (2013) Structural insight into dGTP-dependent activation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase. Nat. Commun. 4, 2722. [DOI] [PubMed] [Google Scholar]
- 47.Brandariz-Nuñez A., Valle-Casuso J. C., White T. E., Laguette N., Benkirane M., Brojatsch J., and Diaz-Griffero F. (2012) Role of SAMHD1 nuclear localization in restriction of HIV-1 and SIVmac. Retrovirology 9, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou P. (2004) Determining protein half-lives. Methods Mol. Biol. 284, 67–77 [DOI] [PubMed] [Google Scholar]
- 49.Jackman M., Kubota Y., den Elzen N., Hagting A., and Pines J. (2002) Cyclin A- and cyclin E-Cdk complexes shuttle between the nucleus and the cytoplasm. Mol. Biol. Cell 13, 1030–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kyei G. B., Cheng X., Ramani R., and Ratner L. (2015) Cyclin L2 is a critical HIV dependency factor in macrophages that controls SAMHD1 abundance. Cell Host Microbe 17, 98–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruiz A., Pauls E., Badia R., Torres-Torronteras J., Riveira-Muñoz E., Clotet B., Martí R., Ballana E., and Esté J. A. (2015) Cyclin D3-dependent control of the dNTP pool and HIV-1 replication in human macrophages. Cell Cycle 14, 1657–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Badia R., Pujantell M., Riveira-Muñoz E., Puig T., Torres-Torronteras J., Martí R., Clotet B., Ampudia R. M., Vives-Pi M., Esté J. A., and Ballana E. (2016) The G1/S specific cyclin D2 is a regulator of HIV-1 restriction in non-proliferating cells. PLoS Pathog. 12, e1005829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu M., Sheppard K. A., Peng C. Y., Yee A. S., and Piwnica-Worms H. (1994) Cyclin A/CDK2 binds directly to E2F-1 and inhibits the DNA-binding activity of E2F-1/DP-1 by phosphorylation. Mol. Cell. Biol. 14, 8420–8431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen J., Saha P., Kornbluth S., Dynlacht B. D., and Dutta A. (1996) Cyclin-binding motifs are essential for the function of p21CIP1. Mol. Cell. Biol. 16, 4673–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dong C., Janas A. M., Wang J. H., Olson W. J., and Wu L. (2007) Characterization of human immunodeficiency virus type 1 replication in immature and mature dendritic cells reveals dissociable cis- and trans-infection. J. Virol. 81, 11352–11362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]