Abstract
Once it enters the host cell, herpes simplex virus type 1 (HSV-1) recruits a series of host cell factors to facilitate its life cycle. Here, we demonstrate that serine/arginine-rich splicing factor 2 (SRSF2), which is an important component of the splicing speckle, mediates HSV-1 replication by regulating viral gene expression at the transcriptional and posttranscriptional levels. Our results indicate that SRSF2 functions as a transcriptional activator by directly binding to infected cell polypeptide 0 (ICP0), infected cell polypeptide 27 (ICP27), and thymidine kinase promoters. Moreover, SRSF2 participates in ICP0 pre-mRNA splicing by recognizing binding sites in ICP0 exon 3. These findings provide insight into the functions of SRSF2 in HSV-1 replication and gene expression.
Keywords: gene expression, herpesvirus, host-pathogen interaction, RNA splicing, transcription factor
Introduction
Herpes simplex virus type 1 (HSV-1) is a human α herpesvirus that is associated with orofacial and genital herpes infections and is related to herpes encephalitis (1). The HSV-1 genome encodes >80 genes that are transcribed by RNA polymerase II (RNAP II)3 (2). Of these viral genes, infected cell polypeptide 0 (ICP0) plays vital roles in facilitating the HSV-1 life cycle. In HSV-1-infected cells, ICP0 prevents the antiviral response triggered by dsDNA by degrading IFI16 (3), PML, and SP100 (4). Additionally, ICP0 inhibits host IRF3 nuclear signaling to prevent the interferon production-mediated antiviral response of the infected cells (5). ICP0 enables efficient viral replication by redistributing host CCND3 to ND10 bodies, which function as precursors of replication compartments (6). ICP0 also participates in HSV-1 reactivation. In the HSV-1 genome, ICP0 dissociates HDAC1 and HDAC2 from the HDAC-RCOR1-REST-KDM1A complex to make the viral DNA accessible for transcription factor binding (7). After infection, HSV-1 uses a series of host cell factors to facilitate its life cycle. Host cell factor 1, which is a cellular transcriptional co-activator, plays a central role in initiating the expression of viral immediate early (IE) genes by interacting with numerous host cell transcription factors, such as virion protein 16 (8) and early growth response protein 1, a protein that mediates IE gene expression by binding to the key regulatory elements near the HSV-1 IE genes (9). Moreover, the DNA methyltransferase DNMT3A has been reported to promote HSV-1 replication by associating with the viral capsid protein VP26 (10).
Serine/arginine-rich splicing factor 2 (SRSF2 or SC35), which is a specific well known serine/arginine-rich (SR) protein family member, is well known as a mediator of genome stability, pre-mRNA splicing, mRNA nuclear export, and translational control (11–15). SRSF2 contains an RNA recognition motif for RNA binding and a domain rich in arginine and serine residues (RS domain) that facilitates its interaction with other SR splicing factors (13).
To date several studies have investigated the roles of SRSF2 in viral infection. In HIV-1 infection, SRSF2 was reported to modulate viral replication by negatively regulating the levels of genomic RNA and HIV-1 structural proteins (16). Additionally, SRSF2 regulates the splicing of the viral factors Tat (17–20) and Rev (21), which are necessary for efficient HIV-1 replication. In human papillomavirus 16 (HPV16), SRSF2 positively regulated the expression and stability of E6E7 RNAs (22). In this report we reveal that the host splicing factor SRSF2 facilitates HSV-1 replication by regulating HSV-1 ICP0, infected cell polypeptide 27 (ICP27), and thymidine kinase (TK) expression. At the transcriptional level, SRSF2 functions as a transcriptional activator by directly binding to the HSV-1 ICP0, ICP27, and TK promoters. At the post-transcriptional level, SRSF2 participates in ICP0 pre-mRNA splicing by co-localizing with ICP0 exon 3. These findings provide insight into the functions of SRSF2 in HSV-1 replication and gene expression and broaden our views concerning the mechanisms through which gene expression is regulated.
Results
SRSF2 Modulates HSV-1 Replication
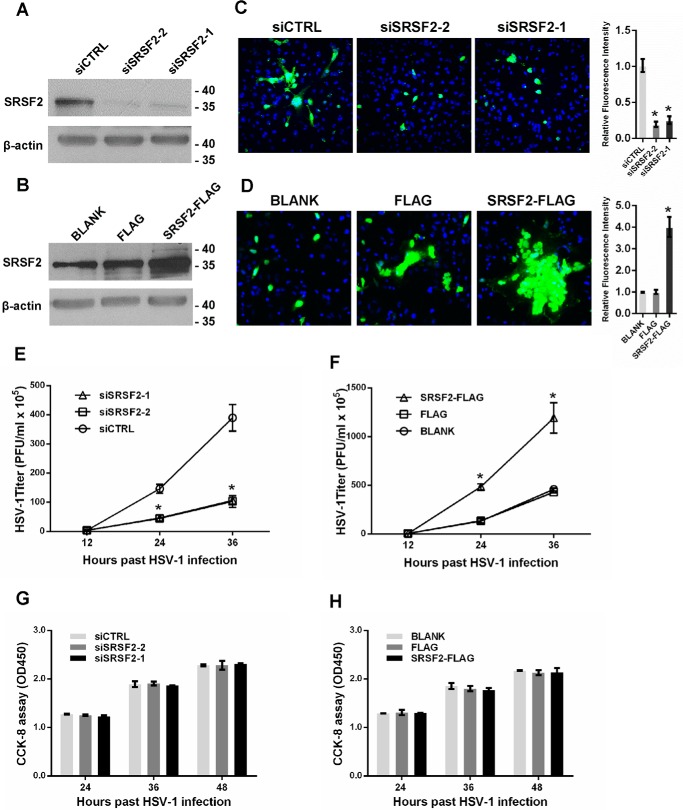
HSV-1 infection has been reported to alter the localization of the SR domain splicing factor SRSF2, resulting in the inhibition of host cell pre-mRNA splicing (23). However, the effects of SRSF2 on the HSV-1 infection process have not been elucidated. To investigate the role of SRSF2 in HSV-1 replication, we first knocked down SRSF2 using SRSF2 siRNA and overexpressed SRSF2 with a pCMV-SRSF2-FLAG construct (Fig. 1, A and B). Then, we examined HSV-1 glycoprotein expression in HeLa cells using fluorescence microscopy. The HSV-1 glycoprotein density and intensity in the SRSF2 knockdown HeLa cells was much lower than that in the cells transfected with negative control siRNA (Fig. 1C). In contrast, increased SRSF2 expression resulted in a much higher density and intensity of the HSV-1 glycoproteins (Fig. 1D), suggesting that SRSF2 affected both HSV-1 replication and spread. Next, cells with down-regulated or up-regulated SRSF2 expression were infected with HSV-1 at a multiplicity of infection (m.o.i.) of 1 for 12, 24, or 36 h. The viral supernatants were collected, and the viral titers were determined using plaque assays. The HSV-1 titers in the culture media from SRSF2-knocked down HeLa cells were markedly lower than the titers in HeLa cells transfected with the negative control siRNA (Fig. 1E). The opposite results were observed in the SRSF2-overexpressing cells (Fig. 1F). To investigate the effect of silencing and overexpression of SC5 on cell cytotoxicity in vitro, a Cell Counting Kit-8 (CCK8) assay was performed in HeLa cells transfected with SRSF2 siRNA or SRSF2-FLAG. The results revealed no difference between the SRSF2 siRNA (Fig. 1G) or SRSF2-FLAG plasmid (Fig. 1H) and control groups 24, 36, and 48 h after the transfections, indicating no significant effect on cell viability or proliferative capacity.
FIGURE 1.
SRSF2 promoted HSV-1 replication. A, HeLa cells were transfected with the SRSF2 siRNA or negative control for 36 h. The SRSF2 expression levels were measured with Western blotting. B, HeLa cells were transfected with SRSF2-FLAG, FLAG, or BLANK for 36 h. The SRSF2 expression levels were measured with Western blotting. C, after 36 h of transfection with SRSF2 siRNA or the negative control, the HeLa cells were infected with HSV-1 at an m.o.i. of 1 for 12 h, fixed, and stained with an anti-HSV-1 glycoprotein antibody (green) and subjected to confocal microscopy analysis. DAPI (blue) was used to stain the nuclei. The right histogram shows the relative fluorescence intensity of the HSV-1 glycoproteins. The fluorescence signal value was measured using ImageJ software and normalized to a single cell. The data are normalized to the control (siCTRL) level and are presented as the mean ± S.D. (*, p < 0.01, Student's t test). D, after 36 h of transfection with SRSF2-FLAG, FLAG, or BLANK, the HeLa cells were infected with HSV-1 at an m.o.i. of 1 for 12 h, fixed, stained with an anti-HSV-1 glycoprotein antibody (green), and subjected to confocal microscopy analysis. DAPI (blue) was used to stain the nuclei. The right histogram shows the relative fluorescence intensity of the HSV-1 glycoproteins. The fluorescence signal value was measured using ImageJ software and normalized to a single cell. The data were normalized to the control level (FLAG) and are presented as the mean ± S.D. (*, p < 0.01, Student's t test). E, after 36 h of transfection with the SRSF2 siRNA or negative control siRNA, HeLa cells were infected with HSV-1 at an m.o.i. of 1. The culture media were collected at the indicated time points after infection and subjected to the plaque assay. The data points represent the mean values determined from three independent experiments. The data are presented as the mean ± S.D. (*, p < 0.01, Student's t test). F, after 36 h of transfection with SRSF2-FLAG, FLAG, or BLANK, the HeLa cells were infected with HSV-1 at an m.o.i. of 1. The culture media were collected at the indicated time points after infection and subjected to the plaque assay. The data points represent mean values determined from three independent experiments. The data are presented as the mean ± S.D. (*, p < 0.01, Student's t test). The cell viability and proliferative ability were determined in HeLa cells transfected with SRSF2 siRNA (G) or SRSF2-FLAG (H) with a CCK8 assay. The data points represent mean values determined from three independent experiments. The data are presented as the mean ± S.D. (no significance, Student's t test).
SRSF2 Regulates Viral Gene Expression
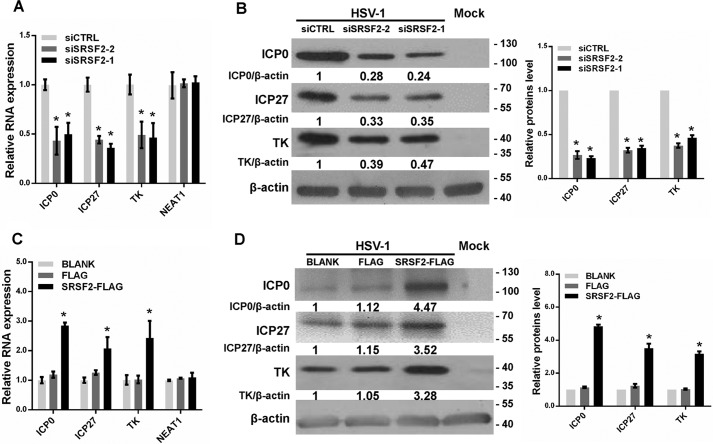
In HSV-1-infected host cells, the IE genes are first activated, promoting the expression of viral proteins to facilitate viral replication. To study the mechanism by which SRSF2 modulates HSV-1 replication, we measured the expression levels of genes required for the appropriate expression of viral early and late gene products, such as HSV-1 ICP0, ICP27, and TK, in HeLa cells transfected with the SRSF2 siRNA. In HSV-1-infected HeLa cells, SRSF2 depletion led to the inhibition of ICP0, ICP27, and TK expression at the mRNA (Fig. 2A) and protein levels (Fig. 2B). Conversely, up-regulation of SRSF2 by SRSF2-FLAG plasmid transfection enhanced ICP0, ICP27, and TK expression at the mRNA (Fig. 2C) and protein levels (Fig. 2D). Also, we measured the expression level of host cellular nuclear paraspeckle assembly transcript 1 (NEAT1), an unrelated non-coding gene, in HeLa cells transfected with the SRSF2 siRNA or SRSF2-FLAG plasmid. The qPCR results showed that SRSF2 had no significant effect on NEAT1 expression (Fig. 2, A and C).
FIGURE 2.
SRSF2 regulated viral gene expression. A, after 36 h of transfection with the SRSF2 siRNA or negative control siRNA, HeLa cells were infected with HSV-1 at a m.o.i. of 1 for 4 h. The ICP0, ICP27, and TK expression levels relative to β-actin expression were determined with qRT-PCR in three independent experiments. An unrelated RNA (NEAT1) was used as the control. The data are presented as the mean ± S.D. (*, p < 0.01, Student's t test). B, after 36 h of transfection with the SRSF2 siRNA or negative control siRNA, HeLa cells were infected with HSV-1 at a m.o.i. of 1 for 8 h. The ICP0, ICP27, and TK expression levels were measured with Western blotting. Protein ratios of the indicated proteins/β-actin were analyzed with ImageJ software, and statistical analysis was conducted with data from three independent experiments (right panel). The data are presented as the mean ± S.D. (*, p < 0.01, Student's t test). C, after 36 h of transfection with SRSF2-FLAG, FLAG, or BLANK, HeLa cells were infected with HSV-1 at a m.o.i. of 1 for 4 h. The ICP0, ICP27, and TK expression levels relative to β-actin expression were determined by qRT-PCR for three independent experiments. An unrelated RNA (NEAT1) was used as the control. The data are presented as the mean ± S.D. (*, p < 0.01, Student's t test). D, after 36 h of transfection with SRSF2-FLAG, FLAG, or BLANK, HeLa cells were infected with HSV-1 at a m.o.i. of 1 for 8 h. The ICP0, ICP27, and TK expression levels were measured with Western blotting. Protein ratios of the indicated proteins/β-actin were analyzed with ImageJ software, and statistical analysis was conducted with data from three independent experiments (right panel). The data are presented as the mean ± S.D. (*p < 0.01, Student's t test).
SRSF2 Enhances Viral Gene Transcription by Binding to Gene Promoters
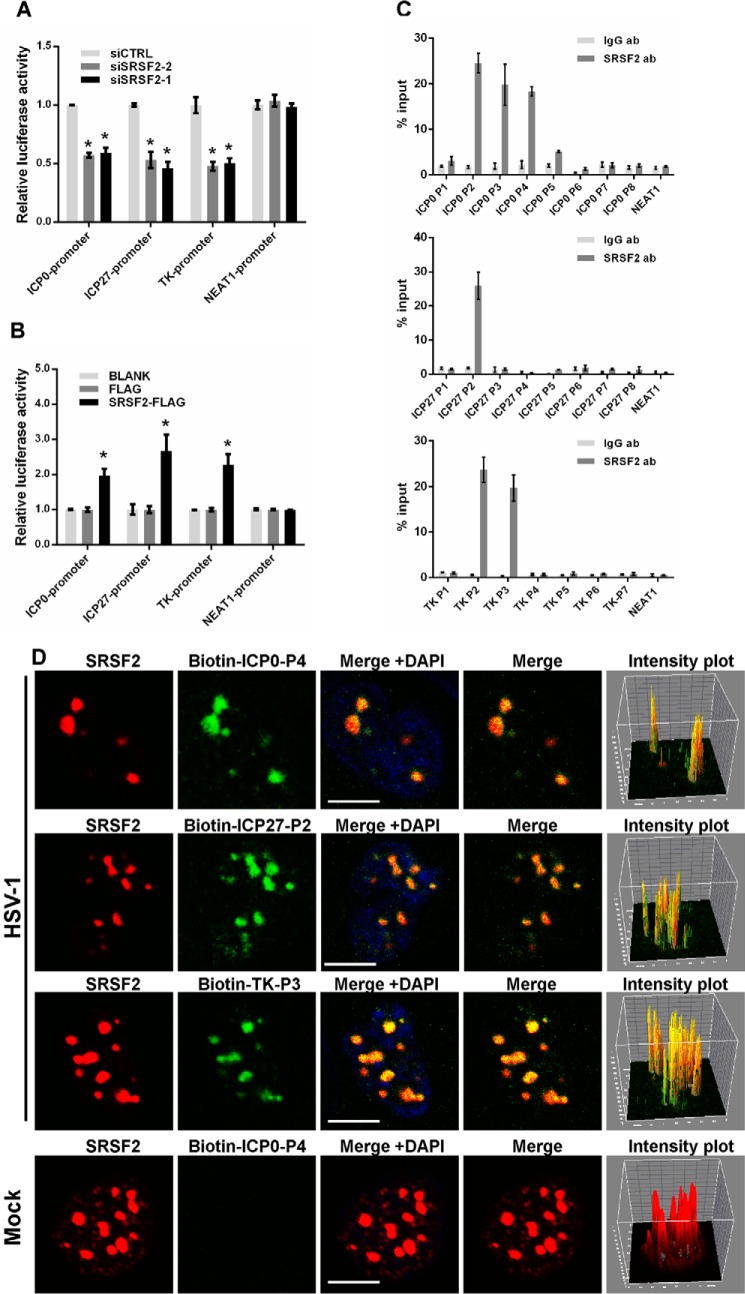
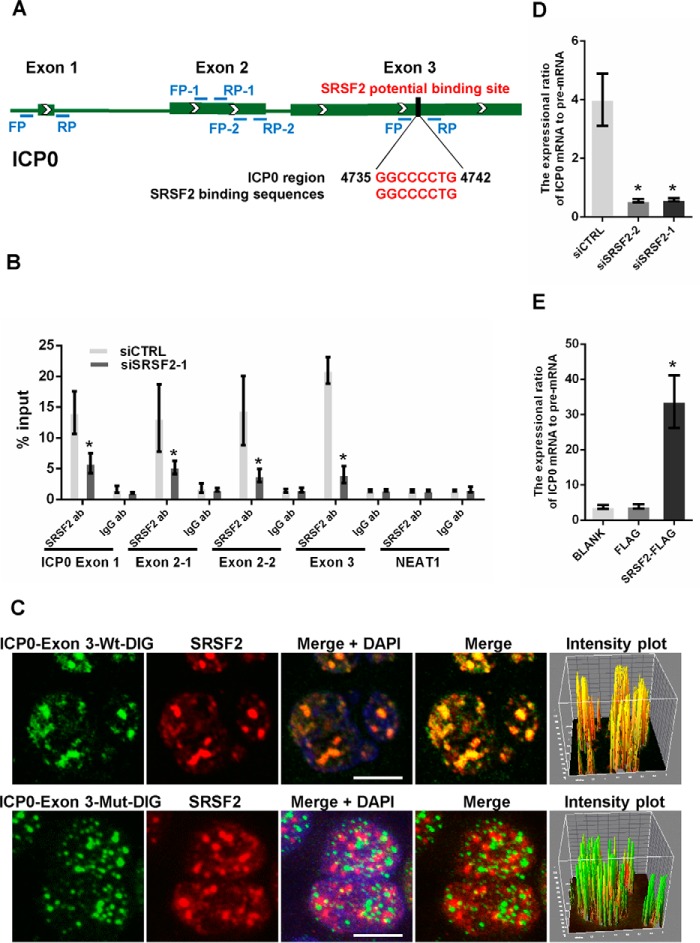
Although SRSF2 is well known as a nuclear speckle component that participates in the process of cellular RNA maturation by binding to RNA, recent reports have indicated that it seems to function as a chromatin regulator (24). Therefore, we performed a luciferase assay to examine whether SRSF2 directly regulated viral gene transcription. We generated two luciferase reporter constructs by inserting the promoter fragments of ICP0 and ICP27 into pGL3-enhancer vectors. To study the effects of SRSF2 on the transcriptional activity of the HSV-1 TK promoter, we directly utilized a pRL-TK reporter vector because this vector contained the same HSV-1 TK gene promoter sequence. The results showed inhibited transcriptional activity of the ICP0, ICP27, and TK promoters in the SRSF2-depleted HeLa cells (Fig. 3A) and increased transcriptional activity in the SRSF2-overexpressing HeLa cells (Fig. 3B). Additionally, we measured the transcription of NEAT1 in HeLa cells transfected with the SRSF2 siRNA or SRSF2-FLAG plasmid as a negative control. To identify the SRSF2 binding sites in the ICP0, ICP27, and TK promoters, we designed sets of primer pairs that recognized the upstream and downstream transcriptional start sites (TSSs) of these genes (Table 1) and performed a chromatin immunoprecipitation (ChIP) assay with anti-SRSF2 antibody or anti-IgG antibody in HSV-1-infected HeLa cells. The NEAT1 promoter was used as a negative control. The results showed that SRSF2 was enriched near the TSSs of ICP0 gene, ICP27 gene, and TK gene (Fig. 3C). Next, we used DNA fluorescence in situ hybridization (FISH) and immunofluorescence microscopy experiments to validate the interaction between SRSF2 and the HSV-1 ICP0, ICP27, and TK promoters. First, we prepared probes by amplifying the P4 region of ICP0, the P2 region of ICP27, and the P3 region of TK; the probes were labeled with biotinylated cytidine bisphosphate at the 3′ end of the DNA. The results confirmed that SRSF2 was associated with the promoters of these genes (Fig. 3D). Additionally, pixel intensity plots were generated for each merged channel (Fig. 3D, right panels). To determine the specificity of this co-localization, we performed DNA-FISH and immunofluorescence microscopy experiments in mock-infected HeLa cells. As shown in Fig. 3D, bottom panels, the probe recognizing the ICP0 promoter fragment detected no signal in mock-infected nuclei.
FIGURE 3.
SRSF2 enhanced ICP0, ICP27, and TK transcriptional activity by binding to their promoters. A, after co-transfection with the SRSF2 siRNA or negative control siRNA and the pGL3 enhancer plasmid containing the ICP0 promoter, ICP27 promoter, or a pRL-TK reporter for 36 h, HeLa cells were infected with HSV-1 at a m.o.i. of 1 for 4 h. The NEAT1 promoter reporter was used as a negative control. The relative transcriptional activities of the ICP0, ICP27, TK, and NEAT1 promoters were determined with a luciferase assay in three independent experiments. The data are presented as the mean ± S.D. (*, p < 0.01, Student's t test). B, after co-transfection with SRSF2-FLAG, FLAG, or BLANK and the pGL3 enhancer plasmid containing the ICP0 promoter, ICP27 promoter, or a pRL-TK reporter vector with the same sequence as the HSV-1 TK gene for 36 h, HeLa cells were infected with HSV-1 for 4 h. The NEAT1 promoter reporter was used as a negative control. The relative transcriptional activity of the ICP0, ICP27, TK, and NEAT1 promoters was analyzed with a luciferase assay in three independent experiments. The data are presented as the mean ± S.D. (*, p < 0.01, Student's t test). C, HeLa cells were infected with HSV-1 for 4 h. ChIP assays were performed with an anti-SRSF2 antibody or an anti-IgG antibody (Ab). -Fold enrichment of the ICP0, ICP27, and TK promoters by anti-SRSF2 relative to the input level was examined with qRT-PCR. The NEAT1 promoter was used as a negative control. D, HeLa cells were infected with HSV-1 at a m.o.i. of 1 for 4 h, fixed, incubated with a biotin-ICP0-P4 fragment (green), a biotin-ICP27-P2 fragment (green), and a biotin-TK-P3 fragment (green) and SRSF2 (red) and subjected to confocal microscopy analysis. The mock-infected nuclei were used as a negative control. The intensity plots for the red and green channels were analyzed with ImageJ software. DAPI (blue) was used to stain the nuclei. Scale bars, 10 μm.
TABLE 1.
Sequences of primers used in the ChIP assay
Primer positions are indicated relative to transcription start site.
| Name | Forward primer | Reverse primer | Primer positions |
|---|---|---|---|
| ICP0-P1 | TAGGGTCAGGGGGTTCCG | GGGGACCCCCACTCATACA | 169/294 |
| ICP0-P2 | GTTCCAGTGTAAGGGTCGGG | ATACGACCCCCATGGAGCC | −13/148 |
| ICP0-P3 | CCGACAGTCTGGTCGCATTT | GGCTCCATGGGGGTCGTAT | −158/7 |
| ICP0-P4 | CCATTGGGGGAATCGTCAC | CTTCTGTGGTGATGCGGAG | −227/−65 |
| ICP0-P5 | GGGCATGCTAATGGGGTTCT | GCAGTGACGATTCCCCCAAT | −380/−206 |
| ICP0-P6 | CAATGAACCCGCATTGGTCC | AGAACCCCATTAGCATGCCC | −493/−361 |
| ICP0-P7 | CTTAATGGGCAACCCCGGTA | CGCCTTCCCGAAGAAACTCA | −621/−432 |
| ICP0-P8 | CTTGTTCCGCTTCCCGGTAT | GAATACCGGGGTTGCCCATT | −707/−599 |
| TK-P1 | GGGCGATTGGTCGTAATCCA | CCCAACGGCGACCTGTATAA | 196/297 |
| TK-P2 | CCGGTATTGTCTCCTTCCGT | CTGCCGGGACGCCCT | 42/191 |
| TK-P3 | AAACGCGGGCGTATTGGT | ACAATACCGGAAGGAACCCG | −107/50 |
| TK-P4 | GCAGGTAGGTCTTCGGGATG | GGCATCTCTGCCCCTTCTTC | −568/−453 |
| TK-P5 | AAAAGCCTAGCAGGTCGGAG | CCTCTCTTCTGGCGCCTAAC | −1459/−1354 |
| TK-P6 | AGAAACTCGGCATACAGGGC | GAGATTCTGGAGCGCGAACA | −1739/−1600 |
| TK-P7 | AAGTGGTCCGGAAGCCAAAA | GAGCCTTCTGATAGCCTCGG | −1982/−1860 |
| ICP27-P1 | CCCACAGTGTGTGGTCGC | TCCGACAAACCATCGGCAG | 301/481 |
| ICP27-P2 | GACGAGGACATGGAAGACCC | CTGGTTGAGGATCGTTGGGG | 138/297 |
| ICP27-P3 | CCAGAGGCCATATCCGACAC | CTGTCCGATTCCAGGTCGTC | −51/115 |
| ICP27-P4 | CGTCCCGTTACCAAGACCAA | CGGCACAGACAAGGACCAAT | −376/−240 |
| ICP27-P5 | CCATCATCCTCTCGGGCATC | GCCCCAAGACAGGACAGTTT | −654/−473 |
| ICP27-P6 | TCACCTTCTTGTACCACCGC | AACACCAGGTGGTGATGGTC | −942/−784 |
| ICP27-P7 | GCCGATGTATGTTTGGCGTC | GCAGGCGGGTAATTTTCGTG | −1131/−1000 |
| ICP27-P8 | TCCGCTGCACCGATGTATTT | CCCGCGATAAGATTGGCGTA | −1460/−1290 |
| NEAT1 | TGTCCCTCGGCTATGTCAGA | GAGGGGACGTGTTTCCTGAG | −920/−769 |
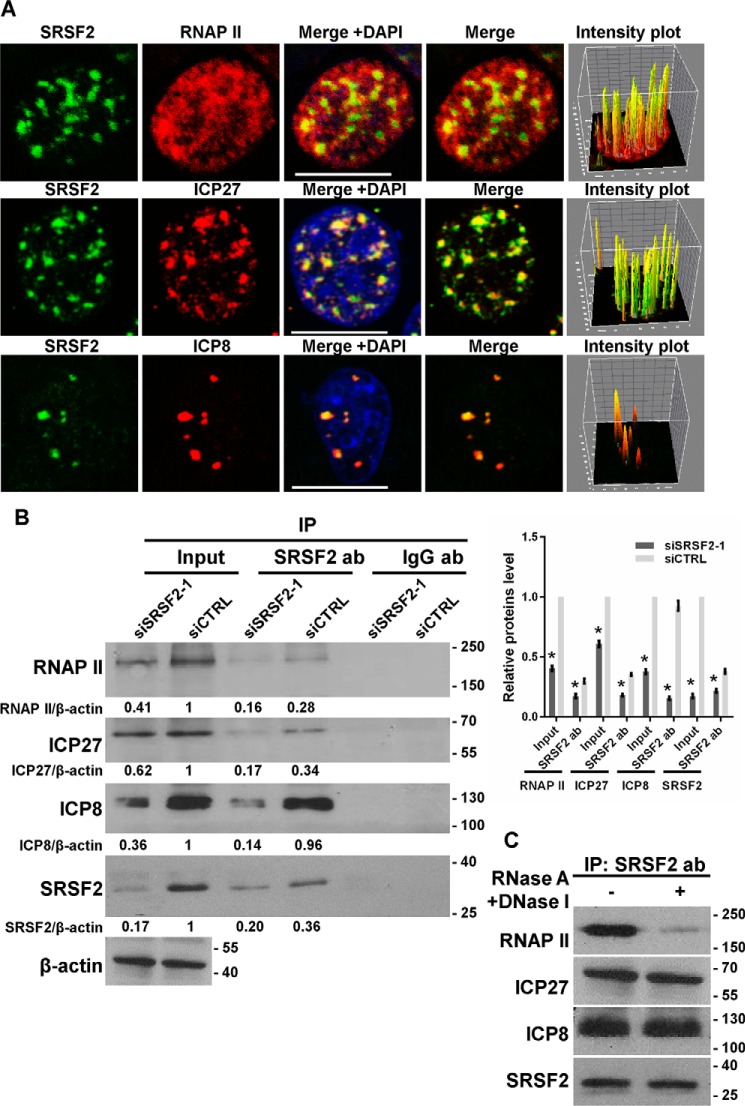
In addition, we tested the co-localization of SRSF2 with Ser-5(P)-RNAP II, ICP27, and infected cell protein 8 (ICP8) using the immunofluorescence assay and pixel intensity analysis (Fig. 4A). Of these proteins, RNAP II widely exists at gene promoters and catalyzes DNA transcription and RNA synthesis, and ICP27 has been shown to recruit RNAP II to viral transcription sites to initiate their transcription (25); ICP8 was the first protein to be identified in HSV-1 replication compartments, which function as sites of viral DNA replication and late gene transcription (26). We also conducted an immunoprecipitation assay to confirm this association. SRSF2 siRNA- or negative-control-transfected cells were infected with HSV-1 at an m.o.i. of 1 for 4 h. Cells lysates were harvested and subjected to the immunoprecipitation assay with the SRSF2 antibody followed by Western blotting using antibodies against RNAP II, ICP27, ICP8, and SRSF2. RNAP II, ICP27, and ICP8 were specifically pulled down in the negative-control-transfected cell lysates. SRSF2 depletion resulted in a significant reduction in the interaction of SRSF2 with RNAP II, ICP27, and ICP8 (Fig. 4B). To remove the influence of the RNA or DNA in these associations, the cell lysate was preincubated with RNase A and DNase I and then subjected to the immunoprecipitation assay with the SRSF2 antibody. The result demonstrated that DNase and RNase treatment significantly reduced the association of SRSF2 with RNAP II without alteration in the association of SRSF2 with ICP8 and ICP27 (Fig. 4C), as suggested earlier (24). Together, these results suggested that SRSF2 activation enhanced the transcriptional activity of the ICP0, ICP27, and TK promoters through the co-localization of SRSF2 with viral gene promoters, RNAP II, ICP27, and ICP8.
FIGURE 4.
SRSF2 associated with RNAP II, ICP27, and ICP8. A, after 4 h of infection with HSV-1, HeLa cells were fixed, stained with antibodies against SRSF2 (green), RNA polymerase II (red), ICP27 (red), and ICP8 (red), and subjected to confocal microscopy analysis. The intensity plots for the red and green channels were analyzed with ImageJ software. DAPI (blue) was used to stain the nuclei. Scale bars, 10 μm. B, after 36 h of transfection with the SRSF2 siRNA or negative control siRNA, HeLa cells were infected with HSV-1 at a m.o.i. of 1 for 4 h. Cell lysates were harvested and subjected to an immunoprecipitation (IP) assay with the anti-SRSF2 antibody or the anti-IgG antibody (Ab). The retrieval of RNA polymerase II, ICP27, SRSF2, and ICP8 by endogenous SRSF2 and IgG was measured by Western blotting. Protein ratios for the indicated proteins/β-actin were analyzed with ImageJ software, and statistical analysis was conducted with data from three independent experiments (right panel). The data are presented as the mean ± S.D. (*, p < 0.01, Student's t test). C, HeLa cells were infected with HSV-1 at a m.o.i. of 1 for 4 h. Cell lysates were harvested with and without DNase and RNase treatment and then subjected to an immunoprecipitation assay with the anti-SRSF2 antibody. The retrieval of RNA polymerase II, ICP27, SRSF2, and ICP8 by endogenous SRSF2 was measured by Western blotting.
SRSF2 Regulates ICP0 Pre-mRNA Splicing
As a nuclear pre-mRNA splicing factor, SRSF2 completes exon inclusion and exclusion via binding to exon splicing enhancers located within pre-mRNA exons (12). In HSV-1, all viral gene transcripts are intronless with the exception of ICP0. Therefore, we analyzed the ICP0 pre-mRNA sequence to search for SRSF2 binding motifs and identified a general consensus SRSF2-binding motif (27) located in exon 3 between 4735 bp and 4742 bp (Fig. 5A). To test whether SRSF2 binds to this site, we performed an RNA immunoprecipitation assay. SRSF2 siRNA- or negative-control-transfected cells were infected with HSV-1 at an m.o.i. of 1 for 4 h. The cell lysates were harvested and subjected to an immunoprecipitation assay with the SRSF2 antibody or IgG antibody followed by qRT-PCR using primers that recognized the ICP0 exon 1, exon 2, and exon 3 regions, including the potential SRSF2-binding site (Fig. 5A). An unrelated RNA (NEAT1) was used as a negative control. To avoid DNA contamination in the qRT-PCR, we eliminated the remaining genomic DNA and then reverse-transcribed cDNA with the kit. The results demonstrated that SRSF2 had the ability to pull down fragments of ICP0 pre-mRNA exon 1, exon 2, and exon 3, whereas SRSF2 knockdown markedly decreased the amount of these fragments retrieved by SRSF2 (Fig. 5B).
FIGURE 5.
SRSF2 regulated ICP0 pre-mRNA splicing. A, schematic of SRSF2 potential binding sites in the ICP0 pre-mRNA sequence. The black box shows the potential binding site, and red characters indicate matching sequences. B, after 36 h of transfection with the SRSF2 siRNAs or negative control, HeLa cells were infected with HSV-1 at a m.o.i. of 1 for 4 h. Cell lysates were harvested and subjected to an RNA immunoprecipitation assay. QRT-PCR was performed to detect the retrieval of ICP0 exon 1, ICP0 exon 2, ICP0 exon 3, and NEAT1 by the anti-SRSF2 antibody or anti-IgG antibody over the input level. The unrelated RNA NEAT1 was used as a negative control. The data are represented as the mean ± S.D. (*, p < 0.01, Student's t test). C, HeLa cells were infected with HSV-1 at a m.o.i. of 1 for 4 h, fixed, incubated with DIG-labeled ICP0 exon 3 fragment (green) or ICP0 exon 3 fragment with a deletion in SRSF2 binding motifs (green) and SRSF2 (red) and subjected to confocal microscopy analysis. The intensity plots for the red and green channels were analyzed with the Image J software. DAPI (blue) was used to stain the nuclei. Scale bars, 10 μm. D, after 36 h of transfection with the SRSF2 siRNAs or negative control, the HeLa cells were infected with HSV-1 at a m.o.i. of 1 for 4 h. The expression ratio of the ICP0 mRNA to the pre-mRNA was determined by qRT-PCR in three independent experiments. The data are represented as the mean ± S.D. (*, p < 0.01, Student's t test). E, after 36 h of transfection with SRSF2-FLAG, FLAG, or BLANK, the HeLa cells were infected with HSV-1 at a m.o.i. of 1 for 4 h. The expression ratio of the ICP0 mRNA to pre-mRNA was determined by qRT-PCR in three independent experiments. The data are represented as the mean ± S.D. (*, p < 0.01, Student's t test).
Next, we prepared probes via transcribing ICP0 exon 3 containing the SRSF2 binding motif (WT) and ICP0 exon 3 with a deletion in the SRSF2 binding motif (Mut). The probes were labeled with digoxigenin (DIG) at the 3′ end of the RNA in vitro and used to perform RNA FISH and immunofluorescence assay experiments. As shown in Fig. 5C, SRSF2 largely colocalized with ICP0 exon 3 containing the SRSF2 binding motif. The intensity plot showed yellow fluorescence, indicating colocalization of SRSF2 and ICP0 exon 3 containing the SRSF2 binding motif (Fig. 5C, upper panels). Conversely, the ICP0 exon 3 with a deletion in the SRSF2 binding motif did not colocalize with SRSF2. The intensity plot showed red and green fluorescence, indicating no colocalization between SRSF2 and ICP0 exon 3 due to the deletion of the SRSF2-binding motif (Fig. 5C, lower panels). These data are the first to show that SRSF2 recognizes and colocalizes with HSV-1 transcripts. Finally, we tested whether the colocalization between SRSF2 and ICP0 exon 3 influenced ICP0 pre-mRNA splicing. Because SRSF2 regulates the transcriptional activity of the ICP0 promoter, the expression difference between the ICP0 intron and the exons regulated by SRSF2 cannot be used as a standard for its effects on ICP0 pre-mRNA splicing. Therefore, we utilized the ratio of the ICP0 mRNA expression level to the pre-mRNA expression level to characterize its splicing ability. As shown in Fig. 5D, the expression ratio of the ICP0 mRNA was 4-fold higher than that of the ICP0 pre-mRNA in HeLa cells transfected with the negative control. However, the ratio declined significantly when SRSF2 expression was decreased with the siRNA. Conversely, overexpression of SRSF2 increased the ratio (Fig. 5E). Taken together, these results prove that SRSF2 regulates ICP0 pre-mRNA splicing by recognizing ICP0 exon 3.
Discussion
SRSF2 is a well known SR protein that is involved in genome stability, alternative splicing, and mRNA export and translation control. In our present study we found that SRSF2 promoted HSV-1 replication by regulating viral gene expression at the transcriptional and post-transcriptional levels. At the transcriptional level, SRSF2 was recruited to the promoters of ICP0, ICP27, and TK with RNAP II, ICP27, and ICP8 to initialize the transcription of these genes. At the post-transcriptional level, SRSF2 recognized the SRSF2 binding sites located in ICP0 pre-mRNA exon 3 to modulate its splicing. As an immediate early protein, ICP0 is synthesized and transported to the nucleus after the earliest stage of HSV-1 infection and plays vital roles in the promotion of viral gene expression (28), the distribution of host cellular nuclear bodies (29), the alteration of host gene expression (30), the disruption of antiviral pathways (4, 31), and the assembly of new virion particles (32). Recently, Workenhe et al. (33) revealed that SRSF2 did not influence ICP0-null HSV-1 replication, suggesting that ICP0 plays key roles in SRSF2-mediated HSV-1 replication. In this study, SRSF2 was revealed to function as a transcriptional activator and splicing factor in ICP0 expression. As a multifunctional viral protein, the SRSF2-mediated induction of ICP0 expression may greatly influence the HSV-1 life cycle.
In our work we demonstrated that SRSF2 could co-localize with the promoters of ICP0, ICP27, and TK to alter their transcriptional activity. First, SRSF2 positively regulated ICP0, ICP27, and TK expression; this regulation was associated with altered transcription. SRSF2 was originally recognized as a pre-mRNA-binding protein. However, Ji X et al. (24) demonstrated that SRSF2 appeared to associate with gene promoters instead of the RNA. Therefore, we hypothesized that SRSF2 might regulate the transcription of viral genes via interacting with their promoters. Based on the ChIP assay results, we concluded that SRSF2 binding sites were located in the TSSs of the ICP0, ICP27, and TK promoters. These results demonstrate for the first time that speckles component SRSF2 co-localizes with HSV-1 DNA, suggesting that the HSV-1 genome is recruited to the nuclear sub-structural speckles to regulate its replication and gene expression. Because HSV-1 genes are transcribed by RNAP II, we tested the relationship between SRSF2 and RNAP II. The immunofluorescence assay and immunoprecipitation assay results confirmed that SRSF2 physically interacts with RNAP II. Additionally, SRSF2 was shown to associate with ICP8, an HSV-1 single-stranded DNA-binding protein that functions as an activator of viral DNA replication and late gene transcription (34). These results revealed the potential ability of SRSF2 to regulate gene expression by affecting viral gene transcription in addition to its traditional roles in pre-mRNA splicing.
Generally, SRSF2 participates in cellular pre-mRNA and alternative splicing by binding to its exon splicing enhancers (12). To screen the ICP0 pre-mRNA sequence, we identified general consensus SRSF2 binding motifs located in exon 3. The RNA immunoprecipitation assay and immunofluorescence assay results confirmed that SRSF2 colocalized with ICP0 pre-mRNA exon 3. To better show its effect on pre-mRNA splicing, we utilized the ratio of the ICP0 mRNA expression level to the pre-mRNA expression level to characterize its splicing ability.
As a traditional RNA-binding protein, SRSF2 bound to the HSV-1 ICP0, ICP27, and TK promoters to initiate their transcription in association with RNAP II and ICP27. Moreover, SRSF2 participated in ICP0 pre-mRNA splicing by recognizing SRSF2 binding motifs located in ICP0 exon 3. Taken together, our study provides useful insight into the functions of SRSF2 in viral infection.
Experimental Procedures
Cell Culture, HSV-1 Infection, and Plaque Assay
HeLa cells (epithelial, human cervix carcinoma cell line; ATCC) were grown in Dulbecco's modified Eagle's medium (Gibco/Invitrogen, 12800-017) containing 10% fetal bovine serum (PAA, A15-101) and 10 units/ml penicillin-streptomycin (Gibco/Invitrogen, 15140-122) in a 5% CO2-humidified incubator at 37 °C. The HeLa cells were infected with the HSV-1 virus SM44 strain at a m.o.i. of 1 pfu per cell. For the plaque assay, Vero cells (Chinese Academy of Sciences) were inoculated with 100 μl of a series of diluted viral supernatants for 1 h. After virus adsorption, the HSV-1-infected cells were overlaid with medium containing 1% human serum for 48 h and stained with 1% crystal violet in 10% formaldehyde for 15 min to visualize the plaques.
Plasmid Construction
Recombinant vectors encoding humanSRSF2 (NM_003016.4) were constructed by PCR-based amplification from the cDNA of human HeLa cells. Then the amplicons were subcloned into the HindIII/XhoI sites of pCMV-N-FLAG (Beyotime, D2722). To generate luciferase reporters for the promoter assay, we cloned the ICP0, ICP27, and NEAT1 promoters into the KpnI/XhoI sites of the pGL3-enhancer reporter. All constructs were confirmed by DNA sequencing. The primers for plasmid construction were as follows: SRSF2-FLAG, 5′-CCCAAGCTTATGGCCCAATGGAATCAGCTA-3′ (sense) and 5′-CCGCTCGAGCATGGGGGAGGTAGCGCA-3′ (antisense); ICP0 promoter, 5′-CGGGGTACCTCTAACGTTACACCCGAGGC-3′ (sense) and 5′-CCGCTCGAGTTCTGTGGTGATGCGGAGAG-3′ (antisense);ICP27 promoter, 5′-CGGGGTACCCCATCATCCTCTCGGGCATC-3′ (sense) and 5′-CCGCTCGAGGTCTTCTGGACGAGACGGG-3′ (antisense); NEAT1 promoter, 5′-CGGGGTACCGGGCCCAGAAACAGCACTAC-3′ (sense) and 5′-CCGCTCGAGAGGACTTTGGACCGTGTAGC-3′ (antisense).
Cell Transfections, RNA Isolation, Reverse Transcription, and qPCR
All the synthetic siRNAs and the negative control (CTRL) were purchased from Shanghai GenePharma Co., Ltd. The siRNA sequences were as follows: SRSF2–1 siRNA, 5′- GUGAGAAGUUGCUUAGAAA-3′ (sense) and 5′- UUUCUAAGCAACUUCUCAC-3′ (antisense); SRSF2–2 siRNA, 5′-CAAACUCUGUACCCAUUAA-3′ (sense) and 5′- UUAAUGGGUACAGAGUUUG-3′ (antisense); negative control siRNA,5′-UUCUCCGAACGUGUCACGU-3′ (sense) and 5′-ACGUGACACGUUCGGAGAA-3′ (antisense). All the siRNAs were transfected using LipofectamineTM 2000 (Invitrogen, 11668-019) according to the manufacturer's protocol. For plasmids, the cells were transiently transfected using Lipofectamine&TM 3000 (Invitrogen, 1656200). Total RNA was isolated using RNA iso Plus (Takara, D9108B) according to the manufacturer's protocol. Real-time qRT-PCR was performed using the ReverTra Ace® qPCR RT Master Mix with genomic DNA remover (TOYOBO, FSQ-301) and the SYBR Green PCR Master Mix (TOYOBO, QPK-201). All mRNA levels were measured and normalized to β-actin mRNA. The primers for RT-PCR analysis were as follows: ICP0, 5′-CCCACTATCAGGTACACCAGCTT-3′ (sense) and 5′-CTGCGCTGCGACACCTT-3′ (antisense); ICP27, 5′-CGATGACTTACTGGCGGGTGT-3′ (sense) and 5′-GCGTCGGTCACGGCATAA-3′ (antisense); ICP0 pre mRNA, 5′-GAGGATGACGACGACCTGGA-3′ (sense) and 5′-GGGGCGCGATCAGGTTA-3′ (antisense); TK, 5′-TGTGTGACACCATTCATTGATGC-3′ (sense) and 5′-TCCTCACATGGGGGAGGTAG-3′ (antisense); NEAT1, 5′-GAGAACCAAAGGGAGGGGTG-3′ (sense) and 5′-TGCTGCGTATGCAAGTCTGA-3′ (antisense); actin-β, 5′-GTACCCAGGCATTGCTGACA-3′ (sense) and 5′-CGCAGCTCAGTAACAGTCCG-3′ (antisense).
Western Blotting
Cells were lysed in ice-cold whole cell extract buffer B (50 mm Tris-HCl, pH 8.0, 4 m urea, and 1% Triton X-100) supplemented with a complete protease inhibitor mixture. The cell extracts were resolved by SDS-PAGE and analyzed by Western blotting. The protein bands were visualized using the ECL blotting detection reagents. The antibodies used for Western blotting included an anti-SRSF2 antibody (Santa Cruz Biotechnology, sc-10252), an anti-HSV-1 ICP0 antibody (Abcam, ab6513), an anti-HSV-1 ICP27 antibody (Abcam, ab31631), an anti-HSV-1 TK antibody (Santa Cruz Biotechnology, sc-28037), an anti-RNAP II antibody (Abcam, ab5131), and an anti-β-actin antibody (Proteintech, 60008-1-Ig).
Cell Counting Kit-8 Assay
To determine the cell viability and proliferative capacity, transfected HeLa cells were plated into 96-well plates at a density of 2 × 104 cells/well. After growth for 24, 36, and 48 h, 10 μl of CCK-8 (DoJinDo, CK04) was added to each well. The plates were incubated for an additional 2 h, and the absorbance was detected at a wavelength of 450 nm.
Luciferase Assay
Luciferase activities were assayed using a Dual-Luciferase Reporter System (Promega, E1960) according to the manufacturer's protocol.
ChIP Assay
The ChIP assay was conducted using Dahl's protocol (35). Briefly, the cells were fixed with 1% formaldehyde and sonicated to shear the DNA to an average fragment size of 200–1000 bp. After centrifugation, the supernatants were incubated with the SRSF2 antibody (Abcam, ab11826) and IgG antibody (Abcam, ab205719). Chromatin DNA was purified with the Dynabeads Protein G kit (Invitrogen, 10004D) and subjected to real-time PCR. The region-specific primers are listed in Table 1.
RNA Immunoprecipitation Assay
The RNA immunoprecipitation assay was performed using the protocol of Peritz et al. (36). Briefly, HSV-1-infected HeLa cell lysates were incubated with the anti-SRSF2 antibody (Abcam, ab11826) and IgG antibody (Abcam, ab205719) at 4 °C overnight. RNA enriched from the immunoprecipitation was retro-transcribed for the real-time PCR. The primers for RT-PCR analysis were as follows: ICP0-exon 1, 5′-TCTCCGCATCACCACAGAAG-3′ (sense) and 5′-GTTCCAGTGTAAGGGTCGGG-3′ (antisense); ICP0-exon 2-1, 5′-CGGATGTCTGGGTGTTTCCC-3′ (sense) and 5′-TCTCGAACAGTTCCGTGTCC-3′ (antisense); ICP0-exon2-2, 5′-TGCATCCCGTGCATGAAAAC-3′ (sense) and 5′-GCTGATTGCCCGTCCAGATA-3′ (antisense); ICP0-exon 3, 5′-GCCAGGAAAACCCCTCCC-3′ (sense) and 5′-AGGAGGAAGAGGCAGAGGAG-3′ (antisense); NEAT1, 5′-GAGAACCAAAGGGAGGGGTG-3′ (sense) and 5′-TGCTGCGTATGCAAGTCTGA-3′ (antisense).
Immunoprecipitation Assay
The immunoprecipitation assaywas performed using the immunoprecipitation protein G Dynabeads® kit (Invitrogen, 10007D) according to the manufacturer's protocol. To digest the RNA or DNA, the cell lysate was preincubated with either 250 μg/ml RNase A (Takara, 2158) or 50 units of DNase I (Takara, 2212) for 10 min.
RNA/DNA FISH and Immunofluorescence Microscopy
To evaluate the influence of SRSF2 on HSV-1 replication, HeLa cells were transfected with SRSF2 siRNA or SRSF2-FLAG plasmid for 36 h. After infection with HSV-1 for 12 h, the cells were fixed with 4% paraformaldehyde for 10 min. After blocking, the cells were incubated with an anti-HSV-1 antibody (Abcam, ab9533). The cells were washed, counterstained with DAPI, and observed with an Olympus FV1000 confocal laser microscope. The fluorescence signal value was measured with Image J software and normalized to a single cell. To validate the interaction between SRSF2 and the ICP0, ICP27, and TK promoter fragments, DNA FISH was performed as previously described in Zhang et al. (37). Briefly, the P4 region of the ICP0 promoter (−380-(−206)), the P2 region of the ICP27 promoter (138–297), and P3 region of the TK gene (-227-(−65)) were amplified and labeled with biotin using the BioNick Labeling System (Invitrogen, 18247-015). HSV-1-infected HeLa cells were incubated with these biotin-labeled promoter regions (10 ng/μl) at 42 °C for 12–16 h. For the co-localization studies, the cells were fixed again and incubated with the anti-SRSF2 antibody (Abcam, ab11826) after DNA-FISH. The cells were washed, counterstained with DAPI, and observed with an Olympus FV1000 confocal laser microscope. The intensity plots for the red and green channels were analyzed with ImageJ software. DNA-FISH and immunofluorescence microscopy experiments in mock-infected nuclei were used as a negative control. To investigate the interaction between SRSF2 and RNAP II, ICP27, or ICP8, HSV-1-infected HeLa cells were fixed and incubated with the anti-SRSF2 antibody (Abcam, ab11826 or Santa Cruz Biotechnology, sc-10252), the anti-RNAP II antibody (Abcam, ab5131), the anti-ICP27 antibody (Santa Cruz Biotechnology, sc-17544), or the anti-ICP8 antibody (Abcam, ab20194). The cells were washed, counterstained with DAPI, and observed with an Olympus FV1000 confocal laser microscope. The intensity plots for the red and green channels were analyzed with ImageJ software. To verify the interaction between SRSF2 and ICP0 exon 3, the ICP0 exon 3 region with or without the potential SRSF2-binding site was transcribed in vitro by T7 RNA polymerase and labeled with DIG) using the DIG Northern Starter Kit (Roche Applied Science, 12039672910). RNA-FISH and immunofluorescence microscopy were performed as previously described in (38). The intensity plots for the red and green channels were analyzed with ImageJ software.
Author Contributions
Z. W. and Y. Z. designed the study and wrote the paper. Q. L., J. L., P. F., W. X., W. Q., F. W. and G. H. finished the experiments. Z. W. and Y. Z. prepared all figures. All authors analyzed the results and approved the final version of the manuscript.
This work was supported by the National Natural Science Foundation of China (31371315), the Ministry of Health, China (2012ZX09102301-019), and Basic Research Fund of Shenzhen (JCYJ20150724173156330). The authors declare that they have no conflicts of interest with the contents of this article.
- RNAP II
- RNA polymerase II
- SRSF2
- serine/arginine-rich splicing factor 2
- SR protein
- serine/arginine-rich protein
- ICP0
- ICP27, and ICP8, infected cell polypeptides 0, 27, and 8, respectively
- TK
- thymidine kinase
- IE genes
- immediate early genes
- CCK8
- cell counting kit-8
- NEAT1
- nuclear paraspeckle assembly transcript 1
- TSS
- transcriptional start site
- qPCR
- quantitative PCR
- m.o.i.
- multiplicity of infection
- DIG
- digoxigenin.
References
- 1.Al-Dujaili L. J., Clerkin P. P., Clement C., McFerrin H. E., Bhattacharjee P. S., Varnell E. D., Kaufman H. E., and Hill J. M. (2011) Ocular herpes simplex virus: how are latency, reactivation, recurrent disease and therapy interrelated? Future Microbiol. 6, 877–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rice S. A., Long M. C., Lam V., Schaffer P. A., and Spencer C. A. (1995) Herpes simplex virus immediate-early protein ICP22 is required for viral modification of host RNA polymerase II and establishment of the normal viral transcription program. J. Virol. 69, 5550–5559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orzalli M. H., DeLuca N. A., and Knipe D. M. (2012) Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc. Natl. Acad. Sci. U.S.A. 109, E3008–E3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu H., and Roizman B. (2003) The degradation of promyelocytic leukemia and Sp100 proteins by herpes simplex virus 1 is mediated by the ubiquitin-conjugating enzyme UbcH5a. Proc. Natl. Acad. Sci. U.S.A. 100, 8963–8968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paladino P., Collins S. E., and Mossman K. L. (2010) Cellular localization of the herpes simplex virus ICP0 protein dictates its ability to block IRF3-mediated innate immune responses. PLoS ONE 5, e10428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalamvoki M., and Roizman B. (2009) ICP0 enables and monitors the function of D cyclins in herpes simplex virus 1 infected cells. Proc. Natl. Acad. Sci. U.S.A. 106, 14576–14580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu H., and Roizman B. (2007) Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc. Natl. Acad. Sci. U.S.A. 104, 17134–17139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Reilly D., Hanscombe O., and O'Hare P. (1997) A single serine residue at position 375 of VP16 is critical for complex assembly with Oct-1 and HCF and is a target of phosphorylation by casein kinase II. EMBO J. 16, 2420–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsia S. C., Graham L. P., Bedadala G. R., Balish M. B., Chen F., and Figliozzi R. W. (2013) Induction of transcription factor early growth response protein 1 during HSV-1 infection promotes viral replication in corneal cells. Br. Microbiol. Res. J. 3, 706–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rowles D. L., Tsai Y. C., Greco T. M., Lin A. E., Li M., Yeh J., and Cristea I. M. (2015) DNA methyltransferase DNMT3A associates with viral proteins and impacts HSV-1 infection. Proteomics 15, 1968–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao R., Sun Y., Ding J. H., Lin S., Rose D. W., Rosenfeld M. G., Fu X. D., and Li X. (2007) Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Mol. Cell. Biol. 27, 5393–5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin S., and Fu X. D. (2007) SR proteins and related factors in alternative splicing. Adv. Exp. Med. Biol. 623, 107–122 [DOI] [PubMed] [Google Scholar]
- 13.Cartegni L., Chew S. L., and Krainer A. R. (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 3, 285–298 [DOI] [PubMed] [Google Scholar]
- 14.Tripathi V., Ellis J. D., Shen Z., Song D. Y., Pan Q., Watt A. T., Freier S. M., Bennett C. F., Sharma A., Bubulya P. A., Blencowe B. J., Prasanth S. G., and Prasanth K. V. (2010) The nuclear retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y., and Steitz J. A. (2005) SRp rises along a messenger's journey. Mol. Cell 17, 613–615 [DOI] [PubMed] [Google Scholar]
- 16.Jacquenet S., Decimo D., Muriaux D., and Darlix J. L. (2005) Dual effect of the SR proteins ASF/SF2, SC35 and 9G8 on HIV-1 RNA splicing and virion production. Retrovirology 2, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erkelenz S., Hillebrand F., Widera M., Theiss S., Fayyaz A., Degrandi D., Pfeffer K., and Schaal H. (2015) Balanced splicing at the Tat-specific HIV-1 3′ ss A3 is critical for HIV-1 replication. Retrovirology 12, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hallay H., Locker N., Ayadi L., Ropers D., Guittet E., and Branlant C. (2006) Biochemical and NMR study on the competition between proteins SC35, SRp40, and heterogeneous nuclear ribonucleoprotein A1 at the HIV-1 Tat exon 2 splicing site. J. Biol. Chem. 281, 37159–37174 [DOI] [PubMed] [Google Scholar]
- 19.Ropers D., Ayadi L., Gattoni R., Jacquenet S., Damier L., Branlant C., and Stévenin J. (2004) Differential effects of the SR proteins 9G8, SC35, ASF/SF2, and SRp40 on the utilization of the A1 to A5 splicing sites of HIV-1 RNA. J. Biol. Chem. 279, 29963–29973 [DOI] [PubMed] [Google Scholar]
- 20.Zahler A. M., Damgaard C. K., Kjems J., and Caputi M. (2004) SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. J. Biol. Chem. 279, 10077–10084 [DOI] [PubMed] [Google Scholar]
- 21.Bøe S. O., Bjørndal B., Røsok B., Szilvay A. M., and Kalland K. H. (1998) Subcellular localization of human immunodeficiency virus type 1 RNAs, Rev, and the splicing factor SC-35. Virology 244, 473–482 [DOI] [PubMed] [Google Scholar]
- 22.McFarlane M., MacDonald A. I., Stevenson A., and Graham S. V. (2015) Human papillomavirus 16 oncoprotein expression is controlled by the cellular splicing factor SRSF2 (SC35). J. Virol. 89, 5276–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandri-Goldin R. M., Hibbard M. K., and Hardwicke M. A. (1995) The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J. Virol. 69, 6063–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji X., Zhou Y., Pandit S., Huang J., Li H., Lin C. Y., Xiao R., Burge C. B., and Fu X. D. (2013) SR proteins collaborate with 7SK and promoter-associated nascent rna to release paused polymerase. Cell 153, 855–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai-Ju J. Q., Li L., Johnson L. A., and Sandri-Goldin R. M. (2006) ICP27 Interacts with the C terminal domain of RNA polymerase II and facilitates its recruitment to herpes simplex Virus 1 transcription sites, where it undergoes proteasomal degradation during infection. J. Virol. 80, 3567–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quinlan M. P., Chen L. B., and Knipe D. M. (1984) The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell. 36, 857–868 [DOI] [PubMed] [Google Scholar]
- 27.Burge C. B., Tuschl T., and Sharp P. A. (1999) Splicing of Precursors to mRNAs by the Spliceosomes. In The RNA World (Gesteland R. F., Cech T. R., and Atkins J. F., eds) pp. 525–560, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 28.Everett R. D. (2000) ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22, 761–770 [DOI] [PubMed] [Google Scholar]
- 29.Lee H. R., Kim D. J., Lee J. M., Choi C. Y., Ahn B. Y., Hayward G. S., and Ahn J. H. (2004) Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J. Virol. 78, 6527–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinnoji R. C., Bedadala G. R., George B., Holland T. C., Hill J. M., and Hsia S. C. (2007) Repressor element-1 silencing transcription factor/neuronal restrictive silencer factor (REST/NRSF) can regulate HSV-1 immediate-early transcription via histone modification. Virol J. 4, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin R., Noyce R. S., Collins S. E., Everett R. D., and Mossman K. L. (2004) The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon stimulated genes. J. Virol. 78, 1675–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sedlackova L., and Rice S. A. (2008) Herpes simplex virus type 1 immediate-early protein ICP27 is required for efficient incorporation of ICP0 and ICP4 into virions. J. Virol. 82, 268–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Workenhe S. T., Ketela T., Moffat J., Cuddington B. P., and Mossman K. L. (2016) Genome wide lentiviral shRNA screen identifies serine/arginine-rich splicing factor 2 as a determinant of oncolytic virus activity in breast cancer cells. Oncogene 35, 2465–2474 [DOI] [PubMed] [Google Scholar]
- 34.Aslani A., Olsson M., and Elias P. (2002) ATP-dependent unwinding of a minimal origin of DNA replication by the origin binding protein and the single strand DNA-binding protein ICP8 from herpes simplex virus type1. J. Biol. Chem. 277, 41204–41212 [DOI] [PubMed] [Google Scholar]
- 35.Dahl J. A., and Collas P. (2008) A rapid micro chromatin immunoprecipitation assay (microChIP). Nat. Protoc. 3, 1032–1045 [DOI] [PubMed] [Google Scholar]
- 36.Peritz T., Zeng F., Kannanayakal T. J., Kilk K., Eiríksdóttir E., Langel U., and Eberwine J. (2006) Immunoprecipitation of mRNA-protein complexes. Nat. Protoc. 1, 577–580 [DOI] [PubMed] [Google Scholar]
- 37.Zhang L. F., Huynh K. D., and Lee J. T. (2007) Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell 129, 693–706 [DOI] [PubMed] [Google Scholar]
- 38.Wang P., Xue Y., Han Y., Lin L., Wu C., Xu S., Jiang Z., Xu J., Liu Q., and Cao X. (2014) The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 344, 310–313 [DOI] [PubMed] [Google Scholar]