Abstract
Protein acetylation is a prevalent posttranslational modification that is regulated by diverse acetyltransferase enzymes. Although histone acetyltransferases (HATs) have been well characterized both structurally and mechanistically, far less is known about non-histone acetyltransferase enzymes. The human ESCO1 and ESCO2 paralogs acetylate the cohesin complex subunit SMC3 to regulate the separation of sister chromatids during mitosis and meiosis. Missense mutations within the acetyltransferase domain of these proteins correlate with diseases, including endometrial cancers and Roberts syndrome. Despite their biological importance, the mechanisms underlying acetylation by the ESCO proteins are not understood. Here, we report the X-ray crystal structure of the highly conserved zinc finger-acetyltransferase moiety of ESCO1 with accompanying structure-based mutagenesis and biochemical characterization. We find that the ESCO1 acetyltransferase core is structurally homologous to the Gcn5 HAT, but contains unique additional features including a zinc finger and an ∼40-residue loop region that appear to play roles in protein stability and SMC3 substrate binding. We identify key residues that play roles in substrate binding and catalysis, and rationalize the functional consequences of disease-associated mutations. Together, these studies reveal the molecular basis for SMC3 acetylation by ESCO1 and have broader implications for understanding the structure/function of non-histone acetyltransferases.
Keywords: acetyltransferase, cell division, chromatin regulation, crystal structure, enzyme structure, gene expression, protein acylation, protein structure, structure-function, X-ray crystallography, cohesin, ESCO1, SMC3, posttranslational modification (PTM)
Introduction
Thousands of proteins are posttranslationally acetylated on lysine residues to mediate diverse biological processes (1). The most well characterized acetylation substrates are histones, which package DNA within the nucleus and help to regulate gene accessibility (2, 3). Extensive characterization of histone acetylation has highlighted the importance of this modification in epigenetic processes, which is catalyzed by histone acetyltransferases (HATs)2 (2, 3). These HATs are categorized into subfamilies (HAT1, Gcn5/PCAF, MYST, p300/CBP, and Rtt109) based on sequence and substrate acetylation properties, all of which share a similar acetyl coenzyme A (Ac-CoA) binding core structure (4–6). More recently, numerous studies have been reported that emphasize the importance of non-histone protein acetylation in a diverse set of other cellular pathways (1), including sister chromatid cohesion (7). During this process, the multi-protein complex cohesin forms a ring around sister chromatid pairs to ensure proper chromosome segregation during mitosis and meiosis (8). In order for cohesin to form and maintain this three-dimensional “ring” structure, multiple acetylation events must occur on the cohesin subunit structural maintenance of chromosomes 3 (SMC3) (8–10). Although the functional consequences of SMC3 acetylation have been extensively investigated, the acetyltransferases that install this modification have not been characterized at the molecular level.
Although several non-histone proteins, such as p53 and FOXO, are also acetylation substrates of HATs, eukaryotes have also evolved a diverse set of non-histone acetyltransferases to help ensure proper posttranslational acetylation throughout the proteome (1, 11–16). In the case of SMC3, acetylation is performed by the establishment of sister chromatid cohesion acetyltransferases (ESCO1 and ESCO2 in human and Eco1 in yeast). The predicted catalytic cores of these enzymes share sequence homology with the Gcn5-related N-acetyltransferase (GNAT) family of proteins (17), but have a unique N-terminal zinc finger that is conserved among all ESCO homologs. Interestingly, other non-histone acetyltransferases, such as α-tubulin acetyltransferase (18–20), also exhibit a GNAT core fold with distinct flanking regions proposed to participate in tubulin-specific substrate activity (19). These findings suggest that ESCO proteins may utilize unique structural features to impart cohesion-specific substrate specificity (7, 21).
In humans and other vertebrates, ESCO1 and ESCO2 both acetylate the head domain of the SMC3 cohesin subunit at Lys-105 and Lys-106, although they appear to have non-redundant activities (22, 23). This is evidenced by the observation that loss of either ESCO protein cannot be rescued by the other variant (22, 23). Notably, mutations in or overexpression of ESCO1 have been linked to endometrial and bladder cancer (24, 25), respectively, and mutations in ESCO2 have been linked to Roberts syndrome, a childhood autosomal recessive disorder that causes mental and physical abnormalities (4, 23, 26, 27). To elucidate the mechanistic details for acetylation by ESCO proteins, we report here the X-ray crystal structure of ESCO1. Together with biochemical and enzymatic studies, this structure reveals unique ESCO1 features that confer SMC3-specific acetylation and generates insight into the pathogenic nature of biologically relevant somatic mutations.
Results
Overall Structure of the ESCO1·Ac-CoA Complex
To obtain a soluble ESCO/Eco1 construct suitable for crystallization trials, we performed recombinant expression trials in Escherichia coli using yeast and human homologs of the enzyme and a variety of constructs thereof. Although the majority of these proteins were poorly expressed, we found that a human ESCO1 construct consisting of the predicted GNAT acetyltransferase domain as well as the invariant N-terminal C2H2 zinc finger was soluble and monomeric (as judged by gel filtration chromatography), and therefore a good candidate for crystallization (Fig. 1, A and B). We were able to obtain crystals of this construct (human ESCO1 residues 599–825) bound to cofactor acetyl-CoA (Ac-CoA) that formed in space group I41 and diffracted to a resolution limit of 2.70 Å. Initial molecular replacement trials were performed using other GNAT proteins as model solutions, but were unable to provide adequate structure solutions. We collected a highly redundant dataset at the zinc peak to take advantage of the natively bound zinc ion and were able to obtain experimental phases using the single wavelength anomalous diffraction method. The structure was subsequently refined to an Rwork of 0.248 and an Rfree of 0.291 and shows one ESCO1 molecule per asymmetric unit (Table 1).
FIGURE 1.
Overall structure of ESCO1. A, schematic diagram of ESCO1 domain architecture. B, sequence alignment with ESCO1 orthologs with GCN5 for reference. Strictly conserved residues are highlighted in black, and highly conserved residues are boxed. C, overall structure, colored by domain, with zinc finger shown in orange, β4-β7 loop insert shown in yellow, and GNAT fold shown in green (as depicted in A) with Ac-CoA shown in sticks and the zinc ion shown as a gray sphere. D, Ac-CoA binding region, with interacting secondary structures labeled. Fo − Fc simulated annealing omit map is shown around the cofactor, contoured at 3.0 σ. E, superimposition of ESCO1 and GCN5 (wheat) bound to histone H3 substrate peptide.
TABLE 1.
Crystallographic statistics for ESCO1
Values in parentheses are for the highest resolution shell. Ramachandran values were calculated by RAMPAGE (46). PDB, Protein Data Bank.
| ESCO1·Ac-CoA crystal | Zn2+ phasing crystal | |
|---|---|---|
| Data collection | ||
| Beamline | BNL X25 | BNL X25 |
| Wavelength, Å | 1.281 | 1.281 |
| Crystal parameters | ||
| Space group | I41 | I41 |
| Cell lengths, Å | 115.6, 115.6, 61.1 | 115.6, 115.6, 61.1 |
| Resolution, Å | 40.85–2.70 (2.75–2.70) | 50.00–2.76 (2.87–2.76) |
| Unique reflections | 11,046 (485) | 10,976 (987) |
| Total observations | 334,558 (10,185) | 329,280 (20,924) |
| Completeness,a % | 98.2 (86.8) | 98.2 (88.7) |
| Redundancy | 30.3 (21.0) | 30.0 (21.2) |
| Rsym | 0.061 (0.624) | 0.062 (0.599) |
| 〈I/σI〉 | 73.3 (2.4) | 71.7 (2.7) |
| Refinement | ||
| Rwork/Rfree | 0.250/0.285 | |
| No. of atoms: total | ||
| Protein | 1370 | |
| Ligand | 51 | |
| Water | 14 | |
| Ion | 1 | |
| Average B-factors, Å2 | 95.0 | |
| Protein | 95.0 | |
| Ligand | 97.0 | |
| Water | 94.0 | |
| Ion | 102.6 | |
| Ramachandran statistics, % | ||
| Favored | 96.6 | |
| Allowed | 3.4 | |
| Outlier | 0.0 | |
| RMS deviations | ||
| Bonds, Å | 0.007 | |
| Angles, ° | 0.996 | |
| MolProbity | ||
| Clash scoreb | 17.34 (87%) | |
| Overall scoreb | 2.05 (97%) | |
| PDB: 5T53 | ||
The ESCO1 structure reveals a catalytic core containing the characteristic α/β motif (β3, β4, β7, α3, and α4) that is common among GNAT proteins (classically designated as motifs A, B, and D) such as Gcn5 (Fig. 1B). An additional conserved feature is the cofactor binding cleft where Ac-CoA is wedged in a groove comprising the β-strands (β7 and β8) and α-helices (α3 and α4) of the GNAT core (Fig. 1, C and D). The N-terminal zinc finger consists of a hairpin loop followed by an α-helix (α1), which fold around the zinc ion, which is ligated by amino acid ligands Cys-619, Cys-622, His-637, and His-641 (Figs. 1B and 2A). Although the zinc finger is tethered to the GNAT region through a loop and β-strand (β1), this region also forms an extensive hydrophobic interface with the α2 helix and β7 strand of the GNAT core. In particular, Phe-640 and Phe-644 of the α1 helix make van der Waals contacts to Met-688, Trp-773, and Phe-775 of the GNAT core, and Gln-643 of the α1 helix makes a hydrogen bond to Asn-691 of the GNAT core (Fig. 2A) (28). Another striking structural feature unique to ESCO1 is an extended “loop insert” (residues 727–766) between the β4 and β7 strands that extends away from the core and contains two short antiparallel β-strands (β5 and β6) that form a hairpin (Fig. 1C). Notably, the segment of the loop connecting these strands (residues 740–753) could not be traced in the electron density map and is presumably disordered.
FIGURE 2.
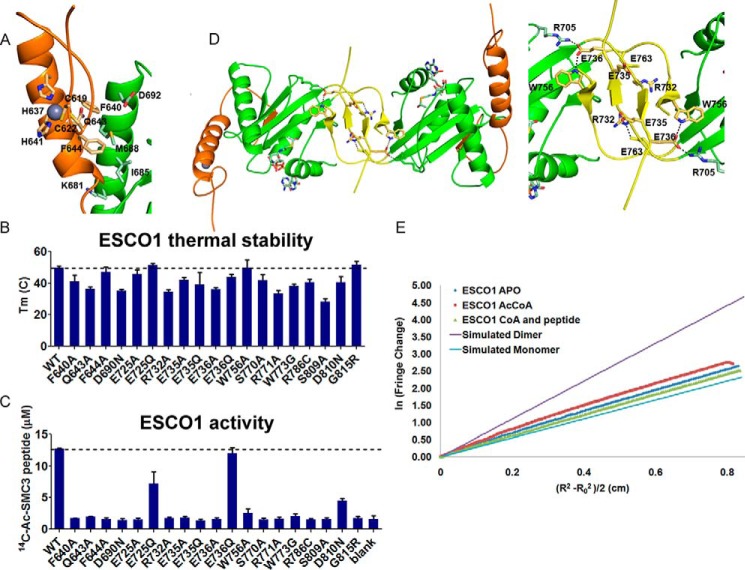
Functional analysis of ESCO1. A, C2H2 zinc finger (orange) with residues involved in zinc ligation or core domain interaction highlighted in wheat. The GNAT α2 core (dark green), C2H2 zinc finger contact residues (light green), and zinc ion (gray) are highlighted. B, results from thermal stability assays of ESCO1 WT and mutants. C, results from enzymatic activity assays of ESCO1 WT and mutants. Error bars are the standard deviation of triplicate assays. The dotted line represents values for ESCO1 WT. Blank represents reaction in the absence of enzyme. D, ESCO1 crystallographic dimer with interface residues shown as sticks in the same color scheme as in Fig. 1. E, analytical ultracentrifugation analysis of ESCO1 with and without Ac-CoA, which is most consistent with the monomer simulation (blue).
A structural alignment of ESCO1·Ac-CoA with the Gcn5·Ac-CoA·histone H3 peptide ternary complex structure reveals that although the core folds superimpose well (Cα RMSD = 3.60 Å), the structural features of Gcn5 that mediate interaction with the H3 substrate peptide (Gcn5 α1 helix and α5-β5 loop) are not conserved in ESCO1 (Fig. 1E). Notably, the aforementioned β5-β6 hairpin from the β4-β7 loop insert of ESCO1 appears to replace one of the substrate peptide binding loops of Gcn5. Variability within this peptide binding cleft is common among GNAT proteins and has previously been shown to be a common mechanism for achieving substrate specificity (29, 30). It is therefore likely that these regions are responsible for cohesin substrate recognition by ESCO1 (Fig. 1E).
ESCO-specific Structural Elements Contribute to Protein Stability and Activity
The packing of the ESCO1 zinc finger against the GNAT core suggests that it may play a role in the stability of the acetyltransferase domain. To test the functional importance of this interaction, three mutants were prepared (F640A, Q643A, and F644A) that we predicted would disrupt the interface based on structural analysis. For these and subsequent functional studies, we employed a larger ESCO1 construct (residues 590–840) than used for crystal structure determination (residues 599–825), which we found to be more stable and active than the construct used for crystallization and X-ray structure determination. We first tested the stability of these proteins using a thermal denaturation assay, which showed that the melting temperature of the wild-type protein (50 °C) was indeed reduced by each these mutations (41, 47, and 36 °C, respectively) (Fig. 2B). Next, we sought to determine the effect of these mutations on catalysis by using a radioactive acetyltransferase assay that monitors transfer of a 14C-labeled acetyl group from Ac-CoA to an SMC3 peptide (residues 97–117). The results from our enzymatic analysis demonstrated that these mutants have severely compromised activity, with F640A, Q643A, and F644A all showing an ∼6-fold decrease in activity at 1 μm protein concentration (Fig. 2C). Together, these findings demonstrate that the zinc finger plays a direct role in ESCO acetyltransferase domain stabilization and an indirect role in positioning of the accompanying active site residues.
We next questioned whether the ESCO-specific hairpin within the β4-β7 loop insert region plays a role in protein function. Interestingly, although this loop is of variable length among ESCO1 homologs, there are several residues that appear to be moderately conserved and therefore may be important for substrate binding and/or catalysis (Fig. 1B). To test this possibility, we implemented the thermal denaturation and catalytic assay on a set of loop mutants to assess their effect on protein stability and activity. We identified three conserved residues in this region (Arg-732, Glu-735, and Glu-736) (Fig. 1B) that are required for optimal protein stability and activity (Fig. 2, B and C). Because this loop is located near the predicted peptide binding cleft, we propose that the mutations cause deficiencies in SMC3 substrate recognition. In either case, ESCO1 provides yet another example of how GNATs exploit distinct structural domains to regulate substrate-specific catalysis.
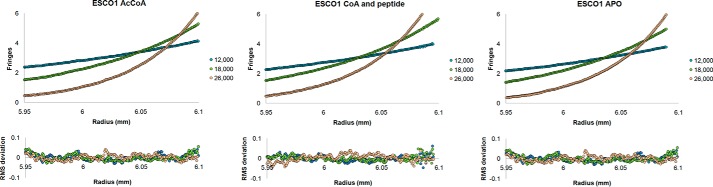
We were also prompted to investigate the oligomeric state of ESCO1 by the observation of a symmetrical dimer in the crystal lattice (Fig. 2D). Specifically, the β5-β6 hairpins from the β4-β7 loop inserts wrap around one another and form an antiparallel β-sheet. This interaction also appears to be supplemented by contacts between residues within these loops (Glu-736 and Trp-756) and the putative cohesion substrate binding site of the adjacent subunit (Fig. 2D). Further analysis of the crystal lattice using the PDBePISA server is also consistent with a dimer, showing that the most stable assembly is a dimer with a buried surface area of 5,300 Å2 and a ΔG of dissociation of 23.9 kcal/mol. To test the potential biological relevance of the crystallographic dimer, we subjected ESCO1 to sedimentation equilibrium analysis in the absence or presence of Ac-CoA. Both of these samples showed a single sedimenting species with calculated molecular masses of 36,700 (with Ac-CoA) and 33,692 (without Ac-CoA), as compared with the theoretical molecular weight of 31,753 (28,950 protein + 2,083 tobacco etch virus (TEV) sequence and His6 tag). We also analyzed the WT enzyme with CoA and the SMC3 peptide and found its sedimentation to also be consistent with a single species corresponding to a size close to that of a monomer 38,401 (theoretical molecular weight protein, CoA and peptide = 34,858) (Figs. 2E and 3). These results are consistent with the initial gel filtration analysis (data not shown), which together suggest that ESCO1 exists predominantly as a monomer in solution.
FIGURE 3.
Sedimentation equilibrium analytical ultracentrifugation of ESCO1. ESCO1 (residues 590–840) was analyzed at 4 °C using interference optics in the Apo, Ac-CoA, and CoA (1 mm) + SMC3 peptide (1 mm) states at three different centrifugation speeds (12,000, 18,000, and 26,000 rpm) and three different protein concentrations (30, 58, and 118 μm). The data for each speed were collected in quadruplicate. The viscosity of the samples was estimated using Sednterp (43, 44), and the most representative runs were included to calculate theoretical molecular masses using the program HeteroAnalysis. The data are summarized in Fig. 2E.
SMC3-specific Acetylation Is Governed by a Unique Active Site Architecture
Because GNAT proteins have been shown to exploit a variety of residue identities and locations to achieve lysine deprotonation, it is difficult to determine which amino acids in ESCO1 are important for catalysis from protein sequence alignments alone (29, 30). Therefore, we used structure-guided analysis of the active site to identify several residues that are positioned to potentially play catalytic roles. We specifically focused on Asp-690, Glu-725, Ser-770, Arg-771, Ser-809, and Asp-810, all of which have the potential to be involved in the proton transfer step of the acetylation mechanism (Fig. 4C) based on their location relative to Ac-CoA. To probe the catalytic contributions made by each of these residues, we prepared a series of point mutants and analyzed their effect on protein stability and catalytic activity.
FIGURE 4.
ESCO1 kinetic assays and substrate binding pocket. A and B, Michaelis-Menten analysis of WT ESCO1 against substrate peptide derived from SMC3 and Ac-CoA co-factor. C, close-up view of the ESCO1 active site highlighting residues (light green) that could play catalytic roles, with Ac-CoA (yellow CPK). D, pH activity profile of ESCO1 WT. E, pH activity profile of mutants R771A (cyan) and D810N (dark blue). Error bars represent standard deviation of triplicate assays.
Although protein stability varied depending on the mutation site, each of the mutants appeared to be properly folded based on their gel filtration elution profiles. Although the steady state catalytic parameters could be determined for the wild-type enzyme (kcat = 0.34 ± 0. 03 min−1, Km, Ac-CoA = 77 ± 14 μm, Km, SMC3 peptide = 53 ± 8 μm), we could not calculate these values for the mutants due to assay detection limits. Therefore, the activity levels were represented as the concentration of product present at the end of each reaction. Surprisingly, all mutations showed low levels of activity (2–9-fold decreases relative to WT), including E725Q, which had no measurable effect on protein stability (Fig. 2, B and C). We next sought to tease out the function of these residues in catalysis, which could be related to substrate binding, lysine deprotonation, or both. Two residues that are particularly well positioned to function as general base residues are Glu-725 and Asp-810. To elucidate the role of these amino acids in catalysis, we calculated the activity of corresponding point mutants at different pH values ranging from 5.5 to 11.0 (Fig. 4, D and E). These assays show that although the E725Q mutant is inactive at all pH values tested, activity for the D810N mutant can be rescued (albeit to a lower degree than WT) as pH values that approach 10, where lysine deprotonation can occur spontaneously. This is strong evidence that Asp-810 contributes to general base catalysis. The possibility that other residues such as Asp-690, Ser-770, Ser-809, and particularly Glu-725, also contribute to lysine deprotonation cannot be ruled out. We also note that Arg-771 is in a position to act as a general acid in the catalytic mechanism. Additionally, these residues could be involved in substrate peptide recognition.
To gain further insight into substrate peptide binding, we analyzed a superposition of the ESCO1·Ac-CoA complex with the ternary Gcn5·CoA·histone H3 peptide complex. We then used this alignment to generate a model for how a substrate peptide might engage ESCO1 (Fig. 5, A and B). When docked onto a surface representation of ESCO1, the substrate lysine (H3K14) extends into a narrow tunnel leading into the active site where the ϵ-amino group is presented to the reactive moiety of Ac-CoA. Additionally, the backbone of the peptide traces down a groove in the enzyme that we predict to be the SMC3 binding cleft. Although there are clear clashes between ESCO1 and the H3 peptide backbone, conformational flexibility in both ESCO1 and the SMC3 peptide would allow the enzyme to accommodate its cognate substrate in a fashion analogous to Gcn5. Moreover, the model suggests that regions of the peptide that are distal to the acetylated lysine residues would come into contact with the β5-β6 loop insert region of ESCO1, consistent with our mutagenesis results. We therefore propose that SMC3 specificity is achieved by an intimate protein-peptide interaction interface that includes the globular domain of the enzyme as well as a more unstructured β4-β7 loop insert region.
FIGURE 5.
Peptide binding and ESCO1-ESCO2 disease-causing mutations. A, surface representation of ESCO1 structure color-coded as in A. B, H3 peptide superimposed onto the structure of ESCO1 using coordinates of the GCN5·CoA H3 peptide structure (PDB ID 1QSN). C, ESCO1 and ESCO2 residues that are mutated in disease are highlighted in light green CPK coloring, and nearby interacting residues are shown.
Implications for Pathogenic ESCO Mutations
The ESCO catalytic domain featured in this study is the site of a number of missense mutations that are linked to diseases such as Roberts syndrome and endometrial cancer. Using a structure-guided approach, we aimed to gain a better understanding of the functional consequences of these mutations. One example of this is the R786C mutation that is found in certain endometrial cancers. This residue lies in the α3 helix of ESCO1 and forms a hydrogen bond to the backbone carbonyl of Lys-781 within the same helix, as well as van der Waals contacts with Phe-711 in the β5 strand and Tyr-662 of the zinc finger (Fig. 5C). This extensive network of interactions within the GNAT core would not be sustained by a mutation to cysteine at this position, which is corroborated by our observation that the R786C mutation shows reduced thermal stability (by 10°) and acetyltransferase activity (by ∼6-fold). Although the ESCO mutations that are classically observed in Roberts syndrome typically occur in ESCO2, the mutation sites are strictly conserved between these proteins. We generated these mutants recombinantly, W773G (W539G in ESCO2) and G815R (G581R in ESCO2), to probe their role in ESCO activity. The structure shows that Trp-773 is a part of the aforementioned hydrophobic network that mediates the interaction between the GNAT core of ESCO1 and the zinc finger (Fig. 5C). This residue is also in a position to make additional hydrophobic contacts with residues Ile-685, Val-689, Leu-693, Val-718, and Cys-781 in the α2 helix, making it a linchpin in the overall fold of the enzyme. Not surprisingly, the W773G mutant shows a large decrease in thermal stability (∼11°) and enzymatic activity (∼7-fold). The other residue that is sensitive to mutation in Roberts syndrome (Gly-815) is located in the α4 helix near a hydrophobic pocket formed by Phe-808, Phe-818, and Pro-881. This residue is also in close proximity (∼5 Å) to the sulfur atom of Ac-CoA. The G815R mutation could potentially disrupt this hydrophobic pocket as well as interfere with Ac-CoA binding through steric clashes. Although we found that this mutant had no effect on protein stability, we did observe an ∼3-fold decrease in activity. This comprehensive analysis of disease-associated ESCO mutants shows that these missense mutations occur throughout the catalytic core at positions that are important for structural stability and enzyme catalysis.
Discussion
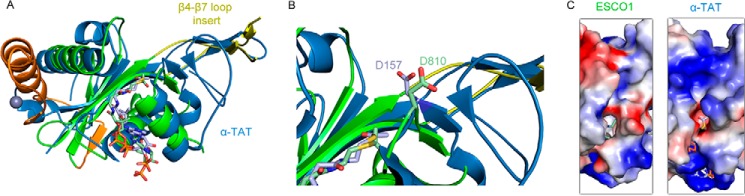
To date, structural and biochemical characterization of lysine side-chain acetylation has been carried out predominantly in the context of HATs and their cognate substrate peptides. These studies have revealed a conserved Ac-CoA binding core with divergent N- and C- terminal flanking regions that have the potential to mediate substrate binding and catalysis. However, the substrate peptide scope of these studies typically does not extend beyond polycationic histone tails. The ESCO1 catalytic domain reported here represents only the second structure of a eukaryotic enzyme that is dedicated to the acetylation of non-histone lysine substrates, the first structure being that of the α-tubulin acetyltransferase, αTAT1 (19, 20). Notably, ESCO1, αTAT1, and Gcn5 all fall into the GNAT family of proteins and, accordingly, follow the same core architectural rules. However, the crystal structure of ESCO1, together with that of αTAT1, shows that non-histone acetyltransferase members of this family diverge from Gcn5 and other HATs in a variety of ways, including variable N- and C-terminal flanking regions, large loop insertions, and substrate peptide binding pocket geometry (Fig. 1E). A structural alignment between ESCO1 and αTAT1 shows that the β4-β7 loop insert region described here is conserved in αTAT, where it has been proposed to result in a wider substrate binding groove than Gcn5 and other HATs (19) (Fig. 6). Our study suggests that this could be a common feature that is exploited by non-histone acetyltransferases to prevent acetylation of histone tails.
FIGURE 6.
Superimposition of ESCO1 and αTAT1. A, overall structural superimposition with αTAT1 shown in blue. B, close-up view of the active sites of ESCO1 and αTAT1 highlighting the putative catalytic general base residue for activity. C, ESCO1 and αTAT1 electrostatic surface potential (calculated in PyMOL (45)) plotted from −65 kiloteslas (red) to +65 kiloteslas (blue) shows the large areas of positive charge on αTAT1 suitable for interacting with negatively charged substrates as compared with ESCO1.
The catalytic mechanism of lysine acetyltransferases typically involves one or more general base residues to initiate deprotonation of the substrate ϵ-amino group, which facilitates the acetyl transfer reaction (29, 30). Our activity assays are consistent with the conclusion that Asp-810 participates as a general base in the ESCO1 catalytic mechanism. Specifically, we found that an isosteric mutation to a residue that cannot serve as a general base at this position (D810N) severely hinders catalytic activity, and that acetylation by this mutant can be partially rescued at pH values where the substrate lysine would naturally exist in the deprotonated state (pH ∼10). Furthermore, in a three-dimensional alignment with αTAT1, this residue aligns very closely to Asp-157, which plays a general base role in tubulin acetylation. Interestingly, the Asp-810 residue in ESCO1 is not strictly conserved among ESCO homologs (Fig. 1B), which suggests that other residues may also have the potential to contribute to substrate lysine deprotonation (19, 20).
Notably, much like αTAT1, the in vitro kcat that we determined for ESCO1 (0.34 ± 0.03 min−1) is on the order of 100-fold slower than that of typical HAT enzymes. We suspect that this does not reflect the rate of this enzyme in vivo, where several extrinsic factors may contribute to catalysis such as other domains of the ESCO1 protein or the intact SMC3 substrate. There are also studies showing that the ESCO proteins can associate with chromatin during cohesin acetylation, which may add another layer of activity regulation (31). Additionally, we note that acetyltransferases typically exist in the context of larger protein complexes in vivo to achieve optimal catalytic activity. This phenomenon is exemplified by several HATs (Sas2, Hat1, and HBO1) and N-terminal acetyltransferases (NatA, NatB, and NatC), which exhibit great enhancement in kcat in the presence of binding partners in vitro and in vivo (4–6, 19, 20, 32–37). We therefore hypothesize that the N-terminal region of ESCO1, as well as the β4-β7 loop insert, may be crucial docking sites for allosteric modulators of acetyltransferase activity. Indeed, recent studies have shown that other proteins (Pds5A and Pds5B) are required for SMC3 acetylation in cells (22, 38).
In the course of preparing this manuscript, another structure, along with limited biochemical characterization of ESCO1 (residues 654–836), was reported by Kouznetsova et al. (39). The ESCO1·Ac-CoA structure reported by Kouznetsova et al. does not contain the zinc finger but contains an extended C-terminal region of ∼10 residues that form a β-strand between β8 and the β4-β7 loop insert region. The β5 and β6 strands are also more extended in the Kouznetsova et al. structure than in the one reported here. Despite these differences, the two structures superimpose well with an RMSD of 0.8 Å2 for all atoms in common. Beyond the similarity of the overall structures, the Kouznetsova et al. study draws two significantly different conclusions then the studies reported here regarding the dimerization state and catalytic mechanism of ESCO1. The Kouznetsova et al. structure contains a cysteine cross-linked ESCO1 dimer in the asymmetric unit cell, and the authors also observe two other ESCO1 dimer configurations in the crystal lattice. Interestingly, analysis of the Kouznetsova et al. crystal lattice using the PDBePISA server indicates that the most stable assembly is a tetramer with a buried surface area of 13,790 Å2 and a ΔG of dissociation of 27.0 kcal/mol. Each of the dimers of the Kouznetsova et al. crystal lattice is mediated by the β4-β7 loop insert regions. Kouznetsova et al. present small angle X-ray scattering data, which they claim is most consistent with the shape of one of the three observed crystallographic dimers, leading the authors to conclude that this dimer is biologically relevant. This conclusion is inconsistent with the analytical ultracentrifugation data, presented here (Figs. 2E and 3), demonstrating that ESCO1 (residues 590–840) is predominantly monomeric, independent of whether Ac-CoA or peptide substrate is bound. The fact that the three Kouznetsova et al. ESCO1 dimers and the crystallographic dimer that we observe are all different suggests that ESCO1 may indeed form dimers under certain circumstances, for example when engaging Smc3 head domain homodimers of cohesion for acetylation, although the nature of that dimer may be influenced by cognate substrate and/or protein cofactor binding (such as Pds5A and/or Pds5B). We also note that the ESCO1 fragments analyzed by Kouznetsova et al. and our study only represent about one-fourth of the intact protein, and it is also possible that other regions of ESCO1 may also influence its oligomerization state.
Kouznetsova et al. (39) also conclude that ESCO1 employs a catalytic mechanism of substrate-assisted catalysis, whereby glutamate residues located at variable distance from autoacetylated ESCO1 lysines and Asp-114 located adjacent to Lys-112 and Lys-113 of yeast Smc3 substrate act as general base residues for catalysis. Such a mechanism has not been observed for any other acetyltransferase, and these findings are also in disagreement with the finding reported here (Fig. 4E) that Asp-810 (and potentially other ESCO1 residues) plays an important role in general base catalysis by ESCO1. We propose that the decrease in lysine acetylation upon alanine substitution of adjacent Glu/Asp residues observed by Kouznetsova et al. disrupts substrate binding as opposed to the catalytic rate of the reaction.
Taken together, the unique ESCO enzyme scaffold and accompanying biochemical characterization described here will be useful in understanding the role of ESCO1 in cohesion establishment, the development of small molecule probes for ESCO1, and the identification of auxiliary components that are important for ESCO protein function.
Experimental Procedures
ESCO1 Expression and Purification
Different N- and C-terminal truncation constructs of ESCO1 were amplified from human cDNA (generous gift from Paul Lieberman) and subcloned into a modified pETDUET vector with an N-terminal TEV protease-cleavable hexahistidine tag. Each construct was then transformed into Rosetta (DE3) pLysS competent E. coli cells. The cells were cultured at 37 °C until they reached an A600 of 0.6–0.8 and induced for protein expression with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. The induced cells were cultured at 18 °C after induction for ∼16 h. The cells were harvested and lysed by sonication in affinity loading buffer containing 25 mm Tris (pH 8), 500 mm NaCl, 5 mm β-mercaptoethanol (β-mercaptoethanol), and 10 μg/ml phenylmethylsulfonyl fluoride. The resulting lysate was clarified by centrifugation and loaded over nickel resin (Thermo Scientific). The resin was washed with >20 resin volumes of wash buffer (loading buffer supplemented with 25 mm imidazole). ESCO1 was eluted using the wash buffer supplemented with 400 mm imidazole. The eluted protein was subjected to TEV proteolysis during dialysis into 25 mm Tris, pH 8.5, 200 mm NaCl, and 10 mm β-mercaptoethanol for 16 h at a final TEV concentration of 6 μg/ml. This solution was passed through Ni-NTA resin to remove the His6-tagged TEV protease as well as any uncut ESCO1. The resin was then washed with approximately 7 column volumes of dialysis buffer supplemented with 25 mm imidazole, which was pooled with the initial flow-through. This solution was dialyzed into buffer for size exclusion chromatography containing 25 mm HEPES, pH 7.0, 200 mm NaCl, and 1 mm tris(2-carboxyethyl)phosphine (TCEP) for ∼16 h. This was concentrated to a volume of 500 μl (10-kDa concentrator; Amicon Ultra, Millipore), and chromatographed on a Superdex 75 prep gel filtration column (GE Healthcare). Peak fractions were concentrated to 10 mg/ml as measured by UV280 for crystallization trials. Single point ESCO1 mutants were generated using the Stratagene QuikChange protocol and purified using the same strategy described above.
ESCO1 Crystallization and Structure Determination
ESCO1 (residues 599–825) at 10 mg/ml was co-crystallized with 1 mm Ac-CoA using hanging drop vapor diffusion in a drop containing a 1:1 v/v mixture of protein to crystallization solution (8% PEG 8000, 0.1 m HEPES, pH 7.5, and 10% ethylene glycol) at 25 °C. Crystals were transferred to a mother liquor supplemented with glycerol in 5% increments (5, 10, and 15%) for 1 min at each concentration and flash-cooled in liquid nitrogen.
Data were collected at beamline X25 at the National Synchrotron Light Source (Brookhaven National Laboratory) and processed using HKL3000 (40). A single crystal was used for two datasets (one high-resolution and one zinc phasing crystal). The crystals survived very well under the beam, and we were able to collect 1,000 frames (in a single wedge) at 0.9° oscillation, with no apparent deterioration in linear Rfactor or diffraction intensity per frame. This coupled with the high symmetry resulted in high redundancy. Initially, we attempted to determine the structure using a library of known acetyltransferase crystal structures as molecular replacement models. This strategy was unable to yield a solution sufficient for model building, and so we next collected a high redundancy dataset at the zinc peak wavelength to exploit the inherent zinc ion for experimental phasing. The Hybrid Substructure Search in Phenix was used to identify a single zinc site in the asymmetric unit. This site was used to generate initial electron density maps in Phaser, which were improved by density modification as implemented in Resolve (performed sequentially in Phenix AutoSol). The resulting maps were of high quality, and manual model building was carried out in Coot. Once the initial model was complete, it was used as a molecular replacement solution for a high-resolution dataset that was subsequently refined using iterative rounds of Phenix (41). Crystallographic refinement and manual model building were carried out in Coot (42). Residues 695–700 were not observed in the electron density and therefore not modeled. Residues 740–754 were initially modeled, but the electron density was too ambiguous to confidently fit the entire loop, so it was omitted from the final structure. Finally, a simulated annealing composite omit map was generated to check for any errors in the final model.
Acetyltransferase Assay
ESCO1 acetyltransferase assays for wild-type and mutant ESCO1 enzymes (residues 590–840) were carried out with 1 μm enzyme at 25 °C for 4 h in 25 mm Tris, pH 8.0, and 50 mm NaCl. For kcat and Km determination for peptide, near saturating concentrations of Ac-CoA (1 mm) were used in all enzymatic reactions, and the substrate peptide (NH2-SLRRVIGAKKDQYFLDKKMVT-COOH; GenScript) concentration was varied from 10 to 1,000 μm. For kcat and Km determination for Ac-CoA, near saturating concentration of peptide (1 mm) was used in all reactions, and the substrate Ac-CoA concentration was varied from 10 to 200 μm. The residues of the peptide used correspond to the SMC3 sequence around Lys-105 and Lys-106. Because this peptide had an overall positive charge, this substrate was compatible with our filter binding assays that involve binding of the radiolabeled peptide to a negatively charged nitrocellulose filter prior to scintillation counting. Specifically, in the assay, radiolabeled [14C]acetyl CoA (4 mCi mmol−1; PerkinElmer Life Sciences) was mixed with the substrate peptide and allowed to incubate with enzyme in a 50-μl reaction volume at 25 °C. To quench the reaction, 20 μl of the reaction mixture was added to P81 paper discs (Whatman), and the paper discs were immediately placed in wash buffer. Washes were then carried out three times in 10 mm HEPES, pH 7.5, with each wash lasting 5 min, to remove un-reacted Ac-CoA. The papers were then dried with acetone and added to 4 ml of scintillation fluid, and the signal was measured with a Packard Tri-Carb 1500 liquid scintillation analyzer. Background control reactions were performed in the absence of enzyme. Reactions were also performed in the absence of the enzyme to ensure that the signal was not dueto autoacetylation of the peptide substrate, and the values were subtracted from each one of the reaction samples. All reactions were performed in triplicate. All radioactive count values were converted to molar units with a standard curve created with known concentrations of radioactive Ac-CoA added to scintillation fluid. Data fitting to the Michaelis-Menten equation was carried out using the GraphPad Prism 5 software.
Thermal Denaturation Assays
Thermal denaturation assays to measure the thermal stability of ESCO1 WT and mutants (residues 590–840) were performed by monitoring fluorescence change using the dye SYPRO Orange (Life Technologies) while heating the protein. The protein sample mixture contained 20 μl of 0.1 mg/ml ESCO1 WT or mutants and 1 μl of 1:10 diluted SYPRO. The temperature was gradually increased from 20 to 95 °C using 1 °C-intervals using a Q-PCR 7900 system. The change in fluorescence was measured using the TAMRA setting in the Q-PCR 7900 system.
Analytical Ultracentrifugation
Sedimentation equilibrium analytical ultracentrifugation experiments with ESCO1 (residues 590–840) were performed at 4 °C with interference optics using a Beckman Optima XL-I analytical ultracentrifuge. The rotor used was a four-hole rotor containing six-channel centerpieces with sapphire windows, spinning at 12,000, 18,000, and 26,000 rpm. Protein samples were analyzed at protein concentrations of 30, 58, and 118 μm in gel filtration buffer (25 mm HEPES, pH 7.0, 200 mm NaCl, 1 mm TCEP) for Apo, 1 mm Ac-CoA, and 1 mm CoA/1 mm SMC3 peptide. The data for each speed were collected in quadruplicate. The viscosity of the samples was estimated using Sednterp (43, 44), and the most representative runs were included to calculate theoretical molecular masses using the program HeteroAnalysis.
Author Contributions
Y. R.-C., A. M., G. P. L., A. S. O., and R. M. designed the research; Y. R.-C., A. M., G. P. L., and A. S. O. performed the research; Y. R.-C. prepared the figures, and Y. R.-C., A. M., G. P. L., A. S. O., and R. M. wrote the manuscript. All authors read and approved the manuscript.
Acknowledgments
We acknowledge support of the proteomics facility at the Wistar Institute (National Institutes of Health Grant P30 CA010815) and the DNA Sequencing Facility at the Perelman School of Medicine, University of Pennsylvania (National Institutes of Health Grant P30 CA016520).
This work was supported by National Institutes of Health Grants R01 GM060293, R35 GM118090, and P01 AG031862 (to R. M.), T32 GM098910 (to G. P. L.), and K12 GM081259 (to Y. R-C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The atomic coordinates and structure factors (code 5T53) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- HAT
- histone acetyltransferase
- Ac-CoA
- acetyl coenzyme A
- GNAT
- Gcn5-related N-acetyltransferase
- RMSD
- root mean square deviation
- PCAF
- P300/CBP-associated factor
- CBP
- CREB-binding protein
- CREB
- cAMP-response element-binding protein
- TEV
- tobacco etch virus
- TCEP
- tris(2-carboxyethyl)phosphine.
References
- 1.Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., and Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 2.Eberharter A., and Becker P. B. (2002) Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 3, 224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang X. J., and Seto E. (2007) HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene 26, 5310–5318 [DOI] [PubMed] [Google Scholar]
- 4.Wang L., Tang Y., Cole P. A., and Marmorstein R. (2008) Structure and chemistry of the p300/CBP and Rtt109 histone acetyltransferases: implications for histone acetyltransferase evolution and function. Curr. Opin. Struct. Biol. 18, 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanner K. G., Langer M. R., and Denu J. M. (2000) Kinetic mechanism of human histone acetyltransferase P/CAF. Biochemistry 39, 11961–11969; Correction Biochemistry (2000) 39, 15652 [DOI] [PubMed] [Google Scholar]
- 6.Tanner K. G., Langer M. R., Kim Y., and Denu J. M. (2000) Kinetic mechanism of the histone acetyltransferase GCN5 from yeast. J. Biol. Chem. 275, 22048–22055 [DOI] [PubMed] [Google Scholar]
- 7.Ivanov D., Schleiffer A., Eisenhaber F., Mechtler K., Haering C. H., and Nasmyth K. (2002) Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr. Biol. 12, 323–328 [DOI] [PubMed] [Google Scholar]
- 8.Jeppsson K., Kanno T., Shirahige K., and Sjögren C. (2014) The maintenance of chromosome structure: positioning and functioning of SMC complexes. Nat. Rev. Mol. Cell. Biol. 15, 601–614 [DOI] [PubMed] [Google Scholar]
- 9.Haering C. H., Löwe J., Hochwagen A., and Nasmyth K. (2002) Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9, 773–788 [DOI] [PubMed] [Google Scholar]
- 10.Haering C. H., Schoffnegger D., Nishino T., Helmhart W., Nasmyth K., and Löwe J. (2004) Structure and stability of cohesin's Smc1-kleisin interaction. Mol. Cell 15, 951–964 [DOI] [PubMed] [Google Scholar]
- 11.Gu W., and Roeder R. G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90, 595–606 [DOI] [PubMed] [Google Scholar]
- 12.Pramanik K. C., Fofaria N. M., Gupta P., and Srivastava S. K. (2014) CBP-mediated FOXO-1 acetylation inhibits pancreatic tumor growth by targeting SirT. Mol. Cancer Ther. 13, 687–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sykes S. M., Mellert H. S., Holbert M. A., Li K., Marmorstein R., Lane W. S., and McMahon S. B. (2006) Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell 24, 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang Y., Luo J., Zhang W., and Gu W. (2006) Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell 24, 827–839 [DOI] [PubMed] [Google Scholar]
- 15.Tang Y., Zhao W., Chen Y., Zhao Y., and Gu W. (2008) Acetylation is indispensable for p53 activation. Cell 133, 612–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li A. G., Piluso L. G., Cai X., Gadd B. J., Ladurner A. G., and Liu X. (2007) An acetylation switch in p53 mediates holo-TFIID recruitment. Mol. Cell 28, 408–421 [DOI] [PubMed] [Google Scholar]
- 17.Neuwald A. F., and Landsman D. (1997) GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 22, 154–155 [DOI] [PubMed] [Google Scholar]
- 18.Akella J. S., Wloga D., Kim J., Starostina N. G., Lyons-Abbott S., Morrissette N. S., Dougan S. T., Kipreos E. T., and Gaertig J. (2010) MEC-17 is an α-tubulin acetyltransferase. Nature 467, 218–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedmann D. R., Aguilar A., Fan J., Nachury M. V., and Marmorstein R. (2012) Structure of the α-tubulin acetyltransferase, αTAT1, and implications for tubulin-specific acetylation. Proc. Natl. Acad. Sci. U.S.A. 109, 19655–19660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shida T., Cueva J. G., Xu Z., Goodman M. B., and Nachury M. V. (2010) The major α-tubulin K40 acetyltransferase αTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc. Natl. Acad. Sci. U.S.A. 107, 21517–21522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tóth A., Ciosk R., Uhlmann F., Galova M., Schleiffer A., and Nasmyth K. (1999) Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13, 320–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minamino M., Ishibashi M., Nakato R., Akiyama K., Tanaka H., Kato Y., Negishi L., Hirota T., Sutani T., Bando M., and Shirahige K. (2015) Esco1 acetylates cohesin via a mechanism different from that of Esco2. Curr. Biol. 25, 1694–1706 [DOI] [PubMed] [Google Scholar]
- 23.Vega H., Waisfisz Q., Gordillo M., Sakai N., Yanagihara I., Yamada M., van Gosliga D., Kayserili H., Xu C., Ozono K., Jabs E. W., Inui K., and Joenje H. (2005) Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat. Genet. 37, 468–470 [DOI] [PubMed] [Google Scholar]
- 24.Price J. C., Pollock L. M., Rudd M. L., Fogoros S. K., Mohamed H., Hanigan C. L., Le Gallo M., NIH Intramural Sequencing Center (NISC) Comparative Sequencing Program, Zhang S., Cruz P., Cherukuri P. F., Hansen N. F., McManus K. J., Godwin A. K., Sgroi D. C., et al. (2014) Sequencing of candidate chromosome instability genes in endometrial cancers reveals somatic mutations in ESCO1, CHTF18, and MRE11A. PLoS ONE 8, e63313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S., Li J., Zhou G., Mu D., Yan J., Xing J., Yao Z., Sheng H., Li D., Lv C., Sun B., Hong Q., and Guo H. (2016) Increased expression of ESCO1 is correlated with poor patient survival and its role in human bladder cancer. Tumour Biol. 37, 5165–5170 [DOI] [PubMed] [Google Scholar]
- 26.Gordillo M., Vega H., and Jabs E. W. (2006) Roberts syndrome. GeneReviews, University of Washington, Seattle, WA [Google Scholar]
- 27.Gordillo M., Vega H., Trainer A. H., Hou F., Sakai N., Luque R., Kayserili H., Basaran S., Skovby F., Hennekam R. C., Uzielli M. L., Schnur R. E., Manouvrier S., Chang S., Blair E., et al. (2008) The molecular mechanism underlying Roberts syndrome involves loss of ESCO2 acetyltransferase activity. Hum. Mol. Genet. 17, 2172–2180 [DOI] [PubMed] [Google Scholar]
- 28.Vetting M. W., S. de Carvalho L. P., Yu M., Hegde S. S., Magnet S., Roderick S. L., and Blanchard J. S. (2005) Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 433, 212–226 [DOI] [PubMed] [Google Scholar]
- 29.Friedmann D. R., and Marmorstein R. (2013) Structure and mechanism of non-histone protein acetyltransferase enzymes. FEBS J. 280, 5570–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marmorstein R., and Zhou M. M. (2014) Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb. Perspect. Biol. 6, a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rahman S., Jones M. J., and Jallepalli P. V. (2015) Cohesin recruits the Esco1 acetyltransferase genome wide to repress transcription and promote cohesion in somatic cells. Proc. Natl. Acad. Sci. U.S.A. 112, 11270–11275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanner K. G., Trievel R. C., Kuo M. H., Howard R. M., Berger S. L., Allis C. D., Marmorstein R., and Denu J. M. (1999) Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J. Biol. Chem. 274, 18157–18160 [DOI] [PubMed] [Google Scholar]
- 33.Lau O. D., Courtney A. D., Vassilev A., Marzilli L. A., Cotter R. J., Nakatani Y., and Cole P. A. (2000) p300/CBP-associated factor histone acetyltransferase processing of a peptide substrate: kinetic analysis of the catalytic mechanism. J. Biol. Chem. 275, 21953–21959 [DOI] [PubMed] [Google Scholar]
- 34.Yan Y., Harper S., Speicher D. W., and Marmorstein R. (2002) The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate. Nat. Struct. Biol. 9, 862–869 [DOI] [PubMed] [Google Scholar]
- 35.Liszczak G., Arnesen T., and Marmorstein R. (2011) Structure of a ternary Naa50p (NAT5/SAN) N-terminal acetyltransferase complex reveals the molecular basis for substrate-specific acetylation. J. Biol. Chem. 286, 37002–37010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liszczak G., Goldberg J. M., Foyn H., Petersson E. J., Arnesen T., and Marmorstein R. (2013) Molecular basis for N-terminal acetylation by the heterodimeric NatA complex. Nat. Struct. Mol. Biol. 20, 1098–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sutton A., Shia W. J., Band D., Kaufman P. D., Osada S., Workman J. L., and Sternglanz R. (2003) Sas4 and Sas5 are required for the histone acetyltransferase activity of Sas2 in the SAS complex. J. Biol. Chem. 278, 16887–16892 [DOI] [PubMed] [Google Scholar]
- 38.Carretero M., Ruiz-Torres M., Rodríguez-Corsino M., Barthelemy I., and Losada A. (2013) Pds5B is required for cohesion establishment and Aurora B accumulation at centromeres. EMBO J. 32, 2938–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kouznetsova E., Kanno T., Karlberg T., Thorsell A. G., Wisniewska M., Kursula P., Sjögren C., and Schüler H. (2016) Sister chromatid cohesion establishment factor ESCO1 operates by substrate-assisted catalysis. Structure 24, 789–796 [DOI] [PubMed] [Google Scholar]
- 40.Minor W., Cymborowski M., Otwinowski Z., and Chruszcz M. (2006) HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 41.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lebowitz J., Lewis M. S., and Schuck P. (2002) Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 11, 2067–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garcia de la Torre J. G. (1992) Sedimentation coefficients of complex biological particles. in Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding S. E., Rowe A. J., and Horton J. C., eds), pp. 333–358, The Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 45.DeLano W. L. (2012) The PyMOL Molecular Graphics System, version 1.5.0.1, Schrödinger, LLC, New York [Google Scholar]
- 46.Lovell S. C., Davis I. W., Arendall W. B. 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., and Richardson D. C. (2003) Structure validation by Cα geometry: φ,ψ and Cβ deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 47.Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 48.Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]