Abstract
mtDNA damage in cardiac myocytes resulting from increased oxidative stress is emerging as an important factor in the pathogenesis of diabetic cardiomyopathy. A prevalent lesion that occurs in mtDNA damage is the formation of 8-hydroxy-2′-deoxyguanosine (8-OHdG), which can cause mutations when not repaired properly by 8-oxoguanine DNA glycosylase (Ogg1). Although the mtDNA repair machinery has been described in cardiac myocytes, the regulation of this repair has been incompletely investigated. Here we report that the hearts of type 1 diabetic mice, despite having increased Ogg1 protein levels, had significantly lower Ogg1 activity than the hearts of control, non-type 1 diabetic mice. In diabetic hearts, we further observed increased levels of 8-OHdG and an increased amount of mtDNA damage. Interestingly, Ogg1 was found to be highly O-GlcNAcylated in diabetic mice compared with controls. In vitro experiments demonstrated that O-GlcNAcylation inhibits Ogg1 activity, which could explain the mtDNA lesion accumulation observed in vivo. Reducing Ogg1 O-GlcNAcylation in vivo by introducing a dominant negative O-GlcNAc transferase mutant (F460A) restored Ogg1 enzymatic activity and, consequently, reduced 8-OHdG and mtDNA damage despite the adverse hyperglycemic milieu. Taken together, our results implicate hyperglycemia-induced O-GlcNAcylation of Ogg1 in increased mtDNA damage and, therefore, provide a new plausible biochemical mechanism for diabetic cardiomyopathy.
Keywords: 8-oxoguanine glycosylase (OGG1), cardiomyopathy, mtDNA; O-linked N-acetylglucosamine (O-GlcNAc), oxidative stress, type 1 diabetes
Introduction
In diabetes mellitus, inadequate glucose regulation and the resultant hyperglycemia are widely regarded as significant contributors to diabetic complications (1). Hyperglycemia leads to overproduction of reactive oxygen species (ROS).4 Because diabetes is accompanied by increased free radical production and/or impaired antioxidant defense capabilities, excessive ROS production has been proposed as the link between high glucose and the pathways responsible for hyperglycemic damage in diabetes (1). ROS are highly reactive molecules that can oxidize essential cellular macromolecules (lipids, proteins, and DNA), thus damaging cell membranes, altering cell metabolism, and increasing mutation rates (1–3).
mtDNA is exposed to high levels of oxidative stress. The proximity of mtDNA to the inner mitochondrial membrane and the oxidative phosphorylation chain makes it highly susceptible to oxidative stress from the electron transport system (4–6). mtDNA is a 16.5-kb double-stranded, circular DNA molecule. It does not contain introns and encodes 13 polypeptides for complexes I, III, IV, and V of the electron transport system as well as 22 tRNAs and two rRNAs (7, 8). Thus, any damage to mtDNA could potentially induce dysfunctional mitochondrial transcripts that could lead to reduced oxidative phosphorylation and decreased mitochondrial function (9). In addition, oxidative mtDNA damage has been reported to be more extensive and longer lasting than that of nuclear DNA damage (10–12).
Base excision repair (6) appears to be the most important mtDNA repair pathway. Base excision repair is initiated when a DNA glycosylase recognizes and removes an improperly modified DNA base. The resulting abasic site is cleaved by endonucleases and/or phosphodiesterases, removing the sugar residue, before DNA polymerase and DNA ligase complete the repair (6). 8-oxoguanine-DNA glycosylase (Ogg1) is the principal DNA glycosylase responsible for repair of the ROS-induced mutagenic DNA lesion 8-hydroxy-2′-deoxyguanosine (8-OHdG) and ring-opened fapyguanine in humans (13). Ogg1 alterations in diabetic models have only been incompletely explored. Oxidative stress-related DNA damage has been proposed as a novel important factor in the pathogenesis of human type 2 diabetes (14) based on the finding that Ogg1 is up-regulated in type 2 diabetic islet cell mitochondria, which is consistent with a rodent model in which hyperglycemia and consequent increased β cell oxidative metabolism lead to DNA damage and induction of Ogg1 expression (14). Moreover, Ogg1 acetylation by p300 significantly increased enzyme activity in both in vitro and in vivo oxidatively stressed cells (13), and phosphorylation by Cdk4 was found to modulate Ogg1 function in in vitro experiments (15), suggesting a complex regulation of Ogg1 DNA repair activity by different posttranslational modifications.
Another common posttranslational modification particularly relevant to diabetes occurs by O-linked N-acetylglucosamine addition (O-GlcNAcylation). O-GlcNAcylation of serine or threonine residues of nuclear, cytoplasmic, and mitochondrial proteins is a dynamic and ubiquitous protein modification (16). This process is emerging as a key regulator of critical biological processes, including nuclear transport, translation and transcription, signal transduction, cytoskeletal reorganization, proteasomal degradation, and apoptosis (17). As glucose is the main precursor for the amino sugar substrates of the hexosamine biosynthesis pathway, which ends in production of UDP-GlcNAc, glucose availability is well accepted to be the key mediator of this protein posttranslational modification (18). Consequently, the elevated protein O-GlcNAcylation level in hearts from diabetic animals has been implicated in glucose toxicity and linked to multiple aspects of cardiomyocyte dysfunction in diabetes (19–25).
GlcNAc residues are monosaccharide units that are linked posttranslationally to the hydroxyl groups of serine and threonine residues of proteins to form O-GlcNAc (26). Protein O-GlcNAcylation levels are regulated by two enzymes that act antagonistically. Uridine diphosphate N-acetylglucosamine polypeptidyl transferase, known as O-GlcNAc transferase (OGT), is the glycosyltransferase that catalyzes transfer of GlcNAc from uridine diphosphate N-acetylglucosamine to acceptor proteins (16, 27). N-acetyl-β-d-glucosaminidase (O-GlcNAcase, OGA) is a glycoside hydrolase that catalyzes cleavage of O-GlcNAc from proteins (28, 29). Both enzymes are ubiquitous and have been shown to be essential for development in vertebrates (30, 31), which underscores their fundamental roles in vital processes.
During the past decade, diabetes-associated O-GlcNAcylationhas been described as either a maladaptive phenomenon that can interfere with protein function (32–34) or, conversely, has been shown to be cardioprotective when acutely activated (21, 35, 36). Moreover, elevated mitochondrial O-GlcNAcylation caused by hyperglycemia, as occurs in diabetes, has been reported to significantly contribute to mitochondrial dysfunction that leads to diabetic cardiomyopathy (19, 20, 23–25, 33, 37), supporting a widely held belief that an excessive increase in O-GlcNAc levels is detrimental to the heart.
Ogg1 O-GlcNAcylation is completely unexplored in either physiological or pathological conditions. Here we report that Ogg1 is highly O-GlcNAcylated in our type 1 diabetes (T1D) murine model and that reducing O-GlcNAcylation with a dominant negative mutant OGT restores Ogg1 activity, likely by competition for Ogg1 binding, which leads to improved mtDNA quality in T1D hearts.
Results
Diabetic Murine Hearts Show Increased Oxidative Stress, mtDNA Damage, and Impaired Ogg1 Activity and O-GlcNAcylation Status
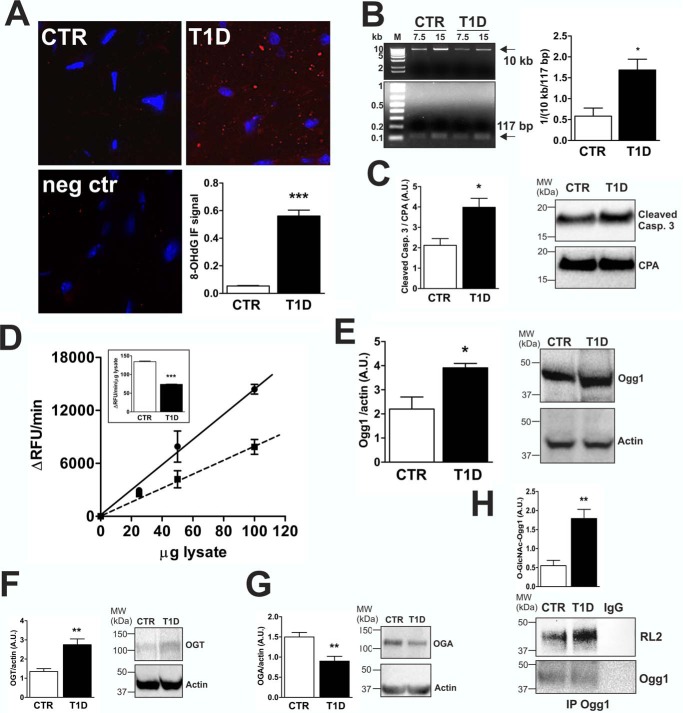
To study whether mtDNA is affectedby hyperglycemia-dependent oxidative stress, we compared 10-week diabetic (T1D) mice (glucose levels in the blood > 500 mg/dl) with control (CTR) mice (glucose levels in the blood < 150 mg/dl). Compared with CTR, we found significantly higher (10-fold) levels of 8-OHdG staining in T1D hearts (Fig. 1A). We then performed an assay to examine mtDNA damage and found a statistically significant increase with T1D. Fig. 1B shows that amplification of a 10-kb fragment from isolated mtDNA was selectively inhibited in T1D versus CTR, indicating that a higher number of lesions were present in mtDNA from diabetic hearts. In addition, we observed increased levels of cleaved caspase 3 in diabetic hearts (Fig. 1C), which indicates an increase in apoptosis. Ogg1 is a key enzyme in mtDNA repair (38). The Ogg1 DNA glycosylase recognizes and hydrolyzes the modified base 8-OHdG, removing the mutagenic 8-OHdG lesion situated opposite cytosine, whereas MutY glycosylase removes adenine from 8-OHdG/A mismatches with its adenine glycosylase activity (39) when 8-OHdG has already induced a mutation. Because we detected increased 8-OHdG levels, which may result in the mtDNA damage registered in T1D hearts, we focused our investigation on determining whether Ogg1, and, thus, the dependent DNA repair process, was somehow affected. We assessed Ogg1 enzymatic activity using a specifically designed fluorescent molecular beacon carrying an oxidized guanosine residue in the stem portion of the molecule annealed to a cytosine residue and compared it with the protein level. We observed a 50% decrease in Ogg1 enzymatic activity in T1D murine heart lysates compared with CTR (Fig. 1D); however, Ogg1 protein levels (detected as a 45-kDa protein) were found to be increased (Fig. 1E). High levels of protein O-GlcNAcylation have been associated with hyperglycemia (32–37). We found that OGT (Fig. 1F) and OGA (Fig. 1G) protein levels were increased and decreased, respectively, in T1D versus CTR hearts; therefore, we checked whether Ogg1 was differently O-GlcNAcylated. Fig. 1H shows that increased Ogg1 O-GlcNAcylation was detected in T1D versus CTR.
FIGURE 1.
Diabetic murine model characterization. T1D mice were studied 10 weeks after STZ treatment. A, IF analysis in heart sections of 8-OHdG levels, a DNA oxidative stress marker. The 8-OHdG IF signal was quantified using Pixel Counter from ImageJ and represents the percentage of red pixels. Negative control (neg ctr) slides were incubated in solution lacking the primary anti-8-OHdG antibody. Images are representative of at least 5 fields/animal (n = 3 CTR and T1D). B, mtDNA damage estimation. 1/(10 kb/117 bp) represents the number of lesions per kilobase within the amplified long mtDNA fragment (10 kb). Data were normalized relative to amplification of a short mtDNA fragment (117 bp). 15 ng of isolated mtDNA was used as a template for the reaction, and, to ensure linearity, 0.5× template (7.5 ng) was included in the experiment. M, DNA molecular weight marker. Data are representative of n = 3 CTR and T1D animals. C, cleaved caspase (Casp.) 3 Western blotting analysis from heart lysates. CPA was used as a loading control. Results are representative of n = 3 CTR and T1D animals. MW, molecular weight; A.U., arbitrary units. D, Ogg1 enzymatic activity from heart lysates. 25, 50, and 100 μg of protein lysate were incubated at 37 °C with 0.5 μm molecular beacon in Ogg1 assay buffer for 30 min. ΔRFU per minute and ΔRFU per minute per microgram of lysate (inset) are reported. Black line, CTR; dashed line, T1D. E, Ogg1 Western blot from 100 μg of total heart lysate. Actin was used as a loading control. F and G, OGT (F) and OGA (G) Western blots from 100 and 200 μg of total heart lysate, respectively. Actin was used as a loading control. H, O-GlcNAcylation status of Ogg1. 500 μg of total heart lysate was immunoprecipitated using anti-Ogg1, separated by SDS-PAGE, and analyzed by Western blotting for O-GlcNAc (RL2) and Ogg1. The O-GlcNacylated Ogg1 band was normalized to Ogg1 detected in the IP. As a control, samples were treated with anti-rabbit IgG. The data in D–H are representative of n = 4 CTR and T1D animals. All data (A–H) are expressed as mean ± S.E. ***, p < 0.001; **, p < 0.01; *, p < 0.05.
O-GlcNAcylation Inhibits Ogg1 Activity in Vitro
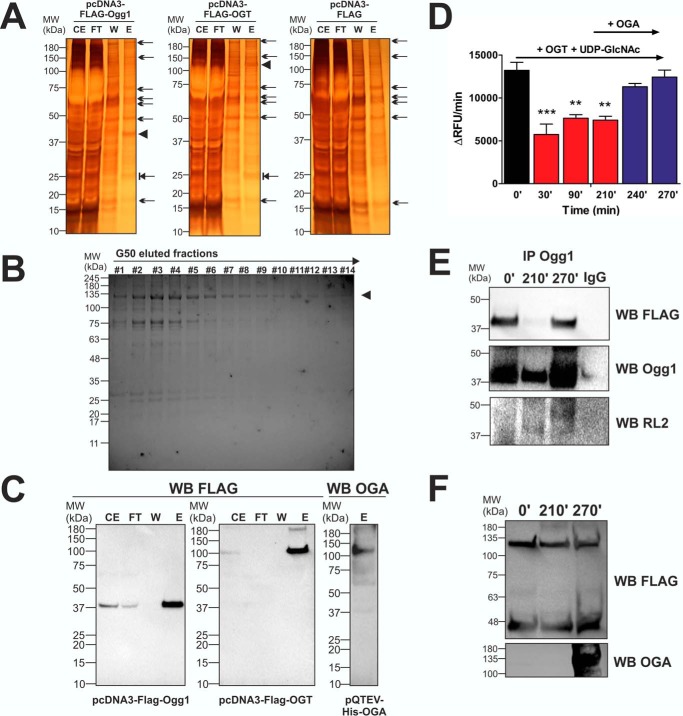
Next we investigated whether Ogg1 activity was affected by O-GlcNAcylation. We expressed and purified a 40-kDa FLAG-tagged Ogg1 and a 120-kDa OGT from HEK 293T cells and a 130-kDa His-tagged OGA from “Rosetta” Escherichia coli (Fig. 2, A–C). In vitro O-GlcNAcylation of Ogg1 was performed by incubating eluted FLAG-Ogg1 and FLAG-OGT at 37 °C in OGT assay buffer in the presence of 50 μm UDP-GlcNAc. Ogg1 activity was then assayed every 30 min by taking 50 μl from the reaction mixture. The data reported in Fig. 2D show an ∼50% decrease in Ogg1 activity after 30 min from the start of the O-GlcNAcylation reaction. We observed no further inhibition at the 90- and 210-min time points, suggesting that OGT activity was sufficient to saturate Ogg1 O-GlcNAcylation sites in the first 30 min. We then introduced recombinant His-OGA, in excess with respect to OGT (0.2 mg/ml versus 0.1 mg/ml), and, after 60 min, we found complete restoration of basal Ogg1 activity. Aliquots of the reaction mixture taken at 0, 210, and 270 min were used to detect the O-GlcNAcylation status of Ogg1. Samples were Ogg1-immunoprecipitated, and Western blots (WB) with anti-O-GlcNAc (RL2), anti-FLAG, or anti-Ogg1 were performed. Fig. 2E shows an increased, ∼40-kDa Ogg1-specific protein O-GlcNAcylation signal (measured with the RL2 antibody) at 210 min compared with 0 min and a decreased signal at 270 min compared with 210 min, suggesting that the observed inhibition of Ogg1 activity was due to the increased O-GlcNAcylation mediated by OGT and that de-O-GlcNAcylating Ogg1 with OGA restored the basal activity. Interestingly, both anti-FLAG and anti-Ogg1 WB show lower Ogg1 levels in the immunoprecipitated sample at 210 min, suggesting that O-GlcNAcylation altered Ogg1 interaction with the Ogg1-specific IgG. As a control, WB analysis of non-Ogg1-precipitated aliquots of reaction mixture showed OGT and Ogg1 at equal levels throughout the experiment and OGA presence only after 210 min of the in vitro O-GlcNAc reaction (Fig. 2F).
FIGURE 2.
In vitro O-GlcNAcylation and de-O-GlcNAcylation of Ogg1. A, recombinant FLAG-Ogg1 and FLAG-OGT were expressed and purified from HEK 293T cells (silver staining). As a control, fractions from pcDNA3-FLAG-transfected cells are reported. Arrows represent nonspecific transgenic products also found in eluted fractions from cells transfected with empty vector. Arrowheads mark the FLAG-Ogg1 and FLAG-OGT recombinant proteins generated and isolated. These proteins represent the most abundant compounds among those eluted. Blocked arrows mark a 25-kDa protein present only in eluted fractions from cells transfected with either pcDNA3-FLAG-Ogg1 or pcDNA3-FLAG-OGT. We may consider this an Ogg1/OGT interactor that does not interfere with subsequent in vitro applications. CE, crude extract; FT, flow-through; W, wash; E, elution; MW, molecular weight. B, recombinant His-OGA was expressed and purified from E. coli “Rosetta.” Fractions collected and stained (Coomassie staining) after the last gel filtration step with G50 resin to remove excess imidazole are reported. Fractions from #6 to #14 were pooled, concentrated, and used for further applications. C, Western blotting analysis of the recombinant FLAG-Ogg1, FLAG-OGT, and His-OGA. D, recombinant FLAG-Ogg1 activity assayed after incubation at time 0 and for 30, 90, and 210 min (0′, 30′, 90′, 210′, respectively) with FLAG-OGT and 50 μm UDP-GlcNAC. To restore initial basal activity, the recombinant His-OGA was added to the reaction mixture, and recombinant FLAG-Ogg1 activity was assayed after 30 min (240′) and 60 min (270′). Data are expressed as mean ± S.E. and were analyzed by comparing all conditions versus activity assayed at time 0. ***, p < 0.001;, **, p < 0.01. E, Western blotting analysis of Ogg1-immunoprecipitated (IP Ogg1) aliquots withdrawn at different O-GlcNAc reaction time points (0′, 210′, and 270′). Recombinant FLAG-Ogg1 (∼40 kDa) was revealed with anti-FLAG and anti-Ogg1, and O-GlcNAc-Ogg1 (∼40 kDa) was revealed with the RL2 antibody. F, Western blots for FLAG-Ogg1 (∼40 kDa), FLAG-OGT (∼120 kDa), and His-OGA (∼130 kDa) levels were performed as loading controls. Aliquots withdrawn at the same time points (0′, 210′, and 270′) were analyzed using anti-FLAG or anti-OGA antibodies.
High-glucose Conditions Replicate the Diabetic Phenotype in Neonatal Cardiac Myocytes (NCM)
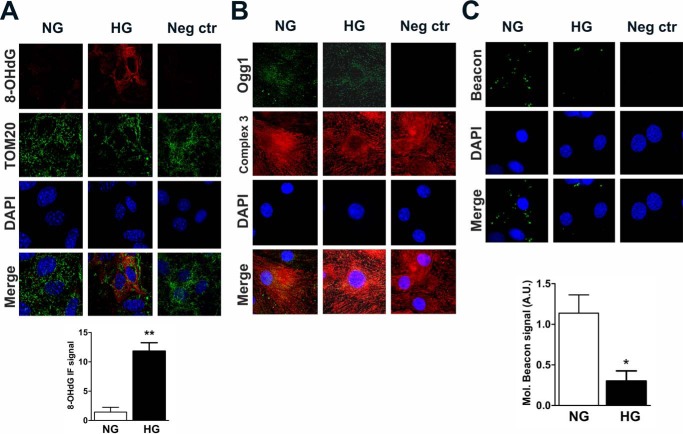
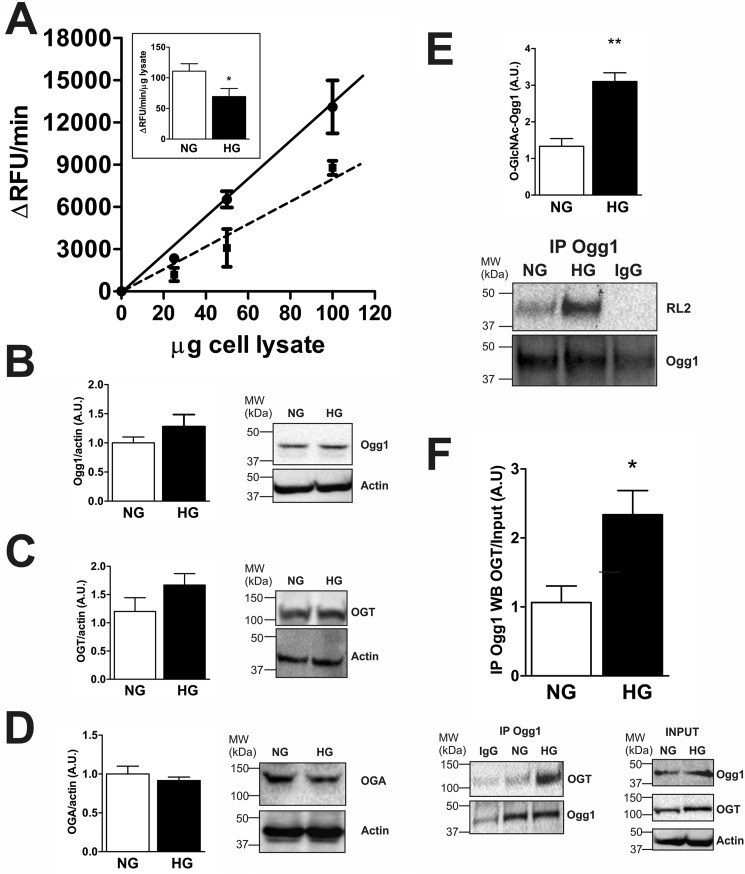
To test methods to reduce Ogg1 O-GlcNAcylation, we used an ex vivo model where NCM primary cultures were exposed to high glucose levels (HG = 25 mm) for 72 h to mimic diabetes-associated hyperglycemia. As in T1D, 8-OHdG levels were significantly increased in HG versus cells cultured with normal glucose levels (NG = 5.5 mm glucose + 19.5 mm mannitol) (Fig. 3A). 8-OHdG also co-localized with the mitochondrial import receptor (TOM20), indicating that the observed DNA oxidation was primarily extranuclear. We then visualized Ogg1 by IF and again observed predominantly extranuclear localization based on closely overlapping signals from Ogg1 and from the mitochondrial marker complex 3 IF (Fig. 3B). Ogg1 localization and in situ activity were examined by visualizing an 8-OHdG-containing molecular beacon following overnight incubation at 37 °C. The images in Fig. 3C demonstrated extranuclear Ogg1 localization and lower activity in HG versus NG. We next measured Ogg1 activity from whole cell lysates (Fig. 4A). Ogg1 activity was confirmed to be ∼50% lower in HG compared with NG lysates even though the Ogg1 protein (detected as a 45-kDa protein) level was slightly increased (Fig. 4B). We then checked whether Ogg1 O-GlcNAcylation was altered in NCM under hyperglycemic conditions, as observed in T1D hearts in vivo, based on an trend toward increased OGT levels (Fig. 4C) and decreased OGA levels (Fig. 4D). Fig. 4E shows that, after 72 h of exposure to HG, Ogg1 was significantly more O-GlcNAcylated than in cells cultured in NG. Moreover, co-immunoprecipitation (IP) of Ogg1 and OGT from lysates revealed that Ogg1 interacts with OGT and, interestingly, that the interaction with OGT was increased in NCM cultured in HG (Fig. 4F). This suggested a possible mechanism through which Ogg1 O-GlcNAcylation increases under hyperglycemic conditions and that we investigated in subsequent studies.
FIGURE 3.
8-OHdG, Ogg1, and in situ Ogg1 activity staining in NCM cultured under hyperglycemic conditions. A and B, co-immunostaining of 8-OHdG with the mitochondrial marker TOM20 (A) and co-immunostaining of Ogg1 with the mitochondrial marker complex 3 (B). As a negative control (Neg ctr), cells were incubated in solution lacking either anti-8OHdG or anti-Ogg1. C, in situ Ogg1 activity fluorescent staining. NCM were seeded on gelatin-coated coverglasses, and after 72 of h treatment, cells were transfected with 1 nm 8-OHdG-containing molecular beacon and incubated overnight at 37 °C. Cells were then fixed in 4% PFA and mounted on a slide with one drop of mounting medium containing DAPI. As a negative control, cells were transfected with the molecular beacon and fixed immediately. 8-OHdG and the molecular beacon signal were quantified with Pixel Counter from ImageJ, and values reported represent the percentage of either red or green pixels. Images are representative of at least 5 fields/condition. A.U., arbitrary units. Data are expressed as mean ± S.E. and represent at least five independent experiments. **, p < 0.01; *, p < 0.05.
FIGURE 4.
Ogg1 activity, Ogg1 O-GlcNAcylation, and Ogg1-OGT interaction levels in NCM cultured under hyperglycemic conditions. A, Ogg1 enzymatic activity from NCM cell lysates. 25, 50, and 100 μg of protein lysate were incubated at 37 °C with 0.5 μm molecular beacon in Ogg1 assay buffer for 30 min. ΔRFU per minute and ΔRFU per minute per microgram of lysate (inset) are reported. Black line, NG; dashed line, HG. B–D, Western blotting of Ogg1 (B), OGT (C), and OGA (D) from NCM cell lysates. Actin was used as a loading control. MW, molecular weight; A.U., arbitrary units. E and F, Ogg1 O-GlcNAcylation status (E) and Ogg1-OGT co-IP assay (F). Ogg1 was immunoprecipitated, and both O-GlcNAcylation and interaction with OGT were detected in the IP with anti-O-GlcNAc (RL2) and anti-OGT, respectively. As control samples were immunoprecipitated with anti-rabbit IgG. OGT and Ogg1 IP signals were normalized to their relative input levels. RL2 signal was normalized to the Ogg1 IP signal. Actin was used as a loading control. All data are expressed as mean ± S.E. and represent at least five independent experiments. **, p < 0.01; *, p < 0.05.
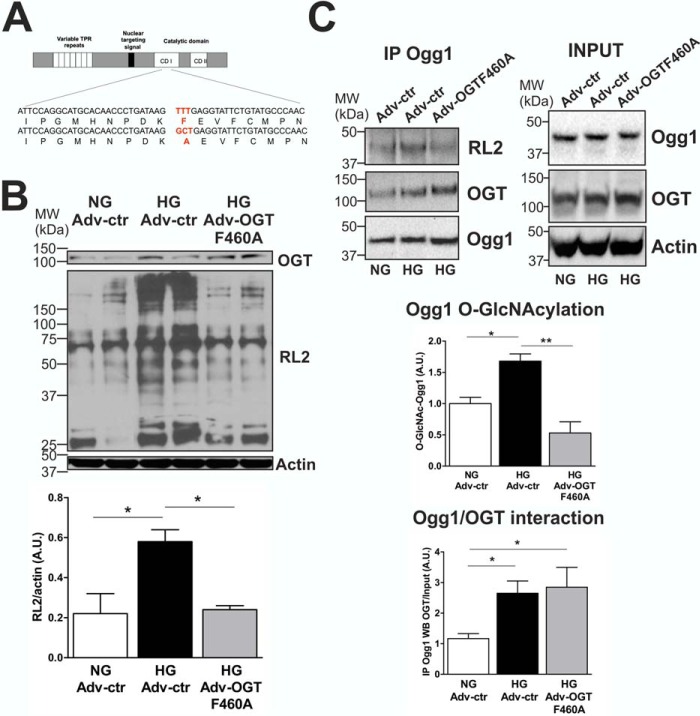
The Dominant Negative OGT Mutant F460A Decreases Ogg1 O-GlcNAcylation in NCM Primary Cultures
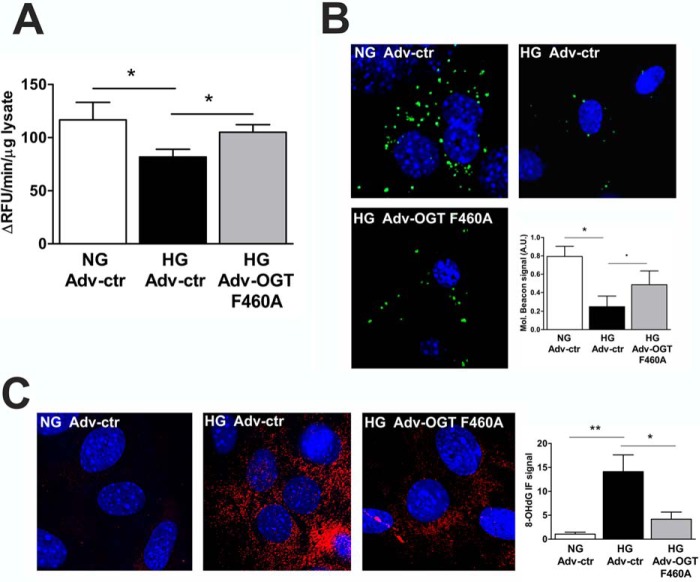
We next decided to introduce a dominant negative OGT mutant (OGT F460A) into our ex vivo hyperglycemia model with the goal of reducing Ogg1 O-GlcNAcylation. OGT catalyzes the addition of a single O-GlcNAc residue to serine or threonine residues of target proteins (16). The catalytic region of OGT consists of two highly conserved domains: CD1 and CD2 (Fig. 5A). It has been shown that Phe-460 in CD1 is an important site for OGT function, as mutation to Ala (F460A) completely abrogates the function of OGT (40). NCM were transduced with an adenovirus encoding the OGT F460A transgene (Adv-OGT F460A) or with an empty vector control (Adv-ctr), and, after 72 h of treatment, total cellular proteins were isolated. HG treatment increased total protein O-GlcNAcylation by 167% compared with NG (Fig. 5B). NCM transduced with Adv-OGT F460A showed increased OGT expression (+40%) and decreased total protein O-GlcNAcylation (−59%) compared with NCM transduced with Adv-ctr and cultured in HG, demonstrating the efficacy of such a treatment in reducing overall O-GlcNAcylation (Fig. 5B). Ogg1 O-GlcNAcylation was then checked, and NCM exposed to HG and transduced with Adv-OGT F460A showed decreased Ogg1-specific O-GlcNAcylation that returned to normal levels. OGT F460A was shown to likely compete with endogenous OGT for interaction with Ogg1, as detected by co-IP experiments (Fig. 5C). Moreover, reducing the O-GlcNAcylation of Ogg1 resulted in increased enzymatic activity both in NCM lysates (Fig. 6A) and in situ (Fig. 6B), which led to lower 8-OHdG levels (Fig. 6C).
FIGURE 5.
OGT F460A is a dominant negative mutant protein used in the NCM hyperglycemia model for decreasing Ogg1 O-GlcNAcylation. A, construction of the OGT dominant negative mutant F460A. The OGT protein is comprised of two domains: the N-terminal TPR domain, which contains a variable number of TPR repeats, and the C-terminal catalytic domain containing two conserved regions (CD 1 and CD 2). The F460A mutation was in the CD 1 region of the OGT catalytic domain, as indicated in red. B, total protein O-GlcNAcylation from NCM cultured in either NG or HG and transduced with either Adv-ctr or Adv-OGT F460A. OGT and actin levels are also reported. MW, molecular weight; A.U., arbitrary units. C, co-IP experiments showing reduced Ogg1-specific O-GlcNAcylation and increased interaction between Ogg1 and OGT F460A. IP signals were normalized to input levels, except for RL2 IP detection, which was normalized to Ogg1 detected in the immunoprecipitate. Actin was used as a loading control. All data are expressed as mean ± S.E., represent at least five independent experiments, and were analyzed by comparing all conditions versus NG Adv-ctr. *, p < 0.05; **, p < 0.01.
FIGURE 6.
NCM transduced with Adv-OGT F460A show increased Ogg1 activity and reduced 8-OHdG levels. A and B, Ogg1 activity assayed in either NCM total lysates (A) or stained in situ (B). A.U., arbitrary units. C, 8-OHdG levels. All data are expressed as mean ± S.E. and are representative of more than four independent experiments for NG Adv-ctr and HG Adv-ctr and of three independent experiments for HG Adv-OGT F460A. **, p < 0.01; *, p < 0.05; p ∼ 0.05 (.).
In Vivo OGT F460A Treatment Restores Ogg1 Activity and Improves mtDNA Quality
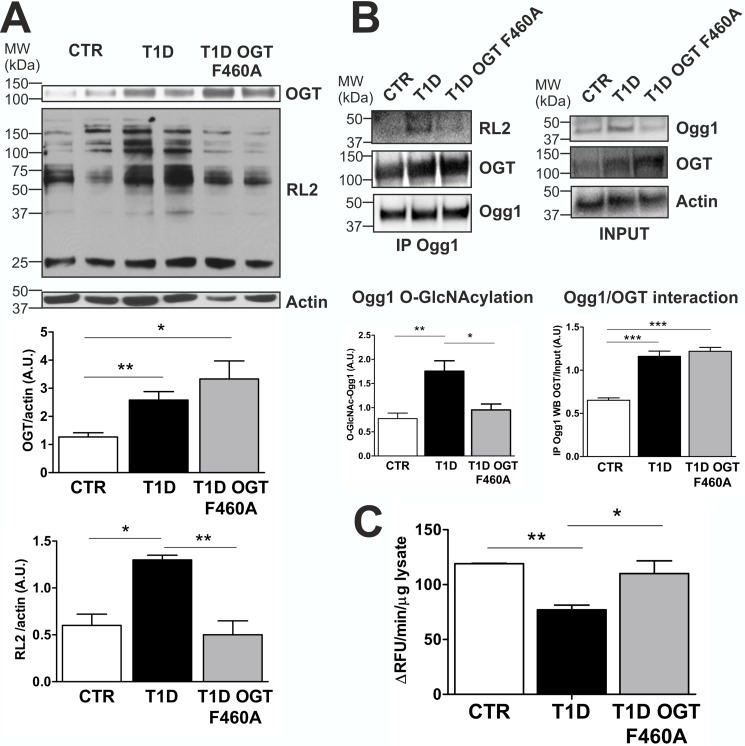
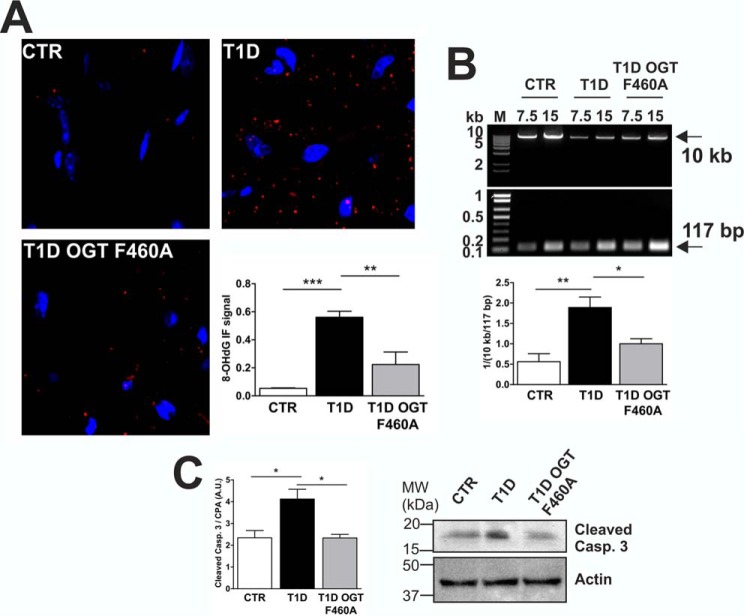
To evaluate OGT F460A treatment in vivo, we used adeno-associated virus serotype 9 (AAV9) to deliver OGT F460A. In T1D murine hearts, total protein O-GlcNAcylation was significantly increased (+115%) compared with CTR. In contrast, diabetic mice expressing OGT F460A displayed decreased (−62%) total protein O-GlcNAcylation, returning O-GlcNAcylation levels to normal (Fig. 7A). OGT F460A treatment was able to specifically reduce in vivo Ogg1 O-GlcNAcylation, presumably by competing with endogenous OGT for Ogg1, as demonstrated by co-IP experiments in which the amount of OGT co-immunoprecipitated with Ogg1 in T1D was not significantly increased despite the increase in the total amount of OGT present, both wild-type and mutant (Fig. 7B). This is in line with what was observed in primary cultures. Reducing Ogg1 O-GlcNAcylation returned enzymatic activity to normal levels (Fig. 7C), leading to reduced levels of 8-OHdG in murine hearts (Fig. 8A). We also found fewer mtDNA lesions associated with lower 8-OHdG (Fig. 8B) and decreased levels of activated cleaved caspase 3 (Fig. 8C).
FIGURE 7.
OGT F460A is a dominant negative mutant protein used in the T1D murine model for decreasing Ogg1 O-GlcNAcylation levels via competitive interaction with Ogg1. A, expression of the OGT dominant negative mutant (F460A) decreases protein O-GlcNAcylation in T1D murine hearts. OGT and Actin WB are also reported. MW, molecular weight; A.U., arbitrary units. B, Ogg1 O-GlcNAcylation status and Ogg1-OGT co-IP assay. Ogg1 was immunoprecipitated, and O-GlcNAc-Ogg1 and OGT were detected in the IP by RL2 OGT WB, respectively. C, Ogg1 activity from total heart lysates. All data are expressed as mean ± S.E. and are representative of at least five CTR, T1D, and T1D transduced with AAV-OGT F460A animals. ***, p < 0.001; **, p < 0.01; *, p < 0.05.
FIGURE 8.
Reduced Ogg1 O-GlcNAcylation in T1D AAV-OGT F460A-transduced mice leads improved mtDNA repair. A, 8-OHdG levels in heart sections. B, mtDNA damage estimation. C, Western blotting analysis of cleaved caspase (Casp.) 3 as a marker of apoptosis. A.U., arbitrary units. Data are expressed as mean ± S.E. and are representative of at least four CTR and T1D and at least three AAV-OGT F460A-transduced T1D animals. M, DNA molecular weight marker; MW, molecular weight. ***, p < 0.001; **, p < 0.01; *, p < 0.05.
Discussion
We investigated whether mtDNA repair processes were influenced by hyperglycemia-dependent oxidative stress and protein O-GlcNAcylation in T1D mice, as no data are current available according to our knowledge. 8-OHdG is one of the most abundant and well characterized DNA lesion generated by oxidative stress (41). 8-OHdG is a miscoding lesion that can cause G:C to T:A or T:A to G:C transversion mutations (42). With age, these transversions accumulate in DNA, particularly in the mitochondrial genome, and the progressive mutagenic changes in DNA sequences have been causally linked to several cancers and neurodegenerative diseases (43). Because mitochondrial endogenous oxidative damage has been reported by Anson et al. (44) to be approximately three times overestimated when analyzed from isolated mitochondria from aged mice, we initially focused our attention on characterizing our T1D murine model by comparing the immunofluorescence levels of 8-OHdG in T1D versus CTR hearts sections, thereby avoiding the artifact that was described. We then observed a correlation between 8-OHdG levels and mtDNA quality. We found that diabetic murine hearts had increased levels of 8-OHdG and an increased number of lesions compared with non-diabetic hearts. 8-OHdG, when chronically accumulated, is known to cause mutations (45). This, together with possible other lesions on mtDNA induced by diabetes, may be the reason why we observed a delayed amplification in our mtDNA damage assay by the polymerase, which may have had problems in amplification of the long fragment over the first crucial cycles of the reaction, as reported for other types of polymerases in vitro (46). As a result of this maladaptive scenario, the subsequent observation of increased apoptosis in diabetic hearts was not surprising.
In T1D, we found increased Ogg1 (as expected because of the impelling need for DNA repair under diabetic conditions). Surprisingly, Ogg1 activity was 50% lower compared with non-diabetic animals. Thus, we explored the possibility that this inverse relationship was due to some maladaptive posttranslational modification resulting in diminished activity in the context of diabetes-associated hyperglycemia.
Our group and others have clearly reported that normal protein O-GlcNAcylation is disrupted in diabetes and that mitochondria are one of the organelles greatly affected by this phenomenon (32, 37). Consistent with a previous report (20), our diabetic mice showed increased OGT and decreasedOGA expression, likely contributing to increased protein O-GlcNAcylation. Therefore, we checked the O-GlcNAcylationstatus of Ogg1 and found it to be increased in T1D versus CTR hearts. The effect of altered Ogg1 O-GlcNAcylation status on Ogg1 activity was investigated in in vitro O-GlcNAcylation and de-O-GlcNAcylation experiments using recombinant Ogg1, OGT, and OGA. Incubation with OGT and 50 μm UDP-GlcNAc rapidly inhibited Ogg1 enzymatic activity, whereas subsequent incubation with OGA restored the originalbasal activity by removing O-GlcNAc. Moreover, the in vitro immunoprecipitations performed to detect the Ogg1 O-GlcNAcylation level over the course of the experiment revealed less Ogg1 in the immunoprecipitated pellets corresponding to samples in which Ogg1 was more highly O-GlcNAcylated. Protein levels were equal in all samples collected during the experiment, and thus the data suggest that O-GlcNAc may change the Ogg1 3D structure, interfering with protein-protein and, possibly, protein-substrate interaction. However, this phenomenon was observed as markedly evident only in our in vitro experiment. Even though we observed less Ogg1 signal in both Ogg1-immunoprecipitated T1D and HG samples relative to their input levels, we cannot exclude that what we observed in vitro was enhanced because of high Ogg1 abundance obtained from the recombinant preparations.
Over the last decade, it has been reported that Ogg1 activity is modulated by acetylation in oxidatively stressed cells (13) and by phosphorylation via interaction with Cdk4 (a serine-threonine kinase) and c-Abl (a tyrosine kinase) (15). Acetylation and Cdk4-mediated phosphorylation were both reported to enhance the rate of 8-OHdG repair by Ogg1, suggesting a complex regulation of the activity of this DNA repair protein. To find evidence that O-GlcNAcylation also negatively modulates Ogg1 activity in vivo, we decided to perform rescue experiments in our T1D mice by reducing the excessive Ogg1 O-GlcNAcylation in an attempt to restore Ogg1 activity and improve mtDNA repair. First, we mimicked diabetes-associated hyperglycemia by culturing NCM for 72 h in HG. Under these acute conditions, we observed, as in T1D mice, increased mtDNA oxidative stress. Ogg1 activity was found to be decreased in HG-cultured NCM cells compared with cells cultured in NG. Under all conditions, Ogg1 and its activity were found to be principally localized outside nuclei and co-localized with either TOM20 or complex 3, confirming the emerging hypothesis that mitochondria are the major site of Ogg1 repair activity under conditions of oxidative stress (38). Moreover, as observed in T1D mice, Ogg1 protein levels did not correlate with activity, as we observed a slight increase in Ogg1 protein, whereas O-GlcNAcylation levels were significantly higher compared with NCM cultured in NG. We further investigated the Ogg1 and OGT interaction by co-immunoprecipitation assays. Cells cultured in HG displayed an increased interaction between Ogg1 and OGT, suggesting that this phenomenon likely contributes to excessive Ogg1 O-GlcNAcylation and, therefore, represents a strategy that could be adapted for lowering Ogg1 O-GlcNAcylation. For this, we used a dominant negative OGT (F460A) that was previously shown to be catalytically inactive (40). NCM transduced with Adv-OGT F460A showed an overall decrease in total protein O-GlcNAcylation and also reduced levels of O-GlcNAcylated Ogg1, likely because of OGT F460A competition with endogenous OGT for interaction with Ogg1. As a result, Ogg1 activity returned to normal, and this led to lower 8-OHdG levels. We therefore administered OGT F460A to T1D mice using an adeno-associated viral vector approach. Mice expressing OGT F460A displayed reduced total and Ogg1-specific O-GlcNAcylation, presumably by competitive interaction for Ogg1 between the mutant and endogenous OGT. This was accompanied by a complete restoration of Ogg1 activity, lower 8-OHdG levels, and improved mtDNA quality.
This work demonstrates for the first time that diabetes-associated and hyperglycemia-dependent protein O-GlcNAcylation impairs the activity of Ogg1, one of the most important mtDNA repair proteins, and that Ogg1-OGT interaction likely contributes, together with imbalanced OGT and OGA levels, to excessive Ogg1 O-GlcNAcylation. Reducing Ogg1 O-GlcNAcylation led to restored enzymatic activity and to improvement in mtDNA repair, thus giving interesting perspectives for a new plausible biochemical mechanism for diabetic cardiomyopathy.
Experimental Procedures
Treatment
Animals
All investigations conformed to the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health (Publication 85-23, revised 1985). This study was conducted in accordance with the guidelines established by the Institutional Animal Care and Use Committee at the University of California, San Diego. In NIH Swiss male mice (25 g, 3 months old), T1D was induced by giving a daily intraperitoneal injection of streptozotocin (STZ) (40 mg/kg) for 5 consecutive days (47). Diabetic status was confirmed by blood glucose levels. Experiments were carried out 10 weeks after STZ injection. In vivo adeno-associated virus transgene delivery in diabetic mice was performed by direct jugular vein injection. AAV9 expressing OGT F460A (AAV-OGT F460A) (6 × 1011 viral particles in 100 μl) was injected 6 weeks after injection with STZ. Experiments were carried out 10 weeks after STZ injection and 4 weeks after AAV-OGT F460A delivery.
NCM Primary Culture
Primary cultures of murine neonatal cardiac myocytes were prepared as described previously (32). Cells (10 × 106 cells/10-cm2 dish; 2 × 106 cells/well of a 6-well plate; 8 × 104 cells/well of a 24-well plate) were plated onto gelatin-coated culture dishes. The plating medium consisted of 4.25:1 Dulbecco's modified Eagle's medium:M199, 10% horse serum, 5% fetal bovine serum, 1% penicillin/streptomycin, and 5.5 mmol/liter d-glucose. Cells were allowed to adhere to the plates for at least 24 h before treatment. Cells were cultured in maintenance medium (4.5:1 Dulbecco's modified Eagle's medium:M199, 2% fetal bovine serum, 1% penicillin/streptomycin/Fungizone) supplemented with either NG (5.5 mm + 19.5 mm mannitol) or HG (25 mm). Cells were also treated with Adv-OGT F460A or Adv-Ctr (19). Cells were infected at a multiplicity of infection of 50/cell for both viruses the day exposure to either NG or HG started. The culture medium was changed daily until cells were harvested after 72 h.
Vectors
Plasmids
pcDNA3-FLAG was constructed by inserting the annealed oligonucleotides 5′-AGCTGC CACCATGGACTACAAAGACGATGACGACAAGG-3′ and 5′-AATTCCTTGTCGTCATC GTCTTTGT AGTCCATGGTGGC-3′ carrying the FLAG sequence into HindIII/EcoRI-cut pCDNA3. This vector was then cut with EcoRI/XbaI, and the 5′-AATTCGTCTCGAGGGATCCCTAAGCGGCCGCT-3′- and 5′-CTAGAGCGGCCGCTTAGGGATCCCT GAGACG-3′-annealed oligonucleotides carrying a multicloning site sequence were inserted. The In-Fusion® HD Cloning Kit (Clontech Laboratories, Inc.) was used to build up pcDNA3-FLAG-Ogg1 and pcDNA3-FLAG-OGT following the procedures described by the manufacturer. XhoI/NotI were used to linearize pcDNA3-FLAG and to digest both murine Ogg1 and rat OGT amplified and purified coding sequences (CDS). The Ogg1 CDS was amplified using 5′-TCGTCTCGAGGATGTTATTCCGTTC-3′ as forward and 5′-TAGAGCGGCCGCCTAGCCCTCTGGCC-3′ as reverse. OGT CDS was amplified using 5′-TCGTCTCGAGGATGGCGTCTTCCGT-3′ as forward and 5′-TAGAGCGGCCGCAAGCTTCAAACCCC-3′ as reverse. High-fidelity KOD Hot Start DNA polymerase (EMD Millipore) was used. pQTEV-His-OGA was made similarly. SalI/NotI were used both to linearize pQTEV-His and to digest human OGA CDS. Constructs were checked by restriction enzyme digestion analysis (XhoI/NotI for pcDNA3-FLAG-/Ogg1, NotI for pcDNA3-FLAG-OGT, and SalI for pQTEV-His-OGA). After confirmation of correct in-frame constructs by sequencing, plasmids were utilized for further experiments.
Adenovirus and Adeno-associated Virus
A dominant negative (F460A) single amino acid point mutation of OGT (TTT⋙GCA) (40) was generated using the QuikChange site-directed mutangenesis kit (Stratagene) with pTrc-HisA-OGT (kindly provided by Dr. G. W. Hart) as template and two primers: 5′-CACAACCCTGATAAGGCTGAGGTATTCTGCTGCC and 3′-GGCATAGCAGAATACCTCAGCCTTA TCAGGGTTGTG. OGT F460A was then cloned into pENTR1A using BamHI and EcoRV restriction enzyme sites and cloned into pAAV-Shuttle using KpnI and EcoRV restriction enzyme sites. The following primers were used to add 5′ BamHI and/or KpnI and 3′ EcoRV to the OGT F460A cDNA: 5′ BamHI, CATGGGATCCATGGCGTCTTCCGTGGG CAACGTG; 5′ KpnI, ACGGGGTACCATGGCGT CTTCCGTGGGCAACGTG; and 3′ EcoRV, CCCGGAT ATCTCAGGCTGACTCAGTGACTTCAAC. The adenovirus expressing OGT F460A was generated using the ViraPowerTM Adenoviral GatewayTM expression kit (Invitrogen). Adeno-associated virus serotype 9 expressing OGT F460A was generated by the University of California, San Diego Vector Development Core. The sequence of each vector was confirmed and shown to produce an open reading frame with the appropriate amino acid change.
Western Blotting and Immunoprecipitations
Total cell or tissue lysates were homogenized in Nonidet P-40 buffer (20 mm Tris, 150 mm NaCl, 0.025 mm O-(2-Acetamido-2-deoxy-d-glucopyranosylidenamino) N-phenylcarbamate(PUGNAc), and 1% Nonidet P-40 (pH 7.4)). 50–200 μg of protein samples were loaded on NuPAGE 4–12% BisTris gels (Invitrogen). Separated proteins were transferred to nitrocellulose membranes that were subsequently blocked in 5% milk/TBS, 0.05% Tween. Anti-Ogg1 (Genetex, GTX20204), anti-O-GlcNAc (RL2) (Thermo Fisher Scientific, MAI-072), anti-OGT (Abcam, Ab177941), anti-OGA (Abcam, Ab124807), anti-FLAG®M2 (Sigma, F1804), anti-cleaved caspase 3 (Cell Signaling Technology, 9661S), anti-Actin (Santa Cruz Biotechnology, SC-1616), and anti-cyclophilin A (CPA) (Abcam, Ab41684) were used as primary antibodies. Anti-mouse IgG-HRP-conjugated (Amersham Biosciences), anti-rabbit IgG-HRP-conjugated (Cell Signaling Technology), and anti-goat IgG-HRP-conjugated (Santa Cruz Biotechnology) were used as secondary antibodies. RL2 and anti-Ogg1 antibodies were validated by Western blotting. An Ogg1-specific siRNA (Ambion, Life Technology, AM16708) was used to knock down Ogg1 in NCM and to demonstrate the specificity of the anti-Ogg1 antibody. High-glucose treatment of NCM for 72 h was used to increase O-GlcNAc levels, and incubation for 2 h at 37 °C with recombinant OGA was used to decrease the O-GlcNAc-specific signal detected by RL2. Blots containing protein from NCM cultured for 72 h in high glucose and incubated with RL2 in the presence of 1 m GlcNAc (MP Biomedicals, catalog no. 100068) inhibited RL2-protein interaction (supplemental Fig. 1). To analyze Ogg1-specific O-GlcNAcylation and the interaction between Ogg1 and OGT, 500 μg of total protein samples from either primary cultures or murine hearts were immunoprecipitated with anti-Ogg1 antibody using the Pierce Crosslink Immunoprecipitation Kit (Thermo Fisher Scientific) following the instructions of the manufacturer. Immunoprecipitates were analyzed by Western blotting using RL2, Ogg1, and OGT antibodies. Images were acquired with the ChemiDocTM MP System (Bio-Rad). Band density was quantified with ImageJ software. Control immunoprecipitations were performed using normal rabbit IgG (Santa Cruz Biotechnology, SC-2027) as a nonspecific IP antibody. Ogg1 and OGT detected in the immunoprecipitated samples were normalized to their corresponding input levels, and inputs were normalized either to Actin or CPA. The Ogg1-specific RL2 signal detected in Ogg1 immunoprecipitated samples was normalized to the Ogg1 signal detected in IP.
8-OHdG IF microscopy
Immediately after excision, murine hearts were rinsed in 1× PBS, bisected transversely, and then the apical half was fixed in 4% paraformaldehyde (PFA) overnight at 4 °C with constant mixing. The following day, fixed tissues were incubated at 4 °C for 2 h in 15% sucrose/1× PBS, followed by subsequent incubation for 2 h in 25% sucrose/1× PBS and 2 h in 1:1 25% sucrose/1× PBS: (optimal cutting temperature compound-Sakura Finetek USA Inc.). Then tissues were embedded in optimal cutting temperature compound and frozen in a dry ice/2-methylbutane bath. Samples were stored at −80 °C until slides were prepared by sectioning tissue blocks with a cryostat. Slides (10 μm) were rinsed once in 1× PBS and then incubated for 30 min at room temperature on a shaking platform in blocking/permeabilization buffer (20 mm glycine, 1% BSA IgG-free, 3% normal goat serum, 0.1% Triton X-100, 0.05% Tween 20, and 1× PBS) before incubation overnight at 4 °C with either anti-8-OHdG (Genetex, GTX41980) or anti-Ogg1 (Genetex, GTX20204) diluted 1:20 in 1:10 blocking/permeabilization buffer:PBS (v/v). Anti-8-OHdG was validated using the Bioxytech® 8-OHdG-EIATM kit (OXIS, catalog no. 21026). Anti-Ogg1 was further validated by transfecting NCM cells with Ogg1-specific siRNA (Ambion, Life Technology, AM16708)for 24 h (supplemental Fig. 2). Anti-TOM20 (Santa Cruz Biotechnology, SC-11415) and anti-UQCRC2 (complex 3) (Abcam, Ab14745), diluted 1:100 and 1:50, respectively, in 1:10 blocking/permeabilization buffer:PBS (v/v), were used in co-localization studies. Slides were incubated with only 1:10 blocking/permeabilization buffer:PBS (v/v) as a control for nonspecific detection of antigens by the secondary antibody. The following day, slides were rinsed three times with 1× PBS/0.05% Tween 20 and then incubated at room temperature for 1 h with either goat anti-mouse IgG secondary antibody Alexa Fluor® 568 conjugate (Thermo Fisher Scientific, A11004) or goat anti-rabbit IgG (H+L) secondary antibody Alexa Fluor® 488 conjugate (Thermo Fisher Scientific, A11034) diluted 1:200 in 1:10 blocking/permeabilization buffer:PBS (v/v). Slides were rinsed five times with 1× PBS/0.05% Tween 20. One drop of mounting medium with DAPI (ProLong® Diamond Antifade Mountant with DAPI, Thermo Fisher Scientific) was then added to the tissue slides before applying coverglasses and sealing the edges with a non-fluorescent nail polish. Images were captured with a Delta Vision deconvolution microscope system (Applied Precision) using a ×100 lens at the University of California, San Diego, School of Medicine Light Microscopy Facility. At least 5 fields/biological sample were imaged, and ∼20 serial optical sections, spaced by 0.2 μm, were acquired. The datasets were deconvolved using SoftWorx software (Applied Precision) on a Silicon Graphics Octane work station. 8-OHdG levels were quantified by counting the percentage of positive (red) pixels compared with total pixels using the Plugin “Color Pixel Counter” in ImageJ. Data were normalized by subtracting the nonspecific signal detected in samples incubated with no primary antibody. NCM cells were immunostained following the same protocol with few modifications. After treatment cells were washed once with 1× PBS and fixed in 4% PFA for 15 min at room temperature. Subsequently, cells were treated the same way as the tissue slides.
Ogg1 Activity Assay from Total Cell and Tissue Lysates
For assessing Ogg1 activity, we used an ad hoc-designed molecular beacon (5′-6-FAM-GCACT[8-OXOdG]AAGCGCCGCACGCCATGTCGACGCGCTTCAGTGC-DABCYL-3′,Sigma) carrying an oxidized guanosine residue in the stem portion of the molecule. The underlined sequence represents the stem-annealed region, whereas the non-underlined sequence represents the loop of the beacon. The 5′-fluorophore 6-FAM (6-carboxyfluorescein) is in close proximity to the 3′ quencher (4′dimethylaminophenilazo) benzoic acid until the beacon is cut by Ogg1, resulting in a fluorescent signal forming the basis of detection of Ogg1 activity. 25, 50, and 100 μg of either total murine heart or NCM lysates were incubated with 0.5 μm molecular beacon in 50 mm HEPES (pH 7.6), 100 mm KCl, 2 mm EDTA, and 2 mm DTT at 37 °C, and the Ogg1-mediated cleavage reaction was followed for 30 min on a Biotek® Synergy 2 multimode reader taking readings every minute of fluorescence emission at 528/20 nm and exciting the samples at 485/20 nm.
In Situ Molecular Beacon Staining
After isolation, NCM cells were seeded on gelatin-coated microscope coverglasses (Thermo Fisher Scientific, 12-454-80, 12CIR-1) and the same protocol described above was followed. After 72 h of either NG or HG treatment, NCM were transfected with 1 nm molecular beacon using Lipofectamine® 2000 (Invitrogen) following the instructions of the manufacturer and incubated at 37 °C overnight. The following day, cells were rinsed once with 1× PBS and fixed in 4% PFA for 15 min. Cells were then washed five times with 1× PBS, and one drop of mounting medium with DAPI (ProLong® Diamond Antifade Mountant with DAPI, Thermo Fisher Scientific) was added. Images and data were acquired as described under “8-OHdG IF Microscopy.” As a negative control, one set of cells was transfected just prior to fixation with 4% PFA.
Recombinant Protein Expression and Isolation
Recombinant FLAG-tagged Ogg1 and OGT were expressed and purified from HEK 293T cells. HEK 293T cells were maintained in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin at 37 °C in 5% CO2. Cells were seeded to be 80% confluent and, ∼12 h later, transfected with pcDNA3-FLAG constructs using Lipofectamine® 2000 (Invitrogen) following the instructions of the manufacturer. 24 h later, cells were harvested and sonicated in OGT assay buffer (50 mm Tris-HCl (pH 7.4), 1 mm DTT, 10 mm KCl, and 12.5 mm MgCl2). Cleared lysates were incubated with anti-FLAG® M2 affinity gel (Sigma) and pre-equilibrated in OGT assay buffer for 2 h at 4 °C with constant mixing. Beads were then washed twice with 1× PBS and once with OGT assay buffer. Bound protein was eluted by incubating the beads four times with OGT assay buffer supplemented with 0.2 mg/ml FLAG peptide (Sigma). The four eluates for each protein were pooled and used for further experiments. Aliquots from each purification step were separated by SDS-PAGE and analyzed for recombinant protein purity using the SilverQuestTM silver staining kit (Invitrogen) following the instructions of the manufacturer. Recombinant His-tagged OGA was expressed and purified from E. coli BL21 RosettaTM (DE3). Bacteria were transformed with pQTEV-OGA purified previously from transformed E. coli DH5α. Rosetta cells carrying pQTEV-OGA were cultured at 37 °C until A600 reached ∼0.4. Recombinant expression was induced by incubating the bacterial culture with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside (Genesee Scientific, Inc.) at room temperature overnight. The next day, bacterial cells were harvested and lysed by sonication in OGT assay buffer supplemented with 200 mm NaCl and 5 mm imidazole. Cleared lysate was incubated for 2 h with nickel-nitrilotriacetic acid-agarose resin (Qiagen) and pre-equilibrated in OGT assay buffer with constant mixing. Beads were then washed with OGT assay buffer supplemented with 200 mm NaCl and 5 mm imidazole until A280 ∼0.1. An imidazole gradient (5–500 mm) was then applied, and fractions where collected. Fractions that exhibited OGA expression by SDS-PAGE and Coomassie staining were pooled and loaded onto a G50 resin to remove the excess imidazole. Eluted fractions were then concentrated using a SpeedVac Concentrator SVC200H (Savant) and pooled, and protein concentration was determined for further experiments.
In Vitro Ogg1 O-GlcNAcylation and De-O-GlcNAcylation
Recombinant FLAG-tagged Ogg1, OGT, and His-tagged OGA were expressed and purified as described above. Approximately 0.1 mg/ml eluted FLAG-Ogg1 and FLAG-OGT were incubated in 1-ml total reaction volume in the presence of OGT assay buffer (50 mm Tris-HCl (pH 7.4), 1 mm DTT, 10 mm KCl, and 12.5 mm MgCl2) and 50 μm UDP-GlcNAc (Sigma) at 37 °C. 50 μl of reaction mixture was collected at 0, 30, 90, and 210 min and used to assess Ogg1 activity. After 3.5-h incubation, 200 μl from a 1 mg/ml solution of recombinant OGA was introduced into the reaction, and after 30 and 60 min (240 and 270 min total), 63 μl was collected for assessing Ogg1 activity. Before running this experiment, the functionality of recombinant His-tagged OGA was confirmed by assessing activity in 50 mm sodium cacodylate (pH 6.4), 3% BSA, 1 μm 4-methylumbellifery GlcNAc. Activity was followed on a Biotek® Synergy 2 multimode reader taking readings every 2 min of fluorescence emission at 460/40 nm and exciting the samples at 360/40 nm (supplemental Fig. 3). FLAG-Ogg1, FLAG-OGT, His-OGA, and O-GlcNac-Ogg1 protein levels were also analyzed by Western blotting. Aliquots of reaction mixture taken at different time points were either immunoprecipitated using anti-Ogg1 or directly analyzed by Western blotting following the procedures described above.
Mitochondrion Preparation Procedures
Immediately after excision, murine hearts were rinsed in MIM buffer (250 mm sucrose, 10 mm HEPES, and 1 mm EDTA (pH 7.2)) and then transferred to 3 ml of MIM buffer supplemented with 1 mg/ml BSA (pH 7.4). Samples were then homogenized in a tissue homogenizer four times for 5 s and then in a Potter homogenizer three times up and down. Homogenized samples were then centrifuged at 600 × g for 10 min to pellet nuclei, cytoskeleton, and unbroken cells. The supernatant fraction was then further centrifuged at 8000 × g for 15 min. The mitochondrial pellet was then gently resuspended using a clean small paintbrush in MIM buffer and centrifuged at 8000 × g for 15 min. Finally, the mitochondrial pellet was gently resuspended in ∼200 μl of MIM buffer, and the protein concentration was determined for further applications. All steps were performed at 4 °C, and all tools were prechilled and kept at 4 °C before and during the isolation procedures.
mtDNA Damage Estimation
Mitochondria from mouse hearts were isolated as described under “Mitochondrion Preparation Procedures.” Isolated mitochondria were resuspended in an appropriate volume of mitochondrial lysis buffer (BioVision Mitochondrial DNA Isolation Kit, K280) and kept for 10 min on ice. Then an appropriate volume of Enzyme B Mix was added, and samples were kept at 50 °C until the solution became clear. Samples were further cleaned by adding an equal volume of phenol/chloroform to the lysed samples. After mixing and centrifuging, the aqueous phase was combined with an equal volume of 100% ethanol. Samples were stored at −20 °C for 20 min and then centrifuged at top speed for 5 min at room temperature. The mtDNA pellet was then washed once with 70% ethanol and centrifuged again at top speed for 5 min at room temperature. The mtDNA pellet was resuspended in Tris-EDTA buffer and quantified by NanoDrop (Thermo Fisher Scientific). mtDNA damage was detected by semiquantitative PCR according to Kovalenko and Santos (48). Briefly, two fragments (117 bp and 10 kb) were amplified from isolated mtDNA using the following primers: 117 bp, 5′-CCCAGCTACTACCAT CATTCAAGT-3′ (forward) and 5′-GATGGTTTGGGAGA TTGGTTGATGT-3′ (reverse); 10 kb, 5′-GAGAGATTT TATGGGTGTAATGCGG-3′ (forward) and 5′-GCCAGCCT GACCCATAGCCATAATAT-3′ (reverse).Because lesions are randomly distributed as a result of oxidative and other kinds of stresses, the amplification of the 10-kb fragment is selectively inhibited. After 20 amplification cycles (thermal protocol for the 117-bp fragment: 2 min at 94 °C, 10 s at 98 °C, 30 s at 57 °C, and 1 min at 68 °C; thermal protocol for the 10-kb fragment: 2 min at 94 °C, 10 s at 98 °C, 30 s at °C, and 10 min at 68 °C; KOD XtremeTM Hot Start DNA Pol from Novagen was used), the PCR mixture was separated on a 1% agarose gel, stained with EtBr, and photographed (ChemiDocTM MP System, Bio-Rad). PCR products were quantified by densitometric analysis using ImageJ. The linearity of the reaction was confirmed by including a control reaction containing 50% template DNA. The ratio between 10- and 117 bp bands gives an estimate of the mtDNA damage, and the inverse of this ratio represents the lesions/10 kb.
Statistical Analysis
All data were analyzed using GraphPad Prism 5 and are presented as mean ± S.E. One-way analysis of variance with appropriate post hoc or unpaired Student's t test was used for comparison between two groups. p < 0.05 was considered to be statistically significant.
Author Contributions
F. C., J. S., B. T. S., D. E. C., and W. H. D. designed the research. F. C., B. T. S., W. H., A. D., J. D. J., and T. D. performed the research. F. C., B. T. S., J. S., and W. H. D. analyzed the data. F. C., B. T. S., J. S., D. E. C., and W. H. D. wrote the paper.
Supplementary Material
This work was supported by National Institutes of Health Grants HL066941 (to W. H. D., PI project 3) and NS047101, and a grant from the P. Robert Majumder Charitable Foundation. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Figs. 1–3.
- ROS
- reactive oxygen species
- Ogg1
- 8-oxoguanine DNA glycosylase
- 8-OHdG
- 8-hydroxy-2′-deoxyguanosine
- OGT
- O-GlcNAc transferase
- OGA
- O-GlcNAcase
- T1D
- type 1 diabetes
- CTR
- control
- WB
- Western blot(s)
- NCM
- neonatal cardiac myocyte(s)
- HG
- high glucose
- NG
- normal glucose
- IP
- immunoprecipitation
- STZ
- streptozotocin
- Adv
- adenovirus
- CDS
- coding sequence(s)
- CPA
- cyclophilin A
- PFA
- paraformaldehyde
- IF
- immunofluorescence
- RFU
- relative fluorescence units.
References
- 1.Rolo A. P., and Palmeira C. M. (2006) Diabetes and mitochondrial function: role of hyperglycemia and oxidative stress. Toxicol. Appl. Pharmacol. 212, 167–178 [DOI] [PubMed] [Google Scholar]
- 2.Halliwell B. (2011) Free radicals and antioxidants: quo vadis? Trends Pharmacol. Sci. 32, 125–130 [DOI] [PubMed] [Google Scholar]
- 3.Redza-Dutordoir M., and Averill-Bates D. A. (2016) Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Biophys. Acta 10.1016/j.bbamcr.2016.09.012 [DOI] [PubMed] [Google Scholar]
- 4.Albring M., Griffith J., and Attardi G. (1977) Association of protein structure of probable membrane derivation with HeLa cell mitochondrial DNA near its origin of replication. Proc. Natl. Acad. Sci. U.S.A. 74, 1348–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costa R. A., Romagna C. D., Pereira J. L., and Souza-Pinto N. C. (2011) The role of mitochondria DNA damage in the cytotoxicity of reactive oxygen species. J. Bioenerg. Biomembr. 43, 25–29 [DOI] [PubMed] [Google Scholar]
- 6.Gredilla R., Bohr V. A., and Stevnsner T. (2010) Mitochondrial DNA repair and association with aging: an update. Exp. Gerontol. 45, 478–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falkenberg M., Larsson N. G., and Gustafsson C. M. (2007) DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 76, 679–699 [DOI] [PubMed] [Google Scholar]
- 8.Taanman J. W. (1999) The mitochondrial genome: structure, transcription, translation and replication. Biochim. Biophys. Acta 1410, 103–123 [DOI] [PubMed] [Google Scholar]
- 9.Ballinger S. W., Patterson C., Yan C. N., Doan R., Burow D. L., Young C. G., Yakes F. M., Van Houten B., Ballinger C. A., Freeman B. A., and Runge M. S. (2000) Hydrogen peroxide- and peroxynitrite-induced mitochondrial DNA damage and dysfunction in vascular endothelial and smooth muscle cells. Circ. Res. 86, 960–966 [DOI] [PubMed] [Google Scholar]
- 10.Giulivi C., Boveris A., and Cadenas E. (1995) Hydroxyl radical generation during mitochondrial electron transfer and the formation of 8-hydroxydesoguanosine in mitochondrial DNA. Arch. Biochem. Biophys. 1995, 909–916 [DOI] [PubMed] [Google Scholar]
- 11.Richter C., Park J. W., and Ames B. N. (1988) Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc. Natl. Acad. Sci. U.S.A. 85, 6465–6467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yakes F. M., and Van Houten B. (1997) Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 21, 514–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhakat K. K., Mokkapati S. K., Boldogh I., Hazra T. K., and Mitra S. (2006) Acetylation of human 8-oxoguanine-DNA glycosylase by p300 and its role in 8-oxoguanine repair in vivo. Mol. Cell Biol. 26, 1654–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyrberg B., Anachkov K. A., Dib S. A., Wang-Rodriguez J., Yoon K. H., and Levine F. (2002) Islet expression of the DNA repair enzyme 8-oxoguanosine DNA glycosylase (Ogg1) in human type 2 diabetes. BMC Endocr. Disord. 2, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu J., Imam S. Z., Hashiguchi K., de Souza-Pinto N. C., and Bohr V. A. (2005) Phosphorylation of human oxoguanine DNA glycosylase (α-OGG1) modulates its function. Nucleic Acids Res. 33, 3271–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreppel L. K., Blomberg M. A., and Hart G. W. (1997) Dynamic glycosylation of nuclear and cytosolic proteins: cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 272, 9308–9315 [DOI] [PubMed] [Google Scholar]
- 17.Ngoh G. A., Facundo H. T., Zafir A., and Jones S. P. (2010) O-GlcNAc signaling in the cardiovascular system. Circ. Res. 107, 171–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macauley M. S., Chan J., Zandberg W. F., He Y., Whitworth G. E., Stubbs K. A., Yuzwa S. A., Bennet A. J., Varki A., Davies G. J., and Vocadlo D. J. (2012) Metabolism of vertebrate amino sugars with N-glycolyl groups: intracellular β-O-linked N-glycolylglucosamine (GlcNGc), UDP-GlcNGc, and the biochemical and structural rationale for the substrate tolerance of β-O-linked β-N-acetylglucosaminidase. J. Biol. Chem. 287, 28882–28897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark R. J., McDonough P. M., Swanson E., Trost S. U., Suzuki M., Fukuda M., and Dillmann W. H. (2003) Diabetes and the accompanying hyperglycemia impairs cardiomyocyte calcium cycling through increased nuclear O-GlcNAcylation. J. Biol. Chem. 278, 44230–44237 [DOI] [PubMed] [Google Scholar]
- 20.Hu Y., Belke D., Suarez J., Swanson E., Clark R., Hoshijima M., and Dillmann W. H. (2005) Adenovirus-mediated overexpression of O-GlcNAcase improves contractile function in the diabetic heart. Circ. Res. 96, 1006–1013 [DOI] [PubMed] [Google Scholar]
- 21.Fülöp N., Zhang Z., Marchase R. B., and Chatham J. C. (2007) Glucosamine cardioprotection in perfused rat hearts associated with increased O-linked N-acetylglucosamine protein modification and altered p38 activation. Am. J. Physiol. Heart. Circ. Physiol. 292, H2227–H2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsh S. A., Dell'Italia L. J., and Chatham J. C. (2011) Activation of the hexosamine biosynthesis pathway and protein O-GlcNAcylation modulate hypertrophic and cell signaling pathways in cardiomyocytes from diabetic mice. Amino Acids 40, 819–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marsh S. A., Powell P. C., Dell'Italia L. J., and Chatham J. C. (2013) Cardiac O-GlcNAcylation blunts autophagic signaling in the diabetic heart. Life Sci. 92, 648–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett C. E., Johnsen V. L., Shearer J., and Belke D. D. (2013) Exercise training mitigates aberrant cardiac protein O-GlcNAcylation in streptozotocin-induced diabetic mice. Life Sci. 92, 657–663 [DOI] [PubMed] [Google Scholar]
- 25.Fricovsky E. S., Suarez J., Ihm S. H., Scott B. T., Suarez-Ramirez J. A., Banerjee I., Torres-Gonzalez M., Wang H., Ellrott I., Maya-Ramos L., Villareal F., and Dillmann W. H. (2012) Excess protein O-GlcNAcylation and the progression of diabetic cardiomyopathy. Am. J. Physiol. Regul. Integr. Comp. Physiol. 307, R689–R699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torres C. R., and Hart G. W. (1984) Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes: evidence for O-linked GlcNAc. J. Biol. Chem. 259, 3308–3317 [PubMed] [Google Scholar]
- 27.Lubas W. A., Frank D. W., Krause M., and Hanover J. A. (1997) Linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J. Biol. Chem. 272, 9316–9324 [DOI] [PubMed] [Google Scholar]
- 28.Dong D. L., and Hart G. W. (1994) Purification and characterization of an O-GlcNAc selective N-acetyl-β-d-glucosaminidase from rat spleen cytosol. J. Biol. Chem. 269, 19321–19330 [PubMed] [Google Scholar]
- 29.Gao Y., Wells L., Comer F. I., Parker G. J., and Hart G. W. (2001) Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic β-N-acetylglucosaminidase from human brain. J. Biol. Chem. 276, 9838–9845 [DOI] [PubMed] [Google Scholar]
- 30.Shafi R., Iyer S. P., Ellies L. G., O'Donnell N., Marek K. W., Chui D, Hart G. W., and Marth J. D. (2000) The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. U.S.A. 97, 5735–5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y. R., Song M., Lee H., Jeon Y., Choi E. J., Jang H. J., Moon H. Y., Byun H. Y., Kim E. K., Kim D. H., Lee M. N., Koh A., Ghim J., Choi J. H., Lee-Kwon W., et al. (2012) O-GlcNAcase is essential for embryonic development and maintenance of genomic stability. Aging Cell 11, 439–448 [DOI] [PubMed] [Google Scholar]
- 32.Hu Y., Suarez J., Fricovsky E., Wang H., Scott B. T., Trauger S. A., Han W., Hu Y., Oyeleye M. O., and Dillmann W. H. (2009) Increased enzymatic O-GlcNAcylation of mitochondrial proteins impairs mitochondrial function in cardiac myocytes exposed to high glucose. J. Biol. Chem. 284, 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gawlowski T., Suarez J., Scott B., Torres-Gonzalez M., Wang H., Schwappacher R., Han X., Yates J. R. 3rd, Hoshijima M., and Dillmann W. (2012) Modulation of dynamin-related protein 1 (DRP1) function by increased O-linked-β-N-acetylglucosamine modification (O-GlcNAc) in cardiac myocytes. J. Biol. Chem. 287, 30024–30034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aguilar H., Fricovsky E., Ihm S., Schimke M., Maya-Ramos L., Aroonsakool N., Ceballos G., Dillmann W., Villarreal F., and Ramirez-Sanchez I. (2014) Role for high-glucose-induced protein O-GlcNAcylation in stimulating cardiac fibroblast collagen synthesis. Am. J. Physiol. Cell Physiol. 306, C794–C804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zachara N. E., O'Donnell N., Cheung W. D., Mercer J. J., Marth J. D., and Hart G. W. (2004) Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress: a survival response of mammalian cells. J. Biol. Chem. 279, 30133–30142 [DOI] [PubMed] [Google Scholar]
- 36.Marsh S. A., Collins H. E., and Chatham J. C. (2014) Protein O-GlcNAcylation and cardiovascular (patho)physiology. J. Biol. Chem. 289, 34449–34456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banerjee P. S., Ma J., and Hart G. W. (2015) Diabetes-associated dysregulation of O-GlcNAcylation in rat cardiac mitochondria. Proc. Natl. Acad. Sci. U.S.A. 112, 6050–6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mirbahai L., Kershaw R. M., Green R. M., Hayden R. E., Meldrum R. A., and Hodges N. J. (2010) Use of a molecular beacon to track the activity of base excision repair protein OGG1 in live cells. DNA Repair 9, 144–152 [DOI] [PubMed] [Google Scholar]
- 39.Nakabeppu Y., Tsuchimoto D., Ichinoe A., Ohno M., Ide Y., Hirano S., Yoshimura D., Tominaga Y., Furuichi M., and Sakumi K. (2004) Biological significance of the defense mechanisms against oxidative damage in nucleic acids caused by reactive oxygen species: from mitochondria to nuclei. Ann. N.Y. Acad. Sci. 1011, 101–111 [DOI] [PubMed] [Google Scholar]
- 40.Lazarus B. D., Roos M. D., and Hanover J. A. (2005) Mutational analysis of the catalytic domain of O-linked N-acetylglucosaminyl transferase. J. Biol. Chem. 280, 35537–35544 [DOI] [PubMed] [Google Scholar]
- 41.Dizdaroglu M., Jaruga P., Birincioglu M., and Rodriguez H. (2002) Free radical-induced damage to DNA: mechanisms and measurement. Free Rad. Biol. Med. 32, 1102–1115 [DOI] [PubMed] [Google Scholar]
- 42.Grollman A. P., and Moriya M. (1993) Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 9, 246–249 [DOI] [PubMed] [Google Scholar]
- 43.Barja G. (2004) Aging in vertebrates, and the effect of caloric restriction: a mitochondrial free radical production-DNA damage mechanism? Biol. Rev. Camb. Philos. Soc. 79, 235–251 [DOI] [PubMed] [Google Scholar]
- 44.Anson R. M., Hudson E., and Bohr V. A. (2000) Mitochondrial endogenous oxidative damage has been overestimated. FASEB J. 14, 355–360 [DOI] [PubMed] [Google Scholar]
- 45.Cooke M. S., Evans M. D., Dizdaroglu M., and Lunec J. (2003) Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 17, 1195–1214 [DOI] [PubMed] [Google Scholar]
- 46.Sikorsky J. A., Primerano D. A., Fenger T. W., Denvir J. (2007) DNA damage reduces Taq DNA polymerase fidelity and PCR amplification efficiency. Biochem. Biophys. Res. Commun. 335, 431–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsueh W., Abel E. D., Breslow J. L., Maeda N., Davis R. C., Fisher E. A., Dansky H., McClain D. A., McIndoe R., Wassef M. K., Rabadán-Diehl C., and Goldberg I. J. (2007) Recipes for creating animal models of diabetic cardiovascular disease. Circ. Res. 100, 1415–1427 [DOI] [PubMed] [Google Scholar]
- 48.Kovalenko Q. A., and Santos J. H. (2009) Analysis of oxidative damage by gene-specific quantitative PCR. Curr. Protoc. Hum. Genet. 10.1002/0471142905.hg1901s62 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.